Introduction
Osteosarcoma (OS) refers to a form of bone cancer
that affects the knee or other ends of the long bones (1). This malignancy is rare among most
demographics, but it is more common among children and adolescents
(2). Although physicians have
used surgery, chemotherapy and radiotherapy to ameliorate the
debilitating symptoms of this bone cancer, the survival rate of OS
is still not satisfactory (3).
Recent progress achieved in molecular genetic research of OS has
improved the understanding of OS development (4). Nonetheless, further research is
needed to identify the underlying molecular mechanism of this tumor
and reduce the survival rate of OS (5).
Circular RNAs (circRNAs) consist of closed-loop
polymeric molecules that are crucial to the expression and
regulation of genes (6). Unlike
RNAs, circRNAs have their 3′ and 5′ terminals joined, and they can
inactivate microRNAs (miRNAs/miRs) by regulating the biological
function of genes in eukaryotes (7). Emerging studies have demonstrated
the crucial roles of circRNAs in OS (8-10). In one previous study, it was
reported that a high level of circ-ITCH in OS cells facilitated the
spread of OS by enhancing EGFR phosphorylation (11). Another previous report documented
that the negative effect of circ_0008792 on OS samples was highly
pronounced (12). However, to
the best of our knowledge, the effects of circ_0032463 on OS cells
has not yet been explored.
In contrast to circRNAs, miRNAs cannot code genes.
However, miRNAs can degrade or inhibit the transcription of mRNAs
by binding to specific regions of the target genes (13). In recent years, researchers have
documented the ability of miRNAs to influence the progression of
various cancers by targeting protein-coding genes and regulating
mRNA expression (14-16). Some miRNAs function as tumor
promoters in OS development, including miR-299-5p, miR-765 and
miR-624-5p (17-19). Other miRNAs can act as tumor
suppressors in OS progression, such as miR-375, miR-224-3p and
miR-627-3p (20-22). As for miR-330-3p, some studies
have reported on the inhibitory role of this miRNA in
carcinogenesis (23,24). A recent study found that
miR-330-3p inhibited tumor progression by suppressing polycomb
complex protein Bmi-1 (Bmi-1) expression in OS (25). Nonetheless, researchers are yet
to confirm whether miR-330-3p could be regulated by circ_0032463 in
OS.
Pinin desmosome associated protein (PNN) is a
protein-coding gene, which is associated with a number of diseases,
such as melanoma and Volkmann ischemic contracture (26). The high expression of PNN has
been found to facilitate ovarian carcinoma (27), colorectal cancer (28) and hepatocellular carcinoma
(29). However, its role in OS
is yet to be confirmed.
The present research aimed to investigate whether
novel circRNA (circ_0032463) could accelerate or inhibit OS
progression. It was hypothesized that circ_0032463 promoted OS by
inhibiting miR-330-3p and upregulating PNN expression. Regardless
of the outcome of the hypothesis, the findings of this study could
provide novel insights into OS treatments.
Materials and methods
Bioinformatics analysis
The GSE96964 dataset (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE96964)
from the Gene Expression Omnibus (GEO) database is a circRNA
expression profile that was selected to screen the upregulated
circRNAs in OS samples with P<0.05 and log fold-change (FC)
>1. GSE16088 (30) and
GSE12865 (31) datasets, also
from the GEO database, are mRNA expression profiles that were
selected to screen the upregulated genes in OS samples with
P<0.05 and logFC >1. The Search Tool for the Retrieval of
Interacting Genes/Proteins (STRING; https://string-db.org/) was used to analyze the key
biological process for the upregulated genes. CircInteractome
(https://circinteractome.nia.nih.gov/index.html) was
utilized to predict the miRNAs sponged by key circRNAs, whereas
starBase (http://starbase.sysu.edu.cn/panCancer.php) and
TargetScan (http://www.targetscan.org/vert_71/) were utilized to
predict the miRNAs that target the key genes.
Tissue collection and cell culture
Tissue samples with and without OS were obtained
from 13 patients who under-went surgery between January 2018 and
December 2019 at The First Affiliated Hospital of Zhengzhou
University (Zhengzhou, China). The inclusion criteria included
patients who were diagnosed with OS and had not received
preoperative radiotherapy, chemotherapy or other adjuvant
treatments, whereas the exclusion criteria included patients who
were diagnosed with other diseases and did not sign the informed
consent. The experimental procedure was approved by the Ethics
Committee of the First Affiliated Hospital of Zhengzhou University
(approval no. ChiECRCT20200309), and informed consent was obtained
from the patients before they could participate in the experiment.
The characteristics of the participants with OS are presented in
Table I. All cell lines used in
this study were obtained from the American Type Culture Collection,
including the human fetal osteoblast cell line (hFOB1.19) and human
OS cell lines (HOS, U2OS, Saos-2 and SOSP-9607). The cells were
maintained in an RPMI-1640 medium (Invitrogen; Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.) and 1% penicillin-streptomycin (Gibco; Thermo
Fisher Scientific, Inc.), and they were incubated in an atmosphere
containing 5% CO2 at 37°C.
 | Table IClinical characteristics of 13
patients with osteosarcoma. |
Table I
Clinical characteristics of 13
patients with osteosarcoma.
|
Characteristics | Number of patients,
n (%) |
|---|
| Total | 13 |
| Sex | |
| Male | 7 (53.85) |
| Female | 6 (46.15) |
| Age, years | |
| <12 | 1 (7.69) |
| 12-29 | 7 (53.85) |
| 30-49 | 3 (23.08) |
| ≥50 | 2 (15.38) |
| Tumor size, cm | |
| ≤12 | 8 (61.54) |
| >12 | 5 (38.46) |
| TNM stage (49) | |
| I-II | 10 (76.92) |
| III-IV | 3 (23.08) |
| Site | |
| Extremities | 9 (69.23) |
| Other | 4 (30.77) |
| Distant
metastasis | |
| Negative | 9 (69.23) |
| Positive | 4 (30.77) |
Cell transfection
To explore the function of circ_0032463, miR-330-3p
and PNN in OS, the OS cells were divided into eight groups: i)
short hairpin RNA (sh-) negative control (NC) group; ii)
sh-circ_0032463 group; iii) miR-mimics group; iv) NC mimics group;
v) miR-inhibitor group; vi) NC inhibitor group; vii)
sh-circ_0032463 + miR-inhibitor group; and viii) sh-PNN +
miR-inhibitor group. The sh-circ_0032463, miR-330-3p inhibitor,
miR-330-3p mimic, sh-PNN and negative control (NC) plasmids were
purchased from Shanghai GenePharma Co., Ltd. The sequences of all
plasmids used in this study are listed in Table SI. The transfections with the
shRNAs, miRNA mimic, inhibitor or corresponding NCs into Saos-2 and
U2OS cells (1×105) were performed using
Lipofectamine® 3000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C for 48 h. Next, 100, 1,000 or 2,500 ng
shRNAs were transfected into the cells seeded into a 96-, 12- or
6-well plate, respectively. Then, 3, 30 or 75 pmol miR-330-3p mimic
or inhibitor were seeded into a 96-, 12- or 6-well plate,
respectively.
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was extracted from the OS tissues and
cells (Hfob1.19, HOS, U2OS, Saos-2 and SOSP-9607) with
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). According to the manufacturer's protocols, the HiScript II Q
RT SuperMix (Vazyme Biotech Co., Ltd.) was employed for circRNA and
PNN mRNA reverse-transcription, the miRNA 1st Strand cDNA Synthesis
kit (Vazyme Biotech Co., Ltd.) was used to prepare miR-330-3p.
Then, RT-qPCR analysis was performed with the SYBR Green PCR Master
Mix kit (Thermo Fisher Scientific, Inc.). The thermocycling
conditions were as follows: Initial denaturation at 95°C for 10
min, 40 cycles of denaturation at 95°C for 10 sec, annealing and
extension at 60°C for 1 min. The 2−ΔΔCq method (32) was then used to analyze the
changes in gene expression. The reference control for mRNA and
circRNA was GAPDH, while that for miRNA was U6. The primer
sequences are presented in Table
II. After incubating 1 mg RNA for 30 min at 37°C, RNase R
(Epicentre; Illumina, Inc.) was utilized to validate the stability
of circRNA. The RNA was then utilized to synthesize cDNA for
RT-qPCR.
 | Table IIPrimer sequences for reverse
transcription-quantitative PCR. |
Table II
Primer sequences for reverse
transcription-quantitative PCR.
| Genes | Primer sequences
(5′→3′) |
|---|
| circ_0032463 | F:
GCCCAGCCTTTGAAGAATTT |
| R:
TTCTGTGGTTGCCATAATCC |
| circ_0097271 | F:
CTGTGGAAACCCTTGGTTGT |
| R:
TCACCTGTGAGAATTGACTGG |
| circ_0113706 | F:
ACACCCTTCTCATGGCAATC |
| R:
CAAATGGCCCAAATCATTGT |
| circ_0111388 | F:
TCCTTATCAACCCCACCAAA |
| R:
TGGCAAGTTCTTTTTCACAGG |
| circ_0012726 | F:
ACTTCTCCACGGTTTCAAGC |
| R:
CTGCAATCCACAGATGAAGC |
| circ_0136823 | F:
AACCCAAGACCCCGAAAG |
| R:
GCAGTGTCAGGCAAAGTCTG |
| circ_0113185 | F:
CATGGGGAAGGCTATGTGAA |
| R:
CCTTCAAGAAGCCAGAGGTG |
| miR-330-3p | F:
CAACTGCCTCTCTGGGCCTG |
| R:
CTGCAGAGAGGCAGCGCTG |
| PNN | F:
GGCTTGTTGATGGGTACCCTT |
| R:
GCTGCGCAAGCTCAACTTTCT |
| GAPDH | F:
CCACATCGCTCAGACACCAT |
| R:
CCAGGCGCCCAATACG |
| U6 | F:
CGCTTCGGCAGCACATATAC |
| R:
TTCACGAATTTGCGTGCAT |
Isolation of cytoplasmic and nuclear
fractions
The separation of the cytoplasmic and nuclear RNA in
the UOS and Saos-2 cells was performed with the PARIS™ kit (Thermo
Fisher Scientific, Inc.). The cells were subsequently gathered and
lysed with the cell fractionation buffer. After centrifugation at
4°C and 10,000 × g for 2 min, the supernatant with cytoplasmic
fraction and the sediment with nuclear fraction were added to the
lysis/binding solution and an equal volume of ethanol. After
filtering the mixture, the elution buffer was used to elute the RNA
from the cytoplasmic and nuclear fractions. U6 snRNA and GAPDH rRNA
were used as the reference controls for nuclear fraction and
cytoplasmic fraction, respectively. The extracted RNA expression of
circ_0032463 and linear gene signal-induced
proliferation-associated 1-like protein 1 (SIPA1L1) was estimated
with RT-qPCR.
BrdU assay
The UOS and Saos-2 cells were seeded into the
96-well plates at a density of 1×105 cells/well. The
BrdU Cell Proliferation Assay kit (Frdbio Bioscience &
Technology) was then used to evaluate the proliferative ability of
the cells transfected for 48 h. This was performed according to the
manufacturer's protocols. After the cell culture medium was
eliminated, the cell fixation solution (100 µl) was
transferred to each well to fix the cells at room temperature for
30 min. Next, the solution was dried, and the cell denaturation
solution (100 µl) was added to each well for denaturation at
20-22°C for 10 min. Following which, the washing solution (300
µl) was used to wash the cells three times. The BrdU
antibody (100 µl) was then added to each well to incubate
the cells for 1 h at 37°C. Subsequently, 100 µl diluted
HRP-labeled goat anti-mouse antibody from the BrdU Cell
Proliferation Assay kit was added to each well, and the mixture was
incubated for another 1 h at 37°C. The TMB substrate solution was
added, and the mixture was incubated at 20-22°C for 10 min.
Finally, 100 µl substrate termination solution was added to
each well, and the optical density at 450 nm was read with a
microplate reader (BioTek Instruments, Inc.).
Cell Counting Kit-8 (CCK-8) assay
A CCK-8 Kit (Dojindo Molecular Technologies, Inc.)
was employed to identify the viability of cells. The OS cells
(1×105) were plated in a 96-well plate. For each period
(0, 24, 48, 72 and 96 h), the CCK-8 solution (10 µl/well)
was added to the OS cells and cultured in the dark for another 2 h.
The optical absorbance at 450 nm was measured with a microplate
reader.
Cell apoptosis detection
A flow cytometry system and Annexin V-FITC/PI
apoptosis detection kit (Shanghai Yeasen Biotechnology Co., Ltd.)
were employed to assess cell apoptosis. The transfected cells
(1×106) were gathered and then cleaned twice with
precooled PBS. This was followed by the addition of binding buffer
(100 µl) to the resuspended cells. The PI staining solution
(10 µl) and Annexin V-FITC (5 µl) were added, and the
cells were incubated in the dark for 10 min. Following which, the
cell apoptosis rate was detected for 1 h using a BD FACSCalibur
flow cytometer (BD Biosciences), and was analyzed using FlowJo
7.6.1 (FlowJo LLC). The percentage of late apoptotic cells is
presented in Q1-UR, whereas the percentage of early apoptotic cells
is presented in Q1-LR.
Transwell assay
The Corning® BioCoat™
Matrigel® Invasion Chambers (Corning, Inc.) were used to
measure cell invasion. The transfected cells (1×105
cells/well) were inoculated in the upper Transwell chambers in
serum-free RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific,
Inc.). RPMI-1640 with 10% FBS, was included in the lower Transwell
chambers to establish a chemical attractant. After 48 h of
incubation at 37°C, cells in the upper chambers that had not
invaded were swabbed away. Invaded cells were then fixed using 4%
paraformaldehyde for 20 min at 22°C. After the fixation, the cells
were treated with 1% crystal violet staining for 15 min at 22°C. An
optical microscope (Olympus Corporation) at ×200 magnification was
used to analyze the invasive abilities of the cells.
Cell adhesion assay
The Cell Adhesion Detection kit (BestBio Science)
was employed to analyze the adhesion ability of U2OS and Saos-2
cells. Briefly, the coating buffer (100 µl) was transferred
to each well in a 96-well plate and was left overnight at 4°C. The
following day, the coating buffer was removed, and the plate was
cleansed three times with a detergent solution. Cells were then
digested with trypsin. Following which, they were washed with PBS
and resuspended with the RPMI-1640 medium. This procedure was
followed by the preparation of the cell suspension. Subsequently,
1×105 cells were inoculated into the 96-well plate in
each well. Five duplicate wells were established as the
experimental group, while five untreated wells served as the
control group. The cells were incubated at 37°C for 30 min. The
culture medium in the control group was not discarded, and the
culture medium in the experimental group was replaced by a fresh
culture medium. After the cell staining solution B (100 µl)
was transferred to each well, the cells were incubated for 1 h at
37°C. A microplate reader was used to record the optical density
value at 450 nm. Finally, the cell adhesion rate was analyzed based
on that OD value.
Dual-luciferase reporter assay
All the wild-type (WT) or mutant (MUT) vectors were
purchased from Shanghai GenePharma Co., Ltd. Briefly, circ_0032463
segment with either WT or MUT biding sites was transformed and
subcloned into the psiCHECK-2 vector (Promega Corporation). The OS
cells (1×105 cells/well in 96-well-plates) were then
co-transfected with MUT or WT circ_0032463 and the miR-330-3p mimic
or mimic NC. After a transfection period of 48 h, the
Dual-Luciferase Reporter kit (Promega Corporation) was employed to
measure the luciferase activity. Similarly, the MUT or WT 3′-UTR
fragment of PNN with or without the predicted miR-330-3p binding
sites was amplified using QuikChange Site-Directed Mutagenesis kit
(Agilent Technologies, Inc.), according to the manufacturer's
protocol, and subcloned into the pmirGLO vector (Promega
Corporation). This step was followed by co-transfection with
miR-330-3p mimic or mimic NC to identify luciferase activities. The
relative luciferase activity was normalized by Renilla
luciferase activity.
RNA Immunoprecipitation Chip (RIP)
assay
This assay was performed with the Magna RIP™
RNA-Binding Protein Immunoprecipitation kit (EMD Millipore),
according to the manufacturer's protocols. 1×107 cells
were then treated with RIP lysis buffer (100 µl) containing
RNase inhibitors and protease inhibitor cocktail. The cell lysates
(200 µl) were then incubated with 5 µg magnetic beads
conjugated to anti-Ago2 (cat. no. 03-110; EMD Millipore) or
anti-IgG (negative control; cat. no. 03-110; EMD Millipore) after
rinsing with washing buffer, and rotated overnight at 4°C. After
treatment with proteinase K, the immunoprecipitated RNAs were
extracted by 500 µl TRIzol® reagent (Invitrogen;
Thermo Fisher Scientific, Inc.), and centrifuged at 4°C and 10,000
× g for 10 min. Finally, RT-qPCR was performed to measure
circ_0032463 and miR-330-3p enrichment.
RNA pull-down
The OS cells (1×107) were treated with
500 µl cell lysis buffer containing 2.5 mM EDTA, 25 mM
Tris-HCl, 70 mM KCl, 0.05% NP-40, RNase inhibitors and protease
inhibitor cocktail. After centrifugation at 4°C and 10,000 × g for
15 min, the OS cell lysates in the supernatant were collected.
Next, biotinylated RNA probes (Bio-miR-330-3p and Bio-NC), along
with Streptavidin magnetic beads (Thermo Fisher Scientific, Inc.),
were incubated overnight at 4°C with the OS cell lysates. This was
performed based on the manufacturer's protocols of the Pierce™
Magnetic RNA Pull-Down kit (Pierce; Thermo Fisher Scientific,
Inc.). Subsequently, the RNA attached to the beads was separated
with washing buffer twice, and the bio-miRNA/mRNA complex was
collected with centrifugation at 5,000 × g for 30 sec to identify
PNN mRNA enrichment using RT-qPCR. The biotinylated RNA probes used
above were designed and synthesized by Guangzhou RiboBio Co.,
Ltd.
Western blotting
Protein samples were extracted from the cells using
RIPA lysis buffer (Beyotime Institute of Biotechnology), and the
protein concentration was measured with a BCA Protein Assay kit
(Bio-Rad Laboratories, Inc.). Protein samples (20 µg) were
separated via 10% SDS-PAGE gel, and subsequently transferred to
PVDF membranes (EMD Millipore) and blocked with 5% non-fat milk for
1 h at room temperature. After blocking, 5% non-fat milk was
removed, and the membranes were incubated with primary antibodies,
including anti-PNN (1:1,000; cat. no. ab244250; Abcam) and
anti-GAPDH (1:1,000; cat. no. ab9485; Abcam), overnight at 4°C. The
following day, the secondary antibody (1:5,000; cat. no. ab6728;
Abcam) was incubated with the membranes at 20-22°C for 1 h. The
protein band was visualized with a chemiluminescent substrate
(Thermo Fisher Scientific, Inc.), and analyzed using ImageJ version
1.46 (National Institutes of Health).
Data analysis
SPSS 22.0 software (IBM Corp.) was employed for
statistical analyses. Three independent experiments were performed,
and the data are presented as the mean ± standard deviation. While
the Student's t-test was used to compare two groups, one-way ANOVA
with Dunnett's or Tukey's post hoc tests was utilized to compare
multiple groups. Pearson's correlation analysis was performed to
determine the correlations between miR-330-3p expression and
circ_0032463 or PNN expression. P<0.05 was considered to
indicate a statistically significant difference.
Results
Identification of
circ_0032463/miR-330-3p/PNN axis in OS
To screen for the key circRNA, the circRNA
expression profile GSE96964 was downloaded from GEO database. With
P<0.05 and logFC >1, seven circRNAs were found to be
upregulated in OS, including hsa_circ_0032463, hsa_circ_0097271,
hsa_circ_0113706, hsa_circ_01113888, hsa_circ_0012726,
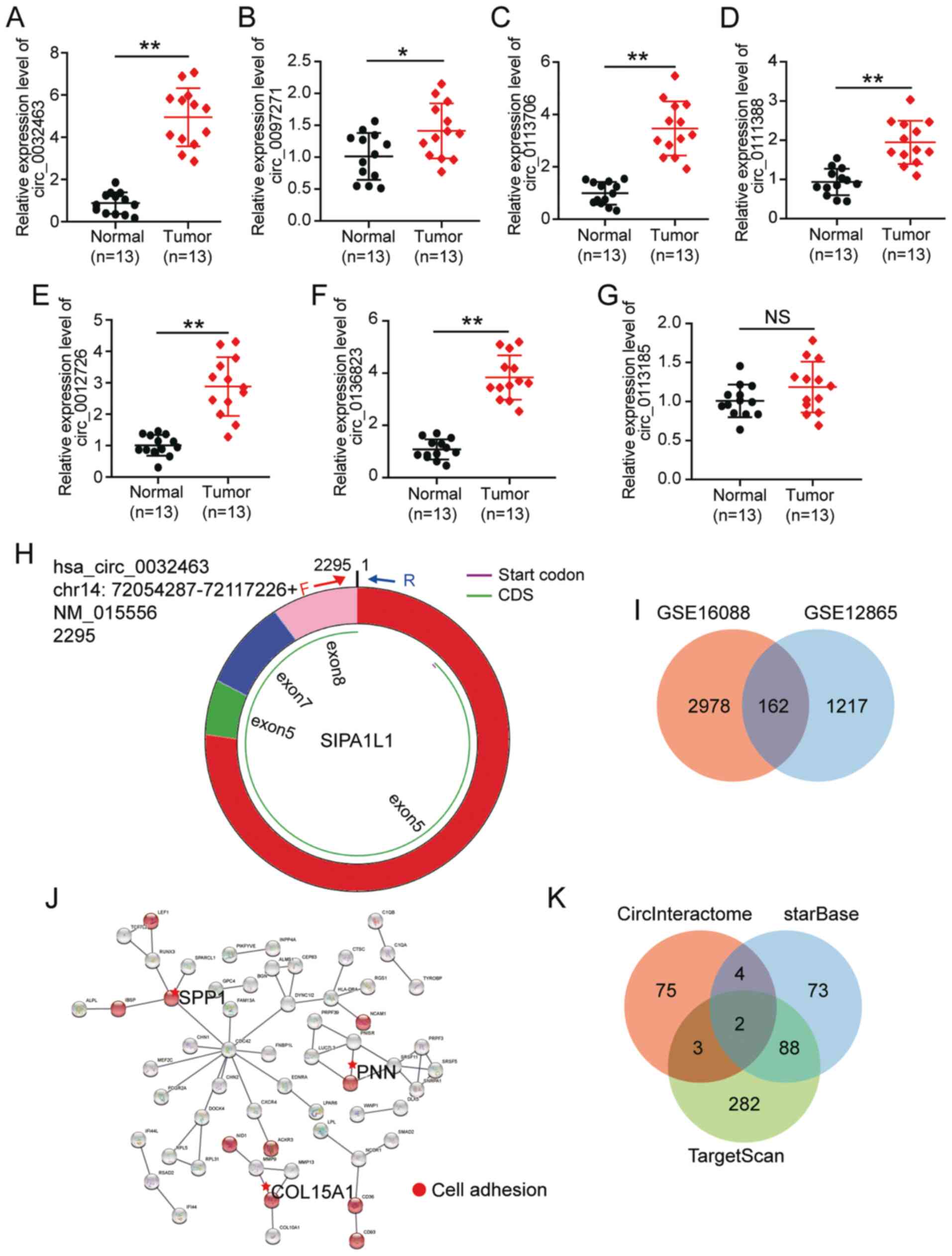
hsa_circ_01368332 and hsa_circ_0113185 (Table III). Then, RT-qPCR analyses
were performed to validate circRNA expression in the 13 tumor
tissues and corresponding normal tissues obtained from
participants. The results showed that hsa_circ_0032463 had the
highest expression in the OS tumor tissues compared with the normal
tissue (Fig. 1A-G). Thus,
hsa_circ_0032463 was chosen as the circRNA of interest. The
structure of circ_0032463 is shown in Fig. 1H. To identify the key gene, two
mRNA expression profiles (GSE16088 and GSE12865) were downloaded
from the GEO database. With P<0.05 and logFC >1, 162
upregulated genes were screened out (Fig. 1I). After uploading 162 genes to
STRING, secreted phosphoprotein, PNN and collagen α-1(XV) chain
were confirmed to be associated with cell adhesion (Fig. 1J). Due to the limited studies on
PNN in OS, PNN was identified as the gene of interest.
CircInteractome was later utilized to predict the miRNAs sponged by
circ_0032463, while starBase and TargetScan were used to predict
the miRNAs that target PNN. Finally, has-miR-330-3p and has-miR-543
were found to be overlapped in each of the database searches
(Fig. 1K). As miR-330-3p has
been identified as a tumor suppressor in OS (33), and has potential roles in the
prognosis of OS (25),
miR-330-3p was chosen as the miRNA of interest.
 | Table IIISeven upregulated circRNAs screened
from GSE96964 dataset with P<0.05 and logFC >1. |
Table III
Seven upregulated circRNAs screened
from GSE96964 dataset with P<0.05 and logFC >1.
| circHome | P-value | LogFC |
|---|
| circ_0032463 | 0.01898124 | 1.6991703 |
| circ_0097271 | 0.00988923 | 1.548159 |
| circ_0113706 | 0.03454026 | 1.3707054 |
| circ_0111388 | 0.03330578 | 1.2384044 |
| circ_0012726 | 0.03208378 | 1.2261671 |
| circ_0136823 | 0.02128171 | 1.1761047 |
| circ_0113185 | 0.04863966 | 1.1112614 |
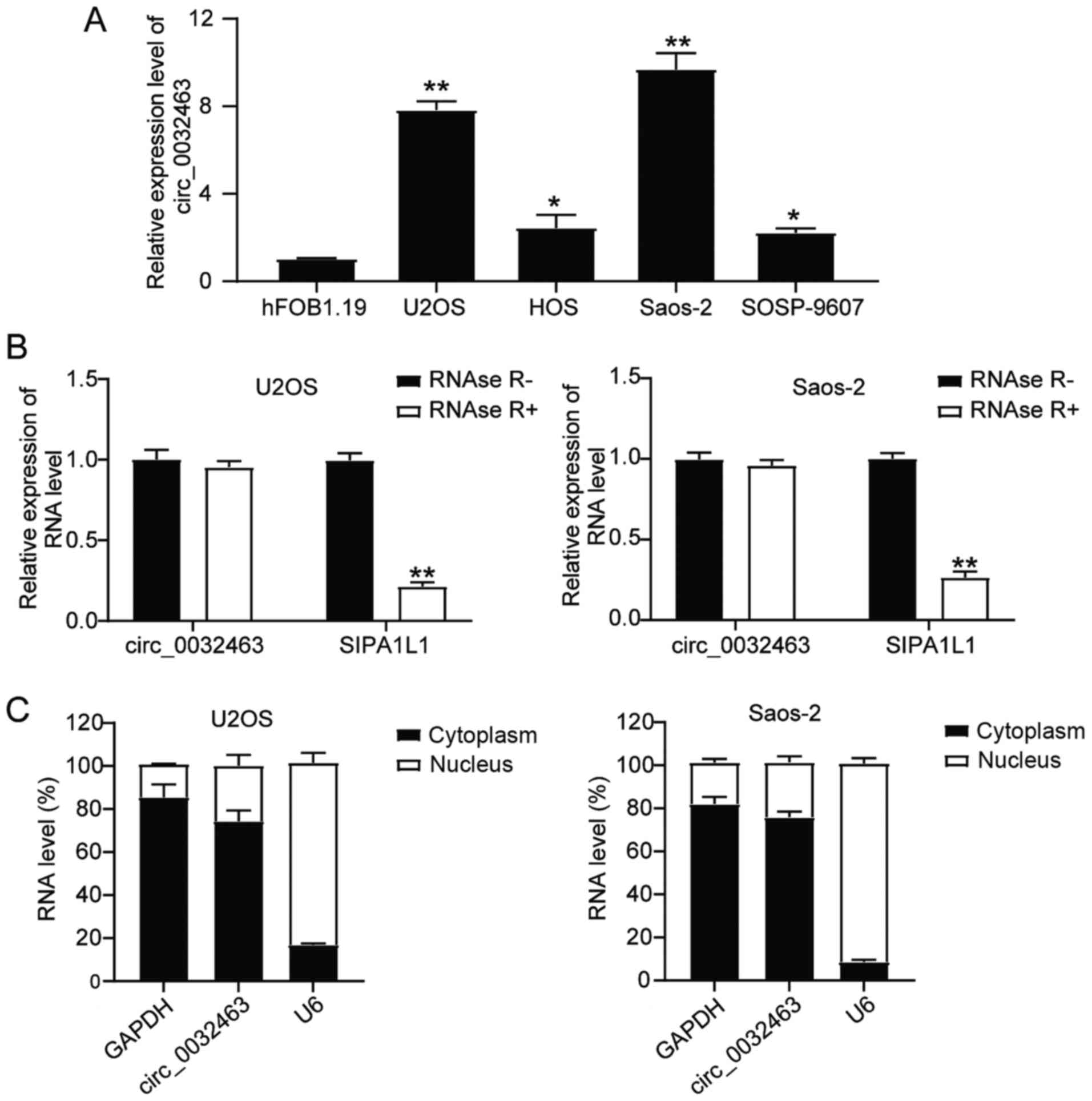
Circ_0032463 is upregulated in OS
The expression of circ_0032463 in the human fetal
osteoblast cell line (hFOB1.19) and OS cell lines (U2OS, HOS,
Saos-2 and SOSP-9607) was measured using RT-qPCR. The enrichment of
circ_0032463 in OS cells was more pronounced than that of hFOB1.19
cells (Fig. 2A). As the
expression of circ_0032463 was higher in Saos-2 and U2OS cells than
that in other OS cells, U2OS and Saos-2 were used in subsequent
experiments. As illustrated in Fig.
2B, the level of the linear form of SIPA1L1 declined
significantly with the exposure to RNase R treatment. However,
circ_0032463 could not be removed with RNase R. After isolating
cytoplasmic and nuclear fractions, circ_0032463 was found to be
enriched in the cytoplasm (Fig.
2C). Overall, the results indicated that circ_0032463 stability
enriched in the cytoplasm was upregulated in OS tissues and
cells.
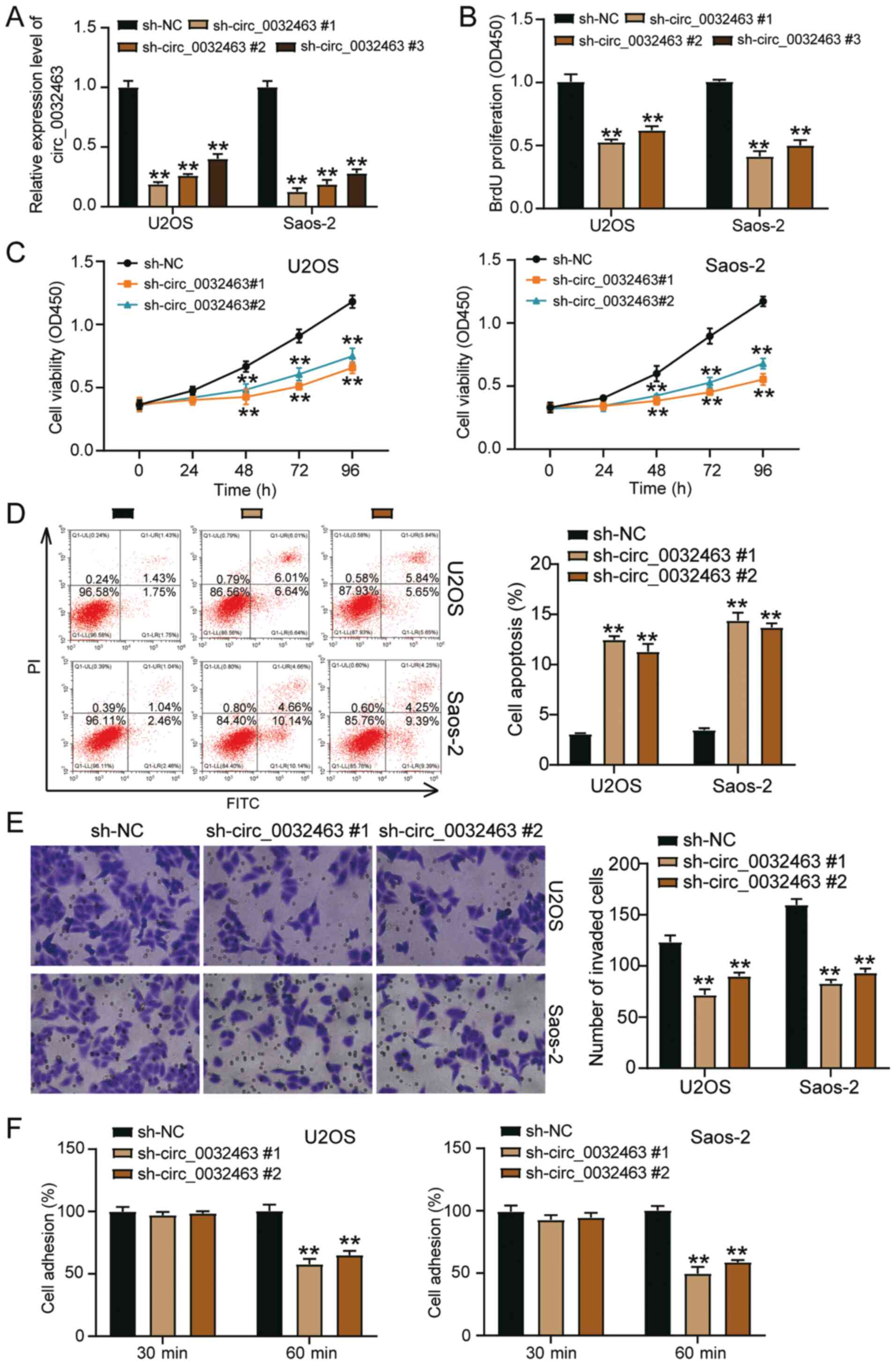
Silencing circ_0032463 suppresses the
progression of OS cells
To explore the function of circ_0032463 in OS cells,
sh-circ_0032463 #1, sh-circ_0032463 #2 and sh-circ_0032463 #3 were
synthesized and transfected into U2OS and Saos-2 cell lines. The
transfection efficiency is shown in Fig. 3A, the suppressive effect of
sh-circ_0032463 #1 and sh-circ_0032463 #2 on circ_0032463
expression was most significant. Hence, they were used in
subsequent experiments. BrdU assay was performed to detect the
proliferative ability of OS cells transfected with sh-circ_0032463
#1 and sh-circ_0032463 #2. As depicted in Fig. 3B, circ_0032463 knockdown
inhibited cell proliferation by 50%. Similar to the outcome of the
BrdU assay, the results of the CCK-8 assay showed that silencing
circ_0032463 suppressed cell viability (Fig. 3C). According to the flow
cytometry results, circ_0032463 knockdown could promote the
apoptosis rate of OS cells (Fig.
3D). The Transwell assay outcomes indicated that silencing
circ_0032463 reduced the number of invaded cells (Fig. 3E). Besides, silencing
circ_0032463 also reduced cell adhesion by 40% compared with the NC
group (Fig. 3F). Overall, these
results confirmed that silencing circ_0032463 could inhibit the
development of OS.
Circ_0032463 is the upstream target of
miR-330-3p
Since miR-330-3p was predicted to be the downstream
target of circ_0032463 by bioinformatics analysis, miR-330-3p
expression in OS was first detected. The results showed that
miR-330-3p expression was lower in OS tissues (Fig. 4A). The result also showed that
miR-330-3p expression was negatively correlated with circ_0032463
expression using Pearson's correlation analysis (Fig. 4B). It was discovered that
miR-330-3p expression in OS cells decreased compared with hFOB1.19
cells (Fig. 4C). CircInteractome
was later employed to predict the two biding sites between
circ_0032463 and miR-330-3p (Fig.
4D). To examine the relationship between miR-330-3p and
circ_0032463, miR-330-3p mimic was constructed and transfected into
U2OS and Saos-2 cells (Fig. 4E).
After performing the dual-luciferase reporter assay, it was found
that the luciferase activity was repressed by ~70% with the
combination of circ_0032463 WT and miR-330-3p mimic, whereas the
luciferase activity in the co-MUT circ_0032463 and miR-330-3p mimic
group remained unchanged compared with the co-MUT circ_0032463 and
NC group (Fig. 4F). Regardless
of whether the binding site (MUT1) or another binding site (MUT2)
was mutated, the luciferase activity decreased slightly (Fig. 4F). The RIP assay data also
indicated that miR-330-3p could bind to circ_0032463 (Fig. 4G). Taken together, the results
confirmed that circ_0032463 could sponge miR-330-3p.
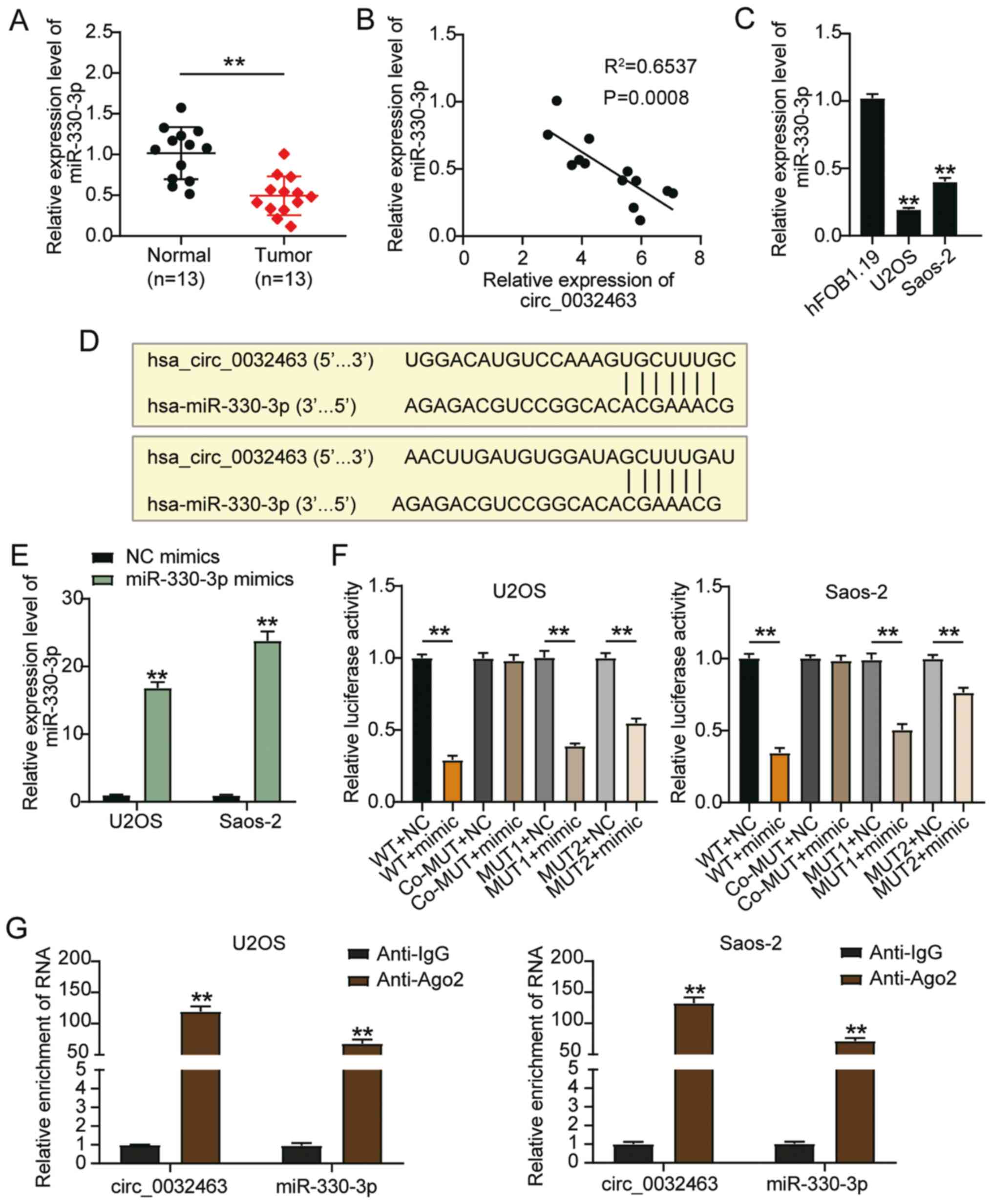 | Figure 4Circ_0032463 may be a sponge for
miR-330-3p in OS progression. (A) miR-330-3p expression in OS
tissues and normal tissues, determined by RT-qPCR.
**P<0.001 vs. normal tissues. (B) The expression of
miR-330-3p had a negative correlation with circ_0032463 expression
in U2OS and Saos-2 cells. (C) miR-330-3p expression in human fetal
osteoblast cell line (hFOB1.19) and OS cell lines (U2OS and
Saos-2), determined by RT-qPCR. **P<0.001 vs.
hFOB1.19 cells. (D) The predicted binding sites between miR-330-3p
and circ_0032463. (E) Transfection efficiency of miR-330-3p mimic
determined by RT-qPCR in U2OS and Saos-2 cells.
**P<0.001 vs. NC mimics. (F) The target relationship
between miR-330-3p and circ_0032463 was verified by dual-luciferase
reporter assay. **P<0.001 vs. NC. (G) Relative
enrichment of circ_0032463 in anti-IgG and anti-Ago2 was detected
by RIP assay. Data are presented as the mean ± SD.
**P<0.001 vs. anti-IgG group. circ, circular RNA;
miR, microRNA; OS, osteosarcoma; NC, negative control; RT-qPCR,
reverse transcription-quantitative PCR; RIP, RNA
Immunoprecipitation Chip; WT, wild-type; MUT, mutant. |
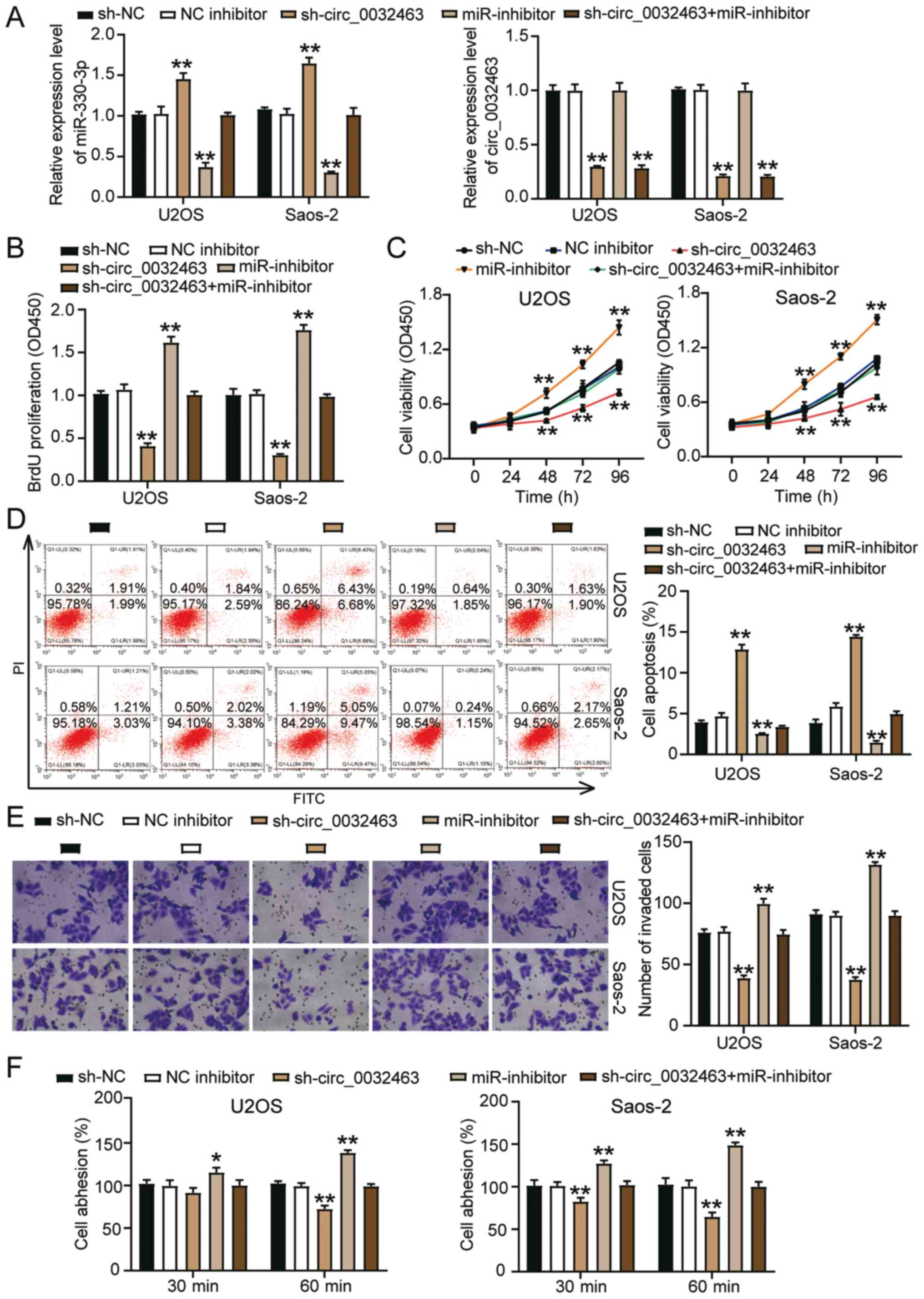
Oncogenic effects of circ_0032463 in OS
cells are relieved by miR-330-3p
To ascertain the interaction between miR-330-3p and
circ_0032463 in OS cells, circ_0032463 shRNA and/or miR-330-3p
inhibitor was introduced into the OS cells. As indicated in
Fig. 5A, sh-circ_0032463
increased miR-330-3p expression by 50%, while the miR-330-3p
inhibitor decreased it by 60%. The expression of miR-330-3p in the
sh-circ_0032463 plus miR-330-3p inhibitor group remained at the
same level compared with the NC group. However, the miR-330-3p
inhibitor could not regulate circ_0032463 expression or reverse the
reduction of circ_0032463 expression that was induced by
sh-circ_0032463 (Fig. 5A). BrdU
assay data showed that the miR-330-3p inhibitor promoted cell
proliferation, and cell proliferation in the sh-circ_0032463 plus
miR-330-3p inhibitor group was similar to that in the NC group
(Fig. 5B). The CCK-8 assay
results demonstrated the miR-330-3p inhibitor promoted the
viability of U2OS and Saos-2 cells (Fig. 5C). Nonetheless, the
co-transfection of sh-circ_0032463 plus miR-330-3p inhibitor could
reverse the promotive effects caused by miR-330-3p inhibitor
(Fig. 5C). The flow cytometry
results indicated that the miR-330-3p inhibitor repressed cell
apoptosis by ~50%; however, these changes could be reversed by
co-transfection with sh-circ_0032463 (Fig. 5D). The miR-330-3p inhibitor
increased the invasive ability of U2OS and Saos-2 cells by 30%, but
this change could be restored by silencing circ_0032463 (Fig. 5E). In addition, the cell adhesion
assay outcomes indicated that circ_0032463 knockdown restricted the
promotion of cell adhesion caused by the miR-330-3p inhibitor
(Fig. 5F). Overall, these
results demonstrated that the oncogenic role of circ_0032463 in OS
cells was exerted via miR-330-3p.
miR-330-3p targets PNN in OS cells
PNN was highly expressed more in OS tissues than in
normal tissues (Fig. 6A).
Besides, PNN expression was negatively correlated to miR-330-3p
expression in OS tissues (Fig.
6B). Similarly, PNN mRNA expression in hFOB1.19 and OS cell
lines measured by RT-qPCR indicated that PNN expression in OS cells
was higher than that in hFOB1.19 cells (Fig. 6C). The binding sites predicted
between miR-330-3p and PNN by TargetScan are illustrated in
Fig. 6D. The results of the
dual-luciferase reporter assay confirmed that the co-transfection
of the miR-330-3p mimic and PNN WT vectors inhibited the luciferase
activity; however, no difference was observed in the PNN MUT group
in both U2OS and Saos-2 cells (Fig.
6E). This relationship was further determined using the RNA
pull-down assay, and experimental analysis showed that PNN could be
pulled down by miR-330-3p (Fig.
6F). Moreover, PNN expression was decreased in U2OS and Saos-2
cells transfected with sh-circ_0032463 compared with the sh-NC
group, and this could be reversed by the miR-330-3p inhibitor
(Fig. 6G). Overall, results
indicated that miR-330-3p could target PNN in OS cells.
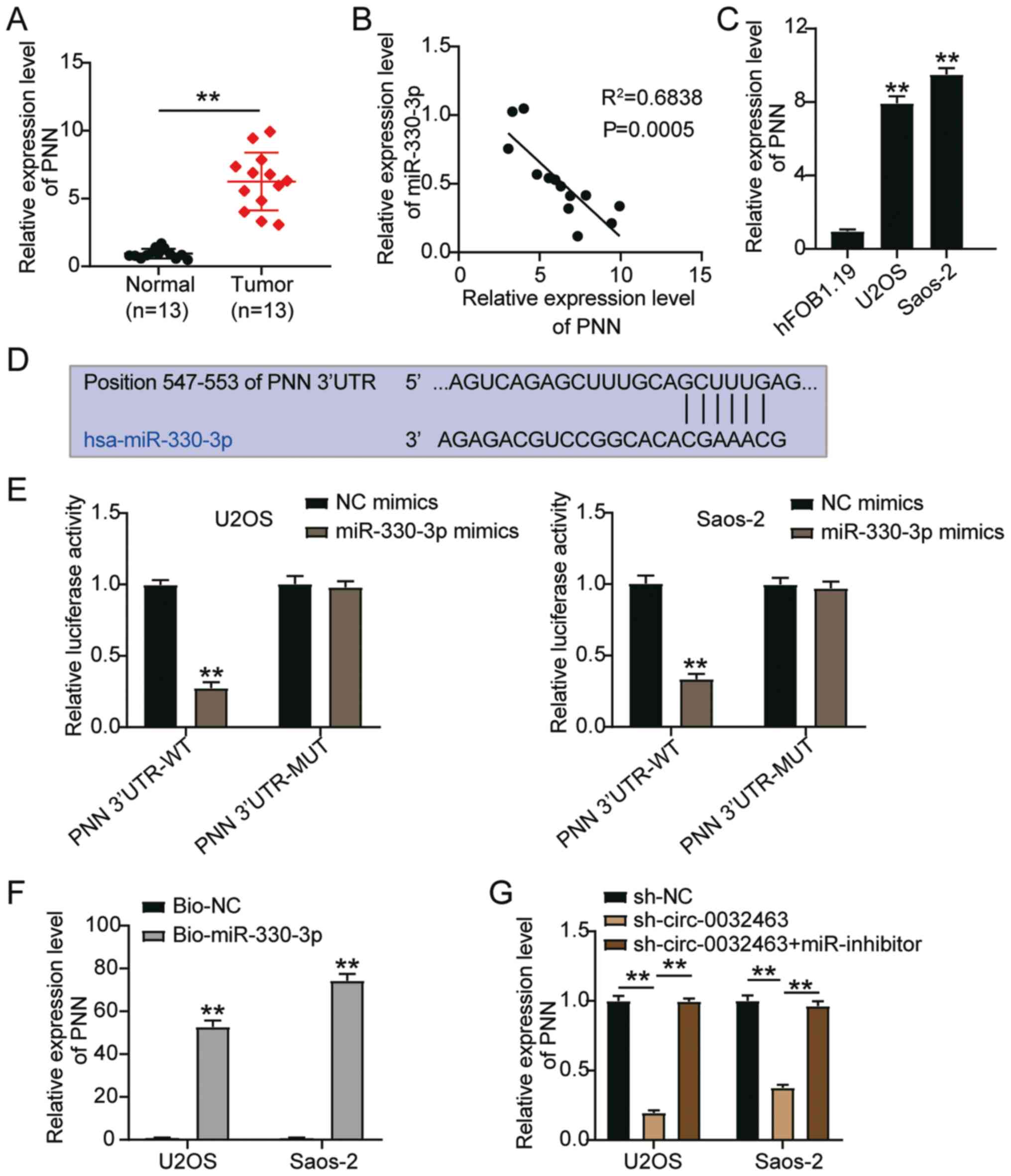 | Figure 6miR-330-3p targets PNN in OS cells.
(A) PNN mRNA expression in OS tissues and normal tissues,
determined by RT-qPCR. **P<0.001 vs. normal tissues.
(B) The correlation analysis between miR-330-3p expression and PNN
expression in OS. (C) PNN mRNA expression in human fetal osteoblast
cell line (hFOB1.19) and OS cell lines (U2OS and Saos-2),
determined by RT-qPCR. **P<0.001 vs. hFOB1.19 cells.
(D) The predicted binding sites between miR-330-3p and PNN. (E) The
target relationship between miR-330-3p and PNN was verified by a
dual-luciferase reporter assay. **P<0.001 vs. NC
mimics. (F) Relative enrichment of PNN in Bio-miR-330-3p and Bio-NC
was detected by a RNA pull-down assay. **P<0.001 vs.
Bio-NC group. (G) Relative expression of PNN in U2OS and Saos-2
cells transfected with sh-NC, sh-circ_0032463 #1 or sh-circ_0032463
#1 + miR-330-3p inhibitor. Data are presented as the mean ± SD.
**P<0.001. circ, circular RNA; miR, microRNA; sh-,
short hairpin RNA; NC, negative control; PNN, Pinin desmosome
associated protein; OS, osteosarcoma; RT-qPCR, reverse
transcription-quantitative PCR; WT, wild-type; MUT, mutant; UTR,
untranslated region. |
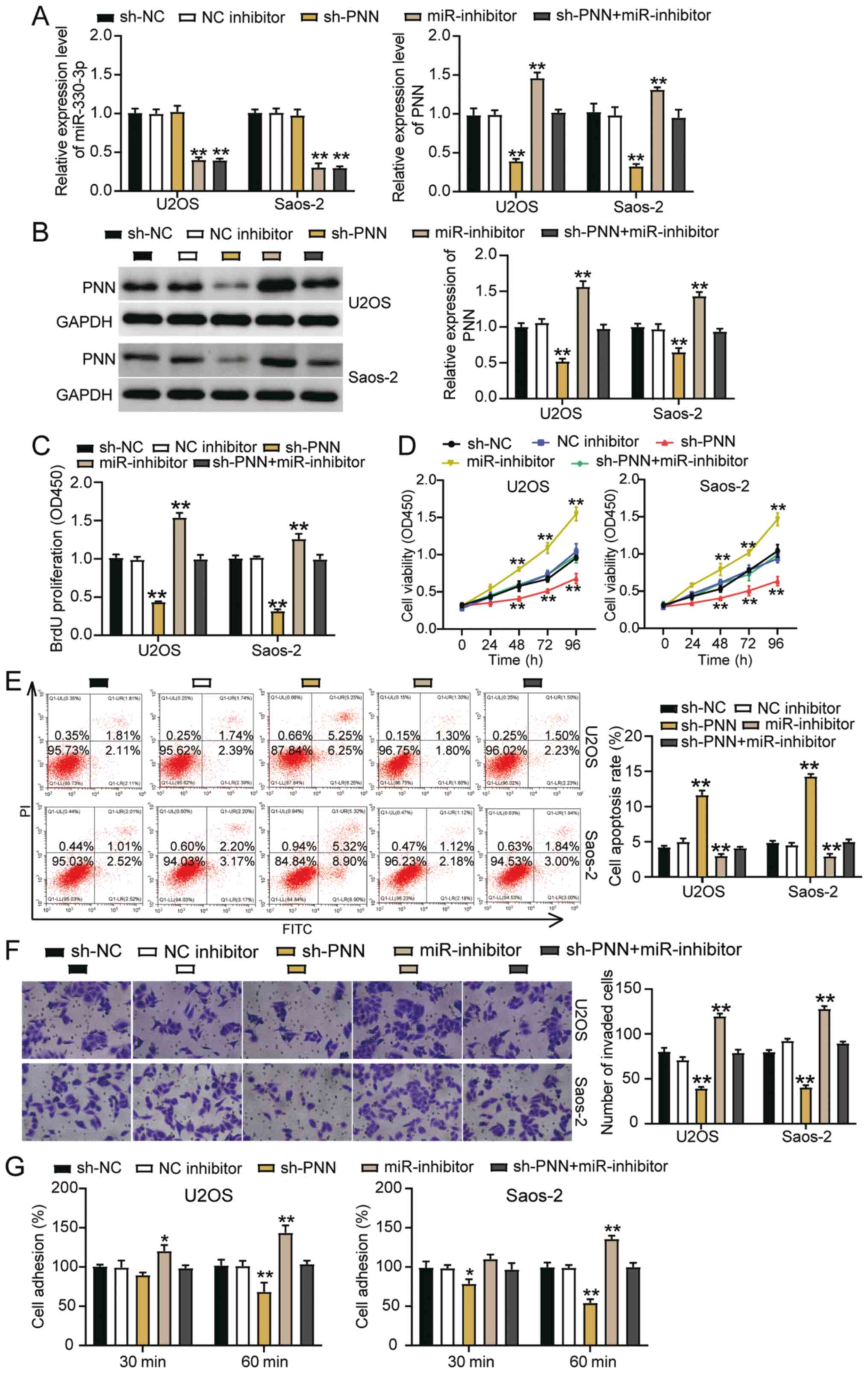
miR-330-3p inhibits OS cell activities by
suppressing PNN
Rescue experiments were carried out to verify
whether miR-330-3p could regulate PNN in SaoS2 and U2OS cells. PNN
knockdown reduced the mRNA expression of PNN by 70% compared with
the sh-NC group, while the miR-330-3p inhibitor increased it by
almost 50% compared with the NC inhibitor group; nonetheless, the
effect could be restored by the co-transfection group (Fig. 7A). Furthermore, sh-PNN did not
affect the expression of miR-330-3p or reverse its downregulation
induced by miR-330-3p inhibitor. Besides, the protein levels of PNN
were similar to the mRNA levels (Fig. 7B). The BrdU assay investigations
revealed that silencing PNN decreased cell proliferation by 50%
compared with the NC group; however, this change could be restored
by co-transfecting the miR-330-3p inhibitor with sh-PNN (Fig. 7C). Similarly, the CCK-8 assay
findings revealed that silencing PNN inhibited cell viability;
however, this effect could be abrogated by co-transfecting the
miR-330-3p inhibitor with sh-PNN (Fig. 7D). The flow cytometry data showed
that sh-PNN could increase the apoptosis of OS cells compared with
the NC group; nonetheless, this facilitation could be reversed by
co-transfection with the miR-330-3p inhibitor (Fig. 7E). Furthermore, >40% reduction
of cell invasion and 40% reduction of cell adhesion induced by
silencing PNN could be abolished by co-transfecting the miR-330-3p
inhibitor with sh-PNN (Fig. 7F and
G). Overall, these outcomes indicated that miR-330-3p could
inhibit the activity of OS cells by suppressing PNN.
Discussion
Adolescents are the most vulnerable groups to OS
(34). CircRNAs have been
reported to be aberrantly expressed in OS and have various roles in
either promoting (35-37) or inhibiting (12,38) the progression of OS
tumorigenesis. The present study discovered a novel circRNA
(circ_0032463) that was upregulated in OS, indicating its potential
role in promoting OS progression. After performing cytological
experiments, circ_0032463 was observed to exert positive effects on
cell proliferation, viability, invasion and adhesion. However, it
inhibited cell apoptosis by acting as a sponge for miR-330-3p,
thereby upregulating PNN in Saos-2 and U2OS cells.
Previous research has indicated the significant role
of circRNAs in various cancer types (39,40). The present study focused on the
role of circRNAs in OS and examined the regulatory mechanism
underlying the development of OS from an epigenetic perspective.
Evidence in the literature has suggested that circRNAs can play
crucial roles in OS development (35,41,42). However, to the best of our
knowledge, the present study is the first to demonstrate the role
of circ_0032463 in OS cells. After bioinformatics analysis in the
present study, it was found that circ_0032463 was upregulated in OS
samples, suggesting that circ_0032463 had a promotive effect in OS.
Further experimental investigations showed that while the knockdown
of circ_0032463 could repress cell proliferation, viability,
invasion and adhesion, it could induce cell apoptosis of OS cells.
This result suggested that silencing circ_0032463 could suppress OS
progression.
Furthermore, circRNAs are often investigated as the
key member of competing endogenous (ce)RNA networks, known as the
'circRNA-miRNA-mRNA axis' (12).
The 'circRNA-miRNA-mRNA axis' theory speculates that circRNAs
mainly sponge miRNAs to regulate the downstream target genes of
miRNAs (43). With this in mind,
the present study examined how circ_0032463 could influence the
development of OS through a miRNA-mediated mechanism. After
performing bioinformatics analysis and cytological experiments, it
was found that miR-330-3p was the downstream miRNA sponged by
circ_0032463 in OS cells. Several studies have documented the
suppressive abilities of miR-330-3p in several malignancies,
including gastric cancer, liver cancer and melanoma (23,24,44). At present, only one previous
study has reported on the role of miR-330-3p in OS, which
demonstrated that this miRNA was downregulated and played an
antitumor role by targeting Bmi-1 (25). The present findings were
consistent with the results of this previous study, as we showed
that miR-330-3p suppressed OS progression. However, the inhibitory
effect of miR-330-3p on OS could be restored by circ_0032463. The
present study also revealed that PNN was the target of miR-330-3p
in OS, which was different from the outcomes of previous
studies.
PNN, a desmosome-associated molecule, has been
demonstrated to be overexpressed and associated with the poor
prognosis of numerous types of cancer (27-29). For example, Tang et al
(45) revealed that PNN
contributed to the epithelial-mesenchymal transition in
nasopharyngeal carcinoma by regulating the activity of E-cadherin.
The present study speculated that PNN might act as a tumor
suppressor in OS based on the information obtained in the
literature (45,46). After bioinformatics analyses and
in vitro experiments were performed, the present study
discovered that PNN was upregulated in OS. It was also found to
induce the proliferation, invasion and adhesion of OS cells.
Besides, these results indicated that the enhancing effect of PNN
in OS could be restored by miR-330-3p.
Although the role of the circ_0032463/miR-330-3p/PNN
axis in OS was illustrated, the present study has a few
limitations. First, only in vitro experiments were
performed. For this reason, in vivo experiments should be
conducted in the future to confirm the validity of these results.
Moreover, PNN has been found to be associated with cell adhesion
(47). Also, a previous study
demonstrated that the knockdown of PNN repressed E-cadherin and
DSG2, which in turn reduced epithelial adhesion (48). This could indicate that the
interaction of PNN with other relevant factors, such as E-cadherin
and DSG2, could affect OS cell adhesion. This area of research
should be explored in the future.
In conclusion, the present research demonstrated
that by sponging miR-330-3p to upregulate PNN, circ_0032463 could
act as a tumor promoter in OS cells. This study also confirmed the
role of the circ_0032463/miR-330-3p/PNN axis in OS development.
This knowledge could be useful in the diagnosis, prognosis and
treatment of OS.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
GQ performed the experiments and data analysis. XW
conceived and designed the study, and wrote the paper. GQ reviewed
and edited the manuscript. GQ and XW confirm the authenticity of
all the raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The First Affiliated Hospital of Zhengzhou University
(Zhengzhou, China). Informed consent was obtained from the patients
before they could participate in the experiment.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
No funding was received.
References
|
1
|
Rickel K, Fang F and Tao J: Molecular
genetics of osteosarcoma. Bone. 102:69–79. 2017. View Article : Google Scholar :
|
|
2
|
Savage SA and Mirabello L: Using
epidemiology and genomics to understand osteosarcoma etiology.
Sarcoma. 2011:5481512011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Isakoff MS, Bielack SS, Meltzer P and
Gorlick R: Osteosarcoma: Current treatment and a collaborative
pathway to success. J Clin Oncol. 33:3029–3035. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Otoukesh B, Boddouhi B, Moghtadaei M,
Kaghazian P and Kaghazian M: Novel molecular insights and new
therapeutic strategies in osteosarcoma. Cancer Cell Int.
18:1582018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bishop MW, Janeway KA and Gorlick R:
Future directions in the treatment of osteosarcoma. Curr Opin
Pediatr. 28:26–33. 2016. View Article : Google Scholar :
|
|
6
|
Kristensen LS, Andersen MS, Stagsted LVW,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nature Rev Genet. 20:675–691.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kristensen LS, Hansen TB, Venø MT and
Kjems J: Circular RNAs in cancer: opportunities and challenges in
the field. Oncogene. 37:555–565. 2018. View Article : Google Scholar :
|
|
9
|
Wang C, Ren M, Zhao X, Wang A and Wang J:
Emerging roles of circular RNAs in osteosarcoma. Med Sci Monit.
24:7043–7050. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang Y and Li J, Wang Y, Jing J and Li J:
The roles of circular RNAs in osteosarcoma. Med Sci Monit.
25:6378–6382. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li H, Lan M, Liao X, Tang Z and Yang C:
Circular RNA cir-ITCH promotes osteosarcoma migration and invasion
through cir-ITCH/miR-7/EGFR pathway. Technol Cancer Res Treat. Jan
21–2020.Epub ahead of print. View Article : Google Scholar
|
|
12
|
Chen L, Shan Y, Zhang H, Wang H and Chen
Y: Up-regulation of Hsa_circ_0008792 inhibits osteosarcoma cell
invasion and migration and promotes apoptosis by regulating
Hsa-miR-711/ZFP1. OncoTargets Ther. 13:2173–2181. 2020. View Article : Google Scholar
|
|
13
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liang M, Huang G, Liu Z, Wang Q, Yu Z, Liu
Z, Lin H, Li M, Zhou X and Zheng Y: Elevated levels of
hsa_circ_006100 in gastric cancer promote cell growth and
metastasis via miR-195/GPRC5A signalling. Cell Prolif.
52:e126612019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rong X, Gao W, Yang X and Guo J:
Downregulation of hsa_circ_0007534 restricts the proliferation and
invasion of cervical cancer through regulating miR-498/BMI-1
signaling. Life Sci. 235:1167852019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yao J, Zhang C, Chen Y and Gao S:
Downregulation of circular RNA circ-LDLRAD3 suppresses pancreatic
cancer progression through miR-137-3p/PTN axis. Life Sci.
239:1168712019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang CL, Li LB, She C, Xie Y, Ge DW and
Dong QR: MiR-299-5p targets cell cycle to promote cell
proliferation and progression of osteosarcoma. Eur Rev Med
Pharmacol Sci. 22:2606–2613. 2018.PubMed/NCBI
|
|
18
|
Lv DB, Zhang JY, Gao K, Yu ZH, Sheng WC,
Yang G and Gao YZ: MicroRNA-765 targets MTUS1 to promote the
progression of osteosarcoma via mediating ERK/EMT pathway. Eur Rev
Med Pharmacol Sci. 23:4618–4628. 2019.PubMed/NCBI
|
|
19
|
Loughlin V and Beniwal JS: Post-traumatic
brachial artery aneurysm and arteriovenous fistulae. J Cardiovasc
Surg (Torino). 29:570–571. 1988.
|
|
20
|
Liu G, Huang K, Jie Z, Wu Y, Chen J, Chen
Z, Fang X and Shen S: CircFAT1 sponges miR-375 to promote the
expression of Yes-associated protein 1 in osteosarcoma cells. Mol
Cancer. 17:1702018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu R, Feng F, Yu X, Liu Z and Lao L:
LncRNA SNHG4 promotes tumour growth by sponging miR-224-3p and
predicts poor survival and recurrence in human osteosarcoma. Cell
Proliferation. 51:e125152018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
He M, Shen P, Qiu C and Wang J: miR-627-3p
inhibits osteosarcoma cell proliferation and metastasis by
targeting PTN. Aging. 11:5744–5756. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jin Z, Jia B, Tan L and Liu Y: miR-330-3p
suppresses liver cancer cell migration by targeting MAP2K1. Oncol
Lett. 18:314–320. 2019.PubMed/NCBI
|
|
24
|
Guan A, Wang H, Li X, Xie H, Wang R, Zhu Y
and Li R: MiR-330-3p inhibits gastric cancer progression through
targeting MSI1. Am J Transl Res. 8:4802–4811. 2016.PubMed/NCBI
|
|
25
|
Zheng Z, Bao F, Chen X, Huang H and Zhang
X: MicroRNA-330-3p expression indicates good prognosis and
suppresses cell proliferation by targeting bmi-1 in osteosarcoma.
Cell Physiol Biochem. 46:442–450. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mini E, Lapucci A, Perrone G, D'Aurizio R,
Napoli C, Brugia M, Landini I, Tassi R, Picariello L, Simi L, et
al: RNA sequencing reveals PNN and KCNQ1OT1 as predictive
biomarkers of clinical outcome in stage III colorectal cancer
patients treated with adjuvant chemotherapy. Int J Cancer.
145:2580–2593. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang Y, Kwok JS, Choi PW, Liu M, Yang J,
Singh J, Ng SK, Welch WR, Muto MG, Tsui SK, et al: Pinin interacts
with C-terminal binding proteins for RNA alternative splicing and
epithelial cell identity of human ovarian cancer cells. Oncotarget.
7:11397–11411. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wei Z, Ma W, Qi X, Zhu X, Wang Y, Xu Z,
Luo J, Wang D, Guo W, Li X, et al: Pinin facilitated proliferation
and metastasis of colorectal cancer through activating EGFR/ERK
signaling pathway. Oncotarget. 7:29429–29439. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang X, Sun D, Dong C, Tian Y, Gao Z and
Wang L: Pinin associates with prognosis of hepatocellular carcinoma
through promoting cell proliferation and suppressing glucose
deprivation-induced apoptosis. Oncotarget. 7:39694–39704. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Paoloni M, Davis S, Lana S, Withrow S,
Sangiorgi L, Picci P, Hewitt S, Triche T, Meltzer P and Khanna C:
Canine tumor cross-species genomics uncovers targets linked to
osteosarcoma progression. BMC Genomics. 10:6252009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sadikovic B, Yoshimoto M, Chilton-MacNeill
S, Thorner P, Squire JA and Zielenska M: Identification of
interactive networks of gene expression associated with
osteosarcoma oncogenesis by integrated molecular profiling. Human
Mol Genet. 18:1962–1975. 2009. View Article : Google Scholar
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
33
|
Wang H, Liu L and Fang S: MicroRNA-330-5p
inhibits osteosarcoma cell growth and invasion by targeting the
proto-oncogene survivin. Mol Med Rep. 20:2236–2244. 2019.PubMed/NCBI
|
|
34
|
Caudill JS and Arndt CA: Diagnosis and
management of bone malignancy in adolescence. Adolesc Med State Art
Rev. 18:62–78. ix2007.
|
|
35
|
Zhang H, Yan J, Lang X and Zhuang Y:
Expression of circ_001569 is upregulated in osteosarcoma and
promotes cell proliferation and cisplatin resistance by activating
the Wnt/β-catenin signaling pathway. Oncol Lett. 16:5856–5862.
2018.PubMed/NCBI
|
|
36
|
Zhang Z, Pu F, Wang B, Wu Q, Liu J and
Shao Z: Hsa_ circ_0000285 functions as a competitive endogenous RNA
to promote osteosarcoma progression by sponging hsa-miRNA-599. Gene
therapy. 27:186–195. 2020. View Article : Google Scholar
|
|
37
|
Liu X, Zhong Y, Li J and Shan A: Circular
RNA circ-NT5C2 acts as an oncogene in osteosarcoma proliferation
and metastasis through targeting miR-448. Oncotarget.
8:114829–114838. 2017. View Article : Google Scholar
|
|
38
|
Wu Z, Shi W and Jiang C: Overexpressing
circular RNA hsa_circ_0002052 impairs osteosarcoma progression via
inhibiting Wnt/β-catenin pathway by regulating miR-1205/APC2 axis.
Biochem Biophys Res Commun. 502:465–471. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Qian L, Yu S, Chen Z, Meng Z, Huang S and
Wang P: The emerging role of circRNAs and their clinical
significance in human cancers. Biochimica Biophys Acta Rev Cancer.
1870:247–260. 2018. View Article : Google Scholar
|
|
40
|
Chen BJ, Byrne FL, Takenaka K, Modesitt
SC, Olzomer EM, Mills JD, Farrell R, Hoehn KL and Janitz M:
Analysis of the circular RNA transcriptome in endometrial cancer.
Oncotarget. 9:5786–5796. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Nie WB, Zhao LM, Guo R, Wang MX and Ye FG:
Circular RNA circ-NT5C2 acts as a potential novel biomarker for
prognosis of osteosarcoma. Eur Rev Med Pharmacol Sci. 22:6239–6244.
2018.PubMed/NCBI
|
|
42
|
Wang L, Du ZG, Huang H, Li FS, Li GS and
Xu SN: Circ-0003998 promotes cell proliferative ability and
invasiveness by binding to miR-197-3p in osteosarcoma. Eur Rev Med
Pharmacol Sci. 23:10638–10646. 2019.PubMed/NCBI
|
|
43
|
Dong W, Bi J, Liu H, Yan D, He Q, Zhou Q,
Wang Q, Xie R, Su Y, Yang M, et al: Circular RNA ACVR2A suppresses
bladder cancer cells proliferation and metastasis through
miR-626/EYA4 axis. Mol Cancer. 18:952019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yao Y, Zuo J and Wei Y: Targeting of TRX2
by miR-330-3p in melanoma inhibits proliferation. Biomed
Pharmacother. 107:1020–1029. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tang T, Yang L, Cao Y, Wang M, Zhang S,
Gong Z, Xiong F, He Y, Zhou Y, Liao Q, et al: LncRNA AATBC
regulates Pinin to promote metastasis in nasopharyngeal carcinoma.
Mol Oncol. 14:2251–2270. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wei Z, Ma W, Qi X, Zhu X, Wang Y, Xu Z,
Luo J, Wang D, Guo W, Li X, et al: Pinin facilitated proliferation
and metastasis of colorectal cancer through activating EGFR/ERK
signaling pathway. Oncotarget. 7:29429–29439. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Alpatov R, Munguba GC, Caton P, Joo JH,
Shi Y, Shi Y, Hunt ME and Sugrue SP: Nuclear speckle-associated
protein Pnn/DRS binds to the transcriptional corepressor CtBP and
relieves CtBP-mediated repression of the E-cadherin gene. Mol Cell
Biol. 24:10223–10235. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Joo JH, Alpatov R, Munguba GC, Jackson MR,
Hunt ME and Sugrue SP: Reduction of Pnn by RNAi induces loss of
cell-cell adhesion between human corneal epithelial cells. Mol
Vision. 11:133–142. 2005.
|
|
49
|
Ritter J and Bielack SS: Osteosarcoma. Ann
Oncol. 21(Suppl 7): vii320–vii325. 2010. View Article : Google Scholar : PubMed/NCBI
|