Introduction
Severe combined immunodeficiency (SCID) is a rare
genetic disease with severe deficiency in both cellular and humoral
immunity, resulting in fatal and recurrent opportunistic infections
(1-5). X-linked SCID (X-SCID) accounts for
40-60% of SCID cases (1,2), resulting from mutations in the
interleukin-2 receptor subunit γ (IL2RG) gene on the X
chromosome (3,6). IL2RG is a common receptor of
interleukins (IL) 2, IL4, IL7, IL9, IL15 and IL21, playing an
essential role in lymphocyte development and function (7,8).
Mutations in IL2RG cause the absence of the protein or
altered protein structures, resulting in impaired interactions
between the receptor complexes and cytokine ligands, ultimately
leading to severe immune deficiency (9). Without the reconstitution of the
immune system by hematopoietic stem cell transplantation or gene
therapy, X-SCID patients usually die within the first year of life
due to recurrent infections (10,11).
Typical X-SCID is characterized by the absence of T
cells, a low number of NK cells and a normal to high number of
dysfunctional B cells (T−B+NK−
phenotype) (5). It was found that
~40% of SCID have maternal engraftment of T lymphocytes due to the
patient's unsuccessful recognition and rejection of the maternal
blood cells (12). It is
hypothesized that maternal T lymphocytes may promote near-term
survival in several SCID cases, but SCID patients with and without
the maternal engraftment succumb without appropriate reconstitution
of the immune system (13,14).
The immunophenotype of engrafted maternal T cells
can be diverse, with most cases exhibiting a mature
(CD45RO+) phenotype (15). Functional regulatory T cells are
detected within maternal engrafted cells (16) and a massive maternal T cell
expansion in response to EBV has been reported (17). However, the engrafted T cells are
generally functionally defective, with a restricted T-cell receptor
repertoire and limited or no proliferative response to mitogens
(12,14,18). Some SCID patients with engrafted
maternal T cells are asymptomatic, while others present
graft-versus-host disease (GVHD) (14,18,19). Maternal engraftment of B and NK
cells are also observed in several cases (17,20-22). Maternal engrafted B cells can
cause the monoclonal IgA or IgG1 gammopathy in SCID (20,22), while the function of the maternal
NK cells have not been explored in depth (17,22). To the best of the authors'
knowledge, there have been no reported cases about maternal
engraftment of NKT cells. Overall, the functional status of
engrafted maternal cells remains largely undefined due to the
difficulty of efficiently separating and characterizing maternal
cells and autologous counterparts with the limited number of
cells.
The present study applied single-cell RNA sequencing
(scRNA-seq) to study the transcriptome landscape of BMMNCs from an
X-SCID patient with engrafted maternal cells. A novel analytic
strategy was designed to discriminate between maternal and
autologous cells. The present study characterized the functional
status of autologous and maternal engrafted T and NK-like cells in
this X-SCID patient with single-cell resolution. It was
hypothesized that the scRNA-seq data from BMMNCs in this X-SCID
patient would provide valuable resources for investigating the
essential characteristics of heterogeneous immune cells and
potentially guide the development of advanced therapies.
Materials and methods
Collection of clinical samples
Peripheral blood (PB), bone marrow (BM), hair
follicles and mouth swabs from the X-SCID patient (13-months-old)
and PB from the patient's parents were obtained with written
informed consent from the patient's parents according to procedures
approved by the Guangzhou Women and Children's Medical Center
(approval number: 105A01). The PB and BM were processed within two
hours of sample collection. Then ~30 hair follicles were collected
in a clean 1.5 ml Eppendorf tube and four mouth swabs collected
separately in sealed tubes were frozen at -80°C prior to DNA
extraction.
Captured exome sequencing
DNA was extracted from the patient's PB using the
QIAamp DNA Blood Mini kit (Qiagen GmbH), following DNA
fragmentation, end-repair, A-tailing and ligation to adapters to
generate sequencing libraries. Exome-derived DNA fragments within
the library were heat denatured and hybridized with biotin probes
targeting immune genes (P039-Exome; MyGenostics) and the libraries
hybridized with probes were enriched via biotin binding to
streptavidin beads (Thermo Fisher Scientific, Inc.) for 45 min at
room temperature (RT). After rinsing, the beads were incubated in
0.1 mol/l HCl at RT to denature the DNA, allowing the hybridized
DNA fragments eluting from the beads. The enriched DNA fragments
were neutralized with equal volume of 0.1 mol/l CH3COOH,
purified with P-30 column (Bio-Rad Laboratories, Inc.), and
amplified for exome sequencing.
Sanger sequencing
To verify the mutation at the IL2RG locus,
Sanger sequencing was performed using the DNA extracted from the PB
of the patient and his parents across the mutation site (primers:
GTG AGA CCC TGC CTC AAA AG and CAG CAC ATA TTT GCC ACA CC).
Whole-genome sequencing (WGS)
The patient's hair follicles and mouth swabs were
digested with 0.1 mg/ml proteinase K (Thermo Fisher Scientific,
Inc.) in 100 µl 150 mM NaCl with 10 mM EDTA and 1% SDS
overnight at 55°C. The solution was then extracted twice with equal
volume of phenol-chloroform-isoamyl alcohol (Thermo Fisher
Scientific, Inc.), once with equal volume of chloroform. The
solution was then added with 20 µl 7.5 mol/l ammonium
acetate and 300 µl absolute ethanol to precipitated DNA. DNA
from the PB of the patient's parents was prepared using the QIAamp
DNA Blood Mini kit (Qiagen GmbH). The libraries were generated
using NEBNext Ultra DNA Library Prep kit for Illumina (New England
BioLabs, Inc.).
Chromosomal microarray
DNA extracted from the patient PB was subjected to
chromosomal microarray (CMA) analysis using an Affymetrix Cytoscan
750K Microarray kit (Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions. This assay covers 550,000 markers
to detect copy number variations and 200,000 high-performing SNP
probes. The data of the Cytoscan 750K Microarray were analyzed
using Chromosome Analysis Suite version 2.0 (Affymetrix; Thermo
Fisher Scientific, Inc.).
Next-generation sequencing and data
processing
Exome sequencing libraries and WGS libraries were
sequenced using the NextSeq 500 system (Illumina, Inc.) in
paired-end 150 bp mode. The raw sequenced reads were filtered for
low quality reads and adapter regions using Trimmomatic v.0.39
(23) with the adapter sequences
(Read 1: AGA TCG GAA GAG CAC ACG TCT GAA CTC CAG TCA; Read 2: AGA
TCG GAA GAG CGT CGT GTA GGG AAA GAG TGT). Cleaned high quality
sequencing reads (Q30) were then aligned to the human reference
genome (GRCh38) using the Burrows Wheeler Aligner (24) (BWA, version 0.7.15) with default
parameters and then sorted using the SAMtools (25) (version 1.10) software. The
'MarkDuplicates' function embedded in Picard (version 1.107,
http://broadinstitute.github.io/picard/) was applied
to mark and discard PCR duplicates. The results were visualized and
displayed using the IGV (26-28) genome browser.
Short tandem repeat (STR) marker
detection
DNA extracted from the patient's PB and mouth swabs
was subjected to genetic screening at 16 STR loci, covering 14
pairs of chromosomes across the genome. DNA from the patient's
mother PB was used as a control.
Fluorescence in situ hybridization
(FISH)
To verify the mixed cell status of the patient PB,
FISH experiments were performed using a Vysis CEP X
SpectrumOrange/Y SpectrumGreen Labeled Fluorescent DNA probe kit
(Abbott Pharmaceutical Co. Ltd.) with the patient PB, according to
the manufacturer's instructions. Briefly, nuclei isolated from the
PB were fixed, denatured and hybridized with the DXZ1 probes
(targeting to the X chromosome) and DYZ3 probes (targeting to the Y
chromosome). After staining with DAPI at RT for 10 min, nuclei were
imaged using a fluorescence microscope. Images of 400 interphase
nuclei were evaluated and quantified.
Single-cell RNA sequencing
The BM collected from the patient was treated with
6% Hespan to remove red cells. Mononuclear cells were then enriched
by Ficoll gradient centrifugation at RT for 30 min at 800 × g.
BMMNCs were rinsed twice and resuspended in PBS (Ca2+
free, Mg2+ free) supplemented with 0.04% BSA. Cells were
then filtered with a 40-µm strainer and stored on ice until
processing. Viability (>95%), concentration (1,400
cells/µl), debris (almost none) and aggregation (almost
none) ratios were determined using an automated cell counter
(Nexcelom Bioscience LLC). According to the manufacturer's manual,
the single-cell suspension was loaded onto ChIP B (10× Genomics)
with GEM beads from Chromium Single Cell 3' Reagent kit V3 (10×
Genomics). In total, two libraries were constructed, each with
~16,000 cells loaded to obtain 10,000 cells. cDNA was amplified for
11 cycles and about 40 ng of purified cDNA (SPRIselect beads, 0.6×)
was input for each library construction (12 cycles of index PCR,
0.6× and 0.8× SPRIselect beads for size-selection). Concentrations
of cDNA and libraries were measured with Qubit (Thermo Fisher
Scientific, Inc.) and their size distribution was determined using
Qsep (BiOptic Inc.). Each library was sequenced in the 2×150 bp
mode (NovaSeq; Illumina, Inc.) for 100 Gb of data.
Collection of public scRNA-seq
datasets
To compare the immune cell features between the
X-SCID patient and healthy individuals, the scRNA-seq data of
BMMNCs from eight healthy donors (four males and four females) from
the HCA data portal (https://data.humancellatlas.org/explore/projects/cc95ff89-2e68-4a08-a234-480eca21ce79)
and peripheral blood mononuclear cells (PBMCs) from a 29-year-old
healthy male donor from the 10× Genomics website (https://support.10xgenomics.com/single-cell-gene-expression/datasets/3.0.0/pbmc_10k_v3)
were collected.
Single-cell RNA-seq data processing
Clean sequencing reads of each sample were mapped to
the human reference genome (GRCh38) by STAR with Cell Ranger
pipelines (version 3.1.0; 10× Genomics) and the feature-barcode
matrices were generated using the 'cellranger count' command with
default parameters. The pre-filtered gene expression matrices were
analyzed using R (v.3.6.0, https://www.r-project.org/) software (29) with the Seurat package (v.3.2.3,
https://satijalab.org/seurat/) (30). Briefly, genes expressed in >3
cells of the data and cells with >200 genes detected were
extracted for further analyses. Low-quality cells were removed when
>10% of unique molecular identifiers (UMIs; >20% for the PBMC
data) were derived from the mitochondria. Gene expression matrices
were then normalized and scaled by the 'SCTransform' function.
Multiple dataset integration and batch
effect correction
To remove the batch effects across different samples
and different versions of reagent chemicals, the integration
methods adopted by the Seurat package (v.3.2.3) were employed to
assemble multiple scRNA-seq datasets into an integrated and
unbatched one. Briefly, a total of 3,000 features with high
cell-to-cell variation were identified by the
'SelectIntegrationFeatures' function. 'Anchors' between individual
datasets were computed with the 'FindIntegrationAnchors' function
and then import to the 'IntegrateData' function to create a
batch-corrected expression matrix of all cells.
Dimensional reduction and unsupervised
clustering analysis
To reduce the dimensionality of the integrated
datasets, the 'RunPCA' function in Seurat was applied with default
parameters on linear-transformation scaled data generated by the
'SCTransform' function. Next, the 'ElbowPlot' function was applied
to identify the true dimensionality of the dataset. Finally, cells
were clustered using the 'FindNeighbors' and 'FindClusters'
functions with a resolution of 0.6 and 30 clusters were obtained.
Nonlinear dimensional reduction was conducted by the 'RunUMAP'
function with dims parameter set to 50 for visualizing the
clustering result in 2D space.
Cluster marker analysis and cell type
annotation
To annotate the cell type of each cluster, the
SingleR package (31) was used to
compare the transcriptome of every single cell to the reference
immune datasets (NovershternHematopoieticData) (32) to determine cellular identity. To
verify the annotation, the expression patterns of the canonical
immune cell markers were plotted for manual inspection. The
'FindAllMarkers' function in Seurat was also used to find the top
markers for each identified cluster. Finally, 28 out of the 30
distinct cell clusters were assigned to well-known immune cell
types and two were assigned as unknown (Unk 1 and Unk 2) due to the
lack of distinct features of known cell types.
Discrimination of the maternal and
autologous cells
As the XIST gene was transcribed exclusively
in cells derived from females, the present study first defined that
the cells expressing XIST (XIST>0, XIST_ON) were from
maternal engraftment, whereas those with null XIST
expression (XIST=0, XIST_OFF) were autologous. Meanwhile, due to
the patient containing exclusive de novo IL2RG c.677C>T
mutation, maternal and autologous cells should be detected on the
variant locus as C (consistent with the reference genome, IL2RG_C)
and T (variant, IL2RG_T), respectively. Last, based on the
expression level of XIST and the sequence in the mutation
site, the maternal (XIST_ON with IL2RG_C) and autologous (XIST_OFF
with IL2RG_T) cells were distinguished.
Classification of the NK and NKT
cells
As CD3 genes (except for the CD247 gene) were
expressed only in NKT but not NK cells, the NK-like cells were
defined with at least one of the CD3 genes (CD3E,
CD3D and CD3G) as NKT and the rest as NK cells.
Differentially expressed genes and
functional enrichment
Due to various versions of reagent chemicals, the
comparison of BMMNC scRNA-seq data between the X-SCID patient and
the normal BMMNCs resulted in highly biased differentiated
expression pattern, although integrated analyses of these data sets
achieved satisfactory results in cell clustering. Therefore, to
conduct unbiased differential expression analysis, the scRNA-seq
data of the X-SCID patient BMMNCs were compared with a scRNA-seq
data set using the same reagent chemical version with PBMCs from a
healthy donor. The 'FindMarkers' function in Seurat R package was
applied with parameter 'test.use='wilcox', logfc.threshold=0.25,
min. pct=0.1' and the Benjamini-Hochberg method was used to
estimate the false discovery rate (FDR). Differentially expressed
genes (DEGs) were filtered using a minimum average log2 (fold
change) of 0.25 and a maximum FDR value of 0.05 for the comparison
between the patient's and the healthy donor's cells and with a
maximum P-value of 0.05 for the comparison between autologous and
maternal cells. Gene Ontology (GO) and Kyoto Encyclopedia of Genes
and Genomes (KEGG) enrichment of the DEGs was conducted using the
'enrichGO' and 'enrichKEGG' function of the clusterProfiler
(v.3.14.3) R package (33). Gene
sets were derived from the GO Biological Process Ontology (34,35). The Hiplot website (https://hiplot-academic.com/), a comprehensive web
platform for scientific data visualization, was used to visualize
the enrichment results (36).
Calculation of exhaustion module
scores
The present study calculated the exhaustion scores
in cell cluster with six well-defined exhaustion-related markers
(PDCD1, HAVCR2, LAG3, CTLA4,
TIGIT, TOX) (37-42) to evaluate the potential exhaustion
states. The 'AddModuleScore' function in Seurat at single cell
level was used to implement the method with default setting.
Single-cell gene set enrichment
To score pathway activities in individual cells,
single-cell gene set enrichment was conducted using the
'AUCell_buildRankings' and 'AUCell_calcAUC' function with default
parameters from the R package AUCell (43). Gene sets were selected from the
MSigDB database (44) and the
fraction of the top-ranking genes within the gene sets in each cell
was represented as the AUC value.
Flow cytometry
PBMCs of the patient (13-months-old) and his mother
were stained with viability dye (eFluor780; cat. no. 65-0865-14;
Invitrogen; Thermo Fisher Scientific, Inc.) and antibodies against
CD3 (PE/Cy7; cat. no. 300420; BioLegend, Inc.), CD56 (APC; cat. no.
17-0566-42; Invitrogen; Thermo Fisher Scientific, Inc.) and CD16
(FITC; cat. no. 11-0168-42; Invitrogen; Thermo Fisher Scientific,
Inc.) or antibodies against CD3 (PE/Cy7; cat. no. 300420;
BioLegend, Inc.), TIGIT (PE; cat. no. 12-9500-42; Invitrogen;
Thermo Fisher Scientific, Inc.) and CD279 (PD-1, FITC; cat. no.
367412) with the eBioscience intracellular fixation and
permeabilization buffer set (cat. no. 88-8824-00; Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
instruction. Briefly, 1 million PBMCs were fixed in 100 µl
IC Fixation Buffer for 30 min at RT and rinsed twice with 200
µl 1X permeabilization buffer. The cells were then
resuspended in 50 µl 1X permeabilization buffer with the
corresponding primary antibodies (1:100) for 30 min at RT protected
from light. The stained cells were analyzed with a 5-laser Cytek
Aurora flow cytometer (Cytek Biosciences). The data were analyzed
with FlowJo X (v.10.0.7 R2; FlowJo LLC). The low proportion of NK
cells would not alter the profile of the total CD3+ T
cells.
Statistical analysis
The DEGs were identified using the wilcox test by
'FindMarkers' function in Seurat R package (30), and the Benjamini-Hochberg method
was used to estimate the FDR. The GO and KEGG functional enrichment
of DEGs were conducted using the one-sided version of fisher's
exact test by 'enrichGO' and 'enrichKEGG' function in
cluster-Profiler R package (33).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Clinical characteristics of the
patient
A male infant with prolonged fever and recurrent
infections since 3-months-old was hospitalized 4 times from
4-months-old to 13-months-old. On the first admission (at
4-months-old), Pneumocystis jirovecii, Aspergillus
and Mycobacterium tuberculosis were detected in the
patient's bronchial alveolar lavage fluid and
Cytomegalovirus (CMV) and Pneumocystis jirovecii in
his PB. T-SPOT (5-6 months) and Streptococcus pneumoniae and
Haemophilus influenzae (7 months) presented later in the
patient's PB (Table I). Pulmonary
imaging continuously found multiple patchy and cable/patchy
high-density shadows (at 4 months, 5-6 months and 13 months-old)
and pneumonia (7 months-old) in the patient (Table I). Since routine blood tests
continuously detected a high number of white blood cells
(14.9-42.9×109 cells/l) and lymphocytes
(11.18-28.74×109 cells/l) in the patient (Table I), the infant was considered to
have persistent and recurrent infections. Peripheral immunological
profiles revealed that the patient exhibited a
T+B+NK+ immunophenotype, with
normal to high number of CD3+ T cells (1,619 or 5,943
cells/µl) and CD8+ T cells (1,371 or 3,619
cells/µl), normal number of CD19+ B cells (307 or
814 cells/µl) and low to normal number of CD4+ T
cells (200 or 2,172 cells/µl) and CD56+ NK cells
(105 or 215 cells/µl; Table
I). The patient had low to normal levels of IgG but he was
administered with IgG injections every month from 4-months-old.
Moreover, IgA was always undetectable in the patient and the IgM
was also almost absent at the age of 7 months (Table I), indicating the loss of function
of most B cells. Genetic screening using the patient's PB (at 5-6
months) detected a c.677C>T variant in exon 5 of the
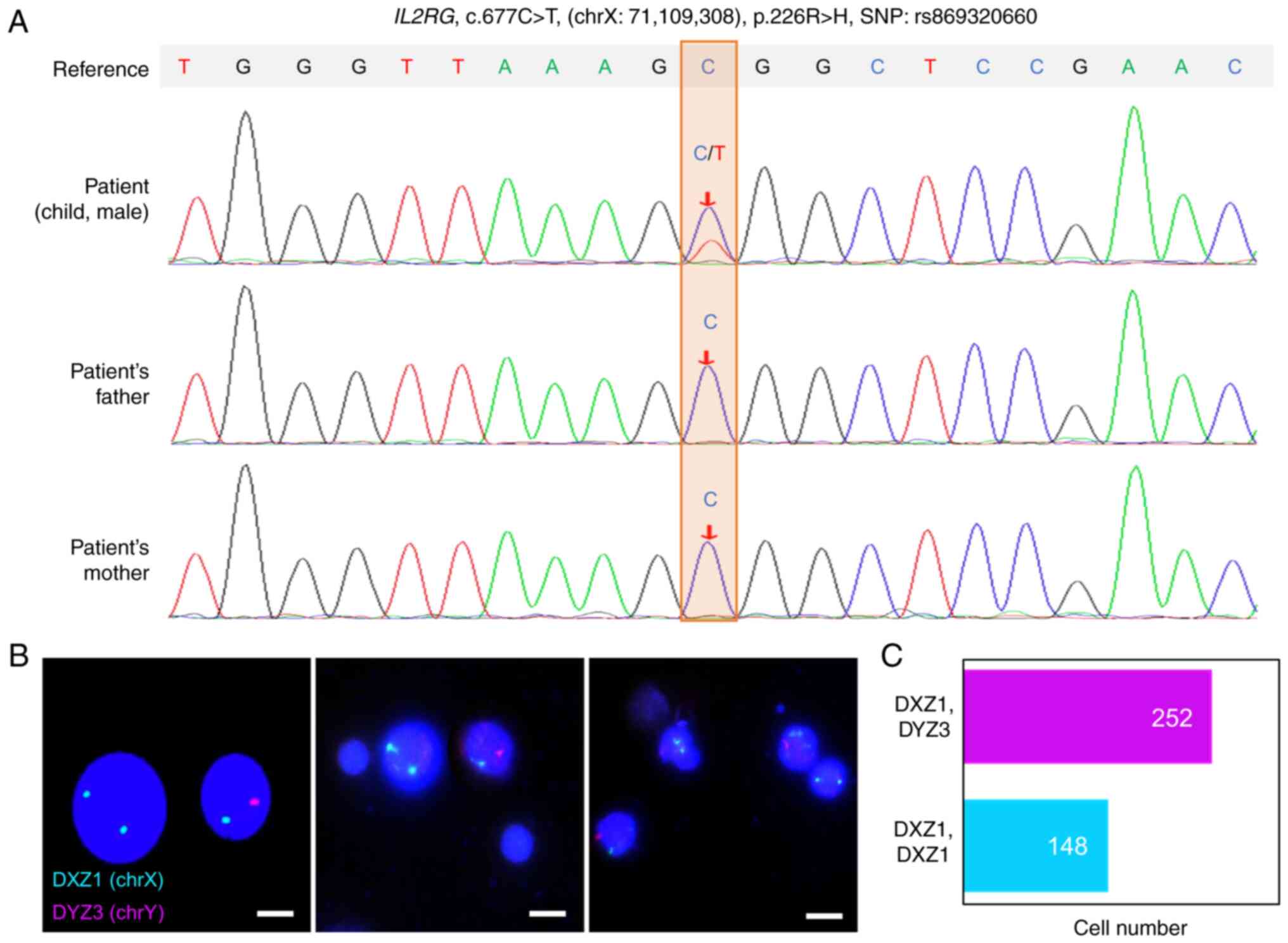
IL2RG gene (Figs. 1A and
S1A). This is a variant
documented as X-SCID-related (ClinVar ID: 225196, https://www.ncbi.nlm.nih.gov/clinvar/;
SNPdb: rs869320660, https://www.ncbi.nlm.nih.gov/snp/) (45). Therefore, the patient was
diagnosed as X-SCID. Phytohemagglutinin (PHA) stimulation failed to
induce the patient's PB cells (at 13 months) into mitosis,
irrespective of cell recovery (data not shown). The patient lacked
signs of GVHD, such as chronic eczematous dermatitis, marked
increase of liver enzyme levels and abnormal bone marrow smear. He
received broad-spectrum antibiotics, antifungal and
anti-tuberculous therapy. However, he had no chance of
hematopoietic stem cell transplantation or gene therapy and
succumbed eventually at the age of 15 months old.
 | Table IClinical immunologic
characterizations of the male X-SCID patient. |
Table I
Clinical immunologic
characterizations of the male X-SCID patient.
| Age
| Normal value |
|---|
| 4 months | 5-6 months | 7 months | 13 months |
|---|
| Infections | Pneumonia | Pneumonia, acute
hematogenous disseminated tuberculosis | Acute hematogenous
disseminated tuberculosis | Pneumonia,
diarrhea, chronic hematogenous disseminated tuberculosis | - |
| Treatment | IVIG,
cephalosporins, azithromycin, caspofungin, sulfamethoxazole,
Ganciclovir | IVIG,
cephaosporins, sulfamethoxazole, voriconazole, Isoniazid,
rifampicin, pyrazinamide | IVIG, meropenem,
isoniazid, Rifampicin, Pyrazinamide | IVIG, isoniazid,
rifampicin, pyrazinamide | - |
| Blood Routine
Examination | | | | | |
| White blood cells
(×109/l) | 15 | 23.6 | 42.9 | 14.9 | 5-12 |
| Lymphocytes
(×109/l) | 12.75 | 17.46 | 28.74 | 11.18 | 1.55-4.8 |
| Monocytes
(×109/l) | 1.05 | 1.18 | 2.57 | 0.45 | 0.55-0.96 |
| Neutrophils
(×109/l) | 1.2 | 4.72 | 9.87 | 3.28 | 2-7.2 |
| Peripheral
immunological profiles | | | | | |
| Serum IgG
(g/l) | 3.75a | ND | 6.84a | 0.55 | 3.60-9.2 |
| Serum IgM
(g/l) | 0.45 | ND | 0.06 | 0.05 | 0.38-1.26 |
| Serum IgA
(g/l) | <0.07 | ND | <0.07 | <0.07 | 0.08-0.56 |
| Serum IgE
(IU/ml) | 7 | ND | <5 | 5 | 0-15 |
| CD3+
Abs (cells/µl) | 1,619 | ND | ND | 5,943 | 805-4,459 |
|
CD3+CD4+ Abs
(cells/µl) | 200 | ND | ND | 2,172 | 345-2,350 |
|
CD3+CD8+ Abs
(cells/µl) | 1,371 | ND | ND | 3,619 | 314-2,080 |
| CD19+
Abs (cells/µl) | 307 | ND | ND | 814 | 240-1,317 |
|
CD16/56+ Abs
(cells/µl) | 105 | ND | ND | 215 | 210-1,514 |
| Laboratory test
results | | | | | |
| Virus nucleic acid
test |
Cytomegalovirus | ND | ND | ND | - |
| Blood culture | - | ND | Streptococcus
pneumoniae, Haemophilus influenzae | - | |
| Mycobacterium
tuberculosis DNA test of blood | Negative | T-SPOT
(Positive) | ND | ND | - |
| Blood culture | Negative | Negative | ND | ND | - |
| PMseq-High
throughput gene test of blood | Pneumocystis
jirovecii | ND | ND | ND | - |
| PMseq-High
throughput gene test of BALF | Pneumocystis
jirovecii, Aspergillus, Mycobacterium tuberculosis | ND | ND | ND | - |
| CT scan | Patchy and cable
strip high density shadow | Multiple patchy and
patchy high-density shadows | Pneumonia | Multiple patchy and
patchy high-density shadows | - |
| Captured-exome
sequencing of blood | - | IL2RG gene
c.677C>T (exon 5) | - | - | - |
| Vaccines | At birth: BCG
vaccine, first dose of hepatitis B vaccine |
Genetic screening of IL2RG mutation in
the X-SCID patient
The c.677C>T variant in IL2RG was detected
heterozygous in the PB of the patient (Figs. 1A and S1A). Intriguingly, whole genome
sequencing re-detected the variant but hemizygously in the
patient's hair follicles and oral pharyngeal cells (Fig. S1B and C). Therefore, it was
suspected the patient harbored maternal engrafted cells, a
phenomenon frequently occurred in the X-SCID. This hypothesis was
confirmed by chromosome microarray analysis (Fig. S1D) and FISH (Fig. 1B), with ~1/3 of the patient's
PBMCs presenting XX genotype (Fig.
1C). STR genetic marker testing also confirmed a maternal
origin of the engrafted cells (Table
SI). Collectively, a variant in the IL2RG gene was found
in an X-SCID patient with maternal cell engraftment. A reported
case with the same c.677C>T variant in IL2RG exhibited
low number of T cells phenotype (45), in contrast to the patient in the
present study.
Single-cell transcriptional landscapes of
BMMNCs in the X-SCID patient
Single-cell RNA sequencing was performed with BMMNCs
from the patient (at 13-months-old) to systematically evaluate the
states and functions of all types of X-SCID immune cells, including
undifferentiated hematopoietic cells. This obtained ~4,000 UMIs and
1,500 genes for each cell (Table
SII), indicating sufficient coverage and representative
transcripts for subsequent analyses.
The sequencing data were comprehensively analyzed in
combination with public available scRNA-seq data of BMMNCs and
PBMCs from healthy donors and 230,525 cells passed quality control
for unsupervised clustering (Fig.
S2A and Tables SII and SIII). Cells from different individuals
were well overlapped (Fig. S2B),
suggesting the proper elimination of potential batch effects among
the different samples. Overall, 30 distinct cell clusters were
obtained (Fig. 2A and Table SIV). Each cluster was annotated
according to the most variably expressed genes (Fig. S2C and Table SV) and canonical
cell-type markers (Fig. 2B and C
and Table SVI). The cluster
identity was further verified with reference datasets of human
immune cells (31) (Fig. S2D). Finally, 28 clusters were
assigned with well-defined immune cell identities, including B
cells, T cells, NK-like cells, monocytes, dendritic cells, platelet
cells, erythrocytes and progenitor and stem cells (Fig. 2A). T and NK-like cell clusters
were further manually annotated based on the expression of
well-known T and NK cell markers (Fig. S3).
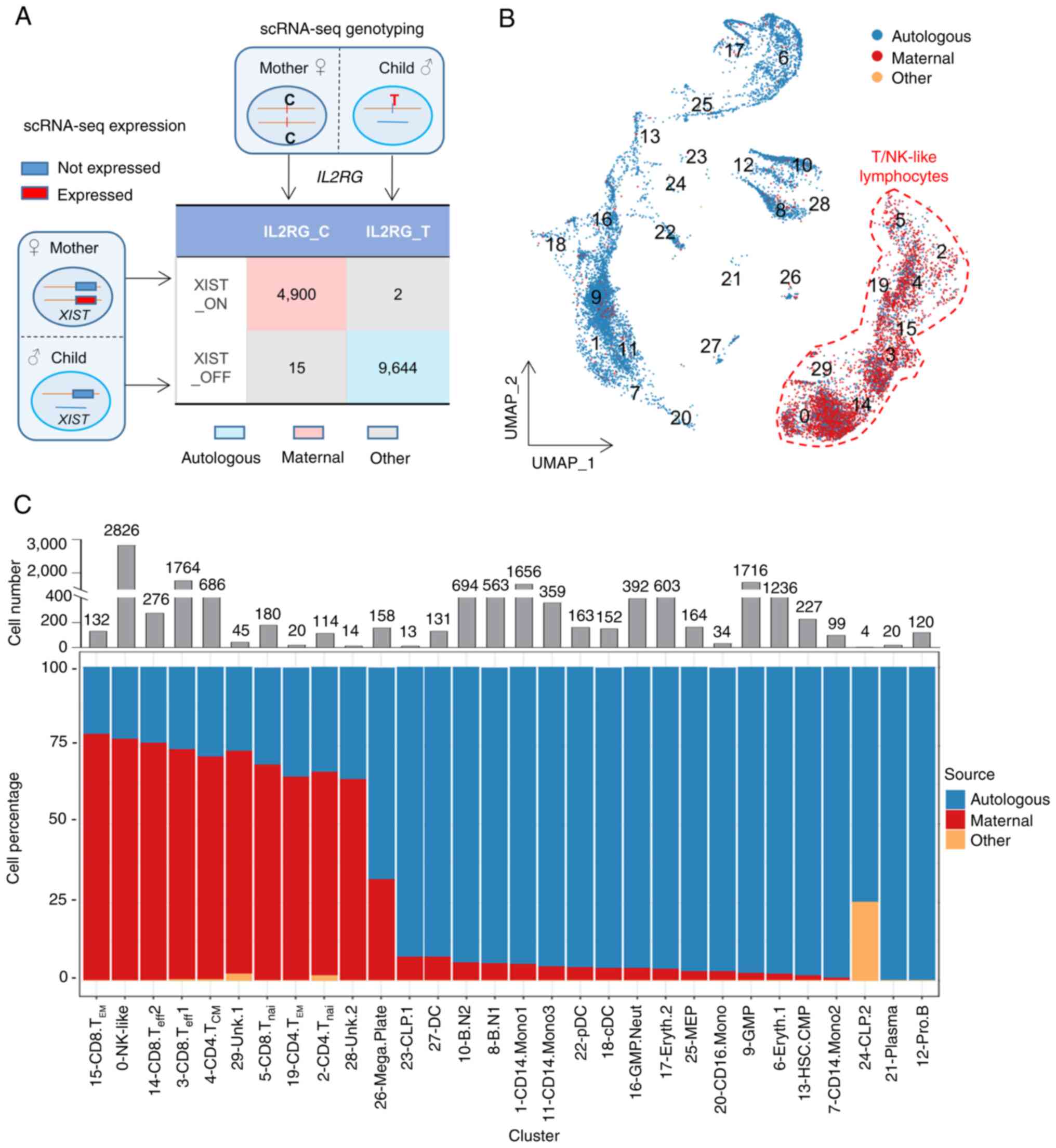
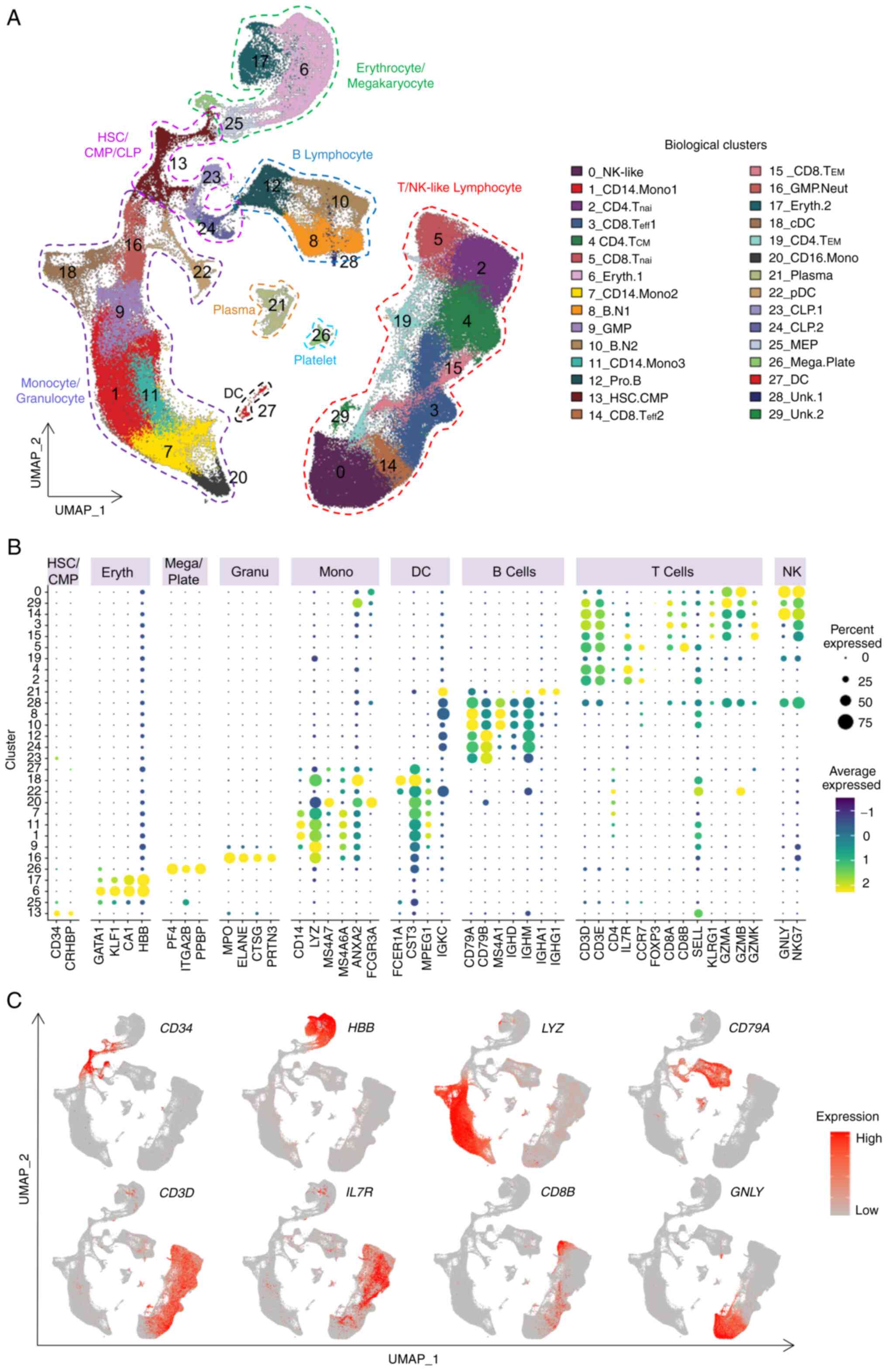 | Figure 2Single-cell transcriptome landscape
of the X-SCID patient BMMNCs. (A) BMMNCs from this X-SCID patient
and BMMNCs and PBMCs from healthy donors were projected into the
UMAP analysis showing 30 cell clusters. Each dot denotes an
individual cell and colors represent the cluster origins. The
annotation of each cluster is listed on the right. The cell number
in each cell cluster from each sample is listed in Table SIV. (B) Dot plot representing the
relative average expression of representative marker genes (x-axis)
across all clusters (y-axis). As indicated on the legend, the dot
size denotes the percentage of cells in a cluster expressing the
gene. Dot color represents the relative average expression level.
(C) Single-cell transcription levels of representative genes
illustrated in the combined UMAP plot. The transcription levels are
color-coded: Gray, not expressed; red, expressed. X-SCID, X-linked
severe combined immunodeficiency; BMMNCs, bone marrow mononuclear
cells; PBMCs, peripheral blood mononuclear cells; UMAP, uniform
manifold approximation and projection; HSC, hemopoietic stem cell;
CMP, common myeloid progenitor; CLP, common lymphoid progenitor;
GMP, granulocyte-macrophage progenitor; MEP,
megakaryocyte-erythroid progenitor; Eryth, erythrocytes; Neut,
neutrophilic; Mega, megakaryocyte; Plate, platelet; Mono,
monocytes; cDC, conventional dendritic cell; pDC, plasmacytoid
dendritic cell; NK-like, natural killer like cells; nai, naive;
B.N, naïve B cells; eff, effector; CM, central memory; EM, effector
memory; Pro, progenitor. Unk, unknown. |
Discrimination of cell sources in the
X-SCID patient at the single-cell level
As the maternal cells did not contain IL2RG
mutation, but autologous cells did, it was hypothesized that cells
of the two origins would be at different immune states in this
X-SCID patient. The expression profile with single-cell precision
provided an unprecedented opportunity to discriminate and
characterize separately the maternal engrafted cells and autologous
cells in this X-SCID patient. To this end, a novel strategy was
designed to discriminate the cell sources of the patient BMMNCs
based on the expression of XIST and the existence of the
IL2RG variant (Figs. 3A
and S4A). XIST is a highly
transcribed gene which mediates X chromosome inactivation (46,47), thus it is expressed exclusively in
cells derived from females. The present study defined that the
cells expressing XIST (XIST_ON) were from maternal
engraftment, whereas those with null XIST expression
(XIST_OFF) were autologous (Fig. S4B
and Table SVII). The expression patterns of XIST in all
samples confirmed the accuracy of this criteria (Fig. S4C). Meanwhile, due to the patient
exclusive de novo IL2RG c.677C>T mutation (Figs. 1A and S1C), maternal and autologous cells
should be detected on the variant locus as C (consistent with the
reference genome, IL2RG_C) and T (variant, IL2RG_T), respectively
(Fig. S4D). The high consistency
between the XIST_ON (XIST_OFF) category and the IL2RG_C (IL2RG_T)
subpopulation (Fig. 3A) confirmed
that both strategies efficiently and accurately discriminate the
cell sources.
To achieve on optimal discrimination of the cell
sources, maternal engrafted cells were determined to be
simultaneously XIST_ON and IL2RG_C (4,900 cells), while autologous
cells were determined to be both XIST_OFF and IL2RG_T (9,644 cells)
(Fig. 3A), with the cell origin
ratio consistent with that in PBMCs (148 XX:252 XY, Fig. 1C). A total of 17 cells failed in
the cell source identification (defined as 'Other'), probably due
to an inadequate sequencing depth and/or sequencing errors, which
were excluded in the subsequent analyses (Figs. 3, S4E and Table SVIII). The maternal
engrafted cells were primarily from the T and NK-like cell groups
but merely negligible in the remaining cell clusters (Fig. 3B, C and Table SIX). The effector memory T cells
(Cluster 15) harbored the highest ratio (78.79%) of maternal cells
(Fig. 3C and Table SIX), which might contribute to
the maintenance of the maternal T cells in the patient. Notably, it
was found that the common lymphoid progenitor cells (Cluster 23 and
24) were almost exclusively autologous (Fig. 3C), similar with the finding that
maternal HSCs were not engrafted into the patient bone marrow
(22). Meanwhile, progenitor B
cells (Cluster 12) and plasma cells (Cluster 21) were also mainly
from autologous sources and thus were not analyzed further.
Cell status across maternal and
autologous T and NK-like cells in the X-SCID patient
In general, the T cells detected in X-SCID patients
are mostly from maternal engraftment (12,16,22). In the present study, the
co-existence of both maternal and autologous T and NK-like cells in
this X-SCID patient together with the single-cell level
discrimination of cell sources provided a unique opportunity to
separately elucidate the functional status of T and NK-like cells
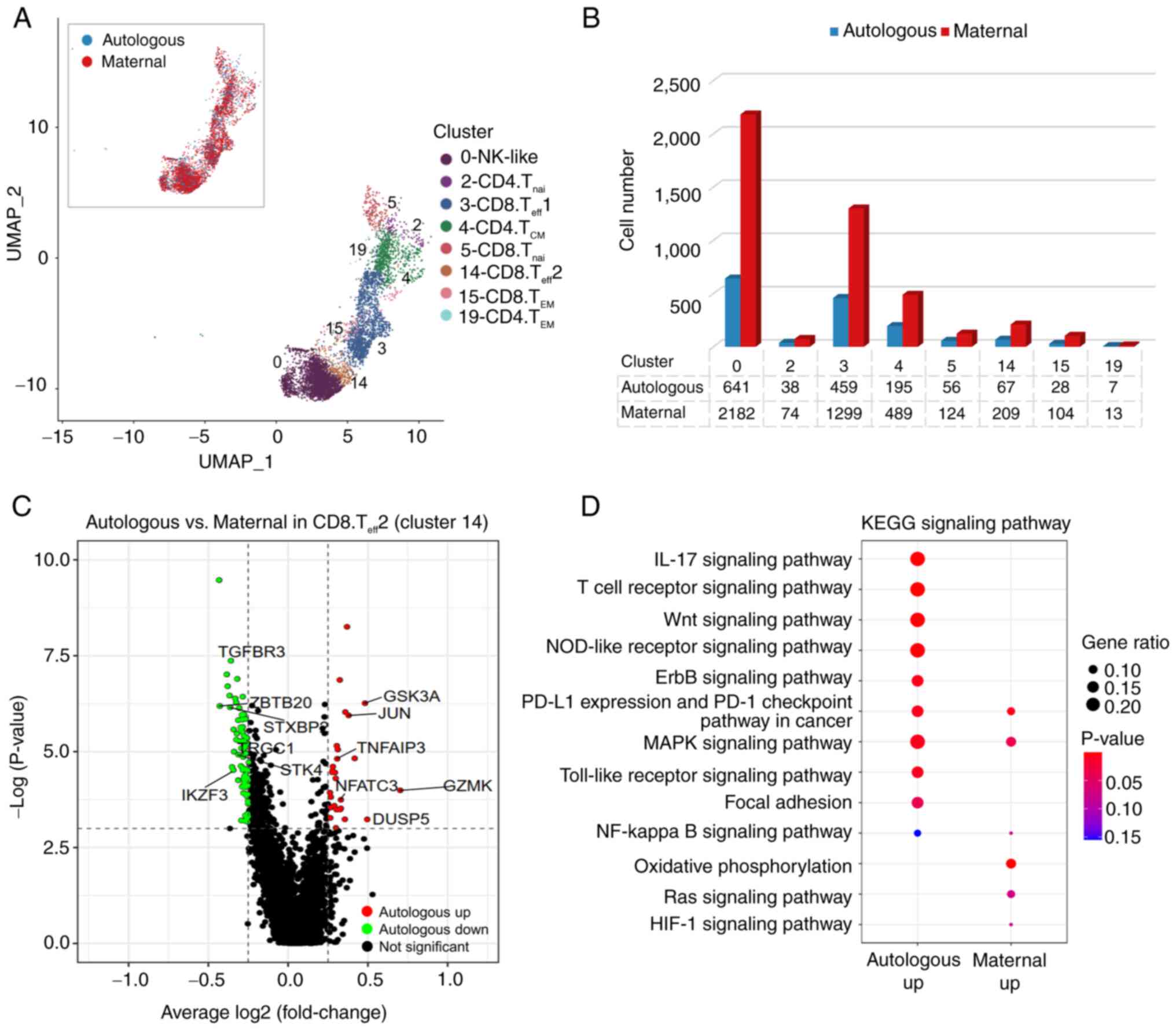
from both sources in the same individual. Cell source
discrimination revealed that maternal cells were more abundant than
autologous ones within the T and NK-like cells (Fig. 4A and B and Table SIX). Differential analyses in
clusters with >50 cells in each origin (Cluster 0, 3, 4, 5 and
14) detected the XIST gene as the most significantly DEG in
all maternal engrafted cells (Fig.
S5), confirming the efficiency of the cell origin separation.
Multiple genes were differentially expressed in a CD8+
effector T cell population (Cluster 14), including maternally
enhanced IKZF3 and TGFBR3 and autologously enriched
GZMK, TNFAIP3, JUN and NFATC3 (Fig. 4C and Table SX). GO enrichment analyses of the
DEGs revealed that the autologously upregulated genes in Cluster 14
were enriched in multiple immunity-related signaling pathways,
especially the NOD-like receptor signaling pathway and Toll-like
receptor signaling pathway (Fig.
4D), indicating a function against infections of the patient
autologous cells. Notably, cells of both origins showed an
enrichment in the PD-L1 expression and PD-1 checkpoint pathway
(Fig. 4D). Other clusters showed
similar transcriptome between cells of the two sources, except for
some DEGs in the CD8+ naïve cells (Fig. S5A). Taken together, the majority
of maternal engrafted cells in the patient exhibited a similar cell
status to that of autologous counterparts, with differentially
expressed genes mainly in a CD8+ effector T cell
population.
Characterization of T cells in the X-SCID
patient BMMNCs
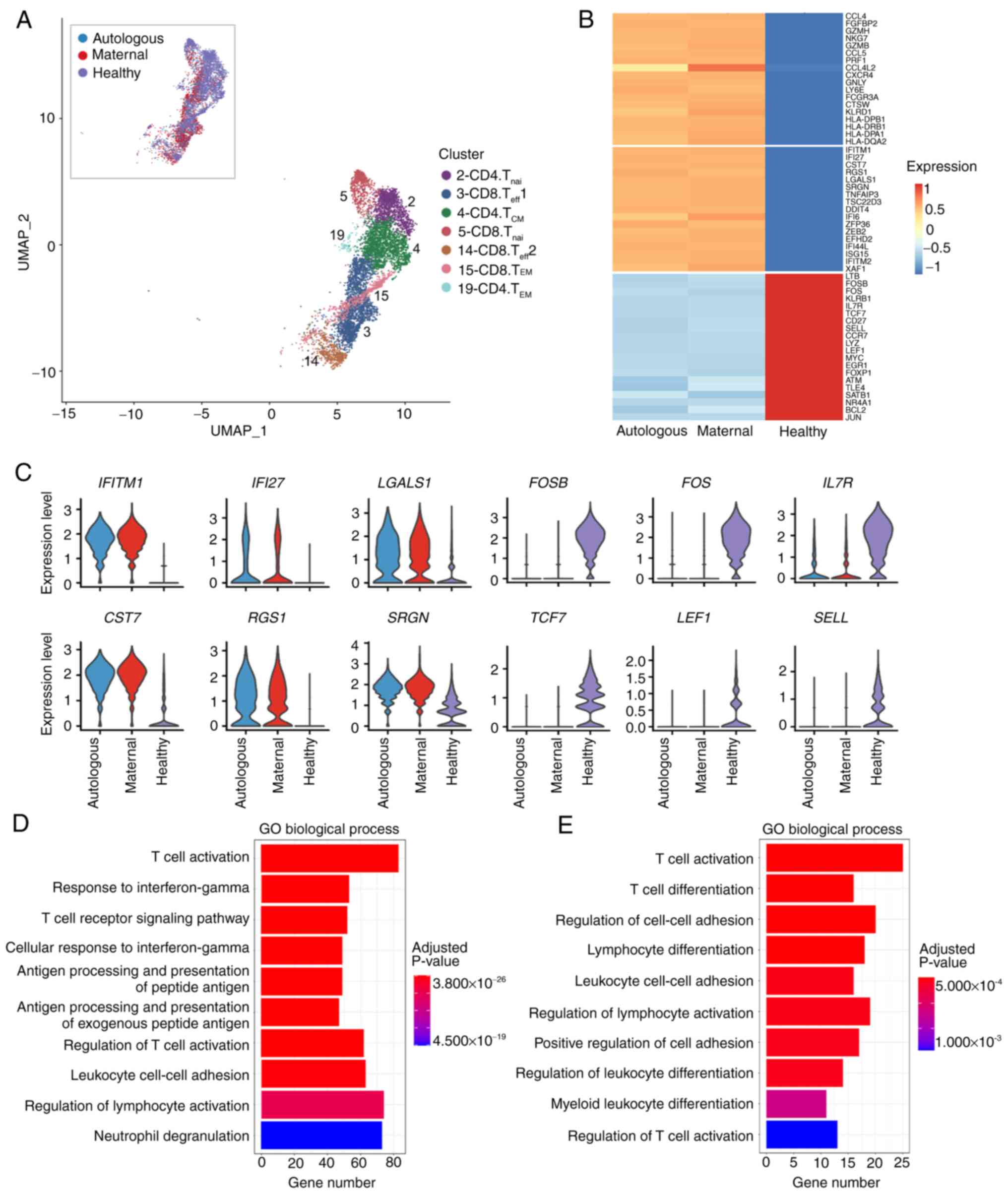
T and NK-like cells in this X-SCID patient were
primarily (65.0-78.79%) of maternal source (Fig. 3C and Table SIX). To further characterized the
status and function of the maternal and autologous T cells in the
patient, the cells in T cell clusters were compared with >50
cells in each cell origin respectively with the counterpart from
the scRNA-seq data of a healthy donor collected in the 10× Genomics
dataset (Figs. 5A and S6A). The DEG patterns were similar
across cell origins and cell clusters in the patient (Fig. S6A and B and Tables SXI and SXII).
Fig. 5B presents the average
expression levels of the DEGs common in cells of both origins and
in all clusters, except for SELL which was maternally
highest-expressed in Cluster 14 (Fig. S6B and Table SXIII). A number of
genes related to antigen presentation and inflammatory effects
(HLA-DPA1, HLA-DPB1, GZMB and PRF) were
higher expressed in both autologous and maternal cells (Fig. 5B), reflecting the immune reaction
of the patient to fight against severe and continuous infections.
Accordingly, type I interferon response genes, such as
IFITM1 and IFI27, tended to be expressed at higher
levels in the patient (Fig. 5C).
However, a number of transcription factors (TFs) that are important
for T cell proliferation, differentiation and maturation (FOS,
TCF7, LEF1 and MYC) (48-51) were significantly attenuated in the
patient (Fig. 5B and C),
accounting for the fact that T cells were not properly
differentiated and matured in the X-SCID patients (52).
To comprehensively understand the nature of the T
cell status and function, GO enrichment analyses of the DEGs
revealed that the patient-upregulated genes were enriched in
biological processes such as T cell activation, response to
interferon-γ and antigen processing and presentation, reflecting an
active immune response in the patient (Fig. 5D). The patient-downregulated genes
were prone to processes of T cell activation and differentiation,
regulation of cell-cell adhesion and regulation of lymphocyte
activation (Fig. 5E), indicating
T cell dysregulation in the activation and differentiation in this
X-SCID patient.
Characterization of NK-like cells in the
X-SCID patient BMMNCs
The NK-like cells contained two cell types, NK and
NKT cells, which were similar in the expression pattern of CD56
(NCAM1) and CD16 (FCGR3A), but differed in the
expression of CD3 (Fig. S7A).
Flow cytometry results also showed the existence of the NK
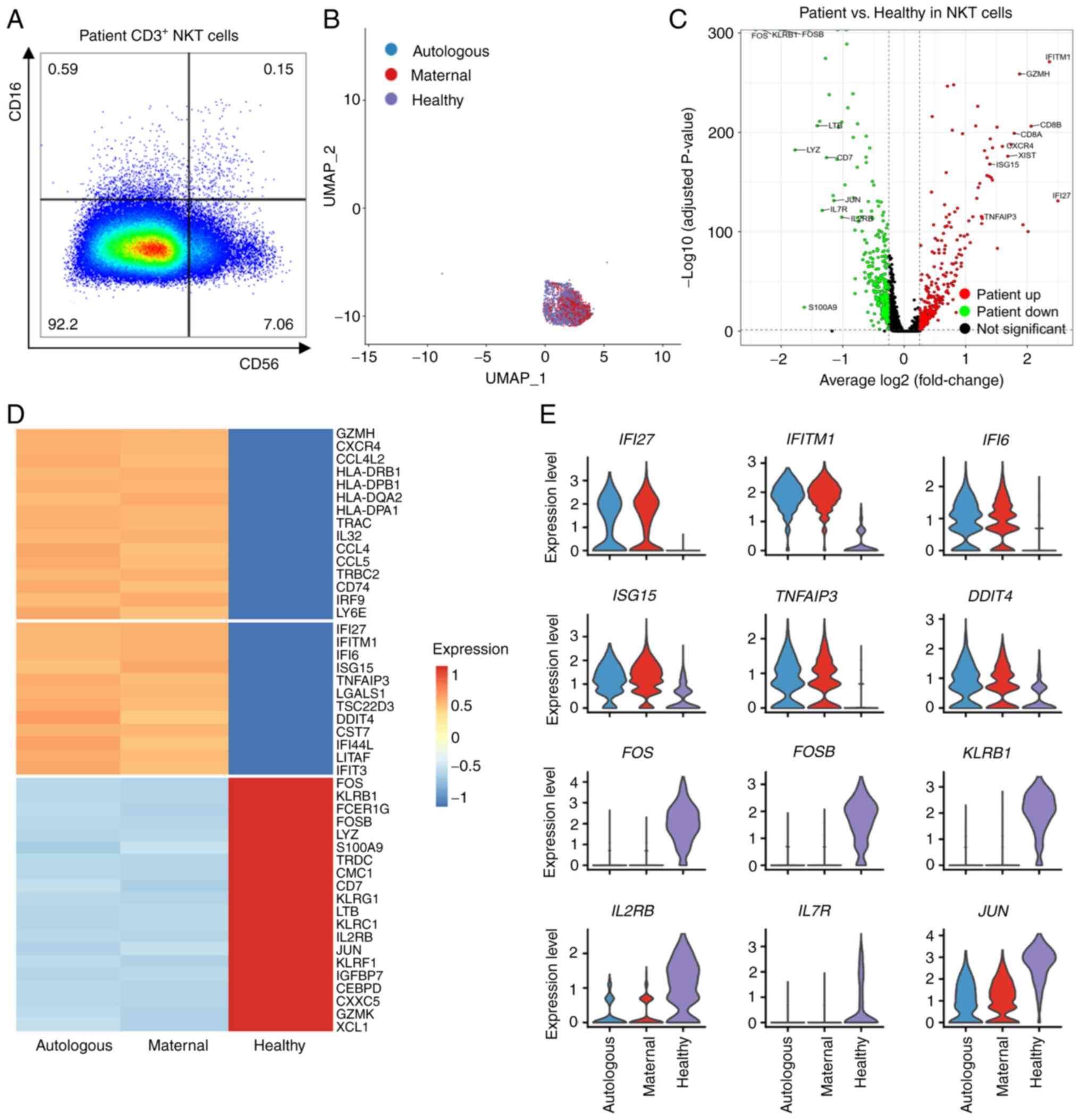
(Fig. S7B) and NKT (Figs. 6A and S7C) cells in the PB of the patient and
his mother. The NK-like cells in Cluster 0 were then classified
into NK [45 (autologous)/110 (maternal), 1.1% of 14,561 (total) and
NKT [596 (autologous)/2,072 (maternal), 18.3% of 14,561 (total)];
Figs. 6B, S7D and Table SXIV]. Moreover, the
expression of CD16 and CD56 in the maternal cells confirmed the
existence of maternal engrafted NKT (Fig. S7E) and NK (Fig. S7F) cells in the patient. As
neither NKT (Fig. S7G) nor NK
(Fig. S7H) cells presented DEGs
other than XIST between autologous and maternal cells, cells
of the two origins were combined in each cluster to compare with
the counterpart cells from the normal donor [520 (NKT)/237 (NK),
4.7% (NKT)/2.2% (NK) of 10,964 (total); Figs. 6B, S7D and Table SXIV]. Nevertheless, the
expression level of DEGs in autologous, maternal and healthy NKT
were showed separately (Fig. 6C and
D and Table SXV) and NK
(Fig. S8A and B and Table SXVI)
cells.
A number of DEGs were detected in the patient's NKT
and NK cells when compared with the normal counterpart. The
CD8A up-regulation (Fig.
6C) in the patient's NKT may reflect an increase of
CD8+ NKT cells due to infections (53). As in T cells, a number of genes
related to antigen presentation and inflammatory effects
(HLA-DPA1, HLA-DPB1, GZMH and CXCR4)
and type I interferon responses (IFI27, IFITM1 and
IFI6) were more abundant in the patient's NKT (Fig. 6C-E) and NK (Fig. S8A-C) cells. This suggest that both
NKT and NK cells exerted a function to defense against persistent
infections in the patient. However, the expression levels of some
effector molecules (e.g. KLRG1, FCER1G, KLRB1
and LYZ) decreased dramatically in the patient's NKT
(Fig. 6D and E) and NK (Fig. S8B and C) cells, suggesting an
insufficient function of these immune cells. The expression of TFs
such as FOS, FOSB, JUN and CEBPD also
decreased obviously in the patient's NKT (Fig. 6D and E) and NK (Fig. S8B and C) cells. Notably, there
were only 45 autologous NK cells, thus the findings within this
cell type require further confirmation. Taken together, both
maternal and autologous NKT, and probably also NK cells, in this
X-SCID patient were dysfunctional, which may also be related to the
X-SCID clinical symptoms in the patient.
GO enrichment analyses of the DEGs revealed that
the genes upregulated in the patient's NKT cells were enriched in
biological processes of type I interferon signaling pathway,
antigen processing and presentation and virus defense response,
consistent with the immune reaction against the infections.
Meanwhile, the patient's NKT-downregulated genes were more
frequently detected in biological processes such as T cell
activation, response to IL-12 and neutrophil activation (Fig. S9), reflecting the inability of NKT
cells to exert complete immune function. Overall, the data
suggested the patient's NKT cells were fighting against the
infections, but the dysfunctional cell state confined their
capability to efficiently eliminate the infections, which could
also facilitate recurrent and continuous infections in the
patient.
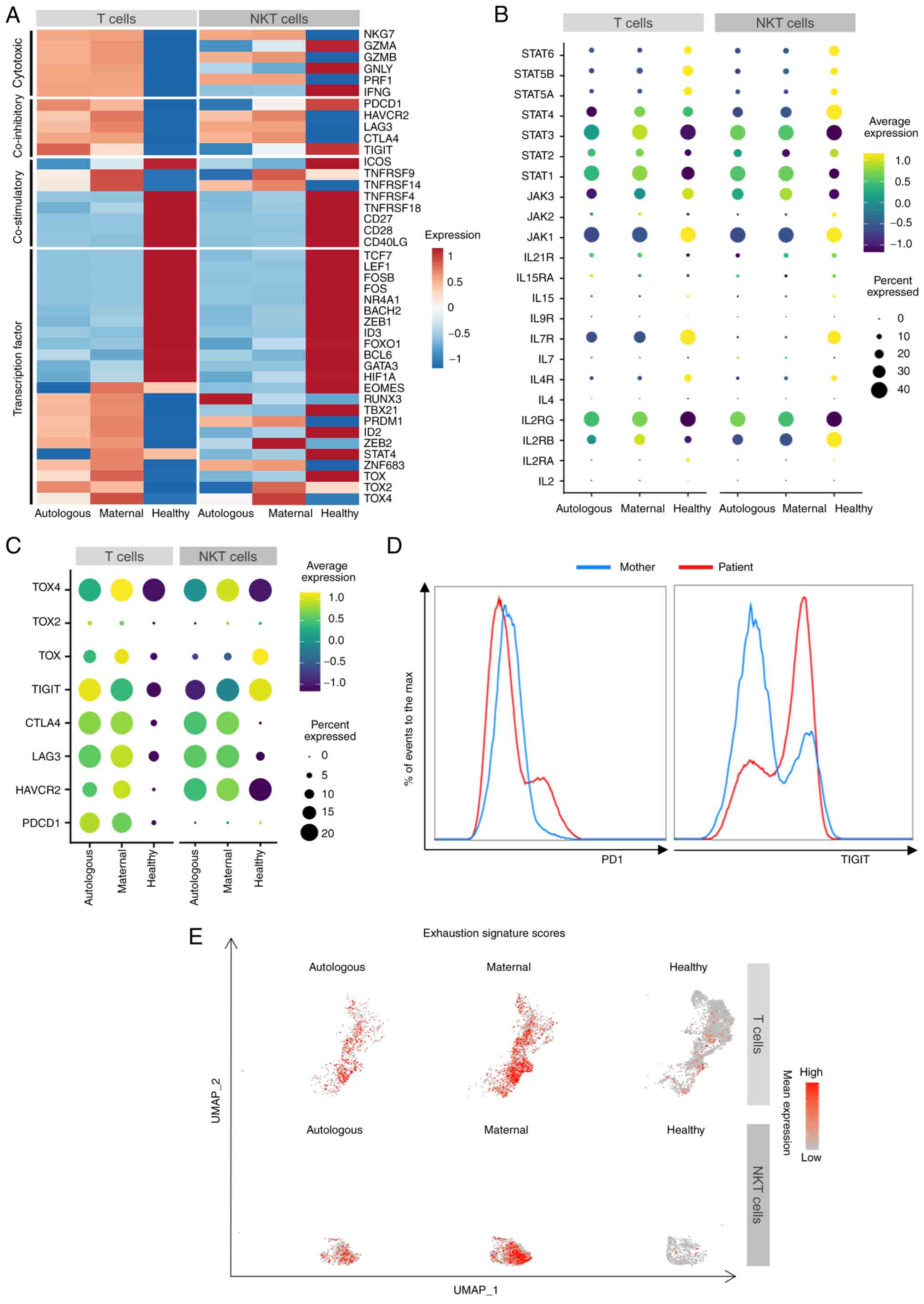
Exhausted-like T and NKT lymphocytes in
the X-SCID patient
To examine the dysfunctional status of T and NKT
cells in this X-SCID patient, some key immune signatures relative
to immunity activity were next explored. The analyses confirmed
that the patient specifically increased expression of a number of
cytotoxic cytokines (NKG7, GZMA, GZMB,
GNLY, PRF1 and IFNG), with exceptions of
IFNG, GZMA and GNLY in the patient's NKT cells
(Fig. 7A). However,
co-stimulatory factors including TNFRSF4, TNFRSF18,
CD27, CD28 and CD40LG were notably decreased
in the patient's cells (Fig. 7A).
Thus, the patient's T and NKT cells were probably incapable of
being sufficiently activated. To this end, the patient's T and NKT
cells expressed a lower level of a number of effector molecules
like LYZ (Figs. 5B and
6D) and were unable to
efficiently eliminate the infections.
Next, the present study explored how T and NKT
cells became dysfunctional in the patient. Multiple key
transcription factors, such as FOS, JUN, TCF7 and
LEF1, are involved in lymphocyte proliferation,
differentiation and immune responses (48-51), but they were notably downregulated
in the patient's T and NKT cells (Fig. 7A), which may contribute to the
inability of peripheral blood cells to proliferate upon PHA
stimulation. As BCL2 expression is regulated by FOS
and JUN (54,55), the lower level of BCL2 in
the patient's T cells (Fig. 5B)
could be attributed to the attenuated FOS and JUN
expression in these cells. As FOS and JUN are
regulated via the JAK-STAT signaling pathway (48,49), the expression pattern of
JAK-STAT-related genes were examined and it was found that
IL7R, JAK1, STAT5A, STAT5B and
STAT6 were downregulated, while IL2RG, STAT1
and STAT3 were upregulated in the patient's T and NKT cells
from both sources (Fig. 7B).
IL2RB and STAT2 were increased in the patient's T but
decreased in his NKT cells. By contrast, JAK3 expression was
decreased in the patient's T but increased in NKT cells.
STAT4 levels were lower in the patient's T and NKT cells,
except for the higher levels in maternal T cells (Fig. 7B). Moreover, IL2RG ligands,
especially IL2 secreted by activated T cells, were expressed at a
negligible level (Fig. 7B). These
abnormalities indicated a dysregulation of the JAK-STAT pathway,
which impaired the expression of key transcription factors
mediating lymphocyte proliferation, differentiation and immune
responses, leading to the dysfunction of T and NKT cells of both
origins in the patient.
Further analyses revealed that the patient's T and
NKT cells of both origins enhanced the expression of multiple
exhaustion-related inhibitory genes, such as HAVCR2 and
CTLA4, and especially LAG3 (37-39,56) and the transcriptional factor
TOX4 (Fig. 7A and C).
Moreover, TIGIT and PDCD1 expression levels were
higher in the patient's T cells (Fig.
7A and C), as confirmed with by flow cytometry of the patient's
peripheral blood (Fig. 7D).
Notably, the expression levels of some inhibitory molecules varied
in the maternal and autologous T and NKT cells in the patient. For
example, PDCD1 was slightly more transcribed in autologous T
cells. TIGIT was higher in autologous T cells, but lower in
autologous NKT cells. In addition, the enrichment score of
canonical exhaustion markers were higher in the maternal T and NKT
cells (Fig. 7E). Taken together,
the T and NKT cells were at an exhausted-like dysfunction status
post a long-term recurrent pathogen infection in this X-SCID
patient.
Discussion
The variant c.677C>T (p.R226H) on the
IL2RG gene was documented in the ClinVar database (Variation
ID: 225196) as a pathogenic variation causing X-SCID. Several
studies have reported patients with this particular variant
presenting with low number of T cells and
B+NK− or B+NK+
phenotype, but did not describe the maternal engraftment in detail
(57-63), although it is a general phenomenon
in SCID patients (12). The
functional status of the engrafted cells remains largely unclear
since the traditional cell separation methods, such as flow
cytometry and magnetic bead separation, were not able to separate
the cell origins effectively. Moreover, since the maternal
engrafted cells may consist of more than one cell type and/or cell
state (12,16,17,22), separation and analysis of cells
merely based on cell sources (depending on HLA) (12) would confine the systematics
evaluation of the cell status of multiple cell types.
The present study applied the state-of-art
single-cell technology to study systematically the transcriptome
landscape of BMMNCs from a 13-months-old male X-SCID patient with
maternal engraftment caused by a de novo c.677C>T
mutation in the IL2RG gene. This allowed the present study
to discriminate the maternal engrafted and autologous cell origins
and characterize their functions/states with single-cell
resolution. It first tried to use published approaches like
'Souporcell' (64) to determine
the cell origin based on SNPs, but shortly realized the SNPs
between the autologous and maternal cells were heterozygous and
represented a low density in genome with sparse read coverage,
which confined the efficiency of cell separation. The present study
thus designed a novel analytical strategy to effectively
distinguish the cell origins of BMMNCs in this X-SCID patient based
on the exclusive maternal expression of the XIST gene and
the autologously specific de novo mutation on IL2RG
and successfully achieved high confidence in cell source
discrimination.
The present study detected both autologous and
maternal engrafted T, NKT and NK cells in this X-SCID patient.
Usually, engraftment of the maternal T cells is the most frequent
and was detected in 62% of the B− SCID and in 50% of the
B+ SCID (12). These T
cells were generally defective in immunity function and limited or
no proliferative response to mitogens (12,14,18), in consistent with the absent cell
proliferation of the patient's T cells upon PHA. The exclusively
autologous HSCs and CLP cells in the patient probably gave rise to
the patient's autologous T cells. The higher expression of
EOMES in maternal T cells (Fig. 7A) and the highest maternal cell
ratio in the effector memory T cell cluster indicated that the
maternal T cells were maintained in the patient probably due to the
engraftment of memory T cells, but not naive T cells, as described
in the previous studies (14,65).
Previous studies typically characterized SCID based
on the effects of classical T, B and NK cells (1,3,15-17,20-22), but did not focus on NKT cells. The
present study found NKT dysfunction in cytotoxicity and immune
regulation in this X-SCID patient. Further study with more cases is
required to verify if the NKT dysfunction exists overall in X-SCID
patients and to understand whether the NKT dysfunction is related
to the X-SCID clinical symptoms. The present study also
characterized the NK cells in this X-SCID patient, which was not
discussed in depth in previous studies (17,22), although the findings in this cell
type require further investigation due to the low cell number in
the patient.
The co-existence of both maternal and autologous T,
NKT and NK cells in the patient allowed the present study to
explore comprehensively the status and function of both normal and
IL2RG-mutated cells of these immune cell types in the same
microenvironment. When compared with the autologous counterpart,
maternal T cells expressed lower level of GZMK and
JUN in Cluster 14 (Fig.
4C) and CD8+ naïve T cells also exhibited
differentially expressed genes (Fig.
S5A). The two clusters consisted of a small proportion of the
total cells, so most of the maternal cells were at a similar cell
status to the autologous ones.
The comprehensive comparison with public scRNA-seq
data from healthy donors revealed that both maternal engrafted and
autologous T, NKT and NK cells in the patient increased the
expression of a number of cytokines and type I interferon
responsive genes, suggesting that both maternal and autologous
cells fight against recurrent infections in the patient. However,
possibly due to the deregulated JAK-STAT pathway (66) in both maternal and autologous
cells, the expression of multiple transcription factors mediating
the differentiation and activation of T, NKT and NK lymphocytes
(FOS, JUN, TCF7 and MYC) (48-51) was substantially decreased in this
X-SCID patient's cells of both origins, consistent with the
abnormalities of T, NKT and NK cells previous reported in the
X-SCID patients (1,67). Due to the necessity of
co-stimulatory molecules for lymphocyte cell proliferation,
differentiation, maintenance and activation (68), the deregulation of CD27,
CD28 and CD40LG in this X-SCID patient's cells from
both origins may have contributed to his T cell dysfunction and
non-proliferation upon PHA. Notably, the exhaustion-related
inhibitory genes HAVCR2, CTLA4 and especially
LAG3 (37-39) were upregulated in the patient's T,
NKT and NK cells from both cell origins. TIGIT and
PDCD1 were higher in the patient's T cells at both
expression and protein levels (Fig.
7A, C and D). This indicated that the T and NK-like cells in
this X-SCID patient may have developed an exhausted-like status. In
consistence with this result, Okano et al (22) reported a higher PD1 protein level
in an X-SCID patients with respiratory syncytial virus infection.
This is in accordance with reports that both the maternal engrafted
cells and autologous cells in SCID are similarly dysfunctional
(13,19). Studies with more patients should
help to verify if the exhaustion-like state of these immune cells
is a common phenomenon in X-SCID patients.
Why were both maternal and autologous cells at a
similar cell status? One scenario was that the compromised immunity
of the patient was unable to efficiently eliminate foreign viruses
and bacteria, resulting in prolonged and recurrent infections and
chronic inflammation that may finally exhaust the T and NKT cells.
The BMMNC sample in this study was collected when the patient 13
months old, close to the mortality of the patient at 15 months.
Maternal engrafted and autologous cells at this time point had both
already become exhausted-like due to the recurrent pneumonia and
acute hematogenous disseminated tuberculosis, with the maternal
cells even more exhausted (Fig. 7C
and D). The exhausted-like T cells in the patient limited the
expression of IL2 (Fig.
7B) as in T cell exhaustion defined in other situations
(69). Although the maternal T
cells in the patient carried the normal IL2RG gene, there
might be insufficient IL2 ligands produced in the repressed immune
state to activate the normal receptor. In this situation, even
though the maternal cells exhibit slightly higher level of
IL2RG, JAK3 and STAT1-4, the normal IL2RG in
the maternal cells could not exert its function to completely
activate the JAK-STAT signaling pathway.
Except for the mutated IL2RG in the
autologous cells, the restricted TCR repertoire in maternal T cells
(16,70,71) might also contribute to the
compromised immunity in the patient. This was supported by the
finding in the present study that the gene sets related to TCR
diversity were depleted in the patient's T cells (Fig. S6C). Analyses of the TCR repertoire
in maternal and autologous T cells using single-cell TCR-seq may
help to clarify this possibility. However, 3' scRNA-seq rather than
5' applied in this study impeded the access to the TCR sequences at
the 5' end of the TCR genes. Another concern is that due to the low
case frequency of X-SCID, the present study collected only this
reported case in the past two year. Integrated analyses of larger
cohorts of X-SCID patients of different phenotypes prior to and
post infections with the 5' scRNA-seq could provide further
insights into the common cell status of T and NKT cells and their
development in the X-SCID patients.
In conclusion, the present study comprehensively
profiled the immune landscape of BMMNCs in an X-SCID patient with
maternal engraftment at the single-cell transcriptome level. With
the state-of-the-art single-cell sequencing technology, it
effectively discriminated the maternal cells from the X-SCID
patient autologous ones. The present study revealed the dysfunction
of T and NKT cells of both origins contributing to X-SCID symptoms
and identified multiple immune signatures involved in T and NKT
lymphocyte dysfunction and exhaustion. These results might provide
new insights into the mechanisms underlying X-SCID pathogenesis and
could contribute to the early diagnosis of SCID patients.
Supplementary Data
Availability of data and materials
The single-cell RNA sequencing data of the X-SCID
patient in this study has been deposited in the GSA database
(Genome Sequence Archive in BIG Data Center, Beijing Institute of
Genomics, Chinese Academy of Sciences) under the accession number
HRA000811 (accessible at http://bigd.big.ac.cn/gsa-human). The bulk WGS and
captured exome sequencing data were also deposited in the GSA
database, under the accession number HRA001493. All other data
generated or analyzed during this study are included in this
published article. Supplementary Information is available from the
corresponding authors on reasonable request. The scripts generated
during this study are available at the GitHub repository
(https://github.com/dongwei1220/X-SCID_scRNA-seq).
Authors' contributions
GL, XW and ZT designed and supervised the study. WD
performed the data analysis and interpretation. HF and XW collected
the clinical samples, XW and XZ prepared the scRNA-seq libraries.
WL, SZ, QQ and XW performed the FACS experiment. WD, XW and WL
wrote the manuscript and other co-authors critically reviewed and
modified the manuscript. LG and XW confirm the authenticity of all
the raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Informed consents for the performed studies were
obtained from the patient's parents, in accordance with the
principles of the ethics committee of the Guangzhou Women and
Children's Medical Center (approval number: 105A01).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by grants from the National
Natural Science Foundation of China (grant no. 31900415), Research
Foundation of Guangzhou Women and Children's Medical Center for
Clinical Doctor (grant no. 1600071) and Guangzhou Basic Research
Plan of Basic and Applied Basic Research Foundation (grant no.
202201011221).
References
|
1
|
Fischer A: Severe combined
immunodeficiencies (SCID). Clin Exp Immunol. 122:143–149. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Notarangelo LD: Primary
immunodeficiencies. J Allergy Clin Immunol. 125(2 Suppl 2):
S182–S194. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cirillo E, Giardino G, Gallo V, D'Assante
R, Grasso F, Romano R, Lillo CD, Galasso G and Pignata C: Severe
combined immunodeficiency-an update. Ann N Y Acad Sci. 1356:90–106.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fazlollahi MR, Pourpak Z, Hamidieh AA,
Movahedi M, Houshmand M, Badalzadeh M, Nourizadeh M, Mahloujirad M,
Arshi S, Nabavi M, et al: Clinical, laboratory, and molecular
findings for 63 patients with severe combined immunodeficiency: A
decade s experience. J Investig Allergol Clin Immunol. 27:299–304.
2017. View Article : Google Scholar
|
|
5
|
Tangye SG, Al-Herz W, Bousfiha A, Chatila
T, Cunningham-Rundles C, Etzioni A, Franco JL, Holland SM, Klein C,
Morio T, et al: Human inborn errors of immunity: 2019 update on the
classification from the international union of immunological
societies expert committee. J Clin Immunol. 40:24–64. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Noguchi M, Yi H, Rosenblatt HM, Filipovich
AH, Adelstein S, Modi WS, McBride OW and Leonard WJ: Interleukin-2
receptor gamma chain mutation results in X-linked severe combined
immunodeficiency in humans. Cell. 73:147–157. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Russell SM, Johnston JA, Noguchi M,
Kawamura M, Bacon CM, Friedmann M, Berg M, McVicar DW, Witthuhn BA
and Silvennoinen O: Interaction of IL-2R beta and gamma c chains
with Jak1 and Jak3: Implications for XSCID and XCID. Science.
266:1042–1045. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kovanen PE and Leonard WJ: Critical roles
of the gamma (c)-dependent cytokines interleukins 2, 4, 7, 9, 15
and 21, and their signaling pathways. Immunol Rev. 202:67–83. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Niemela JE, Puck JM, Fischer RE, Fleisher
TA and Hsu AP: Efficient detection of thirty-seven new IL2RG
mutations in human X-linked severe combined immunodeficiency. Clin
Immunol. 95:33–38. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chong HJ, Maurer S and Heimall J: What to
do with an abnormal newborn screen for severe combined immune
deficiency. Immunol Allergy Clin North Am. 39:535–546. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Myers LA, Patel DD, Puck JM and Buckley
RH: Hematopoietic stem cell transplantation for severe combined
immunodeficiency in the neonatal period leads to superior thymic
output and improved survival. Blood. 99:872–878. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Muller SM, Ege M, Pottharst A, Schulz AS,
Schwarz K and Friedrich W: Transplacentally acquired maternal T
lymphocytes in severe combined immunodeficiency: A study of 121
patients. Blood. 98:1847–1851. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kellermayer R, Hsu AP, Stankovics J,
Balogh P, Hadzsiev K, Vojcek A, Maródi L, Kajtár P, Kosztolányi G
and Puck JM: A novel IL2RG mutation associated with maternal T
lymphocyte engraftment in a patient with severe combined
immunodeficiency. J Hum Genet. 51:495–497. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tezcan I, Ersoy F, Sanal O, Turul T, Uckan
D, Balci S, Hicsonmez G, Prieur M, Caillat-Zucmann S, Le Deist F
and De Saint Basile G: Long-term survival in severe combined immune
deficiency: The role of persistent maternal engraftment. J Pediatr.
146:137–140. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
van der Burg M and Gennery AR: Educational
paper. The expanding clinical and immunological spectrum of severe
combined immunodeficiency. Eur J Pediatr. 170:561–571. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lev A, Simon AJ, Trakhtenbrot L, Goldstein
I, Nagar M, Stepensky P, Rechavi G, Amariglio N and Somech R:
Characterizing T cells in SCID patients presenting with reactive or
residual T lymphocytes. Clin Dev Immunol. 2012:2614702012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Touzot F, Dal-Cortivo L, Verkarre V, Lim
A, Crucis-Armengaud A, Moshous D, Héritier S, Frange P, Kaltenbach
S, Blanche S, et al: Massive expansion of maternal T cells in
response to EBV infection in a patient with SCID-Xl. Blood.
120:1957–1959. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Denianke KS, Frieden IJ, Cowan MJ,
Williams ML and McCalmont TH: Cutaneous manifestations of maternal
engraftment in patients with severe combined immunodeficiency: A
clinicopathologic study. Bone Marrow Transplant. 28:227–233. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Thompson LF, O'Connor RD and Bastian JF:
Phenotype and function of engrafted maternal T cells in patients
with severe combined immunodeficiency. J Immunol. 133:2513–2517.
1984. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kobrynski LJ and Abramowsky C: Monoclonal
IgA gammopathy due to maternal B cells in an infant with severe
combined immunodeficiency (SCID) prior to hematopoietic stem cell
transplantation. J Pediatr Hematol Oncol. 28:53–56. 2006.PubMed/NCBI
|
|
21
|
Morinishi Y, Imai K, Nakagawa N, Sato H,
Horiuchi K, Ohtsuka Y, Kaneda Y, Taga T, Hisakawa H, Miyaji R, et
al: Identification of severe combined immunodeficiency by T-cell
receptor excision circles quantification using neonatal guthrie
cards. J Pediatr. 155:829–833. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Okano T, Nishikawa T, Watanabe E, Watanabe
T, Takashima T, Yeh TW, Yamashita M, Tanaka-Kubota M, Miyamoto S,
Mitsuiki N, et al: Maternal T and B cell engraftment in two cases
of X-linked severe combined immunodeficiency with IgG1 gammopathy.
Clin Immunol. 183:112–120. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bolger AM, Lohse M and Usadel B:
Trimmomatic: A flexible trimmer for Illumina sequence data.
Bioinformatics. 30:2114–2120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li H and Durbin R: Fast and accurate short
read alignment with burrows-wheeler transform. Bioinformatics.
25:1754–1760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li H, Handsaker B, Wysoker A, Fennell T,
Ruan J, Homer N, Marth G, Abecasis G and Durbin R; 1000 Genome
Project Data Processing Subgroup: The sequence alignment/map format
and SAMtools. Bioinformatics. 25:2078–2079. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Robinson JT, Thorvaldsdottir H, Wenger AM,
Zehir A and Mesirov JP: Variant review with the integrative
genomics viewer. Cancer Res. 77:e31–e34. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Robinson JT, Thorvaldsdottir H, Winckler
W, Guttman M, Lander ES, Getz G and Mesirov JP: Integrative
genomics viewer. Nat Biotechnol. 29:24–26. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Thorvaldsdottir H, Robinson JT and Mesirov
JP: Integrative genomics viewer (IGV): High-performance genomics
data visualization and exploration. Brief Bioinform. 14:178–192.
2013. View Article : Google Scholar :
|
|
29
|
R Core Team: 2020, R: A language and
environment for statistical computing. R Foundation for Statistical
Computing; Vienna, Austria: URL https://www.R-project.org/.
|
|
30
|
Stuart T, Butler A, Hoffman P, Hafemeister
C, Papalexi E, Mauck WM III, Hao Y, Stoeckius M, Smibert P and
Satija R: Comprehensive integration of single-cell data. Cell.
177:1888–1902 e21. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Aran D, Looney AP, Liu L, Wu E, Fong V,
Hsu A, Chak S, Naikawadi RP, Wolters PJ, Abate AR, et al:
Reference-based analysis of lung single-cell sequencing reveals a
transitional profibrotic macrophage. Nat Immunol. 20:163–172. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Novershtern N, Subramanian A, Lawton LN,
Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay
T, et al: Densely interconnected transcriptional circuits control
cell states in human hematopoiesis. Cell. 144:296–309. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gene Ontology Consortium: The gene
ontology resource: Enriching a GOld mine. Nucleic Acids Res.
49:D325–D334. 2021. View Article : Google Scholar :
|
|
36
|
Li J, Miao B, Wang S, Dong W, Xu H, Si C,
Wang W, Duan S, Lou J, Bao Z, et al: Hiplot: A comprehensive and
easy-to-use web service for boosting publication-ready biomedical
data visualization. Brief Bioinform. 23:bbac2612022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Catakovic K, Klieser E, Neureiter D and
Geisberger R: T cell exhaustion: From pathophysiological basics to
tumor immunotherapy. Cell Commun Signal. 15:12017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Huang YH, Zhu C, Kondo Y, Anderson AC,
Gandhi A, Russell A, Dougan SK, Petersen BS, Melum E, Pertel T, et
al: CEACAM1 regulates TIM-3-mediated tolerance and exhaustion.
Nature. 517:386–390. 2015. View Article : Google Scholar
|
|
39
|
Ruffo E, Wu RC, Bruno TC, Workman CJ and
Vignali DAA: Lymphocyte-activation gene 3 (LAG3): The next immune
checkpoint receptor. Semin Immunol. 42:1013052019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Khan O, Giles JR, McDonald S, Manne S,
Ngiow SF, Patel KP, Werner MT, Huang AC, Alexander KA, Wu JE, et
al: TOX transcriptionally and epigenetically programs CD8 (+) T
cell exhaustion. Nature. 571:211–218. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kim K, Park S, Park SY, Kim G, Park SM,
Cho JW, Kim DH, Park YM, Koh YW, Kim HR, et al: Single-cell
transcriptome analysis reveals TOX as a promoting factor for T cell
exhaustion and a predictor for anti-PD-1 responses in human cancer.
Genome Med. 12:222020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Seo H, Chen J, Gonzalez-Avalos E,
Samaniego-Castruita D, Das A, Wang YH, López-Moyado IF, Georges RO,
Zhang W, Onodera A, et al: TOX and TOX2 transcription factors
cooperate with NR4A transcription factors to impose CD8 (+) T cell
exhaustion. Proc Natl Acad Sci USA. 116:12410–12415. 2019.
View Article : Google Scholar
|
|
43
|
Aibar S, Gonzalez-Blas CB, Moerman T,
Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine JC,
Geurts P, Aerts J, et al: SCENIC: Single-cell regulatory network
inference and clustering. Nat Methods. 14:1083–1086. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liberzon A, Birger C, Thorvaldsdottir H,
Ghandi M, Mesirov JP and Tamayo P: The molecular signatures
database (MSigDB) hallmark gene set collection. Cell Syst.
1:417–425. 2015. View Article : Google Scholar
|
|
45
|
Pepper AE, Buckley RH, Small TN and Puck
JM: Two mutational hotspots in the interleukin-2 receptor gamma
chain gene causing human X-linked severe combined immunodeficiency.
Am J Hum Genet. 57:564–571. 1995.PubMed/NCBI
|
|
46
|
Brown CJ, Ballabio A, Rupert JL,
Lafreniere RG, Grompe M, Tonlorenzi R and Willard HF: A gene from
the region of the human X inactivation centre is expressed
exclusively from the inactive X chromosome. Nature. 349:38–44.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Brown CJ, Hendrich BD, Rupert JL,
Lafreniere RG, Xing Y, Lawrence J and Willard HF: The human XIST
gene: Analysis of a 17 kb inactive X-specific RNA that contains
conserved repeats and is highly localized within the nucleus. Cell.
71:527–542. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hatakeyama M, Kawahara A, Mori H, Shibuya
H and Taniguchi T: c-fos gene induction by interleukin 2:
Identification of the critical cytoplasmic regions within the
interleukin 2 receptor beta chain. Proc Natl Acad Sci USA.
89:2022–2026. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kawahara A, Minami Y, Miyazaki T, Ihle JN
and Taniguchi T: Critical role of the interleukin 2 (IL-2) receptor
gamma-chain-associated Jak3 in the IL-2-induced c-fos and c-myc,
but not bcl-2, gene induction. Proc Natl Acad Sci USA.
92:8724–8728. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wu JQ, Seay M, Schulz VP, Hariharan M,
Tuck D, Lian J, Du J, Shi M, Ye Z, Gerstein M, et al: Tcf7 is an
important regulator of the switch of self-renewal and
differentiation in a multipotential hematopoietic cell line. PLoS
Genet. 8:e10025652012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Xing S, Li F, Zeng Z, Zhao Y, Yu S, Shan
Q, Li Y, Phillips FC, Maina PK, Qi HH, et al: Tcf1 and Lef1
transcription factors establish CD8 (+) T cell identity through
intrinsic HDAC activity. Nat Immunol. 17:695–703. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Decaluwe H, Taillardet M, Corcuff E,
Munitic I, Law HK, Rocha B, Rivière Y and Di Santo JP: Gamma (c)
deficiency precludes CD8+ T cell memory despite formation of potent
T cell effectors. Proc Natl Acad Sci USA. 107:9311–9216. 2010.
View Article : Google Scholar
|
|
53
|
He Y, Xiao R, Ji X, Li L, Chen L, Xiong J,
Xiao W, Wang Y, Zhang L, Zhou R, et al: EBV promotes human CD8 NKT
cell development. PLoS Pathog. 6:e10009152010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zenz R, Eferl R, Scheinecker C, Redlich K,
Smolen J, Schonthaler HB, Kenner L, Tschachler E and Wagner EF:
Activator protein 1 (Fos/Jun) functions in inflammatory bone and
skin disease. Arthritis Res Ther. 10:2012008. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wagner EF and Eferl R: Fos/AP-1 proteins
in bone and the immune system. Immunol Rev. 208:126–140. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Jin HT, Anderson AC, Tan WG, West EE, Ha
SJ, Araki K, Freeman GJ, Kuchroo VK and Ahmed R: Cooperation of
Tim-3 and PD-1 in CD8 T-cell exhaustion during chronic viral
infection. Proc Natl Acad Sci USA. 107:14733–1478. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Allenspach E, Rawlings DJ, Petrovic A,
Chen K, Adam MP, Everman DB, Mirzaa GM, Pagon RA, Wallace SE, Bean
LJH, et al: X-linked severe combined immunodeficiency. GeneReviews
((R) Seattle (WA): 1993
|
|
58
|
Recher M, Berglund LJ, Avery DT, Cowan MJ,
Gennery AR, Smart J, Peake J, Wong M, Pai SY, Baxi S, et al: IL-21
is the primary common gamma chain-binding cytokine required for
human B-cell differentiation in vivo. Blood. 118:6824–6835. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Lee PP, Chan KW, Chen TX, Jiang LP, Wang
XC, Zeng HS, Chen XY, Liew WK, Chen J, Chu KM, et al: Molecular
diagnosis of severe combined immunodeficiency-identification of
IL2RG, JAK3, IL7R, DCLRE1C, RAG1, and RAG2 mutations in a cohort of
Chinese and Southeast Asian children. J Clin Immunol. 31:281–296.
2011. View Article : Google Scholar
|
|
60
|
Shibata F, Toma T, Wada T, Inoue M, Tone
Y, Ohta K, Kasahara Y, Sano F, Kimura M, Ikeno M, et al: Skin
infiltration of CD56 (bright) CD16 (-) natural killer cells in a
case of X-SCID with Omenn syndrome-like manifestations. Eur J
Haematol. 79:81–85. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Randles LG, Lappalainen I, Fowler SB,
Moore B, Hamill SJ and Clarke J: Using model proteins to quantify
the effects of pathogenic mutations in Ig-like proteins. J Biol
Chem. 281:24216–24226. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Notarangelo LD, Giliani S, Mazza C, Mella
P, Savoldi G, Rodriguez-Perez C, Mazzolari E, Fiorini M, Duse M,
Plebani A, et al: Of genes and phenotypes: The immunological and
molecular spectrum of combined immune deficiency. Defects of the
gamma (c)-JAK3 signaling pathway as a model. Immunol Rev.
178:39–48. 2000. View Article : Google Scholar
|
|
63
|
Puck JM, Pepper AE, Henthorn PS, Candotti
F, Isakov J, Whitwam T, Conley ME, Fischer RE, Rosenblatt HM, Small
TN and Buckley RH: Mutation analysis of IL2RG in human X-linked
severe combined immunodeficiency. Blood. 89:1968–1977.
1997.PubMed/NCBI
|
|
64
|
Heaton H, Talman AM, Knights A, Imaz M,
Gaffney DJ, Durbin R, Hemberg M and Lawniczak MKN: Souporcell:
Robust clustering of single-cell RNA-seq data by genotype without
reference genotypes. Nat Methods. 17:615–620. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Palmer K, Green TD, Roberts JL, Sajaroff
E, Cooney M, Parrott R, Chen DF, Reinsmoen NL and Buckley RH:
Unusual clinical and immunologic manifestations of transplacentally
acquired maternal T cells in severe combined immunodeficiency. J
Allergy Clin Immunol. 120:423–438. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Leonard WJ, Lin JX and O'Shea JJ: The
gammac family of cytokines: Basic biology to therapeutic
ramifications. Immunity. 50:832–850. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Wiekmeijer AS, Pike-Overzet K, IJspeert H,
Brugman MH, Wolvers-Tettero IL, Lankester AC, Bredius RGM, van
Dongen JJM, Fibbe WE, Langerak AW, et al: Identification of
checkpoints in human T-cell development using severe combined
immunodeficiency stem cells. J Allergy Clin Immunol. 137:517–526
e3. 2016. View Article : Google Scholar
|
|
68
|
Chen L and Flies DB: Molecular mechanisms
of T cell co-stimulation and co-inhibition. Nat Rev Immunol.
13:227–242. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Wherry EJ: T cell exhaustion. Nat Immunol.
12:492–499. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Plebani A, Stringa M, Prigione I,
Facchetti P, Ghiotto F, Airoldi I, Giacchino R, Cristina E, Porta
F, Grossi CE and Pistoia V: Engrafted maternal T cells in human
severe combined immunodeficiency: Evidence for a TH2 phenotype and
a potential role of apoptosis on the restriction of T-cell receptor
variable beta repertoire. J Allergy Clin Immunol. 101:131–134.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Knobloch C, Goldmann SF and Friedrich W:
Limited T cell receptor diversity of transplacentally acquired
maternal T cells in severe combined immunodeficiency. J Immunol.
146:4157–4164. 1991. View Article : Google Scholar : PubMed/NCBI
|