According to the somatic stem cell hypothesis,
mutations or chromosomal rearrangements in dormant stem cells
present in organs may induce the formation of CSCs (11). It has been demonstrated that the
implantation of embryonic stem cells or the induction of
pluripotent stem cells in mice results in cancer (12). Another view on the origin of CSCs
is that cancer cells with genetic instability may generate CSCs
(13). Cancer cells transfected
with Oct3/4, Sox2, Klf4 and c-Myc have been reported to transform
into CSCs (14).
In most scenarios, CSC subpopulations have emerged
following the accumulation of epigenetic and/or genetic alterations
in normal stem cells or cancer cells. However, in 2013, Wang et
al (15) hypothesized that
CSCs develop de novo from the misplaced somatic stem cells and
proposed a new theory of carcinogenesis; the stem cell misplacement
theory. This theory stated that misplaced epithelial stem cells,
which reach the wrong tissue stroma by accident undergo malignant
transformation and become CSCs.
CSCs, also known as tumor-initiating cells or
tumor-propagating cells are a subpopulation of tumor cells that
demonstrate properties similar to normal stem cells (16). Tumor-initiating cells may better
describe these cells; however, we have referred to these cells as
CSCs in this review. At the 2006 meeting of the American
Association for Cancer Research, Reya et al (17) proposed the definition of a cancer
stem cell as a cell within a tumor that possesses the capacity to
self-renew and produce the heterogeneous lineages of cancer cells
that comprise the tumor.
Two main approaches have been used to identify
tumorigenic cells in published studies: One method is termed
'spheroid colony formation; and is considered the most appropriate
in vitro assay to detect the malignant transformation of
cells (25), and the other one is
an in vivo method involving implantation of tumorigenic
cells in immunodeficient mice (26).
The hallmark of stem cells is their dual ability to
self-renew and to generate multiple cell lineages with more
differentiated characteristics (26). Self-renewal is the ability of a CSC
to sustain itself and continue to give rise to cells with equal
abilities of tumorigenicity (27).
CSCs can self-renew through asymmetric cell division in which one
daughter cell possesses stem cell properties (28). Prior to asymmetric division,
unequally distributed cellular components are differentially
enriched at either the apical or basal pole, in which the mitotic
spindle apparatus and centrosomes are unequally aligned (29).
Anticancer drugs have been applied alone or in
combination to prolong life or to alleviate the symptoms of cancer
for decades (30,31). However, these drugs have failed to
completely eradicate cancers. Multidrug resistance (MDR) plays an
important role in preventing drug absorption (32). Various factors can contribute to
MDR, including the existence of CSCs (33). CSCs possess multiple mechanisms of
drug resistance: A high expression of ABC transporters and
anti-apoptotic factors, and the maintenance of a quiescent state to
avoid the induction of apoptosis (34). The ABC transporter family acts by
pumping drugs into the extracellular domain (35). To date, 49 human ABC genes have
been identified and are clustered in seven subfamilies (ABCA-ABCG)
(36). There are three major
transporters correlated with MDR, including P-glycoprotein
(MDR1/ABCB1), MDR-associated protein (MRP/ABCC1) and breast cancer
resistance protein (BCRP/ABCG2) (37).
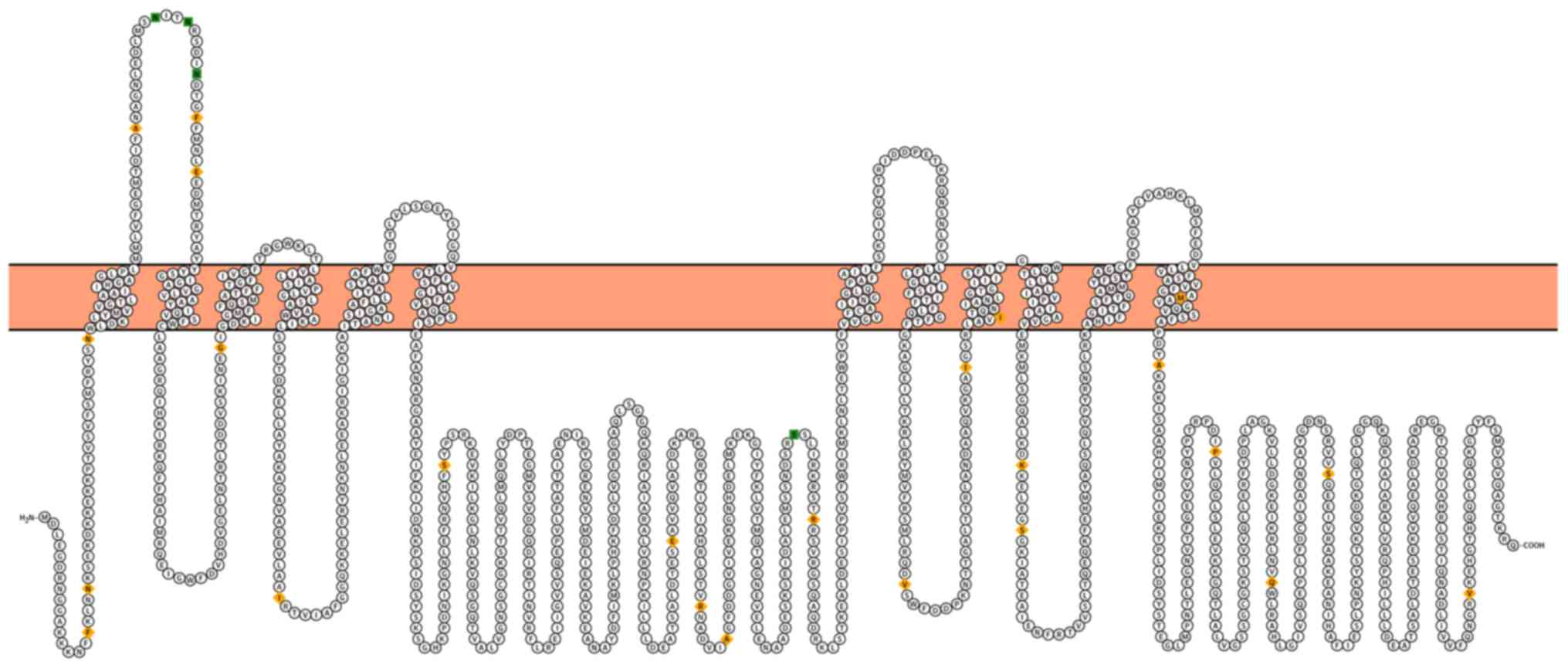
P-glycoprotein (P-gp) is a 170 kDa
phosphoglycoprotein constituting two transmembrane domains and two
cytosolic nucleotide-binding domains (Fig. 2) (38). P-gp overexpression is related to
negative clinical outcomes, including treatment failure, relapse
and survival. An increased P-gp expression has been observed in
breast tumor biopsies treated with conventional chemotherapy
(39). In a previous study on AML,
the relapse rates were associated with elevated P-gp expression
levels (40). A similar
observation was reported in a study on multiple myeloma, in which
6% of patients expressed P-gp at diagnosis, and >43% of patients
exhibited overexpressed P-gp following treatment (41). The patients with osteosarcoma that
did not overexpress P-gp had significantly better relapse-free
rates and improved survival rates of 5 to 14 years (42). P-gp plays a significant role in
transporting a diverse array of molecules, including anionic, and
neutrally charged drugs and toxins (43–45).
MRP1/ABCC1 was the first gene to be identified in
the ABCC subfamily and was cloned from an MDR small cell lung
cancer cell (46). Numerous
studies have demonstrated the upregulation of MRP1 in a variety of
solid tumors, such as those of the lung, breast and prostate
(47–49). MRP1 is potentially an important
target for reversing chemotherapy resistance in many cancers
(50).
Hypoxia is an important factor that affects clinical
outcomes by promoting genetic instability, tumor cell metastasis
and invasiveness (53). The HIF
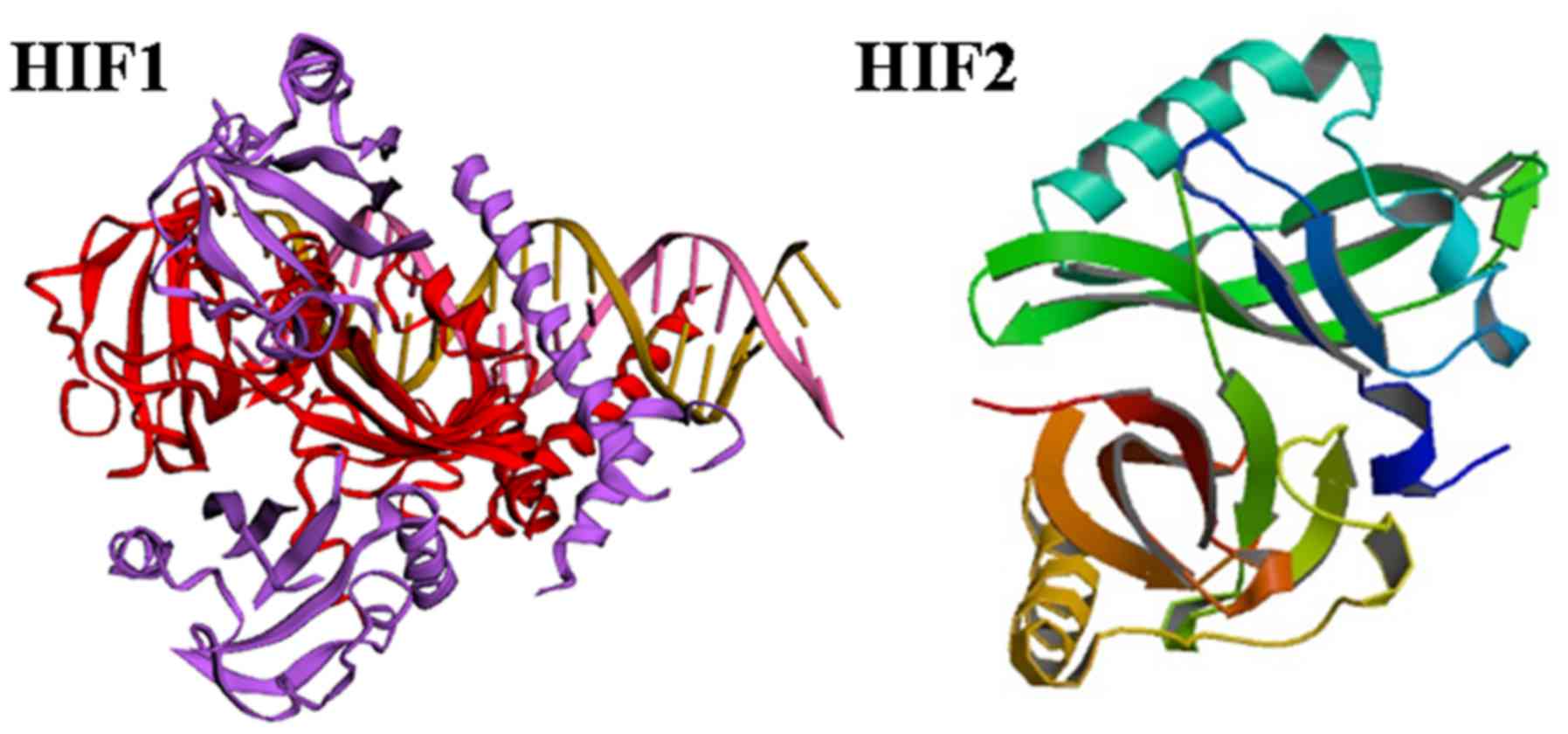
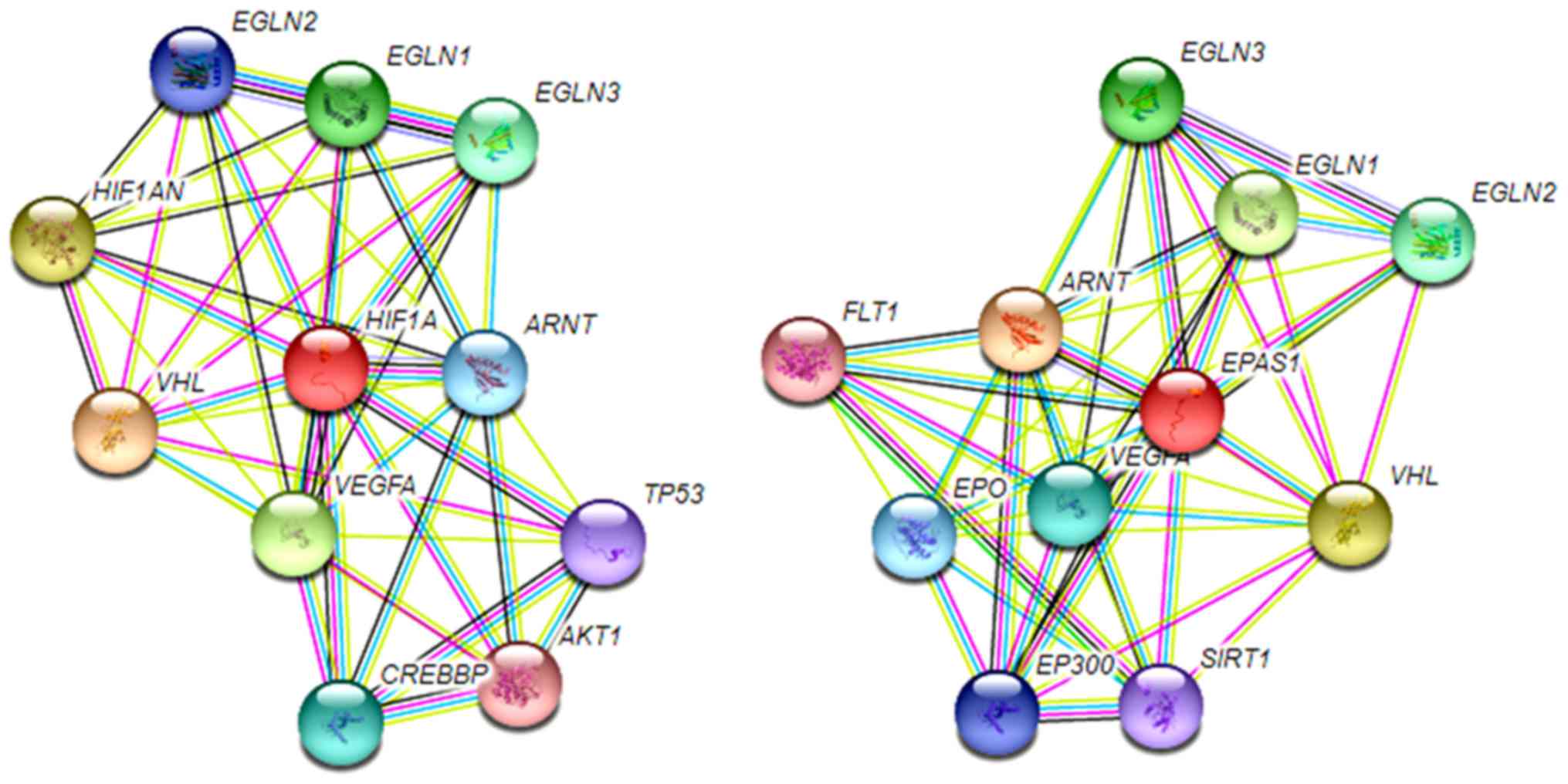
protein is a heterodimeric complex formed by an oxygen-dependent α
subunit and an oxygen-insensitive β subunit (53). The three HIFα subunits (HIF-1α,
HIF-2α and HIF-3α) with a HIF-1β subunit act as key mediators of
cellular adaptation to low oxygen (54). The carboxy-terminal domain of
HIF-1α and HIF-2α consists of domains that regulate its stability
(the oxygen-dependent degradation domain, ODD) and transcriptional
activity (two transactivation domains (TADs), N-TAD and C-TAD
(Fig. 3) (55). Furthermore, both the C- and
N-termini of the α subunits have nuclear localization signals
(N-NLS and C-NLS, respectively) that direct them to the nucleus
(Fig. 3) (56). The stability of HIF-1α and HIF-2α
is regulated by oxygen tension (57). HIF-1α and HIF-2α have been
extensively studied and are ubiquitously expressed in normal tissue
(58). C-TAD regulates most
hypoxia-induced genes, although a subset of genes depended solely
on N-TAD initiation, and N-TAD contributes to target gene
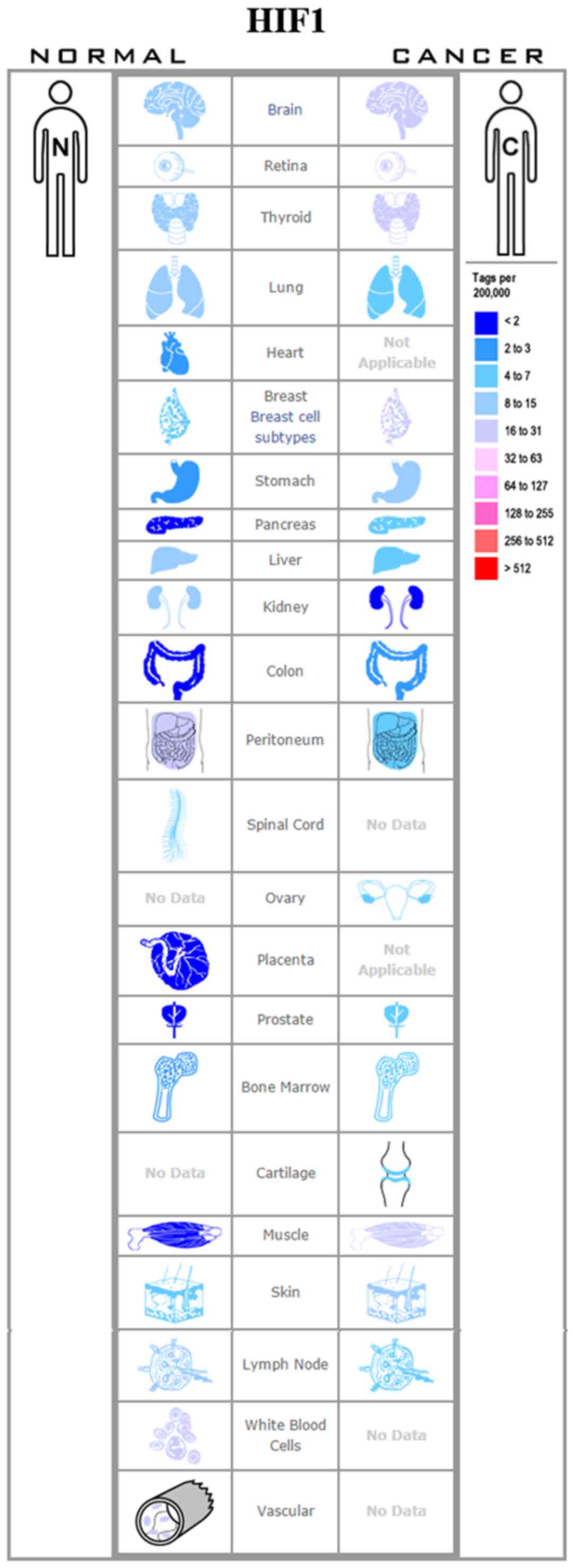
specificity of HIF-1α and HIF-2α (59). An increased HIF-1α or HIF-2α
expression has been observed in many types of cancer, such as
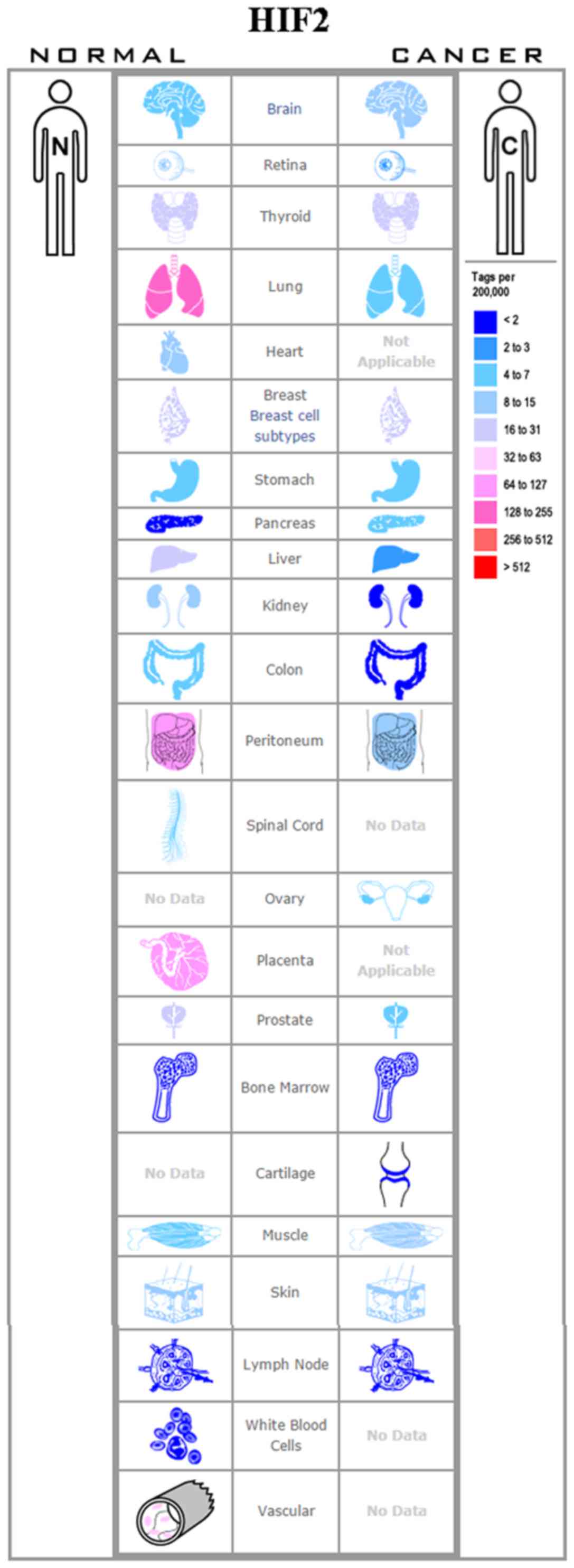
breast (60), colon (61), lung (62), pancreatic (63) and ovarian cancers (65) (Figs.
4 and 5). Upon exposure of the
cells to hypoxia, the HIFα subunits accumulate in the nucleus and
bind to their target genes, such as BNIP3, PGK1, HK1 and TP11
(Fig. 6) (65). The target genes participate in the
proliferation, apoptosis, metabolism and invasion, as well in the
and resistance of cancer cells to therapy (66–68).
Genetic polymorphisms of HIF-1α or HIF-2α in human
cancers have been found in previous studies (69–74).
The single nucleotide polymorphisms 1772C/T (rs11549465) and 1790
G/A (rs11549467) have been shown to be significantly associated
with the overall risk of developing lung, breast, oral, prostate,
cervical and renal cancers (69).
Frank et al (70)
demonstrated a significant association between rs2057482 in HIF-1α
with the risk of rectal cancer. Guo et al (71) found that rs2057482 was associated
with worse clinical outcomes of patients with hepatocellular
carcinoma. Han et al (72)
observed that rs9679290, rs4953346 and rs12617313 of HIF-2α were
associated with the risk of developing renal cell carcinoma.
Yamamoto et al (73)
reported that HIF-2α rs13419896 was associated with a decreased
risk of developing lung cancer. HIF2A rs11125070 and rs4953352 are
associated with the disease-free and overall survival of patients
with colorectal cancer (74).
A non-coding RNA (ncRNA) is a functional RNA
molecule that is not translated into a protein (75). miRNAs are the perfect candidates
for controlling HIF expression during hypoxia (76). These so-called hypoxamiRs
contribute to HIF-1 accumulation and the maintenance of HIF-2 and
HIF-3 (77,78). For example, the hypoxic induction
of miR-18a may allow HIF-1α level decreases and thus contribute to
the HIF switch (79). miR-17,
miR-20a and miR-20b have been reported to be involved in the
HIF-related response during hypoxia in cancer cells (80). lncRNA HIF-1A-AS2 negatively
regulates HIF-1α and is upregulated in non-papillary clear cell
renal carcinomas (81). lncRNA
sONE or NOS3AS regulates the expression of endothelial nitric oxide
synthase (eNOS), under normal oxygen conditions and hypoxic
conditions (82).
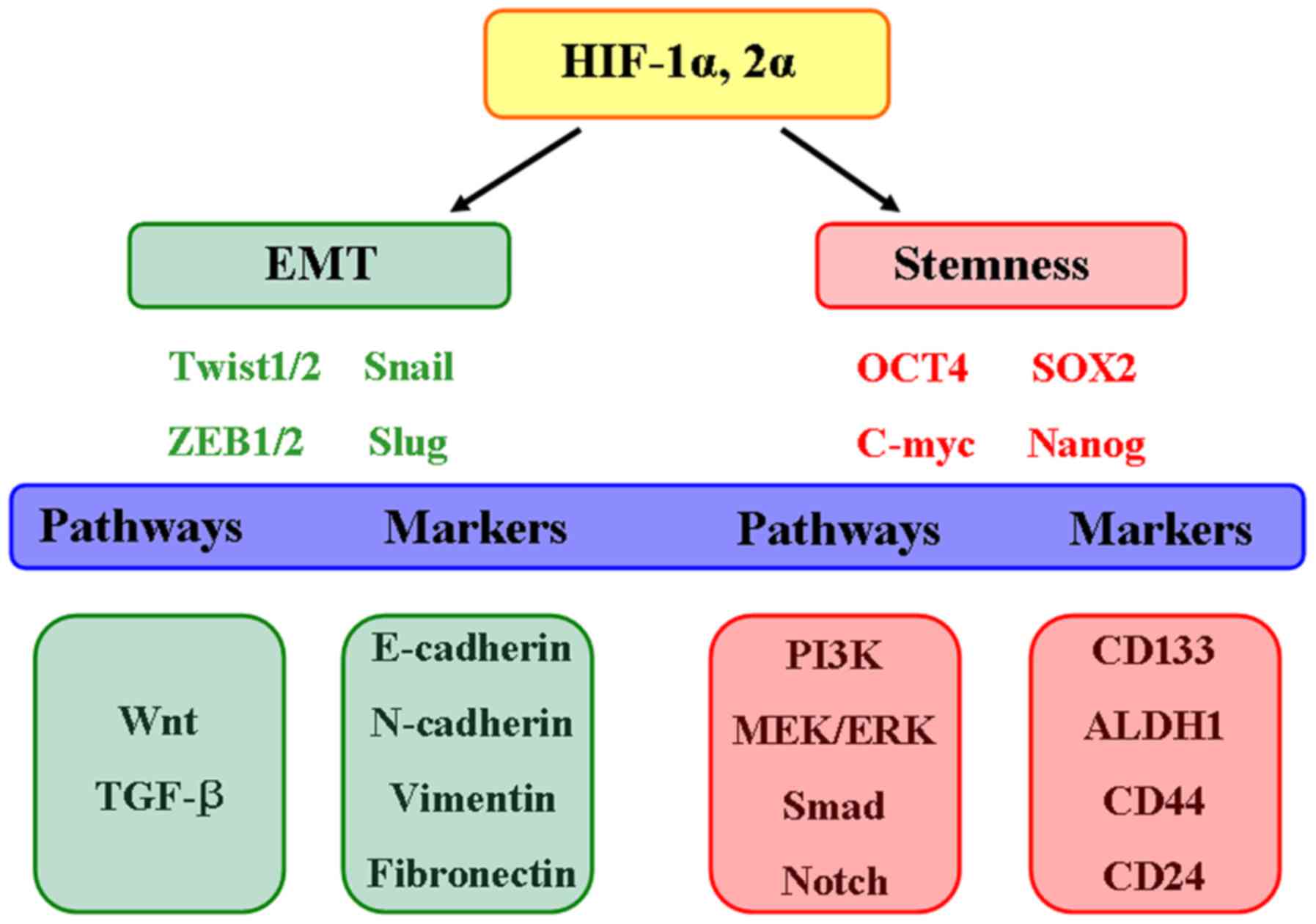
Hypoxia has notable potential to exert significant
effects on the maintenance and evolution of CSCs. Both HIF-1α and
HIF-2α contribute to the regulation of cellular adaptation to
hypoxia and the resistance to cancer therapies (Fig. 7). The simultaneous targeting of the
HIF-1α and HIF-2α pathways may improve clinical responses within
the hypoxic tumor microenvironment. Therefore, the concept of
personalized medicine should be applied in designing clinical
trials for HIF inhibitors.
Not applicable.
No funding was received.
Not applicable.
WWT and YL conceived the study. WWT and GHT
collected the data and wrote the manuscript; WWT prepared the
figures and revised the manuscript. All authors have read and
approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Soni S and Padwad YS: HIF-1 in cancer
therapy: Two decade long story of a transcription factor. Acta
Oncol. 56:503–515. 2017. View Article : Google Scholar
|
|
2
|
Furth J and Kahn M: The transmission of
leukemia of mice with a single cell. Am J Cancer. 31:276–282.
1937.
|
|
3
|
Southam CM and Brunschwig A: Quantitative
studies of auto-transplantation of human cancer. Cancer.
14:971–978. 1961. View Article : Google Scholar
|
|
4
|
Hamburger AW and Salmon SE: Primary
bioassay of human tumor stem cells. Science. 197:461–463. 1977.
View Article : Google Scholar
|
|
5
|
Lapidot T, Sirard C, Vormoor J, Murdoch B,
Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA and
Dick JE: A cell initiating human acute myeloid leukaemia after
transplantation into SCID mice. Nature 3. 67:645–648. 1994.
View Article : Google Scholar
|
|
6
|
Bonnet D and Dick JE: Human acute myeloid
leukemia is organized as a hierarchy that originates from a
primitive hematopoietic cell. Nat Med. 3:730–737. 1997. View Article : Google Scholar
|
|
7
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Singh SK, Hawkins C, Clarke ID, Squire JA,
Bayani J, Hide T, Henkelman RM, Cusimano MD and Dirks PB:
Identification of human brain tumour initiating cells. Nature.
432:396–401. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ponti D, Costa A, Zaffaroni N, Pratesi G,
Petrangolini G, Coradini D, Pilotti S, Pierotti MA and Daidone MG:
Isolation and in vitro propagation of tumorigenic breast cancer
cells with stem/progenitor cell properties. Cancer Res.
65:5506–5511. 2005. View Article : Google Scholar
|
|
10
|
Patrawala L, Calhoun T,
Schneider-Broussard R, Li H, Bhatia B, Tang S, Reilly JG, Chandra
D, Zhou J, Claypool K, et al: Highly purified CD44+
prostate cancer cells from xenograft human tumors are enriched in
tumorigenic and metastatic progenitor cells. Oncogene.
25:1696–1708. 2006. View Article : Google Scholar
|
|
11
|
López J, Valdez-Morales FJ,
Benítez-Bribiesca L, Cerbón M and Carrancá AG: Normal and cancer
stem cells of the human female reproductive system. Reprod Biol
Endocrinol. 11:532013. View Article : Google Scholar :
|
|
12
|
Takahashi K and Yamanaka S: Induction of
pluripotent stem cells from mouse embryonic and adult fibroblast
cultures by defined factors. Cell. 126:663–676. 2006. View Article : Google Scholar
|
|
13
|
Ohnishi K, Semi K, Yamamoto T, Shimizu M,
Tanaka A, Mitsunaga K, Okita K, Osafune K, Arioka Y, Maeda T, et
al: Premature termination of reprogramming in vivo leads to cancer
development through altered epigenetic regulation. Cell.
156:663–677. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Beck B and Blanpain C: Unravelling cancer
stem cell potential. Nat Rev Cancer. 13:727–738. 2013. View Article : Google Scholar
|
|
15
|
Wang RA, Li ZS, Zhang HZ, Zheng PJ, Li QL,
Shi JG, Yan QG, Ye J, Wang JB, Guo Y, et al: Invasive cancers are
not necessarily from preformed in situ tumours - an alternative way
of carcinogenesis from misplaced stem cells. J Cell Mol Med.
17:921–926. 2013. View Article : Google Scholar
|
|
16
|
Clarke MF, Dick JE, Dirks PB, Eaves CJ,
Jamieson CH, Jones DL, Visvader J, Weissman IL and Wahl GM: Cancer
stem cells - perspectives on current status and future directions:
AACR Workshop on cancer stem cells. Cancer Res. 66:9339–9344. 2006.
View Article : Google Scholar
|
|
17
|
Reya T, Morrison SJ, Clarke MF and
Weissman IL: Stem cells, cancer, and cancer stem cells. Nature.
414:105–111. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cabrera MC, Hollingsworth RE and Hurt EM:
Cancer stem cell plasticity and tumor hierarchy. World J Stem
Cells. 7:27–36. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kuroda T, Yasuda S and Sato Y:
Tumorigenicity studies for human pluripotent stem cell-derived
products. Biol Pharm Bull. 36:189–192. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Adorno-Cruz V, Kibria G, Liu X, Doherty M,
Junk DJ, Guan D, Hubert C, Venere M, Mulkearns-Hubert E, Sinyuk M,
et al: Cancer stem cells: Targeting the roots of cancer, seeds of
metastasis, and sources of therapy resistance. Cancer Res.
75:924–929. 2015. View Article : Google Scholar :
|
|
21
|
O'Brien CA, Pollett A, Gallinger S and
Dick JE: A human colon cancer cell capable of initiating tumour
growth in immunodeficient mice. Nature. 445:106–110. 2007.
View Article : Google Scholar
|
|
22
|
Hope KJ, Jin L and Dick JE: Acute myeloid
leukemia originates from a hierarchy of leukemic stem cell classes
that differ in self-renewal capacity. Nat Immunol. 5:738–743. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xia P, Gou WF, Zhao S and Zheng HC:
Crizotinib may be used in Lewis lung carcinoma: A novel use for
crizotinib. Oncol Rep. 30:139–148. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vescovi AL, Galli R and Reynolds BA: Brain
tumour stem cells. Nat Rev Cancer. 6:425–436. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Vargo-Gogola T and Rosen JM: Modelling
breast cancer: One size does not fit all. Nat Rev Cancer.
7:659–672. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tsai RY: Balancing self-renewal against
genome preservation in stem cells: How do they manage to have the
cake and eat it too? Cell Mol Life Sci. 73:1803–1823. 2016.
View Article : Google Scholar :
|
|
27
|
Huntly BJ and Gilliland DG: Leukaemia stem
cells and the evolution of cancer-stem-cell research. Nat Rev
Cancer. 5:311–321. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Daynac M and Petritsch CK: Regulation of
asymmetric cell division in mammalian neural stem and cancer
precursor cells. Results Probl Cell Differ. 61:375–399. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang L, Bu P and Shen X: Asymmetric
division: An antitumor player? Mol Cell Oncol. 3:e11642792016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wu L, Wang G, Liu S, Wei J, Zhang S, Li M,
Zhou G and Wang L: Synthesis and biological evaluation of matrine
derivatives containing benzo-α-pyrone structure as potent anti-lung
cancer agents. Sci Rep. 6:359182016. View Article : Google Scholar
|
|
31
|
Ait-Oudhia S and Mager DE: Array of
translational systems pharmacodynamic models of anti-cancer drugs.
J Pharmacokinet Pharmacodyn. 43:549–565. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
O'Connor R, Clynes M, Dowling P, O'Donovan
N and O'Driscoll L: Drug resistance in cancer - searching for
mechanisms, markers and therapeutic agents. Expert Opin Drug Metab
Toxicol. 3:805–817. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yu Z, Pestell TG, Lisanti MP and Pestell
RG: Cancer stem cells. Int J Biochem Cell Biol. 44:2144–2151. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gottesman MM, Fojo T and Bates SE:
Multidrug resistance in cancer: Role of ATP-dependent transporters.
Nat Rev Cancer. 2:48–58. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ramachandra M, Ambudkar SV, Chen D,
Hrycyna CA, Dey S, Gottesman MM and Pastan I: Human P-glycoprotein
exhibits reduced affinity for substrates during a catalytic
transition state. Biochemistry. 37:5010–5019. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dean M, Hamon Y and Chimini G: The human
ATP-binding cassette (ABC) transporter superfamily. J Lipid Res.
42:1007–1017. 2001.PubMed/NCBI
|
|
37
|
Kort A, van Hoppe S, Sparidans RW,
Wagenaar E, Beijnen JH and Schinkel AH: Brain accumulation of
ponatinib and its active metabolite, N-desmethyl ponatinib, is
limited by P-glycoprotein (P-GP/ABCB1) and breast cancer resistance
protein (BCRP/ABCG2). Mol Pharm. 14:3258–3268. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Eckford PD and Sharom FJ: ABC efflux
pump-based resistance to chemotherapy drugs. Chem Rev.
109:2989–3011. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Trock BJ, Leonessa F and Clarke R:
Multidrug resistance in breast cancer: A meta-analysis of
MDR1/gp170 expression and its possible functional significance. J
Natl Cancer Inst. 89:917–931. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhou DC, Zittoun R and Marie JP:
Expression of multidrug resistance-associated protein (MRP) and
multidrug resistance (MDR1) genes in acute myeloid leukemia.
Leukemia. 9:1661–1666. 1995.PubMed/NCBI
|
|
41
|
Grogan TM, Spier CM, Salmon SE, Matzner M,
Rybski J, Weinstein RS, Scheper RJ and Dalton WS: P-glycoprotein
expression in human plasma cell myeloma: Correlation with prior
chemotherapy. Blood. 81:490–495. 1993.PubMed/NCBI
|
|
42
|
Chan HS, Grogan TM, Haddad G, DeBoer G and
Ling V: P-glycoprotein expression: Critical determinant in the
response to osteosarcoma chemotherapy. J Natl Cancer Inst.
89:1706–1715. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gillet JP and Gottesman MM: Mechanisms of
multidrug resistance in cancer. Methods Mol Biol. 596:47–76. 2010.
View Article : Google Scholar
|
|
44
|
Leslie EM, Deeley RG and Cole SP:
Multidrug resistance proteins: Role of P-glycoprotein, MRP1, MRP2,
and BCRP (ABCG2) in tissue defense. Toxicol Appl Pharmacol.
204:216–237. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Rohwer N and Cramer T: Hypoxia-mediated
drug resistance: Novel insights on the functional interaction of
HIFs and cell death pathways. Drug Resist Updat. 14:191–201. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
da Costa KM, Valente RC, Salustiano EJ,
Gentile LB, Freire-de-Lima L, Mendonça-Previato L and Previato JO:
Functional Characterization of ABCC proteins from trypanosoma cruzi
and their involvement with thiol transport. Front Microbiol.
9:2052018. View Article : Google Scholar :
|
|
47
|
Yin JY, Han LF, Huang Q, Xu XJ, Zhou HH
and Liu ZQ: ABCC1 polymorphism Arg723Gln (2168G> A) is
associated with lung cancer susceptibility in a Chinese population.
Clin Exp Pharmacol Physiol. 38:632–637. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Arumugam P and Song JM: Quantitative
evaluation of ABC transporter-mediated drug resistance based on the
determination of the anticancer activity of camptothecin against
breast cancer stem cells using TIRF. Integr Biol. 8:704–711. 2016.
View Article : Google Scholar
|
|
49
|
Liu C, Li Z, Bi L, Li K, Zhou B, Xu C,
Huang J and Xu K: NOTCH1 signaling promotes chemoresistance via
regulating ABCC1 expression in prostate cancer stem cells. Mol Cell
Biochem. 393:265–270. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Johnson ZL and Chen J: ATP binding enables
substrate release from multidrug resistance protein 1. Cell.
172:81–89.e10. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Castilho L and Reid ME: A review of the JR
blood group system. Immunohematology. 29:63–68. 2013.PubMed/NCBI
|
|
52
|
Fujita K and Ichida K: ABCG2 as a
therapeutic target candidate for gout. Expert Opin Ther Targets.
22:123–129. 2018. View Article : Google Scholar
|
|
53
|
D'Ignazio L, Batie M and Rocha S: Hypoxia
and inflammation in cancer, focus on HIF and NF-κB. Biomedicines.
5:E212017. View Article : Google Scholar
|
|
54
|
Zhang P, Yao Q, Lu L, Li Y, Chen PJ and
Duan C: Hypoxia-inducible factor 3 is an oxygen-dependent
transcription activator and regulates a distinct transcriptional
response to hypoxia. Cell Reports. 6:1110–1121. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang GL, Jiang BH, Rue EA and Semenza GL:
Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS
heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci
USA. 92:5510–5514. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Kallio PJ, Okamoto K, O'Brien S, Carrero
P, Makino Y, Tanaka H and Poellinger L: Signal transduction in
hypoxic cells: Inducible nuclear translocation and recruitment of
the CBP/p300 coactivator by the hypoxia-inducible factor-1alpha.
EMBO J. 17:6573–6586. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Dayan F, Roux D, Brahimi-Horn MC,
Pouyssegur J and Mazure NM: The oxygen sensor factor-inhibiting
hypoxia-inducible factor-1 controls expression of distinct genes
through the bifunctional transcriptional character of
hypoxia-inducible factor-1alpha. Cancer Res. 66:3688–3698. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Jiang BH, Semenza GL, Bauer C and Marti
HH: Hypoxia-inducible factor 1 levels vary exponentially over a
physiologically relevant range of O2 tension. Am J Physiol.
271:C1172–C1180. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Hu CJ, Sataur A, Wang L, Chen H and Simon
MC: The N-terminal transactivation domain confers target gene
specificity of hypoxia-inducible factors HIF-1alpha and HIF-2alpha.
Mol Biol Cell. 18:4528–4542. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Badowska-Kozakiewicz AM, Sobol M and
Patera J: Expression of multidrug resistance protein P-glycoprotein
in correlation with markers of hypoxia (HIF-1α, EPO, EPO-R) in
invasive breast cancer with metastasis to lymph nodes. Arch Med
Sci. 13:1303–1314. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Rodríguez ME, Catrinacio C, Ropolo A,
Rivarola VA and Vaccaro MI: A novel HIF-1α/VMP1-autophagic pathway
induces resistance to photodynamic therapy in colon cancer cells.
Photochem Photobiol Sci. 16:1631–1642. 2017. View Article : Google Scholar
|
|
62
|
Shao JS, Sun J, Wang S, Chung K, Du JT,
Wang J, Qiu XS, Wang EH and Wu GP: HPV16 E6/E7 upregulates HIF-2α
and VEGF by inhibiting LKB1 in lung cancer cells. Tumour Biol.
39:1010428317717137. 2017. View Article : Google Scholar
|
|
63
|
Zhang Q, Lou Y, Zhang J, Fu Q, Wei T, Sun
X, Chen Q, Yang J, Bai X and Liang T: Hypoxia-inducible factor-2α
promotes tumor progression and has crosstalk with Wnt/β-catenin
signaling in pancreatic cancer. Mol Cancer. 16:1192017. View Article : Google Scholar
|
|
64
|
Raspaglio G, Petrillo M, Martinelli E, Li
Puma DD, Mariani M, De Donato M, Filippetti F, Mozzetti S, Prislei
S, Zannoni GF, et al: Sox9 and Hif-2α regulate TUBB3 gene
expression and affect ovarian cancer aggressiveness. Gene.
542:173–181. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Jun JC, Rathore A, Younas H, Gilkes D and
Polotsky VY: Hypoxia-Inducible Factors and Cancer. Curr Sleep Med
Rep. 3:1–10. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Vandyke K, Zeissig MN, Hewett DR, Martin
SK, Mrozik KM, Cheong CM, Diamond P, To LB, Gronthos S, Peet DJ, et
al: HIF-2α promotes dissemination of plasma cells in multiple
myeloma by regulating CXCL12/CXCR4 and CCR1. Cancer Res.
77:5452–5463. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Ma X, Zhang H, Xue X and Shah YM:
Hypoxia-inducible factor 2α (HIF-2α) promotes colon cancer growth
by potentiating Yes-associated protein 1 (YAP1) activity. J Biol
Chem. 292:17046–17056. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Garziera M, Scarabel L and Toffoli G:
Hypoxic modulation of HLA-G expression through the metabolic sensor
HIF-1 in human cancer cells. J Immunol Res. 2017:45875202017.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Anam MT, Ishika A, Hossain MB and Jesmin:
A meta-analysis of hypoxia inducible factor 1-alpha (HIF1A) gene
polymorphisms: Association with cancers. Biomark Res. 3:292015.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Frank B, Hoffmeister M, Klopp N, Illig T,
Chang-Claude J and Brenner H: Single nucleotide polymorphisms in
Wnt signaling and cell death pathway genes and susceptibility to
colorectal cancer. Carcinogenesis. 31:1381–1386. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Guo X, Li D, Chen Y, An J, Wang K, Xu Z,
Chen Z and Xing J: SNP rs2057482 in HIF1A gene predicts clinical
outcome of aggressive hepatocellular carcinoma patients after
surgery. Sci Rep. 5:118462015. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Han SS, Yeager M, Moore LE, Wei MH,
Pfeiffer R, Toure O, Purdue MP, Johansson M, Scelo G, Chung CC, et
al: The chromosome 2p21 region harbors a complex genetic
architecture for association with risk for renal cell carcinoma.
Hum Mol Genet. 21:1190–1200. 2012. View Article : Google Scholar :
|
|
73
|
Yamamoto Y, Kiyohara C, Ogata-Suetsugu S,
Hamada N and Nakanishi Y: Association between genetic polymorphisms
involved in the hypoxia-inducible factor pathway and lung cancer
risk: A case-control study in Japan. Asia Pac J Clin Oncol.
13:234–242. 2017. View Article : Google Scholar
|
|
74
|
Haja Mohideen AM, Hyde A, Squires J, Wang
J, Dicks E, Younghusband B, Parfrey P, Green R and Savas S:
Examining the polymorphisms in the hypoxia pathway genes in
relation to outcome in colorectal cancer. PLoS One. 9:e1135132014.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Autour A, Jeng S C Y, Cawte A D,
Abdolahzadeh A, Galli A, Panchapakesan SSS, Rueda D, Ryckelynck M
and Unrau PJ: Fluorogenic RNA Mango aptamers for imaging small
non-coding RNAs in mammalian cells. Nat Commun. 9:6562018.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Camps C, Saini HK, Mole DR, Choudhry H,
Reczko M, Guerra-Assunção JA, Tian YM, Buffa FM, Harris AL,
Hatzigeorgiou AG, et al: Integrated analysis of microRNA and mRNA
expression and association with HIF binding reveals the complexity
of microRNA expression regulation under hypoxia. Mol Cancer.
13:282014. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Ho JJ, Metcalf JL, Yan MS, Turgeon PJ,
Wang JJ, Chalsev M, Petruzziello-Pellegrini TN, Tsui AK, He JZ,
Dhamko H, et al: Functional importance of Dicer protein in the
adaptive cellular response to hypoxia. J Biol Chem.
287:29003–29020. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
van den Beucken T, Koch E, Chu K,
Rupaimoole R, Prickaerts P, Adriaens M, Voncken JW, Harris AL,
Buffa FM, Haider S, et al: Hypoxia promotes stem cell phenotypes
and poor prognosis through epigenetic regulation of DICER. Nat
Commun. 5:52032014. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Montoya MM, Maul J, Singh PB, Pua HH,
Dahlström F, Wu N, Huang X, Ansel KM and Baumjohann D: A distinct
inhibitory function for miR-18a in Th17 cell differentiation. J
Immunol. 199:559–569. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Li JY, Zhang Y, Zhang WH, Jia S, Kang Y
and Zhu XY: Differential distribution of miR-20a and miR-20b may
underly metastatic heterogeneity of breast cancers. Asian Pac J
Cancer Prev. 13:1901–1906. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Kunej T, Obsteter J, Pogacar Z, Horvat S
and Calin GA: The decalog of long non-coding RNA involvement in
cancer diagnosis and monitoring. Crit Rev Clin Lab Sci. 51:344–357.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Im JH and Muschel RJ: New evidence of
lncRNA role in tumor progression and metastasis. Hepatobiliary Surg
Nutr. 1:55–56. 2012.PubMed/NCBI
|
|
83
|
Schito L and Semenza GL: Hypoxia-inducible
factors: Master regulators of cancer progression. Trends Cancer.
2:758–770. 2016. View Article : Google Scholar
|
|
84
|
Mohlin S, Wigerup C, Jögi A and Påhlman S:
Hypoxia, pseudo-hypoxia and cellular differentiation. Exp Cell Res.
356:192–196. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Shi QY, Zhang SJ, Liu L, Chen QS, Yu LN,
Zhang FJ and Yan M: Sevoflurane promotes the expansion of glioma
stem cells through activation of hypoxia-inducible factors in
vitro. Br J Anaesth. 114:825–830. 2015. View Article : Google Scholar
|
|
86
|
Lee G, Auffinger B, Guo D, Hasan T,
Deheeger M, Tobias AL, Kim JY, Atashi F, Zhang L, Lesniak MS, et
al: Dedifferentiation of glioma cells to glioma stem-like cells by
therapeutic stress-induced HIF signaling in the recurrent GBM
model. Mol Cancer Ther. 15:3064–3076. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Zhang S, Luo X, Wan F and Lei T: The roles
of hypoxia-inducible factors in regulating neural stem cells
migration to glioma stem cells and determinating their fates.
Neurochem Res. 37:2659–2666. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Semenza GL: Regulation of the breast
cancer stem cell phenotype by hypoxia-inducible factors. Clin Sci
(Lond). 129:1037–1045. 2015. View Article : Google Scholar
|
|
89
|
Samanta D, Gilkes DM, Chaturvedi P, Xiang
L and Semenza GL: Hypoxia-inducible factors are required for
chemotherapy resistance of breast cancer stem cells. Proc Natl Acad
Sci USA. 111:E5429–E5438. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Deynoux M, Sunter N, Hérault O and
Mazurier F: Hypoxia and hypoxia-inducible factors in leukemias.
Front Oncol. 6:412016. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Heddleston JM, Wu Q, Rivera M, Minhas S,
Lathia JD, Sloan AE, Iliopoulos O, Hjelmeland AB and Rich JN:
Hypoxia-induced mixed-lineage leukemia 1 regulates glioma stem cell
tumorigenic potential. Cell Death Differ. 19:428–439. 2012.
View Article : Google Scholar
|
|
92
|
Sun JC, He F, Yi W, Wan MH, Li R, Wei X,
Wu R and Niu DL: High expression of HIF-2α and its
anti-radiotherapy effect in lung cancer stem cells. Genet Mol Res.
14:18110–18120. 2015. View Article : Google Scholar
|
|
93
|
Dhatwalia SK, Kumar M and Dhawan DK: Role
of EGCG in containing the progression of lung tumorigenesis - A
multistage targeting approach. Nutr Cancer. 70:334–349. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Vadde R, Vemula S, Jinka R, Merchant N,
Bramhachari PV and Nagaraju GP: Role of hypoxia-inducible factors
(HIF) in the maintenance of stemness and malignancy of colorectal
cancer. Crit Rev Oncol Hematol. 113:22–27. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Thomas S, Harding MA, Smith SC, Overdevest
JB, Nitz MD, Frierson HF, Tomlins SA, Kristiansen G and Theodorescu
D: CD24 is an effector of HIF-1-driven primary tumor growth and
metastasis. Cancer Res. 72:5600–5612. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Bhagat M, Palanichamy JK, Ramalingam P,
Mudassir M, Irshad K, Chosdol K, Sarkar C, Seth P, Goswami S, Sinha
S, et al: HIF-2α mediates a marked increase in migration and
stemness characteristics in a subset of glioma cells under hypoxia
by activating an Oct-4/Sox-2-Mena (INV) axis. Int J Biochem Cell
Biol. 74:60–71. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Johansson E, Grassi ES, Pantazopoulou V,
Tong B, Lindgren D, Berg TJ, Pietras EJ, Axelson H and Pietras A:
CD44 interacts with HIF-2α to modulate the hypoxic phenotype of
perinecrotic and perivascular glioma cells. Cell Reports.
20:1641–1653. 2017. View Article : Google Scholar
|