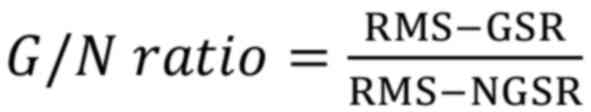Introduction
Tinnitus, a condition that affects 7-25% of the
population worldwide, is a phenomenon in which sound is perceived
in the absence of sound stimuli (1-4).
In 1-2% of the total population, tinnitus may be associated with
debilitating conditions, including insomnia, anxiety, depression,
cognitive dysfunction and stress (1-6).
Although various approaches, such as medical therapy, dietary
supplements, transcranial magnetic/electrical stimulation and sound
therapy, have been devised for the treatment of tinnitus, no
treatment to date has been reported to produce a clear therapeutic
effect (7,8). Therefore, it is crucial to develop
new treatment methods for this condition.
A lack of clarity on the mechanisms underlying the
development of tinnitus makes it difficult to design an effective
treatment strategy. Therefore, the elucidation of the underlying
mechanisms will contribute significantly towards identifying a cure
for tinnitus. Maladaptive auditory-somatosensory plasticity in the
dorsal cochlear nucleus (DCN) after hearing loss has been suggested
as one of the mechanisms promoting the development of tinnitus
(9-11). Using a temporary threshold shift
(TTS) model, our previous study demonstrated that the changes
occurring in the DCN following exposure to noise may play an
important role in the development of tinnitus and a decrease in
auditory projections and subsequent increase in non-auditory
projections via axonal sprouting may be important phenomena
associated with the occurrence of tinnitus (12). Therefore, the identification of
an appropriate checkpoint for these processes may facilitate the
development of a treatment strategy for this condition.
MicroRNAs (miRNAs/miRs) are small non-coding RNAs
that regulate gene expression via translational inhibition or mRNA
degradation (13). As the
expression of most genes is under miRNA control, the majority of
biological processes are regulated by miRNAs to a certain extent.
Therefore, disease development is also hypothesized to be closely
associated with the activities of miRNAs. Indeed, the pathogenesis
of diabetes mellitus, cancer, and cardiovascular and neurological
diseases has been reported to be associated with various miRNAs,
whereas certain miRNAs were recently recognized as therapeutic
targets (14-16). In particular, therapeutic
strategies targeting miRNAs have achieved promising results against
hepatitis C viral infection (16).
Accordingly, the present study was undertaken to
identify miRNAs that may be implicated in the pathogenesis of
tinnitus. miRNA levels were compared in animal models with and
without tinnitus following induction of TTS using microarray
analysis and reverse transcription-quantitative PCR (RT-qPCR).
Additionally, the candidate miRNAs were overexpressed to examine
the differences in the expression of their candidate targets.
Specifically, the expression levels of miR-375-3p in the DCNs of
animals with tinnitus were measured, and the role of miR-375-3p in
tinnitus and the involvement of connective tissue growth factor
(CTG.) were investigated, in the hope that the
identification and targeting of putative miRNAs involved in the
pathogenesis of tinnitus may contribute to the development of novel
approaches to the treatment of this condition.
Materials and methods
Animals
The present study was approved by the Institutional
Animal Care and Use Committee of Chung-Ang University (2016-00092).
All experiments were performed in accordance with the National
Institutes of Health Guide for the Care and Use of Laboratory
Animals (17). All animals were
allowed to acclimate to the laboratory conditions for 1 week prior
to the start of the experiments. The animals were housed in a
temperature- and humidity-controlled room with a 12-h light/dark
cycle, with food and water available ad libitu.. The
experiments were conducted on 12-week-old male Sprague-Dawley rats.
The auditory brainstem response (ABR) recordings, noise exposure
and surgical procedures were performed under anesthesia induced by
intraperitoneal administration of Zoletil (40 mg/kg, Zoletil
50®; Virbac) mixed with xylazine (10 mg/kg,
Rompun®; Bayer-Korea, Ltd.).
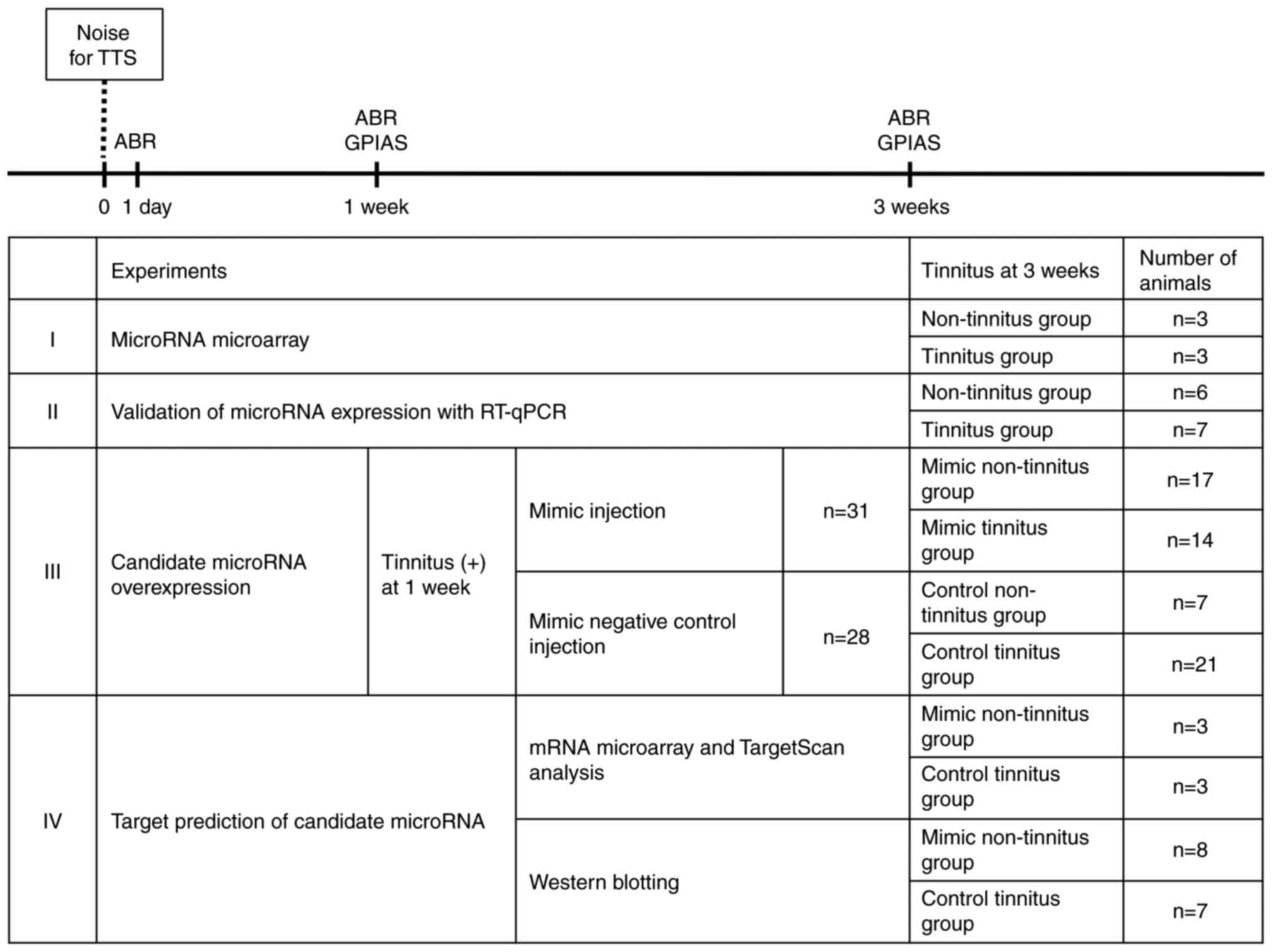
Experimental design
A total of 102 rats were used in the present study.
The study was conducted in four stages (Fig. 1) as follows: i) Identification of
candidate miRNAs involved in the development of tinnitus, using
microarray analysis; ii) validation of miRNA expression using
RT-qPCR; iii) evaluation of the effects of overexpression of the
candidate miRNAs on tinnitus; and iv) target prediction of the
candidate miRNAs. In each stage, the ABR and gap pre-pulse
inhibition of acoustic startle reflex (GPIAS) were recorded in all
animals prior to noise exposure, to confirm that none of the
animals had hearing loss or tinnitus. All rats were exposed to 6-8
kHz narrow-band noise at 110 dB sound pressure level (SPL) for 2 h,
with the left ear plugged and sutured. This noise-exposure protocol
was developed in our previous study and confirmed to induce a TTS
on the right side only (12). To
detect the hearing changes, the ABR was recorded on day 1, and at 1
and 3 weeks post-noise exposure, whereas GPIAS responses were
recorded at 1 and 3 weeks following exposure. The development of
tinnitus was determined based on the recorded GPIAS responses.
A total of 8 rats were used in the first stage of
the experiment. At 3 weeks post-noise exposure, a total of 5 rats
exhibited evidence of tinnitus. Subsequently, 3 rats were randomly
selected each from the tinnitus (n=5) and non-tinnitus (n=3)
groups. The right DCNs were harvested from these rats, following
the protocol outlined in the atlas of Paxinos and Watson (18). The samples were subjected to
miRNA microarray analysis. Based on the results of microarray
analysis, candidate miRNAs were selected. For the second stage of
the experiment, 13 rats were used. The right DCNs were harvested
from these rats at 3 weeks following noise exposure. It was
observed that 7 rats exhibited evidence of tinnitus at this stage.
To validate the candidate miRNAs, RT-qPCR was performed on these
samples. The results were analyzed and the miRNAs that exhibited
significant differences between the tinnitus and non-tinnitus
groups were identified. In the third stage of the experiment, 81
rats were exposed to noise, of which 59 displayed evidence of
tinnitus at 1 week post-exposure. To evaluate the role of the
candidate miRNAs selected with RT-qPCR and miRNA microarray
analyses, a candidate miRNA oligomer (mimic or mimic negative
control) was administered into the lateral ventricles of the 59
rats that manifested evidence of tinnitus. At 3 weeks following
noise exposure, whether the tinnitus persisted was determined using
the GPIAS recordings. In the next stage of the experiment, rats
receiving the candidate miRNA oligomer were divided into the
following four experimental groups based on the type of oligomer
administered and the persistence of tinnitus at 3 weeks following
noise exposure: i) Mimic tinnitus, ii) mimic non-tinnitus, iii)
control tinnitus and iv) control non-tinnitus groups. In the last
stage of the experiment, target prediction analysis for the
candidate miRNAs was performed. Three rats were randomly selected
from each of the mimic non-tinnitus and control tinnitus groups.
mRNA microarray analysis was performed using the three DCN samples
obtained from each group. Based on the results of microarray
analysis and the analysis performed using the TargetScan miRNA
target prediction server (http://www.targetscan.org/cgi-bin/targetscan/vert_72/targetscan.cgi?species=Rat&gid=&mir_sc=miR-375-3p&mir_c=&mir_nc=&sortType=cs&allTxs=&incl_nc=All),
all the potential target genes of the candidate miRNA were
identified. Subsequently, the expression levels of these candidate
target genes were compared between the two experimental groups
using western blotting.
ABR recordings
ABR recordings were performed as described
previously (19). ABR was
measured using SmartEP (version 2.33; Intelligent Hearing Systems)
and high-frequency transducers (HFT9911-20-0035). The ABR signals
detected between the subcutaneous electrodes at the nape of the
neck and the ipsilateral mastoid process were recorded using the
contralateral mastoid process as the return. Tone-pip stimuli of 7,
11 and 15 kHz (duration, 5 msec; cos shaping, 21 Hz) were delivered
in decreasing steps of 5 dB SPL. The responses were band
pass-filtered (100-1,500 Hz), amplified (×100,000) and averaged
over 512 stimuli repetitions at each frequency and sound level. The
lowest stimulus intensity that induced a detectable response was
considered as the threshold, and it was assessed by two
researchers.
Behavioral test for tinnitus
GPIAS recordings were performed as described
previously (12). The recording
system used in the present study consisted of a mesh cage with an
accelerometer (LIS344ALH; STMicroelectronics), an audio amplifier
(PM-5004; Marantz), a full-range loud speaker (TC9FSD13;
Vifa/Peerless, Tymphany), a reference microphone, data acquisition
hardware (NI DAQ-6341; National Instruments Corporation), a
custom-made anechoic noise box, and the LabVIEW (version 2015;
National Instrument)-based custom GUI 3.0 software. This
LabVIEW-based software was used for acoustic stimulation, startle
response acquisition and response analyses.
Acoustic stimulation was carried out using sound
waves of 2 kHz bandwidth and 60 dB SPL; the center frequencies of
7, 11 and 15 kHz were used as the background noise. A broadband
noise burst of 105 dB SPL for a duration of 50 msec served as the
startle stimulus. During each session, 15 gap-conditioned stimuli
and 15 non-gap-conditioned stimuli were presented in a random pair
order. The gap pre-pulse that occurred in each gap-conditioned
stimulus was presented 100 msec before the onset of the startle
stimulus and lasted for 50 msec. The time interval between the
presentation of acoustic stimulations was altered randomly between
17 and 23 sec. The gap-conditioned response/non-gap-conditioned
response (G/N) ratios were calculated according to the following
equation:
where RMS-GSR and RMS-NGSR are the root-mean-squared (RMS) values
of the gap-conditioned startle responses (GSR) and the non-GSR
(NGSR), respectively. The outliers among the measured startle
responses were removed using Grubb's test (
20). The GSR and the NGSR were compared
using the Mann-Whitney U test. The animals were considered to have
no tinnitus if there were significant differences at all
frequencies (P<0.05); otherwise, the rats were considered to
have tinnitus.
miRNA microarray analysis
Microarray analysis of the miRNAs was performed at
BioCore Co., Ltd. using the Affymetrix miRNA 4.0 microarray
(Affymetrix; Thermo Fisher Scientific, Inc.), which contained all
the miRNAs in the miRBase Release 20 database (http://www.mirbase.org/), including 30,434 mature
miRNA probe sets. RNA was prepared as described previously
(21). Briefly, total RNA was
extracted from the DCN samples using the TRI Reagent®
(MRC). The quality and quantity of the RNA were assessed using an
Agilent 2100 BioAnalyzer (Agilent Technologies, Inc.) and a
NanoDrop™ 1000 Spectrophotometer (Thermo Fisher Scientific, Inc.),
respectively. The RNA was labeled using the FlashTag Biotin RNA
Labeling kit (Affymetrix; Thermo Fisher Scientific, Inc.), and the
labeled samples were hybridized to GeneChip miRNA 4.0 microarrays
(Affymetrix; Thermo Fisher Scientific, Inc.). A hybridization
mixture consisting of control oligo B2, 20X hybridization controls
(bioB, bioC, bioD and cre), 27.5% formamide, DMSO, 2X hybridization
buffer and water, was applied to all the samples. Hybridization was
performed in an Affymetrix GeneChip Hybridization Oven 640 at 48°C
and 60 rpm for 16 h. Subsequently, the arrays were stained with
stain cocktails (1 and 2) included in the kit, and washed in
Affymetrix GeneChip Fluidics Station 450, in accordance with the
FS450_0002 fluidics protocol. Following scanning using an
Affymetrix GeneChip Scanner 3000, all the arrays were analyzed
using the Transcriptome Analysis Console™ 4.0 software (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The CEL files
generated were imported into the Gene Expression Workflow in
GeneSpring GX, version 14.9.1 (Agilent Technologies, Inc.).
Background correction, log2 transformation and probe set
summarizing were achieved using the default settings in the
GeneSpring software. Subsequently, principal component analysis was
performed using a covariance dispersion matrix for data quality
control. Unpaired t-tests were performed to compare the individual
gene expression data of the noise-exposed tinnitus group with that
of the non-tinnitus group.
RT-qPCR
Total RNA was extracted from the DCN samples using
the QIAzol Lysis Reagent (Qiagen GmbH) according to the
manufacturer's instructions. The RNA concentration of the samples
was determined using a NanoDrop™ spectrophotometer (Thermo Fisher
Scientific, Inc.). cDNA synthesis was performed using the miScript
II RT kit (Qiagen GmbH), and qPCR was performed with primers for
miR-15b-3p (cat. no. YP00205898), -105 (cat. no. YP00205105),
-221-3p (cat. no. YP00204532), -375-3p (cat. no. YP00204362),
-455-5p (cat. no. YP00204363), -544-5p (cat. no. YP02116293),
-708-5p (cat. no. YP00204490) and -759 (cat. no. YP00206000; all
from Qiagen GmbH) in a Bio-Rad CFX 96 real-time system (Bio-Rad
Laboratories, Inc.) using the miScript SYBR Green PCR kit (Qiagen
GmbH) as follows: Initial heat activation at 95°C for 2 min,
followed by 40 cycles of 95°C for 10 sec and 56°C for 1 min. All
PCR reactions were performed under standard PCR conditions; U6
(cat. no. YP00203907; Qiagen GmbH) was used as the endogenous
control. The relative quantification (RQ) values were calculated
from the quantification cycle (Cq) values using the
2−ΔΔCq method (22).
Validation study using candidate miRNA
oligomers
Based on the results of miRNA microarray and RT-qPCR
analyses, miR-375-3p was selected as the candidate miRNA. To
evaluate its role, miR-375-3p mimic (5′-UUUGUUCGUUCGGCUCGCGUGA-3′)
(Qiagen GmbH) and miR-375-3p mimic negative control (Qiagen GmbH)
were administered to 31 and 28 rats, respectively, that exhibited
evidence of tinnitus at 1 week post-noise exposure. Once the rats
were anesthetized using the method previously described, and placed
in a stereotaxic frame, 5 μ.l of miRNA oligomer (66.67
μ.M) was mixed with 12.5 μ.l of
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) and injected into the right lateral ventricle
(0.8 mm posterior to the bregma, 1.4 mm right lateral to the
midline, to a depth of 3.4 mm from the surface of the skull) at an
infusion rate of 1 μ.l/min. The speed of needle insertion
and withdrawal was maintained at 1.5 mm/min. The site of craniotomy
was closed using bone wax and the scalp was sutured. Two weeks
later (i.e., 3 weeks after noise exposure), the GPIAS responses
were measured to determine whether tinnitus persisted. The rats
were divided into four experimental groups based on the type of
oligomer administered and the persistence of tinnitus, as follows:
i) Mimic tinnitus, ii) mimic non-tinnitus, iii) control tinnitus,
and iv) control non-tinnitus groups.
Target prediction of the candidate
miRNA
To identify the target mRNA regulated by miR-375-3p,
mRNA microarray analysis was performed on the DCN samples from 3
rats randomly selected each from the mimic non-tinnitus and control
tinnitus groups. Microarray analysis of the mRNAs was performed at
BioCore Co., Ltd. Briefly, RNA was isolated and prepared as
detailed above. The cDNAs were generated using the GeneChip Whole
Transcript PLUS Reagent kit (Affymetrix; Thermo Fisher Scientific,
Inc.) and labeled with terminal deoxynucleotidyl transferase (TDT)
using the Affymetrix proprietary DNA Labeling Reagent (Affymetrix;
Thermo Fisher Scientific, Inc.). The labeled samples were
hybridized to the GeneChip RaGene 2.0 ST Array (Affymetrix; Thermo
Fisher Scientific, Inc.). All arrays were scanned using the
Affymetrix GeneChip Scanner 3000, and raw analysis was performed
using the Transcriptome Analysis Console™ software. The CEL files
generated were imported into the Gene Expression Workflow in
GeneSpring GX version 14.9.1 (Agilent Technologies, Inc.).
Subsequently, the microarray data were analyzed as described
above.
Western blot analysis
Western blotting was performed to detect the
expression levels of the candidate target genes obtained using mRNA
microarray and miRNA target prediction server (TargetScan)
analyses. In addition, western blotting was performed to determine
the expression levels of the candidate target genes along with
CTG., which is a previously reported target of miRNA-375-3p
(23). The expression levels of
the proteins were compared between the mimic non-tinnitus and
control tinnitus experimental groups.
Western blotting was carried out on the DCNs, as
described previously (12).
Briefly, the homogenized DCN samples were treated with RIPA lysis
buffer (Biosesang) and a 100X Protease Inhibitor Cocktail
(EDTA-free, lyophilized; cat. no. QTPPI1015; Quartett, Inc.).
Subsequently, the samples were incubated on ice for at least 1 h
and centrifuged at 16,000 × g for 20 min at 4°C (Smart R17 Plus
Micro Centrifuge; Hanil Scientific, Inc.). Following protein
concentration estimation of the supernatants using the BCA assay,
the proteins were denatured at 95°C for 5 min in a 5X SDS-PAGE
loading buffer (cat. no. EBA-1052; Elpis Biotech, Inc.) and
separated on a 10% SDS-PAGE gel. The protein bands were transferred
onto PVDF membranes (Immobilon-P Transfer membrane;
MilliporeSigma); the membranes were then immersed in a blocking
solution [1% bovine serum albumin (cat. no. A0100-010; Gendepot,
Inc.) in Tris-buffered saline with 0.1% Tween-20] for 1 h at room
temperature. Next, the membranes were incubated overnight at 4°C
with primary antibodies against inhibin β-A (INHBA), homeobox A2
(HOXA2), potassium voltage-gated channel subfamily A regulatory
beta subunit 3 (KCNAB3), CTGF, and β-actin, at the following
dilutions: Mouse INHBA (1:200, cat. no. sc-166503; Santa Cruz
Biotechnology, Inc.), rabbit HOXA2 (1:1,000, cat. no. PA568986;
Invitrogen; Thermo Fisher Scientific, Inc.), rabbit KCNAB3
(1:1,000, cat. no. AV35151; Sigma-Aldrich; Merck KGaA), mouse CTGF
(1:1,000, cat. no. sc-101586; Santa Cruz Biotechnology, Inc.), and
mouse β-actin (1:3,000, cat. no. sc-47778; Santa Cruz
Biotechnology, Inc.). On the following day, the membranes were
rinsed and incubated with species-specific horseradish
peroxidase-conjugated secondary antibodies (anti-rabbit: 1:10,000,
cat. no. ADI-SAB-300-J; and anti-mouse: 1:10,000, cat. no.
ADI-SAB-100-J; both from Enzo Life Sciences, Inc.) for 1 h at room
temperature. The protein bands were visualized using ECL solution
(cat. no. WBKLS0500; Immobilon Western Chemiluminescent HRP
Substrate; MilliporeSigma) and analyzed with a chemiluminescence
image analyzer (ChemiDoc; Bio-Rad Laboratories, Inc.).
Statistical analysis
Statistical analysis was performed using the IBM
SPSS 21.0 software (IBM Corp.). The ABR thresholds were assessed
using the two-way repeated measures ANOVA followed by Tukey's post
hoc test. The RQ values obtained from the RT-qPCR and western blot
analyses results were examined using the Mann-Whitney U-test. The
effect of miR-375-3p overexpression was analyzed using the
Pearson's χ2 test. P<0.05 indicated statistically
significant differences.
Results
ABR recordings
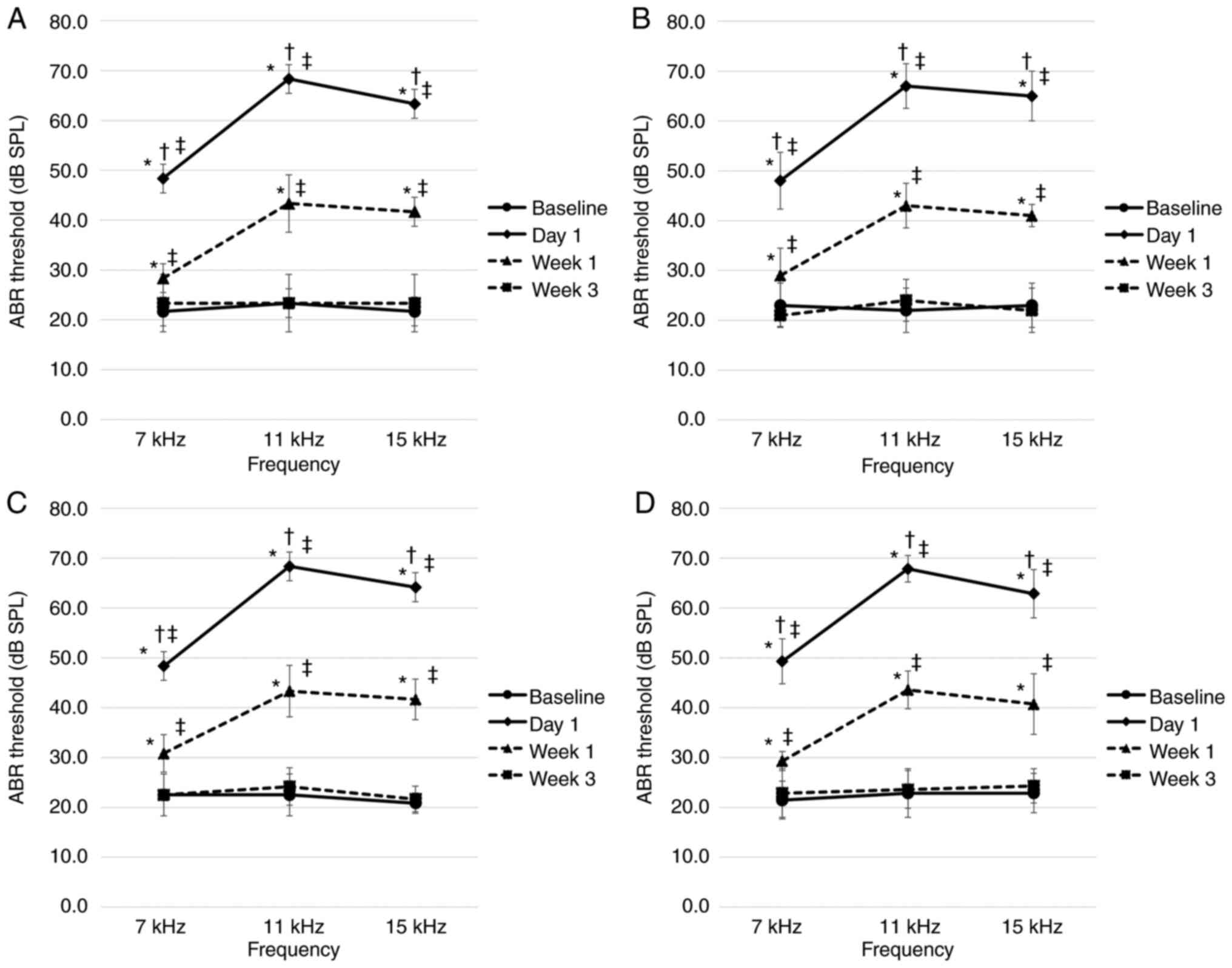
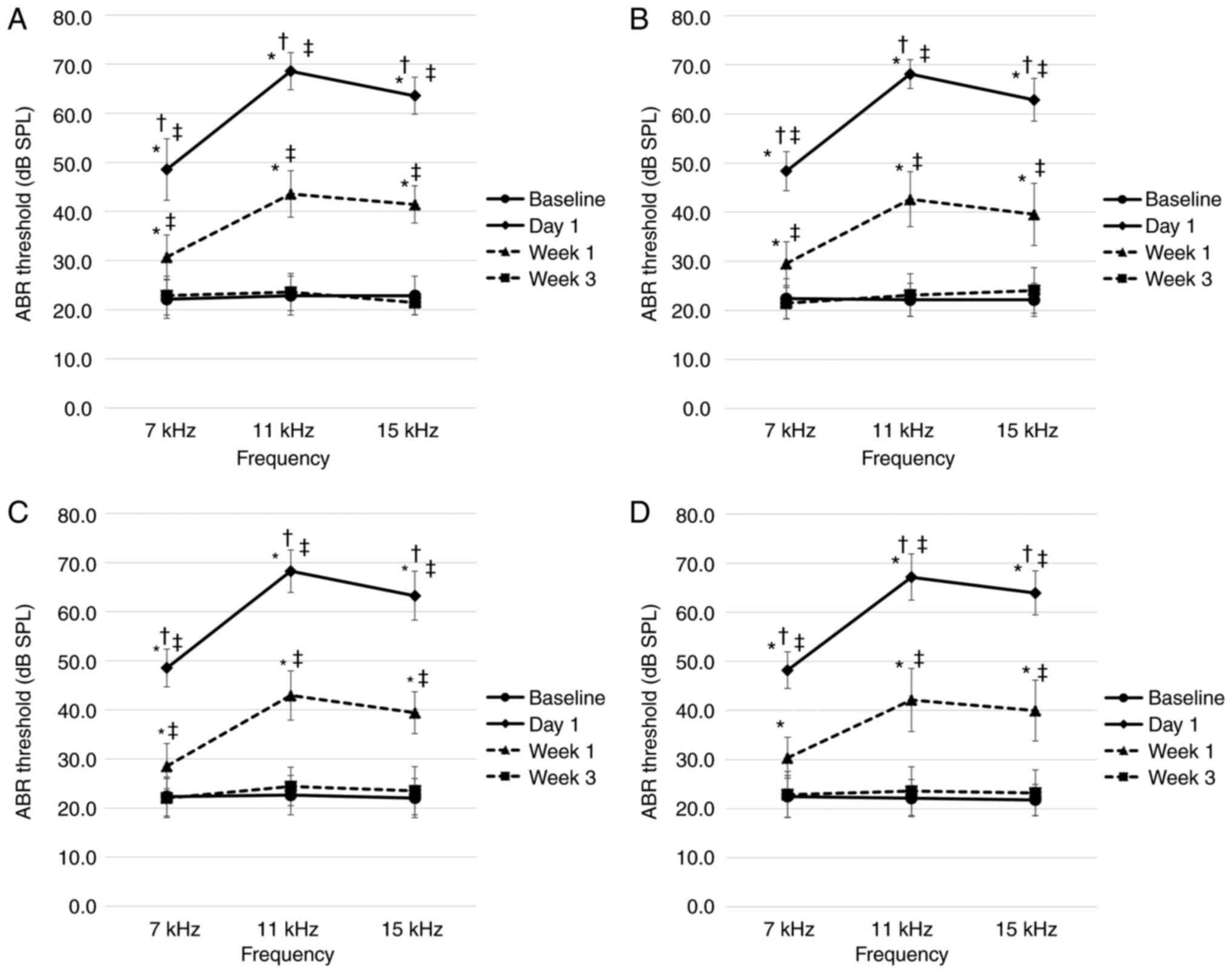
Prior to noise exposure, the ABR thresholds on both
the right and left sides of all rats ranged between 20 and 30 dB
SPL, at all frequencies, with no significant difference detected
between the tinnitus and non-tinnitus groups at all stages. In all
groups and at all frequencies, the ABR thresholds on the right side
on day 1 after noise exposure were significantly higher compared
with those at all other time points. After 1 week of noise
exposure, the ABR threshold was significantly lower compared with
that on day 1, but significantly higher compared with the baseline
threshold and the threshold 3 weeks later. However, there was no
significant difference between the baseline ABR threshold and the
ABR threshold after 3 weeks of exposure to noise. The ABR
thresholds on the left side ranged between 20 and 30 dB SPL. At all
time points post-exposure, the measured ABR thresholds at each
frequency did not differ significantly between the two groups at
the first and second stages during the experiments (Fig. 2). In addition, there were no
significant differences in the ABR thresholds among the four
experimental groups in the third stage (Fig. 3).
Behavioral test for tinnitus
Among individual rats, the pre-exposure G/N ratios
ranged from 30 to 70%. Compared with the no-gap condition, all rats
showed significant decreases in startle responses under the gap
condition at all frequencies (P<0.05). As stated previously, the
GPIAS responses were recorded at 3 weeks following noise exposure.
In the first stage of the experiment, 5 of the 8 rats examined
exhibited no significant decrease in the startle response under the
gap condition at one or more frequencies (P>0.05). Consequently,
these rats manifested behavioral evidence of tinnitus. The
remaining rats exhibited significant decreases in startle responses
under the gap condition at all frequencies; hence, they manifested
no behavioral evidence of tinnitus. In the second stage of the
experiment, 7 of the 13 rats examined exhibited no significant
decrease in startle response under the gap condition at one or more
frequencies (P>0.05). Thus, these rats manifested behavioral
evidence of tinnitus. The remaining rats exhibited significant
decreases in startle responses under the gap condition at all
frequencies, and were accordingly considered to manifest no
behavioral evidence of tinnitus.
Selection of candidate miRNAs based on
microarray analysis
Candidate miRNAs were selected based on the results
obtained from microarray analysis (non-tinnitus group, n=3;
tinnitus group, n=3). First, miRNAs not expressed in humans were
excluded. Subsequently, the remaining miRNAs that satisfied the
following criterion were selected: Log-ratio intensity >0.379 or
<−0.379 (P<0.08) between the tinnitus and non-tinnitus
groups, as determined by the Student's t-test. Using this
criterion, miR-15b-3p, -105, -221-3p, -375-3p, -455-5p, -544-5p,
-708-5p and -759 were selected as candidate miRNAs (Tables I and SI).
 | Table ICandidate microRNAs selected based on
microarray analysis. |
Table I
Candidate microRNAs selected based on
microarray analysis.
| Candidate
microRNAs | Log-ratio | P-value |
|---|
| Log-ratio between
the tinnitus and non-tinnitus groups: |
| >0.379
(P<0.08) | | |
| miR-15b-3p | 0.456 | 0.020 |
| miR-221-3p | 0.500 | 0.033 |
| miR-455-5p | 0.394 | 0.076 |
| miR-544-5p | 0.666 | 0.012 |
| miR-708-5p | 0.773 | 0.043 |
| Log-ratio between
the tinnitus and non-tinnitus groups: |
| <−0.379
(P<0.08) | | |
| miR-105 | −0.519 | 0.030 |
| miR-375-3p | −0.530 | 0.059 |
| miR-759 | −0.661 | 0.058 |
| miR, microRNA. | | |
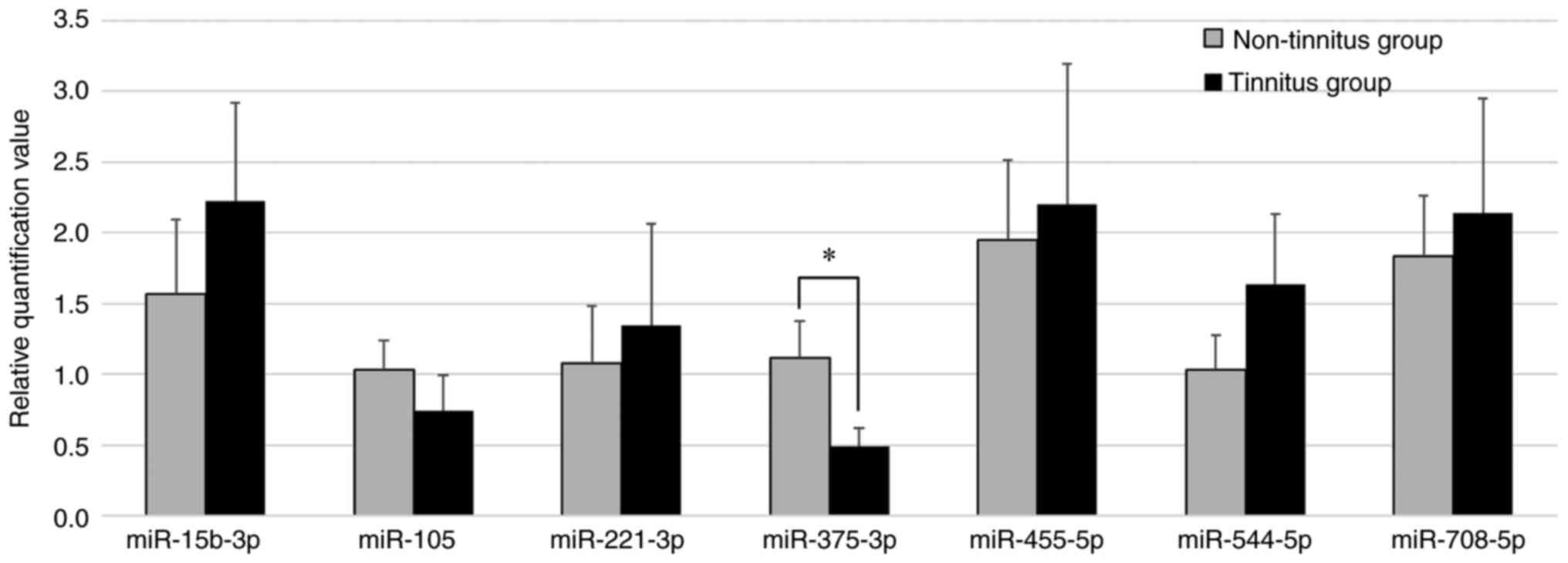
Validation of miRNA expression using
RT-qPCR
To validate the candidate miRNAs, RT-qPCR was
performed (non-tinnitus group, n=6; tinnitus group, n=7) to
identify those miRNAs showing a significant difference in RQ values
in the tinnitus and non-tinnitus groups. It was observed that,
among all the candidate miRNAs, the RQ value of miR-375-3p was
significantly decreased in the tinnitus group compared with that in
the non-tinnitus group (P=0.028), while the RQ values of
miR-15b-3p, -105, -455-5p, -544-5p and -708-5p were not
significantly different (Fig.
4). The Cq value of miR-759 was >35 even with an increased
amount of total RNA as template. As this same observation was made
in multiple replicates of the experiment, miR-759 was excluded from
the candidate miRNAs. Consequently, it was inferred that miR-375-3p
was involved in the development of tinnitus.
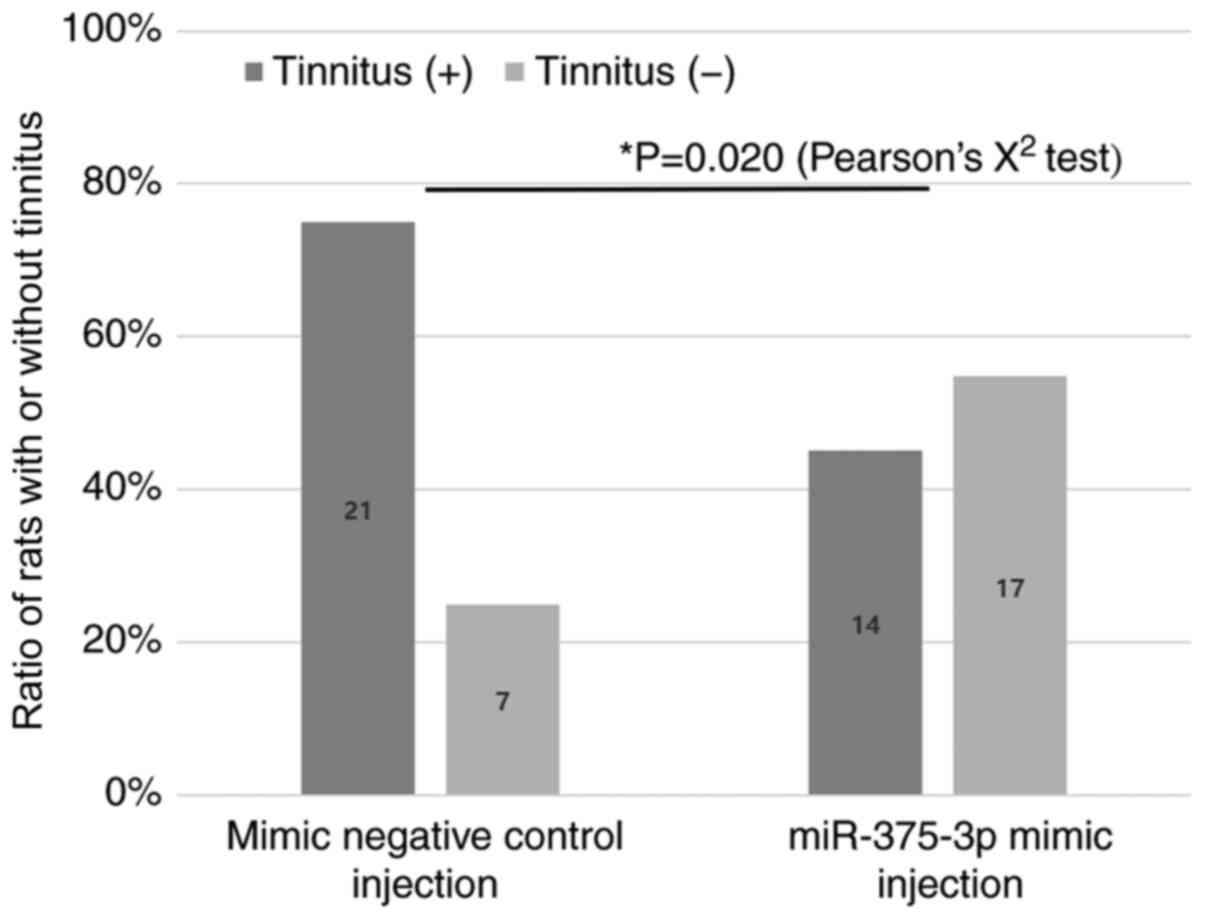
Injection of miR-375-3p oligomers
To evaluate the role of miR-375-3p in the
development of tinnitus, a miR-375-3p mimic or a mimic negative
control was injected into the lateral ventricles of rats with
tinnitus at 1 week post-noise exposure. Two weeks later (i.e., 3
weeks post-noise exposure), GPIAS recordings were performed to
determine whether tinnitus persisted. It was found that tinnitus
persisted in 21 of the 28 rats (75.0%) injected with the miR-375-3p
mimic negative control, and in 14 of the 31 rats (45.2%) injected
with the miR-375-3p mimic. A total of 17 rats (54.8%) exhibited no
tinnitus at 3 weeks post-noise exposure (Fig. 5 and Table II). The effect of miR-375-3p
mimic on preventing the persistence of tinnitus at 3 weeks
post-noise exposure was found to be statistically significant
[P=0.020, odds ratio (OR)=0.275, 95% confidence interval (CI):
0.090-0.833]. These results suggested that a decrease in miR-375-3p
level may play a key role in the persistence of tinnitus.
 | Table IIChanges in GPIAS responses in the
mimic non-tinnitus group wherein tinnitus ceased following
miR-375-3p mimic injection. |
Table II
Changes in GPIAS responses in the
mimic non-tinnitus group wherein tinnitus ceased following
miR-375-3p mimic injection.
| No. | Background noise
(kHz)
|
|---|
6-8
| 10-12
| 14-16
|
|---|
| G/N ratioa | P-value | G/N ratioa | P-value | G/N ratioa | P-value |
|---|
| 1 | | | | | | |
| Baseline | 0.567 | 0.002 | 0.331 | <0.001 | 0.284 | <0.001 |
| Week 1 | 1.100 | 0.793 | 0.360 | 0.001 | 0.559 | 0.223 |
| Week 3 | 0.599 | <0.001 | 0.373 | 0.001 | 0.665 | 0.024 |
| 2 | | | | | | |
| Baseline | 0.477 | 0.001 | 0.571 | 0.028 | 0.249 | <0.001 |
| Week 1 | 0.980 | 0.497 | 0.782 | 0.093 | 0.323 | 0.002 |
| Week 3 | 0.176 | <0.001 | 0.222 | 0.010 | 0.397 | 0.014 |
| 3 | | | | | | |
| Baseline | 0.728 | <0.001 | 0.771 | 0.006 | 0.701 | <0.001 |
| Week 1 | 0.730 | <0.001 | 0.972 | 0.908 | 0.960 | 0.315 |
| Week 3 | 0.785 | 0.006 | 0.785 | 0.003 | 0.842 | 0.015 |
| 4 | | | | | | |
| Baseline | 0.638 | <0.001 | 0.567 | 0.001 | 0.568 | 0.005 |
| Week 1 | 0.984 | 0.627 | 0.744 | 0.010 | 0.972 | 0.576 |
| Week 3 | 0.692 | 0.005 | 0.714 | 0.044 | 0.608 | <0.001 |
| 5 | | | | | | |
| Baseline | 0.537 | 0.001 | 0.295 | 0.001 | 0.296 | <0.001 |
| Week 1 | 0.246 | 0.002 | 0.966 | 0.106 | 0.289 | 0.001 |
| Week 3 | 0.247 | 0.002 | 0.329 | 0.003 | 0.964 | 0.020 |
| 6 | | | | | | |
| Baseline | 0.583 | 0.002 | 0.532 | <0.001 | 0.679 | 0.011 |
| Week 1 | 0.970 | 0.852 | 1.031 | 0.890 | 0.623 | 0.003 |
| Week 3 | 0.734 | 0.033 | 0.692 | 0.005 | 0.602 | 0.004 |
| 7 | | | | | | |
| Baseline | 0.749 | 0.045 | 0.368 | <0.001 | 0.451 | 0.001 |
| Week 1 | 0.401 | <0.001 | 0.703 | 0.014 | 1.015 | 0.633 |
| Week 3 | 0.734 | 0.015 | 0.642 | 0.026 | 0.421 | 0.001 |
| 8 | | | | | | |
| Baseline | 0.121 | <0.001 | 0.357 | <0.001 | 0.443 | 0.015 |
| Week 1 | 1.092 | 0.663 | 0.258 | <0.001 | 0.148 | <0.001 |
| Week 3 | 0.154 | <0.001 | 0.108 | <0.001 | 0.101 | <0.001 |
| 9 | | | | | | |
| Baseline | 0.332 | <0.001 | 0.424 | <0.001 | 0.266 | <0.001 |
| Week 1 | 0.497 | <0.001 | 0.583 | 0.013 | 0.667 | 0.254 |
| Week 3 | 0.480 | <0.001 | 0.354 | <0.001 | 0.284 | 0.013 |
| 10 | | | | | | |
| Baseline | 0.285 | 0.001 | 0.373 | 0.002 | 0.558 | 0.001 |
| Week 1 | 0.378 | <0.001 | 0.765 | 0.178 | 0.990 | 0.760 |
| Week 3 | 0.343 | <0.001 | 0.343 | 0.016 | 0.533 | 0.010 |
| 11 | | | | | | |
| Baseline | 0.624 | 0.006 | 0.642 | 0.029 | 0.526 | 0.001 |
| Week 1 | 1.026 | 0.020 | 1.006 | 0.443 | 0.669 | 0.036 |
| Week 3 | 0.378 | 0.002 | 0.564 | 0.011 | 0.327 | 0.008 |
| 12 | | | | | | |
| Baseline | 0.579 | 0.006 | 0.300 | <0.001 | 0.199 | <0.001 |
| Week 1 | 1.077 | 0.358 | 0.858 | 0.310 | 0.519 | 0.002 |
| Week 3 | 0.616 | 0.001 | 0.555 | 0.007 | 0.448 | 0.040 |
| 13 | | | | | | |
| Baseline | 0.595 | 0.001 | 0.452 | 0.002 | 0.358 | 0.001 |
| Week 1 | 0.589 | 0.001 | 0.561 | 0.012 | 0.812 | 0.101 |
| Week 3 | 0.479 | <0.001 | 0.723 | 0.036 | 0.514 | 0.001 |
| 14 | | | | | | |
| Baseline | 0.453 | <0.001 | 0.669 | 0.038 | 0.668 | 0.012 |
| Week 1 | 0.440 | <0.001 | 0.623 | 0.004 | 0.770 | 0.165 |
| Week 3 | 0.421 | 0.007 | 0.345 | <0.001 | 0.498 | 0.001 |
| 15 | | | | | | |
| Baseline | 0.455 | 0.001 | 0.507 | 0.024 | 0.379 | 0.001 |
| Week 1 | 1.311 | 0.102 | 0.705 | 0.254 | 0.310 | 0.004 |
| Week 3 | 0.518 | 0.024 | 0.502 | 0.021 | 0.433 | 0.025 |
| 16 | | | | | | |
| Baseline | 0.560 | 0.017 | 0.620 | 0.014 | 0.312 | <0.001 |
| Week 1 | 0.286 | 0.047 | 0.596 | 0.141 | 0.532 | 0.006 |
| Week 3 | 0.511 | <0.001 | 0.362 | <0.001 | 0.483 | 0.002 |
| 17 | | | | | | |
| Baseline | 0.594 | 0.005 | 0.486 | <0.001 | 0.611 | 0.021 |
| Week 1 | 1.061 | 0.818 | 0.727 | 0.044 | 0.618 | 0.036 |
| Week 3 | 0.467 | <0.001 | 0.650 | 0.040 | 0.741 | 0.049 |
Target prediction for miR-375-3p
To discern the probable targets regulated by
miR-375-3p, mRNA microarray analysis was performed (Table SII). In the mimic non-tinnitus
group (n=3), tinnitus had ceased after miR-375-3p mimic injection,
whereas in the control tinnitus group (n=3), the rats had
persistent tinnitus after miR-375-3p mimic negative control
injection. The target gene candidates that exhibited decreased
expression in the mimic non-tinnitus group were selected using the
following criteria: Log-ratio <−0.585 and P≤0.25. As an
additional process for target prediction, the miRNA target
prediction program TargetScan was employed (http://www.targetscan.org/cgi-bin/targetscan/vert_72/targetscan.cgi?species=Rat&gid=&mir_sc=miR-375-3p&mir_c=&mir_nc=&sortType=cs&allTxs=&incl_nc=All).
Genes that were predicted as targets for miR-375-3p and whose
target site nucleotide sequences in rats were identical to or
contained one nucleotide difference when compared with the
respective genes in humans were selected from TargetScan.
Collectively, the results from mRNA microarray analysis and
TargetScan revealed three gene targets: KCNAB., INHB.
and HOXA. (Table
III).
 | Table IIICandidate target genes of
microRNA-375-3p selected based on microarray analysis. |
Table III
Candidate target genes of
microRNA-375-3p selected based on microarray analysis.
| Candidate target
genes | Log-ratio between
the mimic non-tinnitus and control tinnitus groups: <−0.585
(P<0.25) | P-value |
|---|
| KCNAB3 | −0.650 | 0.136 |
| INHBA | −0.709 | 0.166 |
| HOXA2 | −0.957 | 0.240 |
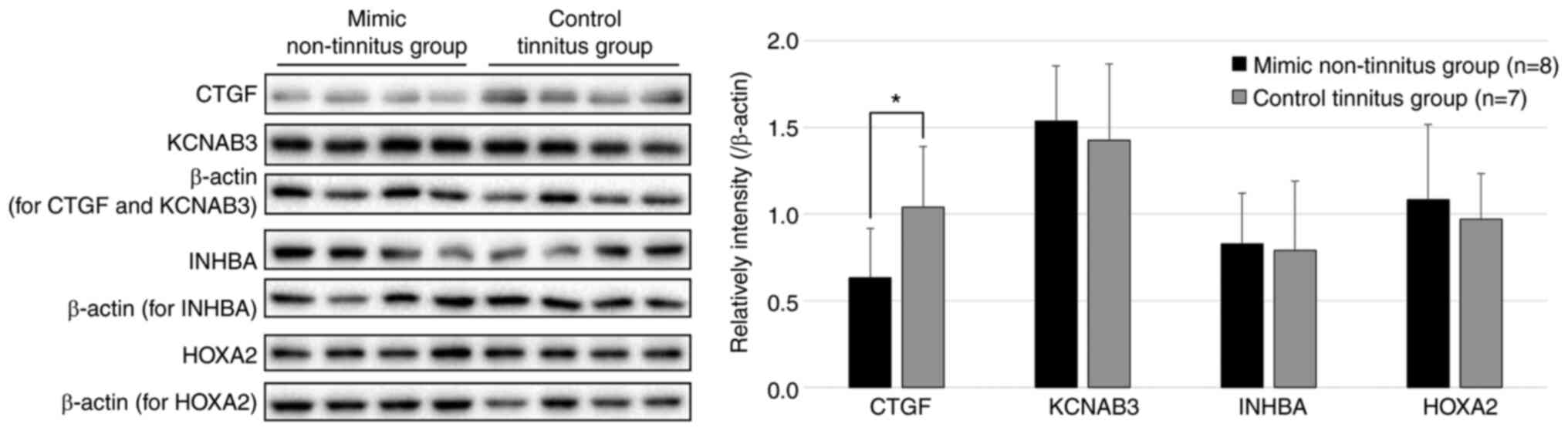
Along with these genes, CTG., a previously
reported target of miR-375-3p (23), was included, and western blotting
was performed to analyze the expression of these potential genes in
the mimic non-tinnitus (n=8) and control tinnitus (n=7)
experimental groups. The western blotting results showed that the
mimic non-tinnitus group, wherein tinnitus ceased after 3 weeks of
noise exposure owing to the injection of the mimic, exhibited a
statistically significant decrease in CTGF levels compared to the
control tinnitus group, in which tinnitus persisted (P=0.028)
(Fig. 6). The other three
proteins did not differ significantly between the groups.
Discussion
Although numerous studies have investigated the
probable causal factors contributing to the development of tinnitus
(9-11), no effective therapeutic methods
have been identified to date for the treatment of this condition.
To address this issue, it is necessary to identify and block the
checkpoints in various pathways associated with the induction of
tinnitus.
It is well known that the majority of genes are
regulated by miRNAs, which are non-coding RNAs that modify gene
expression via post-transcriptional regulation. Moreover, one
single miRNA can regulate the expression of multiple genes, thereby
serving as a checkpoint for disease outbreaks. miRNAs are
particularly abundant in the brain and play important roles in the
development and functioning of neuronal networks, including the
regulation of neurogenesis, synaptogenesis and morphogenesis
(24-26). Various neuronal diseases,
including schizophrenia, autism, fragile X, Rett and Down
syndromes, have also been reported to be associated with aberrant
miRNA expression (27-29). In addition, vestibular
compensation, a change in brainstem function concomitant with a
decrease in sensory input, has been reported to be regulated by
miRNAs (21). Given that
tinnitus is known to be induced by changes in the brainstem in
response to a reduced auditory input (9-11), the developmental mechanism of
tinnitus may have similarities to the vestibular compensation
process. Accordingly, it was hypothesized that one or more miRNAs
may play checkpoint roles in the development of tinnitus and, thus,
we sought to identify these miRNAs.
In our previous study, a tinnitus animal model was
developed to elucidate the mechanisms underlying tinnitus resulting
from noise exposure and inducing TTS in rats (12). Using this model, the DCNs of rats
were collected and the auditory and non-auditory projections were
compared between the tinnitus and the non-tinnitus groups. It was
observed that a decrease in the auditory projections and a
subsequent increase in the non-auditory projections via axonal
sprouting were important phenomena associated with the development
of tinnitus at 3 weeks post-noise exposure. In the present study,
the same model was used in an effort to identify the putative
miRNAs implicated in the regulation of the aforementioned
processes. At 3 weeks post-noise exposure, the DCNs of the rats
were collected to identify miRNAs exhibiting differential
expression in the tinnitus and non-tinnitus groups and the
candidate miRNAs were selected based on microarray analysis.
Through subsequent validation using RT-qPCR, it was found that
miR-375-3p was significantly decreased in the tinnitus group
compared with the non-tinnitus group. miRNAs associated with
tinnitus were identified by comparing the groups with and without
tinnitus, although the same degree of hearing loss was induced in
both groups. If the control group without noise exposure was also
included in the comparisons, the results would have been clearer.
However, even if the same degree of hearing loss occurs due to
exposure to the same noise frequencies, tinnitus only occurs in a
proportion of the cases. Therefore, when conducting tinnitus
research, it is essential to distinguish between the changes
occurring owing to hearing loss and the changes that cause
tinnitus; this may be achieved by comparing tinnitus and
non-tinnitus animal models. The effect of miR-375-3p on tinnitus
development was evaluated by injecting a miR-375-3p mimic or a
mimic negative control. In the mimic negative control-injected
group, tinnitus persisted in 75.0% of the rats. In our previous
study using the same tinnitus animal model, we found that tinnitus
persisted 3 weeks after noise exposure in 81.5% of animals that
developed tinnitus at 1 week after noise exposure (12). Similar results were also obtained
in the present study. However, in the mimic-injected group, the
proportion of animals with persistent tinnitus decreased
significantly to 45.2%. This finding confirmed that the
overexpression of miR-375-3p could reduce the persistence of
tinnitus.
Several researchers have reported the effects of
miR-375 on the brain, although it is found in multiple organs or
tissues (30-32). For example, the expression of
miR-375 was found to be downregulated in several neural injury
models, including models of cerebral ischemia/reperfusion injury
(33-35). However, the overexpression of
miR-375, using an miR-375 mimic, has been shown to provide
significant protection against the ischemia/reperfusion-induced
brain injury by reducing cell apoptosis. A recent study reported
that CTG. is one of the targets of miR-375-3p, and that the
overexpression of miR-375-3p resulted in the downregulation of CTGF
in a brain ischemia/reperfusion injury model (23). CTGF has been demonstrated to
exert pro-apoptotic effects under different conditions, including
carcinoma and brain injury. CTGF levels have been demonstrated to
increase in the brains of rats with traumatic brain injury
(36). The CTGF protein has been
shown to have pro-apoptotic activity, and to promote a reduction in
neuronal survival (37,38).
Our previous study demonstrated that, subsequent to
hearing loss, the auditory projections degrade quickly and more
severely in the tinnitus group compared with the non-tinnitus
group, thereby leading to a significant increase in the
somatosensory projections. This has been identified as an important
process in the development of tinnitus (12). In the present study, it was
observed that miR-375-3p expression decreased in the tinnitus group
compared with the non-tinnitus group. Considering the changes in
miR-375-3p expression following neural injury, it was hypothesized
that the development of tinnitus may be attributed to the decreased
expression of miR-375-3p. In addition, when miR-375-3p was
overexpressed with miR-375-3p mimic injection, the proportion of
animals with sustained tinnitus to those with ceased tinnitus
decreased significantly compared with the control group. It was
inferred that miR-375-3p may prevent the persistence of tinnitus by
attenuating the neural damage following noise exposure. This
mechanism may be involved in the apoptotic process of CTG.,
one of the gene targets of miR-375-3p. A possible scenario is that
the downregulation of CTGF by miR-375-3p is weakened as the
expression of miR-375-3p decreases, leading to more severe neural
damage due to increased CTGF expression and the apoptosis of
auditory neurons. Considering that there was no difference in the
hearing level of the rats in the tinnitus and non-tinnitus groups,
it was hypothesized that there may be differences in the degree of
damage of high-threshold fibers, rather than the low-threshold
fibers, which determine the hearing thresholds (39). On the other hand, it was observed
that tinnitus ceased in some animals administered the mimic
negative control. This is consistent with our previous report,
which demonstrated that 18.5% of animals exhibiting evidence of
tinnitus at 1 week post-noise exposure exhibited no tinnitus at 3
weeks post-noise exposure, even though no specific treatment was
administered, under identical experimental settings (12). This suggests that the
susceptibility of auditory neurons to damage by noise exposure
varies among individuals, and that the individual differences in
miR-375-3p expression may be responsible for this varied
susceptibility. In addition, some animals exhibited persistent
tinnitus even after the administration of miR-375-3p mimic. This
suggests that other factors, besides miR-375-3p expression, may
also be involved in tinnitus development. This discrepancy may also
be due to certain changes in other pathway(s) that cannot be
compensated or reversed by miR-375-3p supplementation, such as the
limbic system, which is associated with the occurrence and
persistence of tinnitus (40).
However, further research is necessary to verify this
hypothesis.
There were certain limitations to the present study
that must be acknowledged. Only male rats were selected in this
study. The rats were ensured to be an identical strain and within a
certain age range to reduce variabilities. Furthermore, ABR
recordings were checked before noise exposure to prove that the
hearing did not differ between the groups. Interestingly, female
rats do not display greater variability during the reproductive
cycle compared with male rats; furthermore, male and female mice
and humans also have similar levels of variability in terms of gene
expression (41-43). Therefore, it is inferred that
using both sexes in this study would not have made a significant
difference in the results, provided no difference in the hearing
levels was confirmed between the tinnitus and non-tinnitus groups.
However, for the results of this experiment to be translated into
treatment strategies in the future, additional studies using female
rats may be helpful.
To the best of our knowledge, no previous studies
have examined the involvement of miRNAs in the development of
tinnitus. In the present study, the expression of miR-375-3p was
found to be reduced in the DCNs of rats with tinnitus, and the
overexpression of miR-375-3p prevented the persistence of tinnitus
by reducing the expression of CTG.. These findings will
contribute significantly to the development of a novel therapeutic
approach to tinnitus, thereby bringing about a significant
breakthrough in the treatment of this potentially debilitating
condition.
Supplementary Data
Availability of data and materials
The raw data of microRNA microarray analysis
generated during the current study are available in the Gene
Expression Omnibus (GEO) (accession numbers: GSE172259),
[https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE172259].
Authors' contributions
MC designed the experiments and study. KHH, HC, KRH
and MC performed the experiments. MC, KHH and SKM have seen and
confirm the authenticity of the raw data. MC and KHH wrote the
manuscript, analyzed and interpreted the data. SKM, YKK and IP
contributed to designing the experiment and revising the
manuscript. All the authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
All procedures for animal care and use were approved
by the Institutional Animal Care and Use Committee of Chung-Ang
University (2016-00092).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by the National Research
Foundation of Korea funded by the Korea government Ministry of
Education (grant no. 2018R1D1A1A02085478) and the Chung-Ang
University Research Grants provided in 2020.
References
|
1
|
Axelsson A and Ringdahl A: Tinnitus-a
study of its prevalence and characteristics. Br J Audiol. 23:53–62.
1989. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hébert S, Canlon B and Hasson D: Emotional
exhaustion as a predictor of tinnitus. Psychother Psychosom.
81:324–326. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Langguth B: A review of tinnitus symptoms
beyond 'ringing in the ears': A call to action. Curr Med Res Opin.
27:1635–1643. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shargorodsky J, Curhan GC and Farwell WR:
Prevalence and characteristics of tinnitus among US adults. Am J
Med. 123:711–718. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jastreboff PJ: Tinnitus retraining
therapy. Prog Brain Res. 166:415–423. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Moller AR: Tinnitus: Presence and future.
Prog Brain Res. 166:3–16. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cima RFF, Mazurek B, Haider H, Kikidis D,
Lapira A, Norena A and Hoare DJ: A multidisciplinary European
guideline for tinnitus: Diagnostics, assessment, and treatment.
HNO. 67(Suppl 1): S10–S42. 2019. View Article : Google Scholar
|
|
8
|
Tunkel DE, Bauer CA, Sun GH, Rosenfeld RM,
Chandrasekhar SS, Cunningham ER Jr, Archer SM, Blakley BW, Carter
JM, Granieri EC, et al: Clinical practice guideline: Tinnitus.
Otolaryngol Head Neck Surg. 151(Suppl 2): S1–S40. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dehmel S, Pradhan S, Koehler S, Bledsoe S
and Shore S: Noise overexposure alters long-term
somatosensory-auditory processing in the dorsal cochlear
nucleus-possible basis for tinnitus-related hyperactivity? J
Neurosci. 32:1660–1671. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Koehler SD and Shore SE: Stimulus
timing-dependent plasticity in dorsal cochlear nucleus is altered
in tinnitus. J Neurosci. 33:19647–19656. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zeng C, Yang Z, Shreve L, Bledsoe S and
Shore S: Somatosensory projections to cochlear nucleus are
upregulated after unilateral deafness. J Neurosci. 32:15791–15801.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Han KH, Mun SK, Sohn S, Piao XY, Park I
and Chang M: Axonal sprouting in the dorsal cochlear nucleus
affects gap-prepulse inhibition following noise exposure. Int J Mol
Med. 44:1473–1483. 2019.PubMed/NCBI
|
|
13
|
Hollins SL and Cairns MJ: MicroRNA: Small
RNA mediators of the brains genomic response to environmental
stress. Prog Neurobiol. 143:61–81. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Campbell K and Booth SA: MicroRNA in
neurodegenerative drug discovery: The way forward? Expert Opin Drug
Discov. 10:9–16. 2015. View Article : Google Scholar
|
|
15
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gebert LF, Rebhan MA, Crivelli SE, Denzler
R, Stoffel M and Hall J: Miravirsen (SPC3649) can inhibit the
biogenesis of miR-122. Nucleic Acids Res. 42:609–621. 2014.
View Article : Google Scholar
|
|
17
|
National Research Council: Guide for the
Care and Use of Laboratory Animals. The National Academies Press
Inc; Washington, DC: 2011
|
|
18
|
Paxinos G and Watson C: The Rat Brain in
Stereotaxic Coordinates. Academic Press Inc; Burlington, MA:
2006
|
|
19
|
Mun SK, Han KH, Baek JT, Ahn SW, Cho HS
and Chang MY: Losartan prevents maladaptive Auditory-Somatosensory
plasticity after hearing loss via transforming growth Factor-β
signaling suppression. Clin Exp Otorhinolaryngol. 12:33–39. 2019.
View Article : Google Scholar
|
|
20
|
Longenecker RJ and Galazyuk AV:
Methodological optimization of tinnitus assessment using prepulse
inhibition of the acoustic startle reflex. Brain Res. 1485:54–62.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chang MY, Park S, Choi JJ, Kim YK, Suh MW,
Lee JH, Oh SH and Park MK: MicroRNAs 218a-5p, 219a-5p, and 221-3p
regulate vestibular compensation. Sci Rep. 7:87012017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expressio. data using real-time quantitative PCR and
the 2(-Delta Delta C(T)). Method Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
23
|
Ou J, Kou L, Liang L and Tang C: MiR-375
attenuates injury of cerebral ischemia/reperfusion via targetting
Ctgf. Biosci Rep. 37:BSR201712422017. View Article : Google Scholar
|
|
24
|
Dugas JC, Cuellar TL, Scholze A, Ason B,
Ibrahim A, Emery B, Zamanian JL, Foo LC, McManus MT and Barres BA:
Dicer1 and miR-219 Are required for normal oligodendrocyte
differentiation and myelination. Neuron. 65:597–611. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Giraldez AJ, Cinalli RM, Glasner ME,
Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP and
Schier AF: MicroRNAs regulate brain morphogenesis in zebrafish.
Science. 308:833–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Schratt GM, Tuebing F, Nigh EA, Kane CG,
Sabatini ME, Kiebler M and Greenberg ME: A brain-specific microRNA
regulates dendritic spine development. Nature. 439:283–289. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Banerjee-Basu S, Larsen E and Muend S:
Common microRNAs target established ASD genes. Front Neurol.
5:2052014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Im HI and Kenny PJ: MicroRNAs in neuronal
function and dysfunction. Trends Neurosci. 35:325–334. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sun E and Shi Y: MicroRNAs: Small
molecules with big roles in neurodevelopment and diseases. Exp
Neurol. 268:46–53. 2015. View Article : Google Scholar
|
|
30
|
Ding L, Xu Y, Zhang W, Deng Y, Si M, Du Y,
Yao H, Liu X, Ke Y, Si J and Zhou T: MiR-375 frequently
downregulated in gastric cancer inhibits cell proliferation by
targeting JAK2. Cell Res. 20:784–793. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
He XX, Chang Y, Meng FY, Wang MY, Xie QH,
Tang F, Li PY, Song YH and Lin JS: MicroRNA-375 targets AEG-1 in
hepatocellular carcinoma and suppresses liver cancer cell growth in
vitro and in vivo. Oncogene. 31:3357–3369. 2012. View Article : Google Scholar
|
|
32
|
Kapsimali M, Kloosterman WP, de Bruijn E,
Rosa F, Plasterk RH and Wilson SW: MicroRNAs show a wide diversity
of expression profiles in the developing and mature central nervous
system. Genome Biol. 8:R1732007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bhinge A, Namboori SC, Bithell A, Soldati
C, Buckley NJ and Stanton LW: MiR-375 is essential for human spinal
motor neuron development and may be involved in motor neuron
degeneration. Stem Cells. 34:124–134. 2016. View Article : Google Scholar
|
|
34
|
Wang C, Pan Y, Cheng B, Chen J and Bai B:
Identification of conserved and novel microRNAs in cerebral
ischemia-reperfusion injury of rat using deep sequencing. J Mol
Neurosci. 54:671–683. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang Y, Dong X, Li Z, Wang W, Tian J and
Chen J: Downregulated RASD1 and upregulated miR-375 are involved in
protective effects of calycosin on cerebral ischemia/reperfusion
rats. J Neurol Sci. 339:144–148. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hertel M, Tretter Y, Alzheimer C and
Werner S: Connective tissue growth factor: A novel player in tissue
reorganization after brain injury? Eur J Neurosci. 12:376–380.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hall-Glenn F and Lyons KM: Roles for CCN2
in normal physiological processes. Cell Mol Life Sci. 68:3209–3217.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Khodosevich K, Lazarini F, von Engelhardt
J, Kaneko H, Lledo PM and Monyer H: Connective tissue growth factor
regulates interneuron survival and information processing in the
olfactory bulb. Neuron. 79:1136–1151. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bharadwaj HM, Masud S, Mehraei G, Verhulst
S and Shinn-Cunningham BG: Individual differences reveal correlates
of hidden hearing deficits. J Neurosci. 35:2161–2172. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Elgoyhen AB, Langguth B, De Ridder D and
Vanneste S: Tinnitus: Perspectives from human neuroimaging. Nat Rev
Neurosci. 16:632–642. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Becker JB, Prendergast BJ and Liang JW:
Female rats are not more variable than male rats: A meta-analysis
of neuroscience studies. Biol Sex Differ. 7:342016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Itoh Y and Arnold AP: Are females more
variable than males in gene expression? Meta-analysis of microarray
datasets. Biol Sex Differ. 6:182015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lauer AM and Schrode KM: Sex bias in basic
and preclinical noise-induced hearing loss research. Noise Health.
19:207–212. 2017. View Article : Google Scholar : PubMed/NCBI
|