As the most frequent malignancy occurring in
females, breast cancer (BC) represents a great threat to women's
health. In 2018, the World Health Organization registered 2,088,849
BC cases and 626,679 BC-associated deaths globally (1). However, the survival rate for
patients with BC has been increasing since the 1990s. One reason
for this is the improvement in systemic treatment concepts
(2,3). Moreover, molecular subtyping has been
implemented in clinical practice as an important tool for
risk-adapted therapy in patients with BC (4-7).
Triple negative breast cancer (TNBC) is classified
as the most aggressive molecular subtype of BC (8). TNBCs are estimated to comprise ~15%
of all cases of BC (9,10), and its occurrence is highest in
younger females (9) and those of
African-American ancestry (8,11).
The prognosis for women diagnosed with TNBC is less favourable than
for those suffering from other intrinsic subtypes (12). Characterised by the absence of
hormone receptors and a lack of Her2neu (ErbB2) gene amplification,
TNBCs are heterogeneous carcinomas (9) and are often poorly differentiated.
Several attempts have been made to further categorise TNBC subtypes
according to distinct gene expression patterns. For example,
Lehmann et al (13)
analysed the gene expression profiles of 587 TNBC cases and
identified six distinct TNBC subtypes: Two basal-like, an
immunomodulatory, a mesenchymal, a mesenchymal stem-like and a
luminal androgen receptor subtype.
Due to the absence of hormone receptors and the lack
of Her2neu gene amplification, therapy for patients with TNBC is
restricted to neoadjuvant chemotherapy (NACT), radiotherapy and
surgery (16). In the case of
NACT, chemotherapy is administered prior to surgery, while in
adjuvant chemotherapy (ACT), surgery precedes the chemotherapy
treatment. NACT is considered to be equivalent to ACT in terms of
clinical outcome, and has become an established treatment in BC
therapy (17-21). Additionally, fewer adverse effects
were observed with NACT compared with ACT (17). The primary endpoint of NACT is
defined as the pathologically determined response of the tumour in
the breast and the axillary lymph nodes. Achieving pathological
complete response (pCR) at the time of surgery, defined as no
residual invasive or non-invasive tumour tissue in the breast and
the axillary lymph nodes, represents an important surrogate marker
for beneficial overall survival in these patients (22-27).
Further advantages of NACT treatment can be seen in the increasing
rate of breast conserving surgery and the evaluation of short-term
surrogate markers (pathological, clinical and molecular) for
outcome prediction (21,27,28).
In this context, it is known that an early tumour response (after
1-2 cycles of NACT) is also a predictive factor for pCR, and offers
the opportunity to distinguish responders from non-responders at a
very early time point of therapy (27). This early determination of poor
response offers the chance to alter treatment regimens and drugs
with the goal of achieving an optimized response up to a pCR.
Therefore, reliable measures to determine early response to NACT
represent an important prerequisite for innovative personalised
treatment concepts for the improvement of the clinical outcome for
patients with non-responding tumours. The NACT regime for patients
with TNBC includes both an anthracycline and a taxane that should
be administered for 18-24 weeks (28). The addition of platinum,
irrespective of breast cancer susceptibility protein status,
increases pCR rates; however, it is accompanied by enhanced
drug-dependent toxicity in patients with TNBC (7). A variety of diagnostic possibilities,
including ultrasound (US), palpation, mammography, magnetic
resonance imaging (MRI) and positron emission tomography/computed
tomography, are available for response monitoring during NACT
(28). Although US measurements
are used routinely in the clinic to assess tumour response to NACT,
few data exist concerning the accuracy of US for the prediction of
pCR (29,30). Morphological reactions of solid
tumours during therapy (for example, fragmentation and tumour
necrosis with or without stromal reaction) may compromise the
accuracy of US in discriminating between responding and
non-responding tumours. A previous study performed by Chagpar et
al (31) investigated the
accuracy of physical examination, US and mammography in predicting
residual tumour size in NACT-treated patients with BC, and found
only a moderate correlation. MRI appears to be an accurate method
to detect residual disease; however, the false-negative rate is
increased in patients receiving chemotherapy. Furthermore, these
methods depend on tumour size reduction, which can, in certain
cases, take months (32).
As an early response to NACT seems to be associated
with achieving pCR and an improved prognosis for patients with
TNBC, prediction of therapy response is particularly important in
this BC subgroup (24,33-35).
A poor prognosis for patients with TNBC improves to a similarly
high level if patients achieve a pCR (34). This underlines the need for more
specific and sensitive predictive methods. Liquid biopsy-based
non-coding RNAs (ncRNAs) in the serum and urine of patients
undergoing NACT as innovative biomarkers may be a promising tool to
predict early therapy response before the current standard imaging
methods are employed.
To pursue this question, the present study examined
the expression of 14 ncRNAs, including 12 microRNAs (miRNA/miRs;
let-7a; let-7e; miR-7, -9, -15a, -17, -18a, -19b, -21, -30b, -222
and -320c), one PIWI-interacting RNA (piRNA; piR-36743) and one
transfer RNA (GlyCCC2) in a triple positive (BT-474) and three TNBC
(BT-20; HS-578T; MDA-MB-231) cell lines treated with the cytostatic
drugs carboplatin, epirubicin, gemcitabine and paclitaxel, both
intra- and extracellularly. Analysis of cell cycle determinants
[cyclin D1, DNA topoisomerase 2α (TOP2α) and targeting protein for
Xklp2 (TPX2)] and RNA metabolic proteins [DEAD-box polypeptide
(DDX)5 and DDX17] were performed at the mRNA and protein level to
investigate drug-dependent effects on the cell cycle. Subsequently,
ncRNA expression was analysed in serum and urine specimens from
patients with TNBC and healthy controls. Additionally, alterations
in ncRNA expression in patients with TNBC prior to NACT
(t0) and immediately prior to the third cycle of therapy
(t1) were investigated.
BC cell lines BT-474 and BT-20 (cat. nos. 300130 and
300131; CLS Cell Lines Service GmbH), HS-578T (cat. no. 86082104;
Sigma-Aldrich; Merck KGaA) and MDA-MB-231 (cat. no.
92020424; Sigma-Aldrich; Merck KGaA) (46) were incubated in DMEM/F12 medium
(Gibco; Thermo Fisher Scientific, Inc.), supplemented with 5% FBS
(Gibco; Thermo Fisher Scientific, Inc.), 1% HEPES buffer (Gibco;
Thermo Fisher Scientific, Inc.) and 1% 100 U/ml
penicillin/streptomycin (Sigma-Aldrich; Merck KGaA). Insulin (2.5%;
Insuman® rapid; Sanofi S.A.) was added to BT-474 cells. Cells were
incubated under humidified conditions at 37°C and 5%
CO2. The cell lines were authenticated using
PCR-single-locus-technology. Mycoplasma testing is performed
regularly.
Female patients with histologically proven, invasive
TNBC as defined by the St. Gallen Consensus 2012 (47), who were subjected to NACT were
included in the present study. The inclusion criteria were as
follows: i) Primary diagnosis of TNBC; and ii) age of >18 years.
The exclusion criteria were as follows: i) Distant metastasis at
the time of diagnosis; and ii) other previous or coexistent
malignancies. The present study was approved by the institutional
ethical review board of the University of Freiburg (permit no.
607/16) and written informed consent was obtained from each
participant. Patients (aged 40-70 years) of the Department of
Gynaecology and Obstetrics at the Medical Center-University of
Freiburg were enrolled between February 2017 and September 2017;
additionally, 20 healthy controls (aged 23-70 years) were enrolled
between March 2016 and August 2016, of whom 10 provided serum
samples, and 10 provided urine samples. All patients received NACT
according to standard treatment protocols [90 mg/m2
epirubicin + 600 mg/m2 cyclophosphamide (EC) ×4 (once
every 3 weeks), followed by 80 mg/m2 paclitaxel (P) ×12 (weekly)]
(28). BC tissue biopsies were
collected prior to NACT, and evaluated via haematoxylin and eosin
(H&E) and Ki67 staining by the Pathology Department of the
Medical Center-University of Freiburg. Histopathological
preparations of BC were graded according to the classification
published by Elston and Ellis (48) on H&E-stained slides, which
includes counting of mitotic cells. Furthermore, the proliferation
index was measured in sections immunohistochemically stained with
the Ki67 antibody clone MIB-1. Any nuclear positivity (nucleoli
excluded) was counted. The percentage of positive nuclei was either
calculated from counting at least 200 tumour cells or estimated
semi-quantitatively, especially for percentages <10 and >30%.
In total, 9 patients with TNBC were enrolled. Patient
characteristics are displayed in Table
I. Blood (2-9 ml) was drawn by physicians at two time points
(t0, prior to NACT; t1, immediately prior to
third cycle of NACT), according to standard protocols. In order to
evaluate the individual response of each patient to NACT, follow-up
by US was performed and interpreted, according to the RECIST
criteria (30). After coagulation
for 20 min, whole blood was centrifuged (10 min, 2,000 × g, 4°C).
Cell-free serum specimens were transferred to a fresh test tube.
Samples exhibiting haemolytic characteristics were excluded. In
total, 20 ml of urine was centrifuged (15 min, 2,000 × g, 4°C) and
the cell-free supernatant was transferred to fresh test tubes.
Serum and urine samples were stored at -20°C. Of the nine serum and
urine samples collected from patients with TNBC, only eight met
quality standards and were further analysed.
Intracellular ncRNA was isolated using an EURx®
GeneMATRIX Universal RNA/miRNA Purification kit (EURx Sp. z o.o.)
according to the manufacturer's protocol. For the isolation of
secreted microvesicular extracellular ncRNA, conditioned cell
culture medium from BC cells was decanted into a 15-ml tube and
centrifuged at 4°C at 8,000 × g for 5 min. In total, 10 ml
supernatant was transferred to a new 15-ml tube. Microvesicles from
liquid samples (conditioned cell culture medium, serum and urine)
were isolated via filtration. RNA was subsequently isolated using a
Norgen Total RNA Purification kit (Norgen Biotek Corp.), according
to the manufacturer's protocol. In total, 20 ml urine and 2 ml
serum were applied. The RNA concentration was determined using a
NanoDrop™ ND1000 (Thermo Fisher Scientific, Inc.) and RNA was
stored at -20°C until further processing.
The targets of the mRNA analysis included cyclin D1,
TOP2α, TPX2, DDX5 and -17. Cq values were normalised to the
housekeeping gene, 5′-aminolevulinate synthase 1 (61).
Proteins from the BC cell lines were isolated from
the flow-through of the RNA-binding columns used to purify RNA by
the addition of 1 ml of isopropanol and subsequent centrifugation
for 30 min at 12,000 × g at 4°C. The protein pellet was washed with
70% ethanol and lysed in protein lysis buffer (1% SDS, 1% triton, 1
mM EDTA, 0.1 M HEPES) for 30 min at 1,000 RPM and 50°C in an
Eppendorf ThermoMixer®. Protein concentrations were determined
using the bicinchoninic acid method as previously described
(62). Western blotting was
conducted as described by Schagger and von Jagow (63). Briefly, 10 μg/lane of
protein was separated on a 10% SDS gel. Proteins were transferred
onto a 0.45-μm PVDF membrane (Sigma-Aldrich; Merck KGaA) and
membranes were blocked with 3% skimmed milk in PBS-Tween 20 at room
temperature for 30 min. Monoclonal mouse anti-human antibodies for
immunodetection were purchased from Santa Cruz Biotechnology, Inc.
The following dilutions were used: Cyclin D1 (1:500; cat. no.
sc-8396); DDX5 (1:5,000; cat. no. sc-365164); DDX17 (1:10,000; cat.
no. sc-398168); p-Actin (1:1,000; cat. no. sc-47778); and
proliferating cell nuclear antigen (PCNA; 1:1,000; cat. no. sc-56).
Membranes were incubated with primary antibodies overnight at 4°C.
A secondary peroxidase-conjugated anti-mouse antibody was purchased
from Jackson ImmunoResearch Europe, Ltd. (cat. no. 115-036-146) and
was incubated at room temperature with the membrane for 1 h
(1:10,000). Bands were visualised using ECL reagent (Santa Cruz
Biotechnology, Inc.). PCNA and p-Actin served as loading
controls.
In the present study, the influence of
chemotherapeutic treatment on the expression levels of 14
BC-related ncRNAs (let-7a/e, miR-7, -9, -15a, -17, -18a, -19b, -21,
-30b, -222 and -320c, piR-36743 and GlyCCC2) was investigated in
three TNBC (BT-20, HS-578T and MDA-MB-231) cell lines and one
triple positive BC (BT-474) cell line, both intra- and
extracellularly.
All examined cell lines expressed all of the 14
investigated ncRNA targets. In addition to intracellular
expression, the vast majority of the panel were detectable as
secreted molecules in microvesicles in cell culture medium, with
the exception of miR-7, -9 and -15a in the medium of MDA-MB-231,
and miR-9 in the medium of HS-578T cells. Chemotherapy treatment of
cells with carboplatin, epirubicin, gemcitabine and paclitaxel
resulted in distinct ncRNA expression alterations depending both on
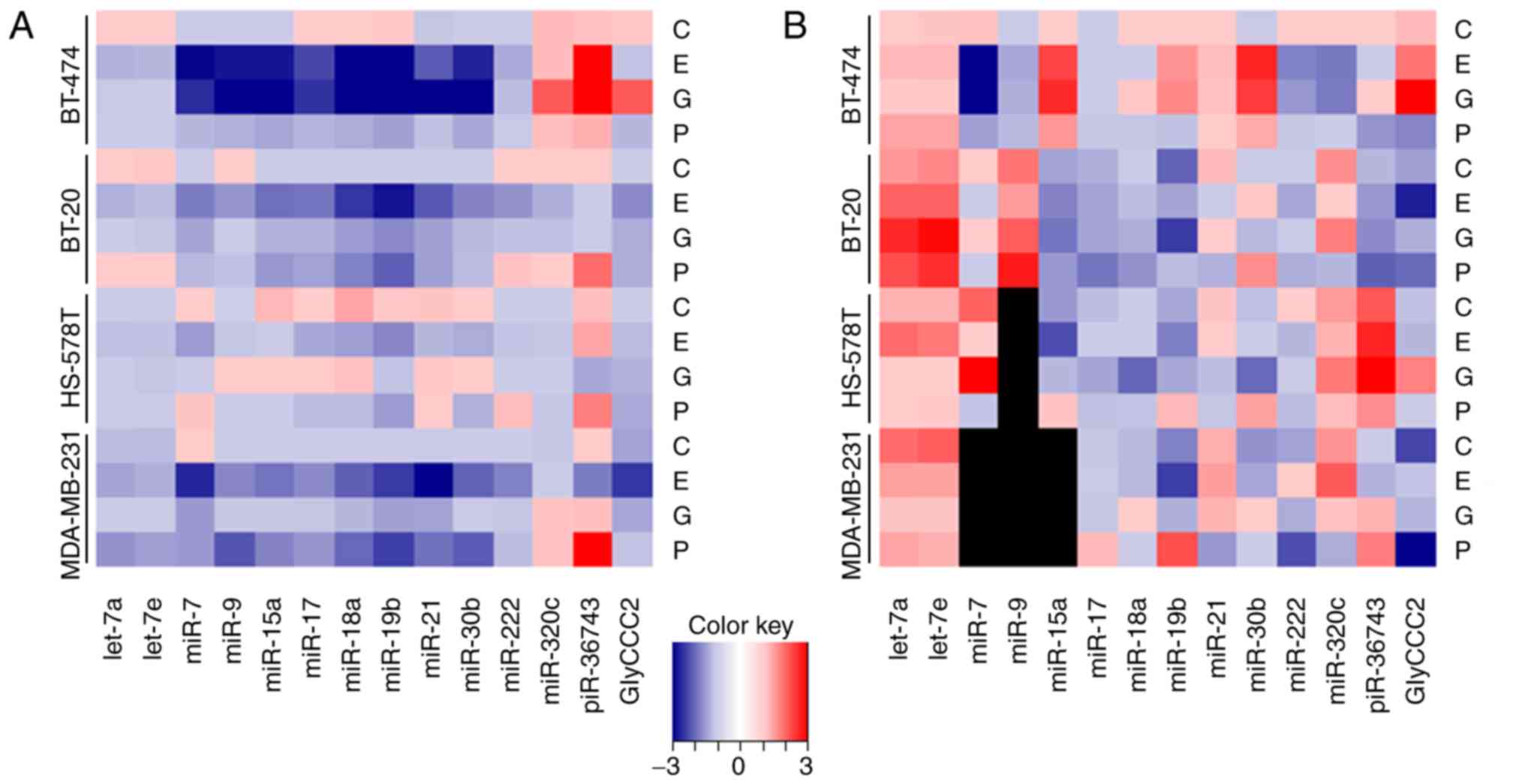
the cytotoxic agent and on the specific BC cell line (Fig. 1).
Multivariable analysis confirmed the inhibitory
effect of epirubicin on ncRNA expression, especially in hormone
receptor positive BT-474 and TNBC cell lines BT-20 and MDA-MB-231.
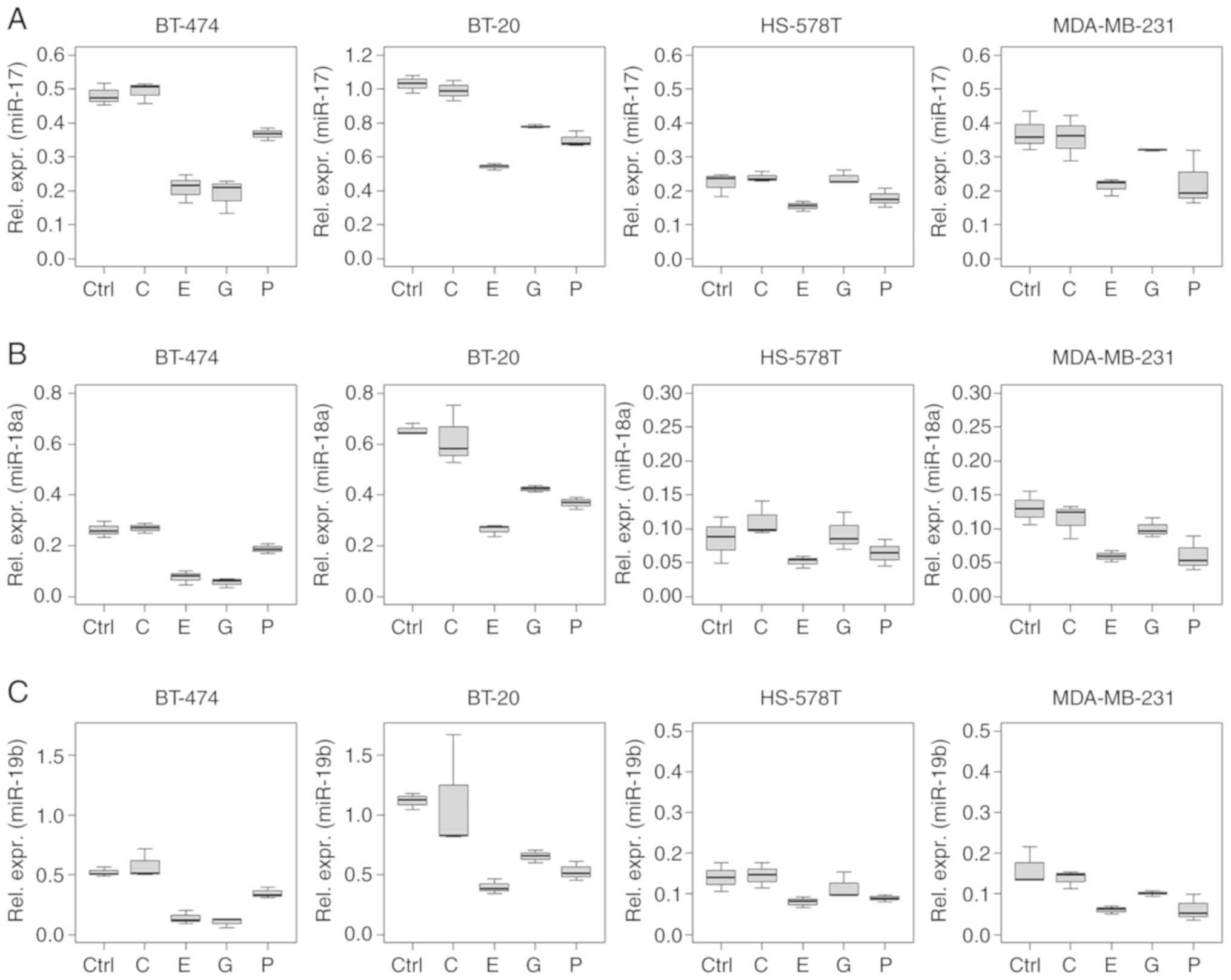
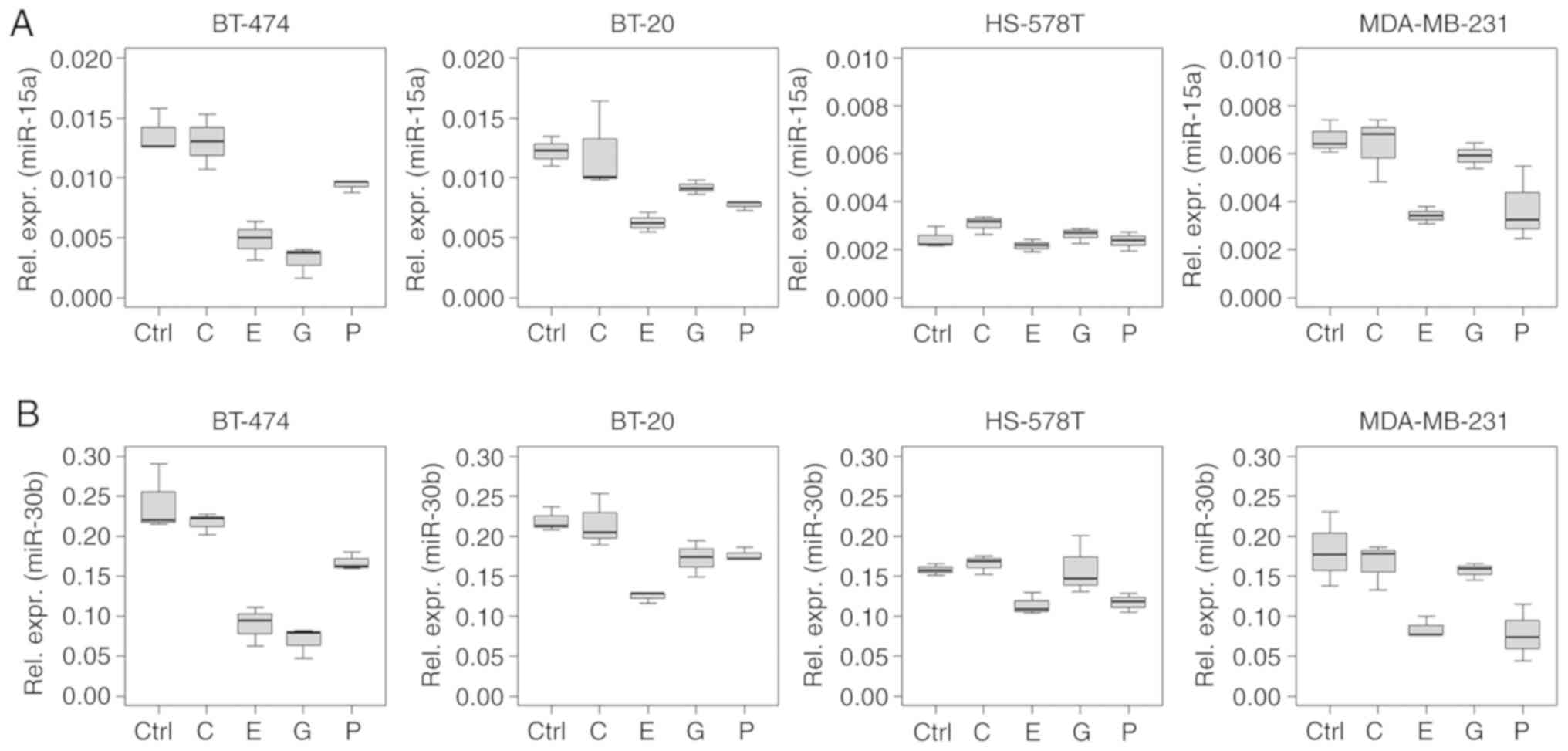
miR-7, -15a, -17, -18a, -19b, -21, -30b and -222 were downregulated
in BT-474, BT-20 and MDA-MB-231 cell lines compared with the
intercept, while reduced expression of miR-9 was restricted to
BT-474 and MDA-MB-231 cells. GlyCCC2 expression was downregulated
in BT-20 and MDA-MB-231 cells. piR-36743 expression decreased
following epirubicin treatment in MDA-MB-231 cells, while an
increase in expression was found in BT-474 cells. In HS-578T, the
effect of epirubicin was limited to miR-17 and -18a expression. All
estimates of multivariable analysis, including confidence intervals
(CIs) and P-values are listed in Table SII.
In a multivariable analysis, gemcitabine treatment
caused major changes in the expression levels of ncRNAs in BT-474
cells, while few detectable alterations were observed in the TNBC
cell lines. Specifically, miR-7, -9, -15a, -17, -18a, -19b, -21 and
-30b were downregulated compared with the intercept, while
miR-320c, piR-36743 and GlyCCC2 were upregulated in BT-474 cells.
In the TNBC cell lines, downregulated expression was detected for
miR-17, -18a and -21 in BT-20, and miR-7 and -21 in MDA-MB-231
cells. No significant effect was detected in HS-578T cells. All
estimates of multivariable analysis, including CIs and P-values are
listed in Table SII.
Intracellularly, the expression levels of the ncRNAs
predominantly decreased following paclitaxel (2.0 μg/ml)
treatment. However, in the cases of miR-7 (HS-578T), -222 (BT-20
and HS-578T), -320c (BT-474, BT-20 and MDA-MB-231) and piR-36743
(all cell lines), upregulation was detected. The levels of secreted
microvesicular ncRNAs were influenced by paclitaxel and differed
among the investigated cell lines (Fig. 1).
Multivariable analysis found that paclitaxel
triggered ncRNA expression fluctuations predominantly in the TNBC
cell lines BT-20 and MDA-MB-231. Compared with the intercept,
miR-17, -18a, -19b and -21 were downregulated in BT-20 cells, while
piR-36743 was upregulated. In MDA-MB-231 cells, the expression
levels of let-7a, let-7e, miR-7, -9, -15a, -17, -18a, 19b, -21,
-30b and -222 were decreased, and piR-36743 was upregulated. In the
HS-578T cell line, no alteration in the investigated ncRNA profiles
was detected, except for the upregulation of piR-36743. Paclitaxel
treatment caused the downregulation of miR-17 in BT-474 cells
compared with the intercept. All estimates of multivariable
analysis, including CIs and P-value are listed in Table SII.
Focusing on the extracellular compartment, no
conclusions can be drawn from the observation of an increase in the
relative levels of ncRNA expression under control conditions
compared with the intracellular expression levels for all targets,
which was mirrored by significantly increased estimates in
multivariable analysis (Table
SIII). This phenomenon cannot be explained by biological
variation but is, at least in part, observed due to different
normalisation methods in intra- and extracellular analysis.
The applied model of multivariable analysis allowed
the estimation of the additional impact on ncRNA expression caused
by three-way interactions with the variables 'chemotherapy', 'cell
line' and 'extracellular compartment'. Most of these influences
were observed in the BT-474 cell line and were less frequent in the
TNBC cell lines. Depending on the estimate, the aforementioned
regulation of the expression of the ncRNAs will be increased
(estimate >1) or alleviated (estimate <1; Table SIII). For the most part, the
three-way interactions attenuated the effects presented in Table SII. This was observed for miR-15a,
-17, -18a, -19b, -21, -30b, -320c and piR-36743 following
gemcitabine treatment, as well as for miR-15a, -17, -30b, -320c and
piR-36743 following epirubicin treatment in BT-474 cells. In the
TNBC cell line HS-578T, the three-way interactions resulted in the
opposite effects on miR-18a, -21 and piR-36743 following
gemcitabine treatment. The same applied to miR-21 and -222
following epirubicin treatment, and for miR-17 and -19b following
paclitaxel treatment in MDA-MB-231 cells (Table SIII).
While lacking significance in the aforementioned
observations, three-way interaction influences estimated by
multivariable analysis added to the effect of piR-36743 upregu-
lation in HS-578T cells following epirubicin treatment (1.310;
P=0.208) by 2.662 (P=0.024). Similarly, let-7a expression following
gemcitabine treatment (0.929; P=0.760) decreased by 0.357 (P=0.036)
in this cell line. Let-7e expression (0.887; P=0.619) further
decreased in the same context by 0.337 (P=0.027). The reduced
GlyCCC2 expression induced by paclitaxel treatment in MDA-MB-231
cells (0.772; P=0.372) was further decreased by a value of 0.277
(P= 0.029). In BT-474 cells, paclitaxel induced the downregulation
of miR-9 (0.752; P=0.233), which was increased by 0.332 (P=0.023).
These effects may indicate that let-7a and -7e, and GlyCCC2 may be
promising liquid biomarker candidates depending on the administered
chemotherapy (Table SIII). In the
case of piR-36743, three-way interaction influences added to
already significant findings as aforementioned. While the
upregulation of piR-36743 following paclitaxel treatment in HS-578T
cells (1.793; P= 0.008) increased by 3.492 (P=0.004), piR-36743
increased in MDA-MB-231 cells (2.922; P<0.001) by 2.428
(P=0.040; Table SIII).
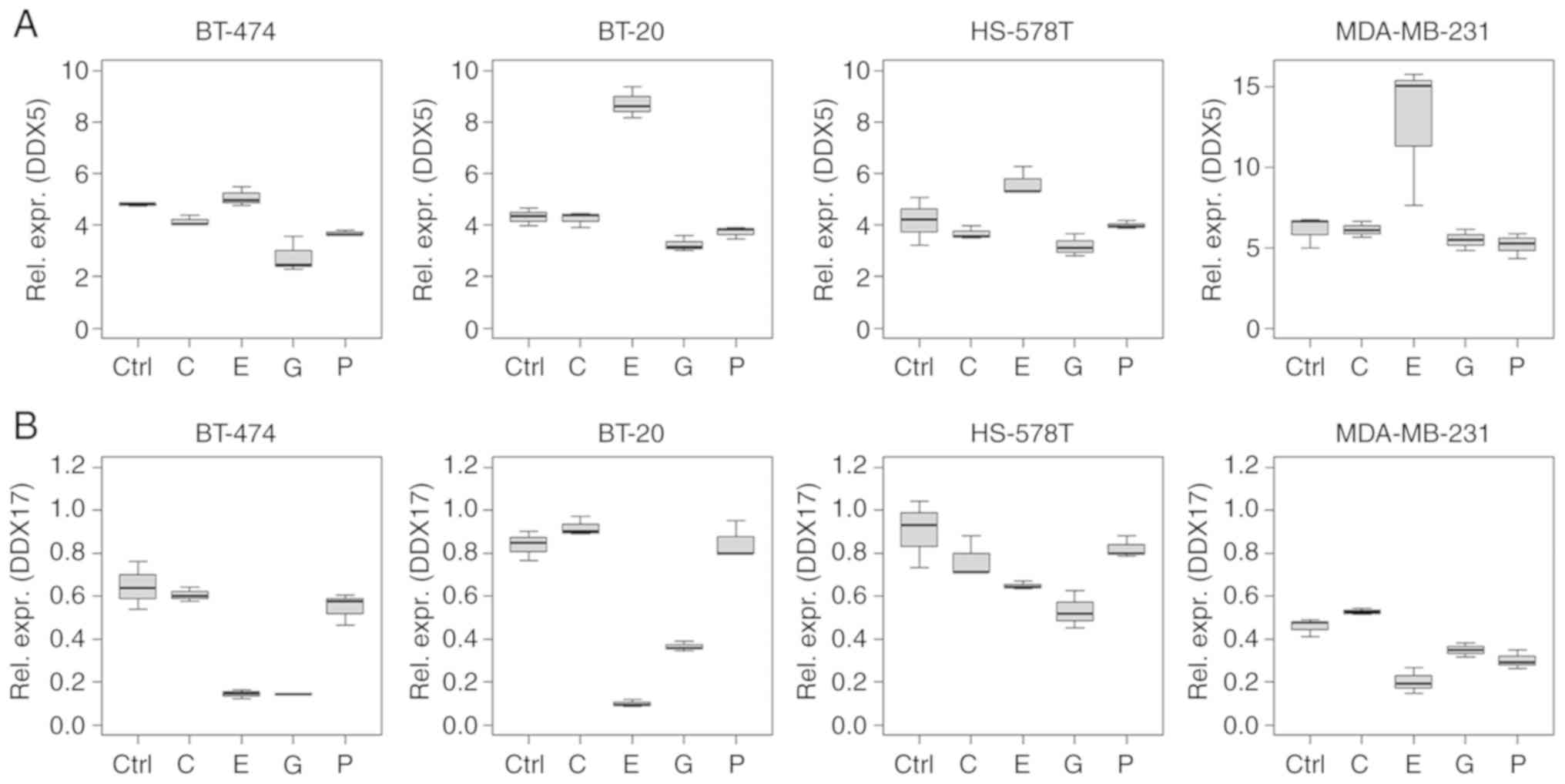
Several ncRNAs analysed in the present study showed
a similar regulation pattern, especially in the intracellular
analysis. This phenomenon was observed for miRNAs of the miR-17~92
cluster (miR-17, -18a and -19b), and for miR-15a and -30b (Figs. 2 and 3).
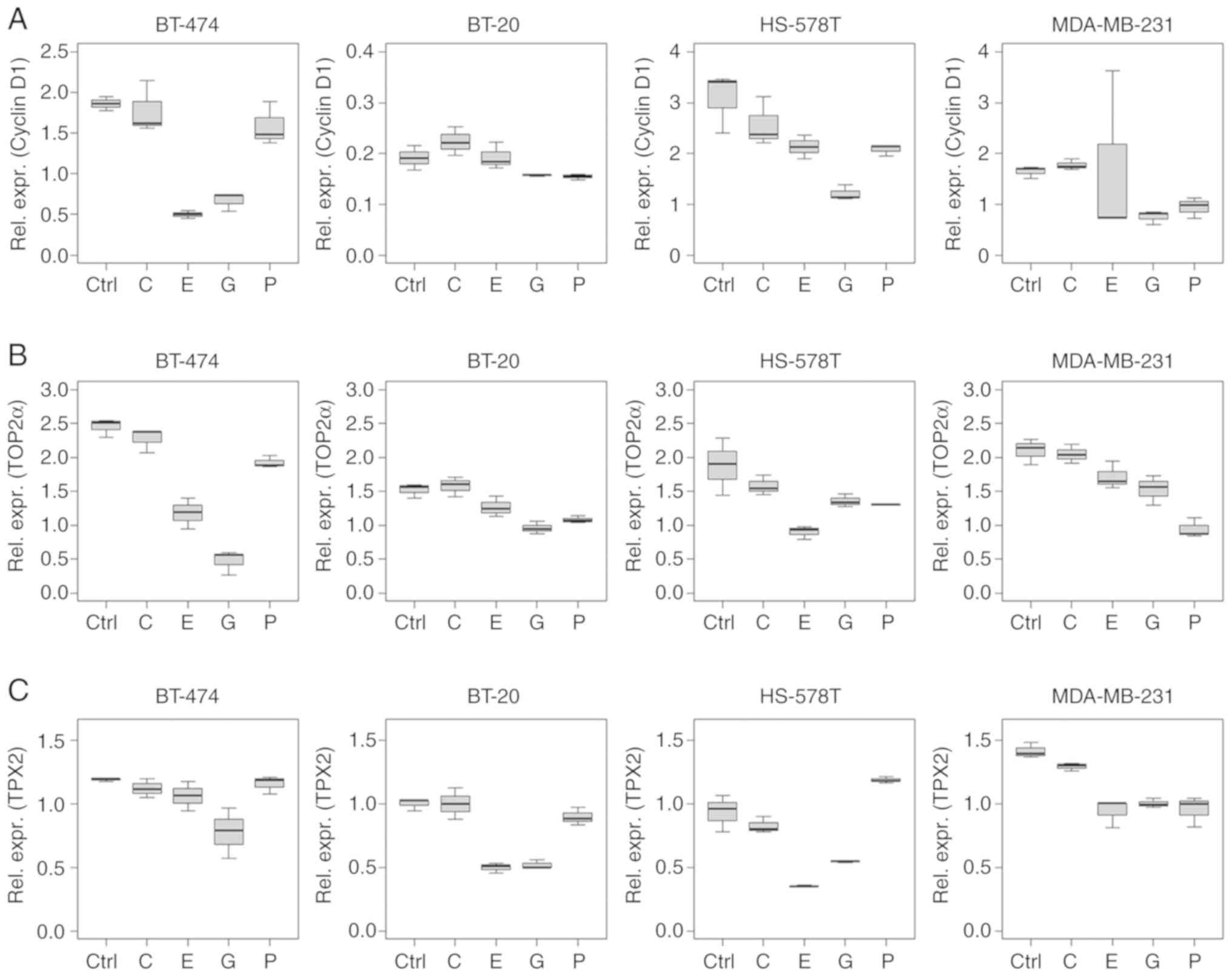
Cyclin D1 mRNA expression levels reduced after
treatment with epirubicin and gemcitabine (Fig. 4A). Multivariable analysis indicated
a decrease in cyclin D1 expression levels in BT-474 and HS-578T
cells (epirubicin, P<0.001 and P=0.010, respectively), and in
BT-474, HS-578T and MDA-MB-231 cells (gemcitabine, P=0.002,
P<0.001 and P=0.016, respectively) compared with the intercept.
A reduction was also observed in paclitaxel-treated HS-578T cells
(P=0.007). Carboplatin did not influence cyclin D1 mRNA expression
in this setting. All estimates of multivariable analysis, including
CIs and P-values are listed in Table
SIV.
TOP2α mRNA levels were reduced following
chemotherapy, except following carboplatin treatment in all the
cell lines tested (Fig. 4B).
P-values, estimates and CIs are listed in Table SIV.
TPX2 mRNA level alterations were similar to those of
cyclin D1, with reductions in expression caused by epirubicin
(BT-20, HS-578T and MDA-MB-231; P<0.001) and gemcitabine (all
cell lines; P<0.001) treatment (Fig. 4C). Paclitaxel treatment reduced
TPX2 levels in BT-474 and MDA-MB-231 cells (P<0.001), but
increased TPX2 expression in HS-578T cells (P=0.001; Table SIV).
While epirubicin treatment induced differential
regulation of DDX5 and -17 mRNA levels, DDX17 was regulated in a
similar manner as the miR-17~92 cluster (Figs. 2 and 5). Specifically, DDX5 mRNA was
upregulated by epirubicin compared with the intercept (BT-20 and
MDA-MB-231, P<0.001) and downregulated in BT-474 by gemcitabine
(P=0.034). By contrast, DDX17 mRNA was downregulated following
epirubicin treatment in all the cell lines tested (all P<0.001).
Downregulation was also observed following gemcitabine treatment in
BT-20, BT-474, HS-578T (P<0.001) and MDA-MB-231 cells (P=0.049),
as well as by paclitaxel in MDA-MB-231cells (P=0.006; Table SIV).
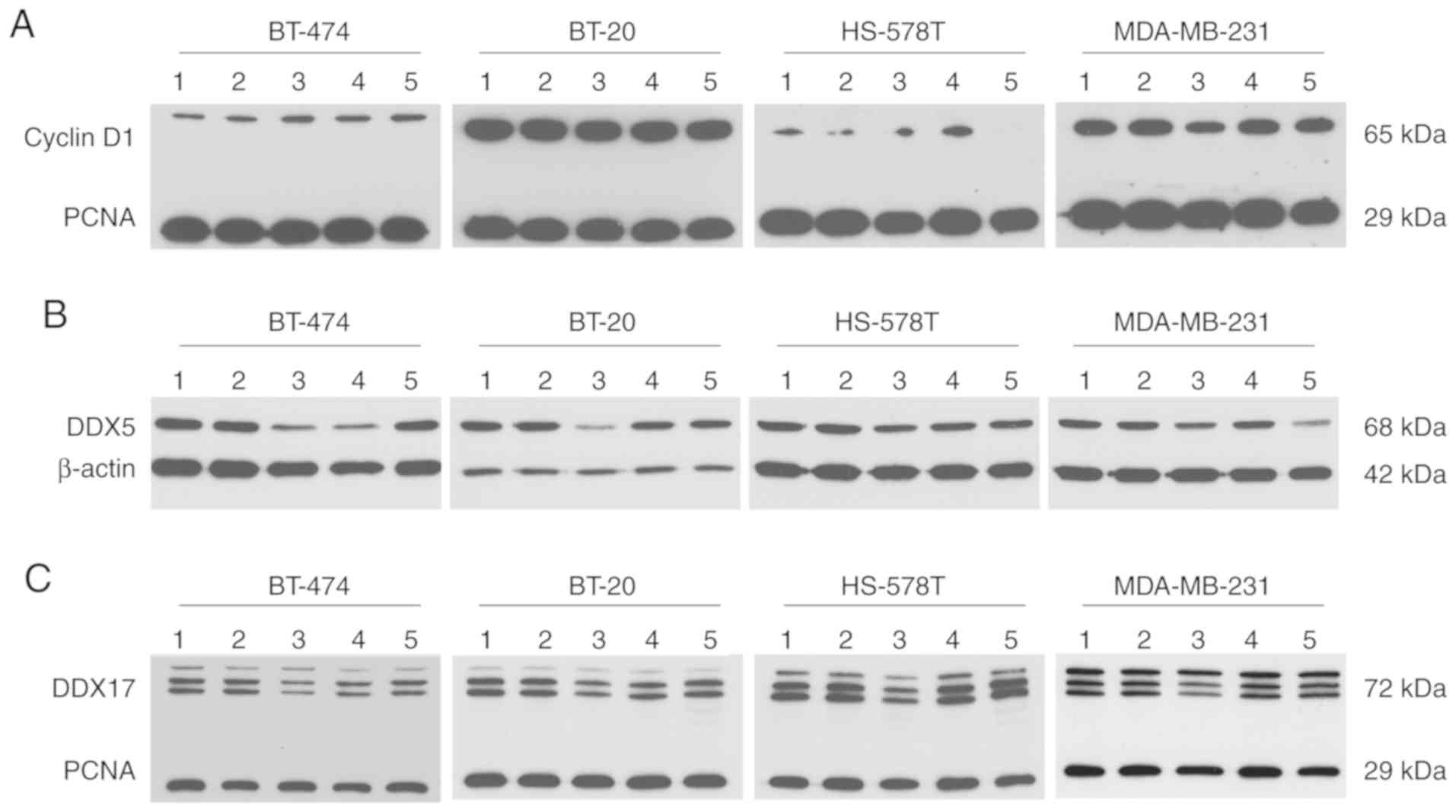
At the protein level, cyclin D1 was detected at a
molecular weight of ~65 kDa. This immunoprecipitated protein may
represent a ubiquitylated form of cyclin D1 and was detected in all
investigated cell lines. The expression of cyclin D1 in HS-578T
cells, however, was present at modest levels. Compared with the
control conditions, no regulation of cyclin D1 was observed, except
for a modest reduction caused by epirubicin treatment in MDA-MB-231
cells (Fig. 6A). Western blotting
revealed differential DDX5 regulation following epirubicin
treatment at the protein level compared with mRNA regulation; DDX5
was downregulated at the protein level. Following gemcitabine
treatment, DDX5 protein levels also declined in BT-474 and HS-578T
cells, while they declined in MDA-MB-231 cells following paclitaxel
treatment. A reduced DDX17 protein level caused by epirubicin is
consistent with reduced DDX17 mRNA. In BT-474 and BT-20 cells,
gemcitabine led to lower DDX17 protein expression; paclitaxel had
the same effect in MDA-MB-231 cells (Fig. 6B and C).
In the present study, no clear pattern was observed
when dividing the TNBC group into early responder and non-responder
[early response as defined by the achievement of a clinical partial
response (cPR) according to RECIST criteria (30) immediately prior to the third cycle
of NACT]. Also, no association was observed regarding pCR as an
outcome (data not shown). However, a trend was discovered in the
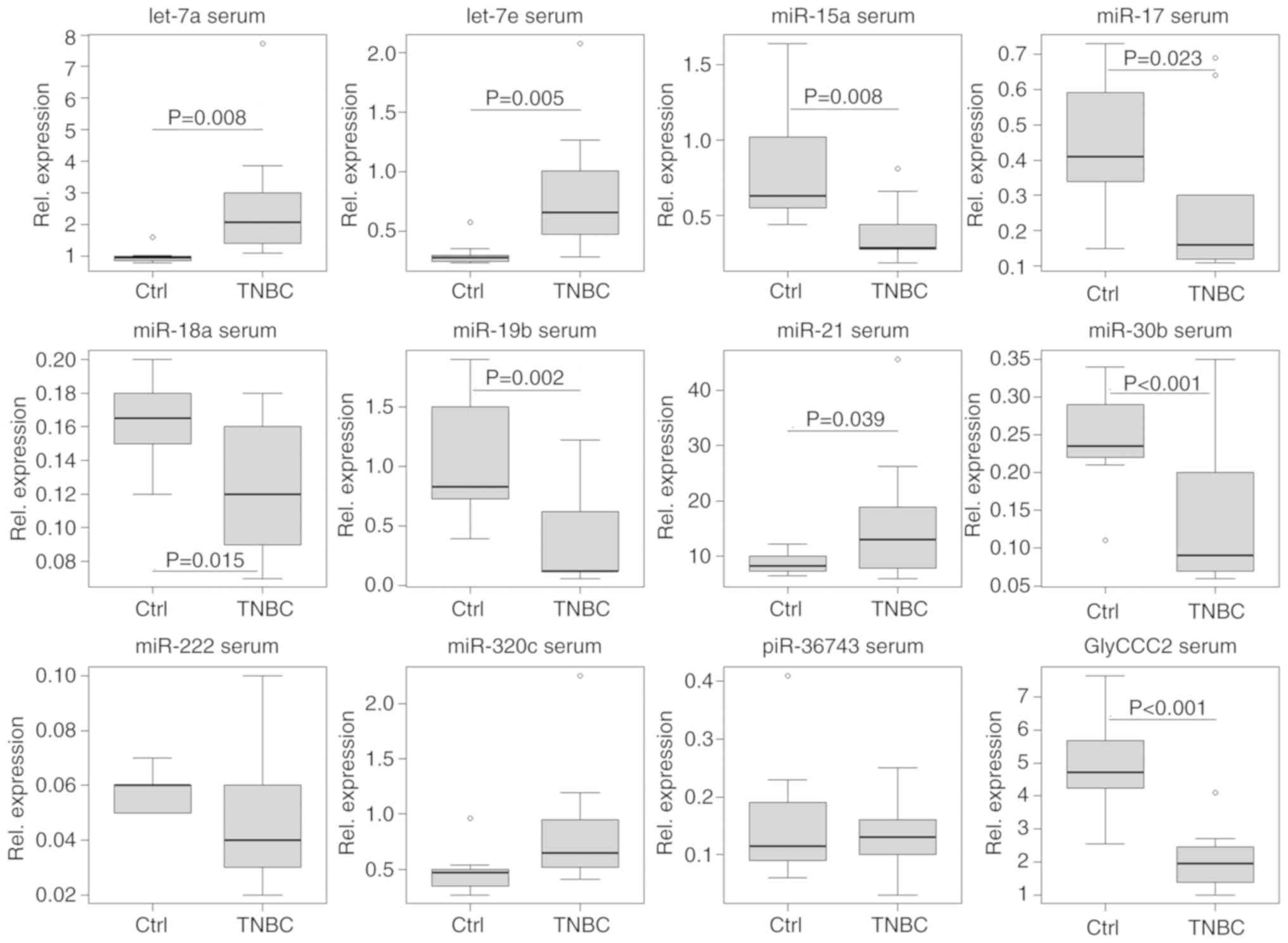
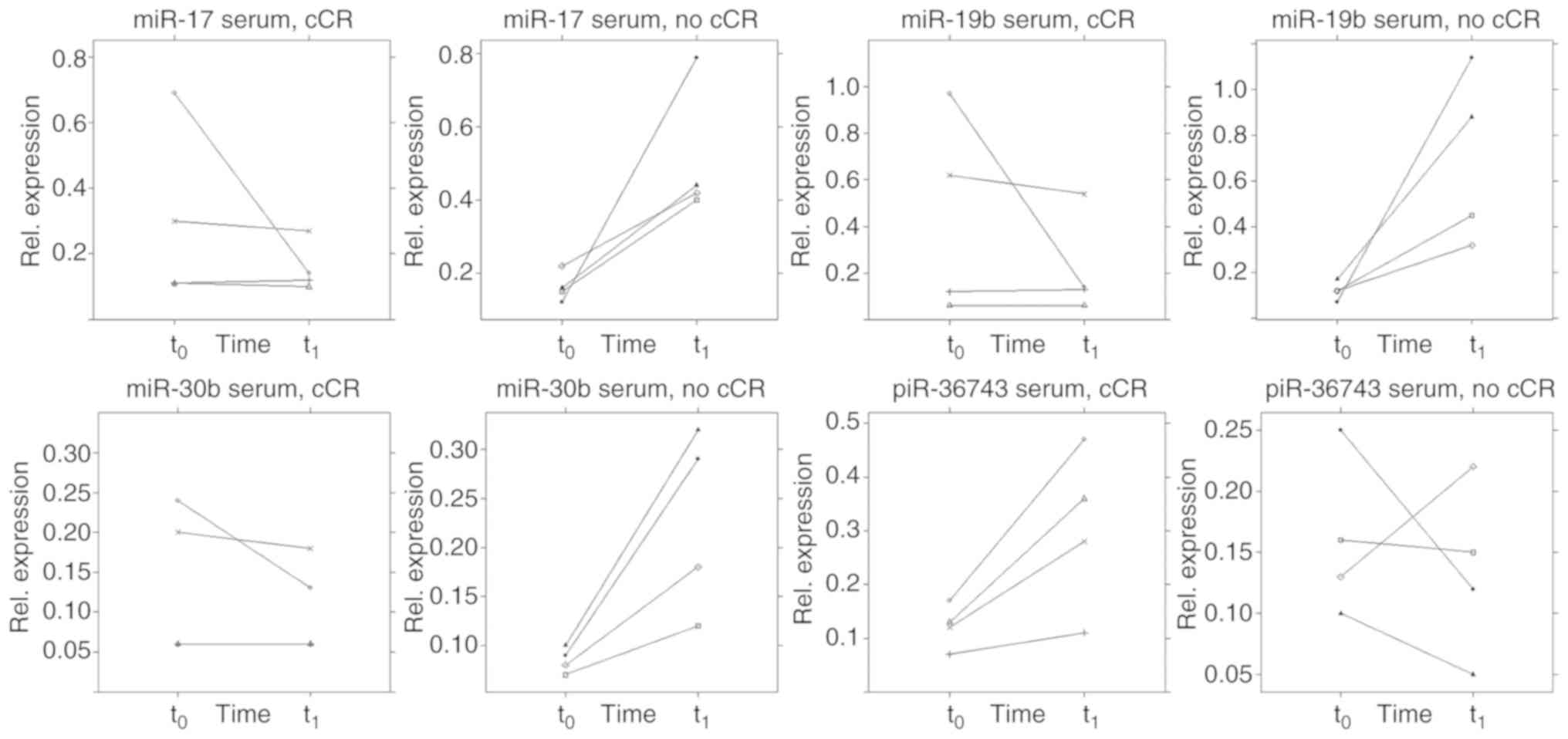
serum of patients receiving a cCR during NACT (Fig. 9). In this respect, those patients
(n=4) showed a different ncRNA regulation pattern immediately prior
to the third cycle of NACT (q) compared with patients (n=4) who did
not achieve a cCR during NACT. A two-tailed paired t-test estimated
a difference from t0 to t1 between the two
groups in terms of serum expression of miR-17 (P=0.029, SE=0.173),
-19b (P=0.030, SE= 0.282), -30b (P=0.011, SE=0.048) and piR-36743
(P=0.028, SE=0.072). However, these results remain preliminary due
the small sample size. By contrast, no specific pattern was
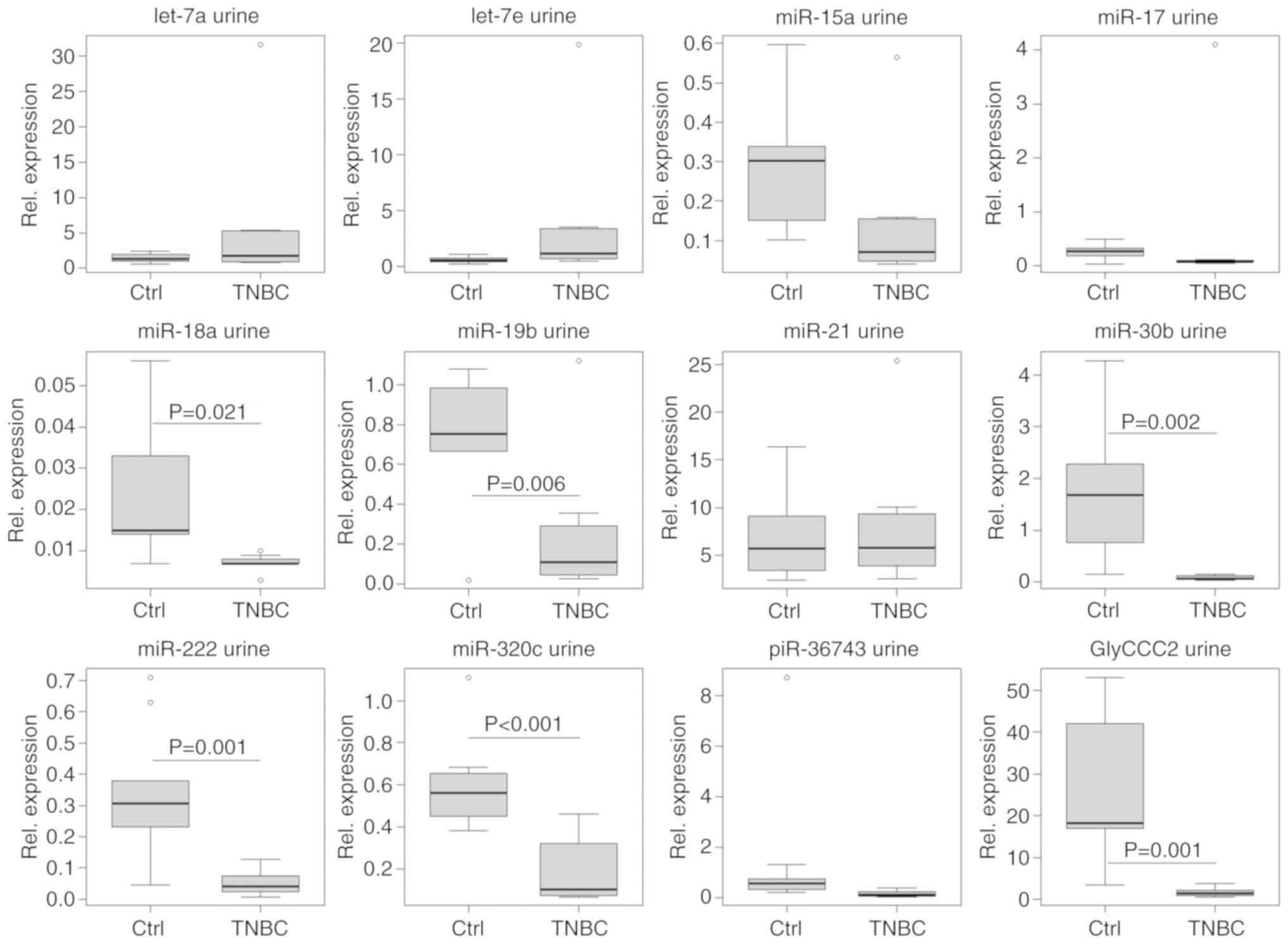
detected within the TNBC group using the urine specimens. A general
rise of ncRNA expression from t0 to t1 was
evident (Fig. 10).
The present study demonstrated that dysregulation of
ncRNAs in TNBC cell lines, serum and urine can be induced by
chemotherapy treatment. In vitro, the influences of
chemotherapeutic treatment with epirubicin, gemcitabine and
paclitaxel on the expression levels of a (TN)BC-related ncRNA panel
were evaluated. Even in the extracellular compartment, influences
were detected, indicating the effects of chemotherapy-driven ncRNA
secretion. The expression profiles differed depending on cell line
and cytostatic drug. In this in vitro model, epirubicin
caused the strongest effect on ncRNA regulation.
Except for miR-7 and -9, the present study reliably
detected all targets in microvesicles from liquid samples
(conditioned cell culture media, serum and urine). As
intracellularly, miR-7 and -9 resulted in distinct expression
alterations caused by chemotherapy treatment, these ncRNAs may
still be involved in therapy response mechanisms. Previous studies
have demonstrated associations between BC therapy response, and the
dysregulation of miR-7 and -9 in cell lines and tissue (65-68).
However, these two miRNAs were not detectable in microvesicles of
liquid samples in the present study, and cannot be recommended as
suitable liquid biopsy marker candidates in this context.
ncRNA expression alterations occurred along with
altered expression of cell cycle markers and, therefore, may be
associated with it. While epirubicin exhibited the most potent
effects on ncRNA regulation, carboplatin treatment did not
significantly influence ncRNA expression in the present study. This
observation is consistent with the present findings on the
influence of chemotherapeutic treatment on cell cycle determinants.
Cyclin D1, TOP2α and TPX2 were not influenced by carboplatin,
indicating limitations of this in vitro model. All cells
were treated with the chemotherapeutic agent for 18 h. Carboplatin
may cause an inhibitory effect on cell proliferation, and possibly
affect ncRNA expression, when treatment time is prolonged (69-72).
In western blotting, cyclin D1 could not be detected in its native
conformation, with a molecular weight of 37 kDa. However, the
product detected was of 65 kDa, and may represent a dimer or a
ubiquitylated form of cyclin D1. Cyclin D1 is degraded by
ubiquitylation and the native protein possesses a short half-life
of <30 min (73,74).
Apart from carboplatin-treated cells, mRNA
expression of cell cycle determinants was reduced in most of the
other samples as aforementioned. Epirubicin and gemcitabine
treatment impacted proliferation and led to ncRNA expression
alterations.
DDX proteins, a family of RNA helicases, are
dysregulated in several types of cancer, including BC (75). Their cellular functions are highly
context-dependent and influenced by posttranslational regulation
(75-77). DDX5 and -17 are closely related and
part of multiprotein complexes. One function of these two proteins
is oestrogen receptor-α co-activation, which may not play an
important role in TNBC; however, the co-activation of p53, RNA
metabolism and processing may be of importance (75,78,79).
Additionally, DDX proteins are involved in the processing of miRNAs
(75). The interplay between DDX
proteins and miRNA dysregulation may be an important feature of
TNBC. In the present study, it was found that DDX5 and -17 mRNA
expression in TNBC cells was influenced by chemotherapy treatment.
The mRNA and protein downregulation observed supports results from
previous studies that described an inhibition of cancer cell
proliferation when simultaneously depleting DDX5 and -17 (80,81).
The findings of the present study suggested an oncogenic function
for these DDX proteins in TNBC cells. However, the mRNA
upregulation of DDX5 triggered by epirubicin is of interest. As
protein levels of DDX5 were reduced after epirubicin treatment, a
feedback loop may be involved.
The widely studied miR-17~92 cluster exerts
oncogenic functions, with its members upregulated in several types
of cancer, including TNBC (82-84).
Furthermore, the expression of miR-17~92 varies in different BC
subtypes (85). In the present
study, it was observed that miRNAs of the miR-17~92 cluster
(miR-17, -18a and -19b), as well as miR-15a and -30b, were
regulated in a similar manner; DDX17 mRNA displayed a similar
expression profile to the miR-17~92 cluster. The present study
demonstrates that the expression levels of miR-17~92 cluster
miRNAs, and miR-15a and -30b, were reduced by epirubicin,
gemcitabine and paclitaxel treatment in the in vitro cell
models. It was also found that the miR-17~92 cluster miRNA
expression levels were reduced in serum and urine samples from
patients with TNBC compared with healthy controls.
piR-36743 was of interest due to the
chemotherapy-driven expression alterations found in the in
vitro models. piR-36743 was upregulated, while most of the
other investigated ncRNAs experienced chemotherapy-dependent
downregulation. The functional background of piR-36743 remains
poorly understood. Hashim et al (86) found that piR-36743 (also known as
DQ598677) was upregulated in BC biopsies compared with healthy
breast tissue.
The comparison of microvesicular ncRNA expression
levels in the urine and serum of patients with TNBC prior to NACT
and immediately prior to the third cycle of therapy provided
preliminary results. Urinary ncRNA expression showed a general
increase from t0 to t1, except for let-7a/e
and miR-17. This observation may be explained by the general
effects of treatment or other factors. In serum, a different
picture was observed. To evaluate the ncRNA panel for their liquid
biopsy biomarker potential in TNBC therapy response evaluation,
patients undergoing NACT were divided into a (early) responder and
non-responder group. However, it is emphasised that due to a small
sample size (8 patients with TNBC), the present study only provided
a preliminary result in this respect. As aforementioned, in an
attempt to visualize ncRNA differences in the serum of patients
with TNBC from t0 to t1 of NACT, no clear
pattern could be distinguished between the early responders and
non-responders, and no association with the achievement of a pCR
could be drawn. However, larger cohorts may shed further light on
this issue and should be analysed to further investigate this
question. It is hoped that the investigated ncRNA types of the
present study may act as potential minimally-invasive biomarkers to
indicate therapy response in TNBC. This hypothesis is supported as
different ncRNA regulation was identified in the serum of patients
with TNBC who achieved a cCR during NACT compared with those who
did not. Therefore, the involvement of these ncRNAs in TNBC is
likely. However, the small patient number in this pilot study is a
limitation. Only studies using bigger cohorts can clarify if this
ncRNA panel may serve as a liquid biopsy tool in predicting therapy
response.
To date, few studies have been published connecting
serum-based ncRNAs to the response of patients with BC to NACT. Liu
et al (87) reported
reduced miR-125b expression levels in responding vs. non-responding
patients with BC before, during and after NACT. Additionally,
miR-21 expression decreased throughout NACT in responding patients.
Furthermore, Gu et al (88)
reported that lower miR-451 expression in serum was associated with
the resistance of patients with BC to NACT. Supporting this,
Al-Khanbashi et al (89)
reported an association between elevated serum levels of miR-451,
and improved clinical and pathological response to NACT of locally
advanced BC.
In the present study, expression analysis was
conducted using relative quantification. As each experimental setup
requires the evaluation of the most suitable reference genes, an
individual normalisation for each matrix was conducted. As
aforementioned, intracellular ncRNA expression was normalised to
the geometric mean of RNU44 and -48, while microvesicular ncRNA in
conditioned cell culture media was normalised to the global mean.
In serum, miR-26b and -191 served as endogenous controls and in
urine, miR-16 and -26b were used as endogenous controls. The most
suitable endogenous controls were assessed using the BestKeeper
software tool (55). However,
different normalisation strategies can complicate the comparability
of results. Nevertheless, it is essential to choose an appropriate
normalisation approach according to each experiment.
Different sample matrices can represent different
physiological and pathophysiological processes. Precise
TNBC-related ncRNA expression profiles can be obtained from tumour
tissue; however, sample acquisition remains invasive. TNBC cells
can serve as a model for tumour tissue. In the case of liquid
biopsies, specimens derived from the blood (for example, serum), an
altered ncRNA profile could result from tumour-derived and secreted
ncRNAs. However, dysregulated ncRNAs may originate from other
malignant or non-malignant cells, or may originate from an immune
response to the tumour. The latter may indirectly indicate the
existence of a malignancy. Immune cells constitutively release
exosomes, which can carry ncRNAs (90). Urine-based ncRNAs are further away
still from the tumour site. It is assumed that urine-based altered
ncRNA levels originate directly from tumour tissue and undergo
glomerular filtration by the kidneys after secretion into the
circulation. In this manner, ncRNAs may be found in urine.
Similarly to serum specimens, urine-based ncRNAs may also be
secreted by immune cells. Furthermore, ncRNAs can enter urine from
the cells of the urinary tract (91).
The present study detected chemotherapy-driven
expression alterations of a (TN)BC-related panel of ncRNAs in
vitro. Furthermore, the vast majority of this panel was
detectable in serum and urine specimens, and could be used to
discriminate between patients with TNBC and healthy controls.
Results regarding the eligibility of this panel in predicting
therapy response in vivo remain preliminary due to the small
sample size. The liquid biopsy potential of the investigated ncRNAs
in the evaluation of the response of patients with TNBC to NACT
should be further investigated in larger cohorts.
The article processing charge was funded by the
open access publication fund of the Albert Ludwigs University
Freiburg. No funding was received for the study.
The datasets analysed during the current study are
available from the corresponding author on reasonable request.
AR, TE, MH and MJ substantially developed the study
design and experimental setup. AR, MJ, TE, KB, DW, JA, MM and SG
contributed to sample collection and processing. AR, MJ, CN, and SG
performed miRNA quantification analysis and data collection. GR
conducted statistical analyses in extenso. AR, MH, KB, GR,
MJ, DW, JA, MM and TE drafted the manuscript or revised it
critically. All authors have read and approved the final
manuscript.
The present study was approved by the institutional
ethical review board of the University of Freiburg (permit no.
607/16) and written informed consent was obtained from each
participant.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
|
1
|
World Health Organization IAfRoC: Cancer
Today. 2018, https://gco.iarc.fr/today/home.
Accessed April 4, 2019.
|
|
2
|
DeSantis C, Ma J, Bryan L and Jemal A:
Breast cancer statistics, 2013. CA Cancer J Clin. 64:52–62. 2014.
View Article : Google Scholar
|
|
3
|
Early Breast Cancer Trialists'
Collaborative Group (EBCTCG): Effects of chemotherapy and hormonal
therapy for early breast cancer on recurrence and 15-year survival:
An overview of the randomised trials. Lancet. 365:1687–1717. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Prat A, Parker JS, Karginova O, Fan C,
Livasy C, Herschkowitz JI, He X and Perou CM: Phenotypic and
molecular characterization of the claudin-low intrinsic subtype of
breast cancer. Breast Cancer Res. 12:R682010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pérou CM, Sprlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sprlie T, Perou CM, Tibshirani R, Aas T,
Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey
SS, et al: Gene expression patterns of breast carcinomas
distinguish tumor subclasses with clinical implications. Proc Natl
Acad Sci USA. 98:10869–10874. 2001. View Article : Google Scholar
|
|
7
|
Coates AS, Winer EP, Goldhirsch A, Gelber
RD, Gnant M, Piccart-Gebhart M, Thürlimann B, Senn HJ and Panel
Members: Tailoring therapies-improving the management of early
breast cancer: St Gallen International Expert Consensus on the
Primary Therapy of Early Breast Cancer 20l5. Ann Oncol.
26:1533–1546. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
American Cancer Society: Breast
CancerFacts &Figures. 2017-2018, American Cancer Society;
Atlanta, GA: 2017, https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-stati-stics/breast-cancer-facts-and-figures/breast-cancer-facts-and-figures-2017-2018.pdf.
Accessed April 02, 2019.
|
|
9
|
Badve S, Dabbs DJ, Schnitt SJ, Baehner FL,
Decker T, Eusebi V, Fox SB, Ichihara S, Jacquemier J, Lakhani SR,
et al: Basal-like and triple-negative breast cancers: A critical
review with an emphasis on the implications for pathologists and
oncologists. Mod Pathol. 24:157–167. 2011. View Article : Google Scholar
|
|
10
|
Plasilova ML, Hayse B, Killelea BK,
Horowitz NR, Chagpar AB and Lannin DR: Features of triple-negative
breast cancer: Analysis of 38,813 cases from the national cancer
database. Medicine (Baltimore). 95:e46142016. View Article : Google Scholar
|
|
11
|
Yeh J, Chun J, Schwartz S, Wang A, Kern E,
Guth AA, Axelrod D, Shapiro R and Schnabel F: Clinical
characteristics in patients with triple negative breast cancer. Int
J Breast Cancer. 2017:17961452017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Anders CK and Carey LA: Biology,
metastatic patterns, and treatment of patients with triple-negative
breast cancer. Clin Breast Cancer. 9(Suppl 2): S73–S81. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lehmann BD, Bauer JA, Chen X, Sanders ME,
Chakravarthy AB, Shyr Y and Pietenpol JA: Identification of human
triple-negative breast cancer subtypes and preclinical models for
selection of targeted therapies. J Clin Invest. 121:2750–2767.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Toft DJ and Cryns VL: Minireview:
Basal-like breast cancer: From molecular profiles to targeted
therapies. Mol Endocrinol. 25:199–211. 2011. View Article : Google Scholar :
|
|
15
|
Bertucci F, Finetti P, Cervera N, Esterni
B, Hermitte F, Viens P and Birnbaum D: How basal are
triple-negative breast cancers? Int J Cancer. 123:236–240. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Harbeck N and Gnant M: Breast cancer.
Lancet. 389:1134–1150. 2017. View Article : Google Scholar
|
|
17
|
Mieog JS, van der Hage JA and van de Velde
CJ: Neoadjuvant chemotherapy for operable breast cancer. Br J Surg.
94:1189–1200. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mieog JS, van der Hage JA and van de Velde
CJ: Preoperative chemotherapy for women with operable breast
cancer. Cochrane Database Syst Rev. CD0050022007.PubMed/NCBI
|
|
19
|
Fisher B, Bryant J, Wolmark N, Mamounas E,
Brown A, Fisher ER, Wickerham DL, Begovic M, DeCillis A, Robidoux
A, et al: Effect of preoperative chemotherapy on the outcome of
women with operable breast cancer. J Clin Oncol. 16:2672–2685.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rastogi P, Anderson SJ, Bear HD, Geyer CE,
Kahlenberg MS, Robidoux A, Margolese RG, Hoehn JL, Vogel VG, Dakhil
SR, et al: Preoperative chemotherapy: Updates of National surgical
adjuvant breast and bowel project protocols B-18 and B-27. J Clin
Oncol. 26:778–785. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
van der Hage JA, van de Velde CJ, Julien
JP, Tubiana-Hulin M, Vandervelden C and Duchateau L: Preoperative
chemotherapy in primary operable breast cancer: Results from the
European organization for research and treatment of cancer trial
10902. J Clin Oncol. 19:4224–4237. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
von Minckwitz G, Untch M, Blohmer JU,
Costa SD, Eidtmann H, Fasching PA, Gerber B, Eiermann W, Hilfrich
J, Huober J, et al: Definition and impact of pathologic complete
response on prognosis after neoadjuvant chemotherapy in various
intrinsic breast cancer subtypes. J Clin Oncol. 30:1796–1804. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Berruti A, Amoroso V, Gallo F, Bertaglia
V, Simoncini E, Pedersini R, Ferrari L, Bottini A, Bruzzi P and
Sormani MP: Pathologic complete response as a potential surrogate
for the clinical outcome in patients with breast cancer after
neoadjuvant therapy: A meta-regression of 29 randomized prospective
studies. J Clin Oncol. 32:3883–3891. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cortazar P, Zhang L, Untch M, Mehta K,
Costantino JP, Wolmark N, Bonnefoi H, Cameron D, Gianni L,
Valagussa P, et al: Pathological complete response and long-term
clinical benefit in breast cancer: The CTNeoBC pooled analysis.
Lancet. 384:164–172. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Loibl S, Volz C, Mau C, Blohmer JU, Costa
SD, Eidtmann H, Fasching PA, Gerber B, Hanusch C, Jackisch C, et
al: Response and prognosis after neoadjuvant chemotherapy in 1,051
patients with infiltrating lobular breast carcinoma. Breast Cancer
Res Treat. 144:153–162. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
von Minckwitz G, Untch M, Nüesch E, Loibl
S, Kaufmann M, Kümmel S, Fasching PA, Eiermann W, Blohmer JU, Costa
SD, et al: Impact of treatment characteristics on response of
different breast cancer phenotypes: Pooled analysis of the German
neo-adjuvant chemotherapy trials. Breast Cancer Res Treat.
125:145–156. 2011. View Article : Google Scholar
|
|
27
|
von Minckwitz G, Blohmer JU, Costa SD,
Denkert C, Eidtmann H, Eiermann W, Gerber B, Hanusch C, Hilfrich J,
Huober J, et al: Response-guided neoadjuvant chemotherapy for
breast cancer. J Clin Oncol. 31:3623–3630. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Diagnosis and treatment of patients with
primary and metastatic breast cancer. http://www.ago-online.de.
Accessed April 2, 2019.
|
|
29
|
Marinovich ML, Houssami N, Macaskill P,
von Minckwitz G, Blohmer JU and Irwig L: Accuracy of ultrasound for
predicting pathologic response during neoadjuvant therapy for
breast cancer. Int J Cancer. 136:2730–2737. 2015. View Article : Google Scholar
|
|
30
|
Eisenhauer EA, Therasse P, Bogaerts J,
Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S,
Mooney M, et al: New response evaluation criteria in solid tumours:
Revised RECIST guideline (version 1.1). Eur J Cancer. 45:228–247.
2009. View Article : Google Scholar
|
|
31
|
Chagpar AB, Middleton LP, Sahin AA,
Dempsey P, Buzdar AU, Mirza AN, Ames FC, Babiera GV, Feig BW, Hunt
KK, et al: Accuracy of physical examination, ultrasonography, and
mammography in predicting residual pathologic tumor size in
patients treated with neoadjuvant chemotherapy. Ann Surg.
243:257–264. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sadeghi-Naini A, Sannachi L, Tadayyon H,
Tran WT, Slodkowska E, Trudeau M, Gandhi S, Pritchard K, Kolios MC
and Czarnota GJ: Chemotherapy-response monitoring of breast cancer
patients using quantitative ultrasound-based Intra-tumour
heterogeneities. Sci Rep. 7:103522017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Amoroso V, Generali D, Buchholz T,
Cristofanilli M, Pedersini R, Curigliano G, Daidone MG, Di Cosimo
S, Dowsett M, Fox S, et al: International expert consensus on
primary systemic therapy in the management of early breast cancer:
Highlights of the fifth symposium on primary systemic therapy in
the management of operable breast cancer, Cremona, Italy 2013. J
Natl Cancer Inst Monogr. 2015:90–96. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liedtke C, Mazouni C, Hess KR, Andrè F,
Tordai A, Mejia JA, Symmans WF, Gonzalez-Angulo AM, Hennessy B,
Green M, et al: Response to neoadjuvant therapy and long-term
survival in patients with triple-negative breast cancer. J Clin
Oncol. 26:1275–1281. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
von Minckwitz G, Blohmer JU, Raab G, Lohr
A, Gerber B, Heinrich G, Eidtmann H, Kaufmann M, Hilfrich J,
Jackisch C, et al: In vivo chemosensitivity-adapted preoperative
chemotherapy in patients with early-stage breast cancer: The
GEPARTRIO pilot study. Ann Oncol. 16:56–63. 2005. View Article : Google Scholar
|
|
36
|
Hombach S and Kretz M: Non-coding RNAs:
Classification, biology and functioning. Adv Exp Med Biol.
937:3–17. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Brosnan CA and Voinnet O: The long and the
short of noncoding RNAs. Curr Opin Cell Biol. 21:416–425. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Anastasiadou E, Jacob LS and Slack FJ:
Non-coding RNA networks in cancer. Nat Rev Cancer. 18:5–18. 2018.
View Article : Google Scholar
|
|
39
|
Izzotti A, Carozzo S, Pulliero A,
Zhabayeva D, Ravetti JL and Bersimbaev R: Extracellular MicroRNA in
liquid biopsy: Applicability in cancer diagnosis and prevention. Am
J Cancer Res. 6:1461–1493. 2016.PubMed/NCBI
|
|
40
|
Qi P, Zhou XY and Du X: Circulating long
non-coding RNAs in cancer: Current status and future perspectives.
Mol Cancer. 15:392016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yang X, Cheng Y, Lu Q, Wei J, Yang H and
Gu M: Detection of stably expressed piRNAs in human blood. Int J
Clin Exp Med. 8:13353–13358. 2015.PubMed/NCBI
|
|
42
|
Fanale D, Castiglia M, Bazan V and Russo
A: Involvement of Non-coding RNAs in Chemo- and radioresistance of
colorectal cancer. Adv Exp Med Biol. 937:207–228. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Magee P, Shi L and Garofalo M: Role of
microRNAs in chemo- resistance. Ann Transl Med. 3:3322015.
|
|
44
|
Wang YW, Zhang W and Ma R: Bioinformatic
identification of chemoresistance-associated microRNAs in breast
cancer based on microarray data. Oncol Rep. 39:1003–1010.
2018.PubMed/NCBI
|
|
45
|
Malhotra A, Jain M, Prakash H, Vasquez KM
and Jain A: The regulatory roles of long non-coding RNAs in the
development of chemoresistance in breast cancer. Oncotarget.
8:110671–110684. 2017. View Article : Google Scholar
|
|
46
|
Kao J, Salari K, Bocanegra M, Choi YL,
Girard L, Gandhi J, Kwei KA, Hernandez-Boussard T, Wang P, Gazdar
AF, et al: Molecular profiling of breast cancer cell lines defines
relevant tumor models and provides a resource for cancer gene
discovery. PLoS One. 4:e61462009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Goldhirsch A, Winer EP, Coates AS, Gelber
RD, Piccart-Gebhart M, Thürlimann B and Senn HJ: Panel members:
Personalizing the treatment of women with early breast cancer:
Highlights of the St Gallen international expert consensus on the
primary therapy of Early breast cancer 2013. Ann Oncol.
24:2206–2223. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Elston CW and Ellis IO: Pathological
prognostic factors in breast cancer. I. The value of histological
grade in breast cancer: Experience from a large study with
long-term follow-up. Histopathology. 19:403–410. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Busk PK: A tool for design of primers for
microRNA-specific quantitative RT-qPCR. BMC Bioinformatics.
15:292014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
51
|
Torres A, Torres K, Wdowiak P, Paszkowski
T and Maciejewski R: Selection and validation of endogenous
controls for microRNA expression studies in endometrioid
endometrial cancer tissues. Gynecol Oncol. 130:588–594. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bockmeyer CL, Säuberlich K, Wittig J, Eßer
M, Roeder SS, Vester U, Hoyer PF, Agustian PA, Zeuschner P, Amann
K, et al: Comparison of different normalization strategies for the
analysis of glomerular microRNAs in IgA nephropathy. Sci Rep.
6:319922016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chen L, Jin Y, Wang L, Sun F, Yang X, Shi
M, Zhan C, Shi Y and Wang Q: Identification of reference genes and
miRNAs for qRT-PCR in human esophageal squamous cell carcinoma. Med
Oncol. 34:22017. View Article : Google Scholar
|
|
54
|
Masè M, Grasso M, Avogaro L, D'Amato E,
Tessarolo F, Graffigna A, Denti MA and Ravelli F: Selection of
reference genes is critical for miRNA expression analysis in human
cardiac tissue. A focus on atrial fibrillation. Sci Rep.
7:411272017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Pfaffl MW, Tichopad A, Prgomet C and
Neuvians TP: Determination of stable housekeeping genes,
differentially regulated target genes and sample integrity:
BestKeeper-Excel-based tool using pair-wise correlations.
Biotechnol Lett. 26:509–515. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Mestdagh P, Van Vlierberghe P, De Weer A,
Muth D, Westermann F, Speleman F and Vandesompele J: A novel and
universal method for microRNA RT-qPCR data normalization. Genome
Biol. 10:R642009. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
D'Haene B, Mestdagh P, Hellemans J and
Vandesompele J: miRNA expression profiling: From reference genes to
global mean normalization. Methods Mol Biol. 822:261–272. 2012.
View Article : Google Scholar
|
|
58
|
Schwarzenbach H, da Silva AM, Calin G and
Pantel K: Data Normalization strategies for MicroRNA
quantification. Clin Chem. 61:1333–1342. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Erbes T, Hirschfeld M, Rücker G, Jaeger M,
Boas J, Iborra S, Mayer S, Gitsch G and Stickeler E: Feasibility of
urinary microRNA detection in breast cancer patients and its
potential as an innovative non-invasive biomarker. BMC Cancer.
15:1932015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Marabita F, de Candia P, Torri A, Tegnér
J, Abrignani S and Rossi RL: Normalization of circulating microRNA
expression data obtained by quantitative real-time RT-PCR. Brief
Bioinform. 17:204–212. 2016. View Article : Google Scholar :
|
|
61
|
Song W, Zhang WH, Zhang H, Li Y, Zhang Y,
Yin W and Yang Q: Validation of housekeeping genes for the
normalization of RT-qPCR expression studies in oral squamous cell
carcinoma cell line treated by 5 kinds of chemotherapy drugs. Cell
Mol Biol (Noisy-le-Grand). 62:29–34. 2016. View Article : Google Scholar
|
|
62
|
Smith PK, Krohn RI, Hermanson GT, Mallia
AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ and
Klenk DC: Measurement of protein using bicinchoninic acid. Anal
Biochem. 150:76–85. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Schägger H and von Jagow G: Tricine-sodium
dodecyl sulfate-polyacrylamide gel electrophoresis for the
separation of proteins in the range from 1 to 100 kDa. Anal
Biochem. 166:368–379. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
R Core Team: R: A language and environment
for statistical computing. R Foundation for Statistical Computing;
Vienna: 2018
|
|
65
|
Gao D, Qi X, Zhang X, Fang K, Guo Z and Li
L: hsa_ circRNA_0006528 as a competing endogenous RNA promotes
human breast cancer progression by sponging miR-75p and activating
the MAPK/ERK signalling pathway. Mol Carcinog. 58:554–564. 2019.
View Article : Google Scholar
|
|
66
|
Gao D, Zhang X, Liu B, Meng D, Fang K, Guo
Z and Li L: Screening circular RNA related to chemotherapeutic
resistance in breast cancer. Epigenomics. 9:1175–1188. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Uhr K, Sieuwerts AM, de Weerd V, Smid M,
Hammerl D, Foekens JA and Martens JWM: Association of microRNA-7
and its binding partner CDR1-AS with the prognosis and prediction
of 1st-line tamoxifen therapy in breast cancer. Sci Rep.
8:96572018. View Article : Google Scholar :
|
|
68
|
Garcia-Vazquez R, Ruiz-García E, Meneses
García A, Astudillo-de la Vega H, Lara-Medina F, Alvarado-Miranda
A, Mal donado-Martínez H, Gonzalez-Barrios JA, Campos-Parra AD,
Rodríguez Cuevas S, et al: A microRNA signature associated with
pathological complete response to novel neoadjuvant therapy regimen
in triple-negative breast cancer. Tumour Biol.
39:10104283177028992017. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Unger FT, Klasen HA, Tchartchian G, de
Wilde RL and Witte I: DNA damage induced by cis- and carboplatin as
indicator for in vitro sensitivity of ovarian carcinoma cells. BMC
Cancer. 9:3592009. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Su WC, Chang SL, Chen TY, Chen JS and Tsao
CJ: Comparison of in vitro growth-inhibitory activity of
carboplatin and cisplatin on leukemic cells and hematopoietic
progenitors: The myelosuppressive activity of carboplatin may be
greater than its antileukemic effect. Jpn. J Clin Oncol.
30:562–567. 2000.
|
|
71
|
Kuittinen T, Rovio P, Staff S, Luukkaala
T, Kallioniemi A, Grénman S, Laurila M and Maenpaa J: Paclitaxel,
carboplatin and 1,25D3 inhibit proliferation of endometrial cancer
cells in vitro. Anticancer Res. 37:6575–6581. 2017.PubMed/NCBI
|
|
72
|
He Q, Liang CH and Lippard SJ: Steroid
hormones induce HMG1 overexpression and sensitize breast cancer
cells to cisplatin and carboplatin. Proc Natl Acad Sci USA.
97:5768–5772. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Diehl JA, Zindy F and Sherr CJ: Inhibition
of cyclin D1 phosphorylation on threonine-286 prevents its rapid
degradation via the ubiquitin-proteasome pathway. Genes Dev.
11:957–972. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Luo H, Zhang J, Dastvan F, Yanagawa B,
Reidy MA, Zhang HM, Yang D, Wilson JE and McManus BM:
Ubiquitin-dependent proteolysis of cyclin D1 is associated with
coxsackievirus-induced cell growth arrest. J Virol. 77:1–9. 2003.
View Article : Google Scholar :
|
|
75
|
Fuller-Pace FV: DEAD box RNA helicase
functions in cancer. RNA Biol. 10:121–132. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Mazurek A, Luo W, Krasnitz A, Hicks J,
Powers RS and Stillman B: DDX5 regulates DNA replication and is
required for cell proliferation in a subset of breast cancer cells.
Cancer Discov. 2:812–825. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Wang D, Huang J and Hu Z: RNA helicase
DDX5 regulates microRNA expression and contributes to cytoskeletal
reorganization in basal breast cancer cells. Mol Cell Proteomics.
11:M111.0119322012. View Article : Google Scholar :
|
|
78
|
Bates GJ, Nicol SM, Wilson BJ, Jacobs AM,
Bourdon JC, Wardrop J, Gregory DJ, Lane DP, Perkins ND and
Fuller-Pace FV: The DEAD box protein p68: A novel transcriptional
coactivator of the p53 tumour suppressor. EMBO J. 24:543–553. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Wortham NC, Ahamed E, Nicol SM, Thomas RS,
Periyasamy M, Jiang J, Ochocka AM, Shousha S, Huson L, Bray SE, et
al: The DEAD-box protein p72 regulates ERalpha-/oestrogen-dependent
transcription and cell growth, and is associated with improved
survival in ERalpha-positive breast cancer. Oncogene. 28:4053–4064.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Shin S, Rossow KL, Grande JP and Janknecht
R: Involvement of RNA helicases p68 and p72 in colon cancer. Cancer
Res. 67:7572–7578. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Jalal C, Uhlmann-Schiffler H and Stahl H:
Redundant role of DEAD box proteins p68 (Ddx5) and p72/p82 (Ddx17)
in ribosome biogenesis and cell proliferation. Nucleic Acids Res.
35:3590–3601. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Concepcion CP, Bonetti C and Ventura A:
The microRNA-17-92family of microRNA clusters in development and
disease. Cancer J. 18:262–267. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Mogilyansky E and Rigoutsos I: The
miR-17/92 cluster: A comprehensive update on its genomics,
genetics, functions and increasingly important and numerous roles
in health and disease. Cell Death Differ. 20:1603–1614. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Farazi TA, Horlings HM, Ten Hoeve JJ,
Mihailovic A, Halfwerk H, Morozov P, Brown M, Hafner M, Reyal F,
van Kouwenhove M, et al: MicroRNA sequence and expression analysis
in breast tumors by deep sequencing. Cancer Res. 71:4443–4453.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Calvano Filho CM, Calvano-Mendes DC,
Carvalho KC, Maciel GA, Ricci MD, Torres AP, Filassi JR and Baracat
EC: Triple-negative and luminal A breast tumors: Differential
expression of miR-18a-5p, and miR-17-5p, and miR-20a-5p. Tumour
Biol. 35:7733–7741. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Hashim A, Rizzo F, Marchese G, Ravo M,
Tarallo R, Nassa G, Giurato G, Santamaria G, Cordella A, Cantarella
C and Weisz A: RNA sequencing identifies specific PIWI-interacting
small non-coding RNA expression patterns in breast cancer.
Oncotarget. 5:9901–9910. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Liu B, Su F, Chen M, Li Y, Qi X, Xiao J,
Li X, Liu X, Liang W, Zhang Y and Zhang J: Serum miR-21 and
miR-125b as markers predicting neoadjuvant chemotherapy response
and prognosis in stage II/III breast cancer. Hum Pathol. 64:44–52.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Gu X, Xue JQ, Han SJ, Qian SY and Zhang
WH: Circulating microRNA-451 as a predictor of resistance to
neoadjuvant chemotherapy in breast cancer. Cancer Biomark.
16:395–403. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Al-Khanbashi M, Caramuta S, Alajmi AM,
Al-Haddabi I, Al-Riyami M, Lui WO and Al-Moundhri MS: Tissue and
Serum miRNA profile in locally advanced breast cancer (LABC) in
response to Neo-adjuvant chemotherapy (NAC) treatment. PLoS One.
11:e01520322016. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Edgar JR: Q&A: What are exosomes,
exactly? BMC Biol. 14:462016. View Article : Google Scholar :
|
|
91
|
Ben-Dov IZ, Whalen VM, Goilav B, Max KE
and Tuschl T: Cell and microvesicle urine microRNA deep sequencing
profiles from healthy individuals: Observations with potential
impact on biomarker studies. PLoS One. 11:e01472492016. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Mansoori B, Mohammadi A, Shirjang S,
Baghbani E and Baradaran B: Micro RNA 34a and Let-7a expression in
human breast cancers is associated with apoptotic expression genes.
Asian Pac J Cancer Prev. 17:1887–1890. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Liu K, Zhang C, Li T, Ding Y, Tu T, Zhou
F, Qi W, Chen H and Sun X: Let-7a inhibits growth and migration of
breast cancer cells by targeting HMGA1. Int J Oncol. 46:2526–2534.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Marques MM, Evangelista AF, Macedo T,
Vieira RADC, Scapulatempo-Neto C, Reis RM, Carvalho AL and Silva
IDCGD: Expression of tumor suppressors miR-195 and let-7a as
potential biomarkers of invasive breast cancer. Clinics (Sao
Paulo). 73:e1842018. View Article : Google Scholar :
|
|
95
|
Huang SK, Luo Q, Peng H, Li J, Zhao M,
Wang J, Gu YY, Li Y, Yuan P, Zhao GH and Huang CZ: A Panel of serum
noncoding RNAs for the diagnosis and monitoring of response to
therapy in patients with breast cancer. Med Sci Monit.
24:2476–2488. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Khalighfard S, Alizadeh AM, Irani S and
Omranipour R: Plasma miR-21, miR-155, miR-10b, and Let-7a as the
potential biomarkers for the monitoring of breast cancer patients.
Sci Rep. 8:179812018. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Mi Y, Liu F, Liang X, Liu S, Huang X, Sang
M and Geng C: Tumor suppressor let-7a inhibits breast cancer cell
proliferation, migration and invasion by targeting MAGE-A1.
Neoplasma. 66:54–62. 2019. View Article : Google Scholar
|
|
98
|
Guo Q, Wen R, Shao B, Li Y, Jin X, Deng H,
Wu J, Su F and Yu F: Combined Let-7a and H19 signature: A
prognostic index of progression-free survival in primary breast
cancer patients. J Breast Cancer. 21:142–149. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Lehmann TP, Korski K, Gryczka R, Ibbs M,
Thieleman A, Grodecka-Gazdecka S and Jagodzinski PP: Relative
levels of let-7a, miR-17, miR-27b, miR-125a, miR-125b and miR-206
as potential molecular markers to evaluate grade, receptor status
and molecular type in breast cancer. Mol Med Rep. 12:4692–4702.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Thakur S, Grover RK, Gupta S, Yadav AK and
Das BC: Identification of Specific miRNA signature in paired sera
and tissue samples of Indian Women with Triple Negative breast
cancer. PLoS One. 11:e01589462016. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Ahram M, Mustafa E, Zaza R, Abu Hammad S,
Alhudhud M, Bawadi R and Zihlif M: Differential expression and
androgen regulation of microRNAs and metalloprotease 13 in breast
cancer cells. Cell Biol Int. 41:1345–1355. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Yu CC, Chen YW, Chiou GY, Tsai LL, Huang
PI, Chang CY, Tseng LM, Chiou SH, Yen SH, Chou MY, et al: MicroRNA
let-7a represses chemoresistance and tumourigenicity in head and
neck cancer via stem-like properties ablation. Oral Oncol.
47:202–210. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Qiu L, Zhang GF, Yu L, Wang HY, Jia XJ and
Wang TJ: Novel oncogenic and chemoresistance-inducing functions of
resistin in ovarian cancer cells require miRNAs-mediated induction
of epithelial-to-mesenchymal transition. Sci Rep. 8:125222018.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Xiao G, Wang X and Yu Y: CXCR4/Let-7a Axis
regulates metastasis and chemoresistance of pancreatic cancer cells
through targeting HMGA2. Cell Physiol Biochem. 43:840–851. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Serguienko A, Grad I, Wennerstrpm AB,
Meza-Zepeda LA, Thiede B, Stratford EW, Myklebost O and Munthe E:
Metabolic reprogramming of metastatic breast cancer and melanoma by
let-7a microRNA. Oncotarget. 6:2451–2465. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Tsang WP and Kwok TT: Let-7a microRNA
suppresses therapeutics-induced cancer cell death by targeting
caspase-3. Apoptosis. 13:1215–1222. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Wu J, Li S, Jia W, Deng H, Chen K, Zhu L,
Yu F and Su F: Reduced let-7a is associated with chemoresistance in
primary breast cancer. PLoS One. 10:e01336432015. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Bhutia YD, Hung SW, Krentz M, Patel D,
Lovin D, Manoharan R, Thomson JM and Govindarajan R: Differential
processing of let-7a precursors influences RRM2 expression and
chemosensitivity in pancreatic cancer: Role of LIN-28 and SET
oncoprotein. PLoS One. 8:e534362013. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Lu L, Schwartz P, Scarampi L, Rutherford
T, Canuto EM, Yu H and Katsaros D: MicroRNA let-7a: A potential
marker for selection of paclitaxel in ovarian cancer management.
Gynecol Oncol. 122:366–371. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Lv K, Liu L, Wang L, Yu J, Liu X, Cheng Y,
Dong M, Teng R, Wu L, Fu P, et al: Lin28 mediates paclitaxel
resistance by modulating p21, Rb and Let-7a miRNA in breast cancer
cells. PLoS One. 7:e400082012. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Yin PT, Pongkulapa T, Cho HY, Han J,
Pasquale NJ, Rabie H, Kim JH, Choi JW and Lee KB: Overcoming
chemoresistance in cancer via combined MicroRNA therapeutics with
anticancer drugs using multifunctional magnetic Core-shell
nanoparticles. ACS Appl Mater Interfaces. 10:26954–26963. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Farré PL, Scalise GD, Duca RB, Dalton GN,
Massillo C, Porretti J, Grana K, Gardner K, De Luca P and De Siervi
A: CTBP1 and metabolic syndrome induce an mRNA and miRNA expression
profile critical for breast cancer progression and metastasis.
Oncotarget. 9:13848–13858. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Aure MR, Leivonen SK, Fleischer T, Zhu Q,
Overgaard J, Alsner J, Tramm T, Louhimo R, Alnaes GI, Perala M, et
al: Individual and combined effects of DNA methylation and copy
number alterations on miRNA expression in breast tumors. Genome
Biol. 14:R1262013. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Lv J, Xia K, Xu P, Sun E, Ma J, Gao S,
Zhou Q, Zhang M, Wang F, Chen F, et al: miRNA expression patterns
in chemoresistant breast cancer tissues. Biomed Pharmacother.
68:935–942. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Oztemur Islakoglu Y, Noyan S, Aydos A and
Gur Dedeoglu B: Meta-microRNA biomarker signatures to classify
breast cancer subtypes. OMICS. 22:709–716. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Shan Y, Liu Y, Zhao L, Liu B, Li Y and Jia
L: MicroRNA-33a and let-7e inhibit human colorectal cancer
progression by targeting ST8SIA1. Int J Biochem Cell Biol.
90:48–58. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Svoboda M, Sana J, Fabian P, Kocakova I,
Gombosova J, Nekvindova J, Radova L, Vyzula R and Slaby O: MicroRNA
expression profile associated with response to neoadjuvant
chemoradiotherapy in locally advanced rectal cancer patients.
Radiat Oncol. 7:1952012. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Cai J, Yang C, Yang Q, Ding H, Jia J, Guo
J, Wang J and Wang Z: Deregulation of let-7e in epithelial ovarian
cancer promotes the development of resistance to cisplatin.
Oncogenesis. 2:e752013. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Sorrentino A, Liu CG, Addario A, Peschle
C, Scambia G and Ferlini C: Role of microRNAs in drug-resistant
ovarian cancer cells. Gynecol Oncol. 111:478–486. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Xiao M, Cai J, Cai L, Jia J, Xie L, Zhu Y,
Huang B, Jin D and Wang Z: Let-7e sensitizes epithelial ovarian
cancer to cisplatin through repressing DNA double strand break
repair. J Ovarian Res. 10:242017. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Block I, Burton M, Sprensen KP, Andersen
L, Larsen MJ, Bak M, Cold S, Thomassen M, Tan Q and Kruse TA:
Association of miR-548c-5p, miR-7-5p, miR-210-3p, miR-128-3p with
recurrence in systemically untreated breast cancer. Oncotarget.
9:9030–9042. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Foekens JA, Sieuwerts AM, Smid M, Look MP,
de Weerd V, Boersma AW, Klijn JG, Wiemer EA and Martens JW: Four
miRNAs associated with aggressiveness of lymph node-negative,
estrogen receptor-positive human breast cancer. Proc Natl Acad Sci
USA. 105:13021–13026. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Gong C, Tan W, Chen K, You N, Zhu S, Liang
G, Xie X, Li Q, Zeng Y, Ouyang N, et al: Prognostic value of a
BCSC-associated MicroRNA signature in hormone receptor-positive
HER2-negative breast cancer. EBioMedicine. 11:199–209. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Raychaudhuri M, Bronger H, Buchner T,
Kiechle M, Weichert W and Avril S: MicroRNAs miR-7 and miR-340
predict response to neoadjuvant chemotherapy in breast cancer.
Breast Cancer Res Treat. 162:511–521. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Shi Y, Ye P and Long X: Differential
expression profiles of the transcriptome in breast cancer cell
lines revealed by next generation sequencing. Cell Physiol Biochem.
44:804–816. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Cui YX, Bradbury R, Flamini V, Wu B,
Jordan N and Jiang WG: MicroRNA-7 suppresses the homing and
migration potential of human endothelial cells to highly metastatic
human breast cancer cells. Br J Cancer. 117:89–101. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Vera-Puente O, Rodriguez-Antolin C,
Salgado-Figueroa A, Michalska P, Pernia O, Reid BM, Rosas R,
Garcia-Guede A, SacristAn S, Jimenez J, et al: MAFG is a potential
therapeutic target to restore chemosensitivity in
cisplatin-resistant cancer cells by increasing reactive oxygen
species. Transl Res. 200:1–17. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Filipits M, Nielsen TO, Rudas M, Greil R,
Stoger H, Jakesz R, Bago-Horvath Z, Dietze O, Regitnig P and
Gruber-Rossipal C: et al The PAM50 risk-of-recurrence score
predicts risk for late distant recurrence after endocrine therapy
in postmenopausal women with endocrine-responsive early breast
cancer. Clin. Cancer Res. 20:1298–1305. 2014.
|
|
129
|
Blume CJ, Hotz-Wagenblatt A, Hüllein J,
Sellner L, Jethwa A, Stolz T, Slabicki M, Lee K, Sharathchandra A,
Benner A, et al: p53-dependent non-coding RNA networks in chronic
lymphocytic leukemia. Leukemia. 29:2015–2023. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Liu DZ, Chang B, Li XD, Zhang QH and Zou
YH: MicroRNA-9 promotes the proliferation, migration, and invasion
of breast cancer cells via down-regulating FOXO1. Clin Transl
Oncol. 19:1133–1140. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Ma L, Young J, Prabhala H, Pan E, Mestdagh
P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S,
et al: miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin
and cancer metastasis. Nat Cell Biol. 12:247–256. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Sporn JC, Katsuta E, Yan L and Takabe K:
Expression of MicroRNA-9 is associated with overall survival in
breast cancer patients. J Surg Res. 233:426–435. 2019. View Article : Google Scholar
|
|
133
|
Orangi E and Motovali-Bashi M: Evaluation
of miRNA-9 and miRNA-34a as potential biomarkers for diagnosis of
breast cancer in Iranian women. Gene. 687:272–279. 2019. View Article : Google Scholar
|
|
134
|
Naorem LD, Muthaiyan M and Venkatesan A:
Identification of dysregulated miRNAs in triple negative breast
cancer: A meta-analysis approach. J Cell Physiol. 234:11768–11779.
2019. View Article : Google Scholar
|
|
135
|
Kia V, Paryan M, Mortazavi Y, Biglari A
and Mohammadi-Yeganeh S: Evaluation of exosomal miR-9 and miR-155
targeting PTEN and DUSP14 in highly metastatic breast cancer and
their effect on low metastatic cells. J Cell Biochem.
120:5666–5676. 2019. View Article : Google Scholar
|
|
136
|
Jang MH, Kim HJ, Gwak JM, Chung YR and
Park SY: Prognostic value of microRNA-9 and microRNA-155 expression
in triple-negative breast cancer. Hum Pathol. 68:69–78. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Cheng CW, Yu JC, Hsieh YH, Liao WL, Shieh
JC, Yao CC, Lee HJ, Chen PM, Wu PE and Shen CY: Increased cellular
levels of MicroRNA-9 and MicroRNA-221 correlate with cancer
stemness and predict poor outcome in human breast cancer. Cell
Physiol Biochem. 48:2205–2218. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
D'Ippolito E, Plantamura I, Bongiovanni L,
Casalini P, Baroni S, Piovan C, Orlandi R, Gualeni AV, Gloghini A,
Rossini A, et al: miR-9 and miR-200 Regulate PDGFRp-mediated
endothelial differentiation of tumor cells in Triple-negative
breast cancer. Cancer Res. 76:5562–5572. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Li X, Pan Q, Wan X, Mao Y, Lu W, Xie X and
Cheng X: Methylation-associated Has-miR-9 deregulation in pacli-
taxel-resistant epithelial ovarian carcinoma. BMC Cancer.
15:5092015. View Article : Google Scholar
|
|
140
|
Pezuk JA, Miller TLA, Bevilacqua JLB, de
Barros ACSD, de Andrade FEM, E Macedo LFA, Aguilar V, Claro ANM,
Camargo AA, Galante PAF and Reis LFL: Measuring plasma levels of
three microRNAs can improve the accuracy for identification of
malignant breast lesions in women with BI-RADS 4 mammography.
Oncotarget. 8:83940–83948. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Kodahl AR, Lyng MB, Binder H, Cold S,
Gravgaard K, Knoop AS and Ditzel HJ: Novel circulating microRNA
signature as a potential non-invasive multi-marker test in
ER-positive early-stage breast cancer: A case control study. Mol
Oncol. 8:874–883. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Yang L, Zhao W, Wei P, Zuo W and Zhu S:
Tumor suppressor p53 induces miR-15a processing to inhibit neuronal
apoptosis inhibitory protein (NAIP) in the apoptotic response DNA
damage in breast cancer cell. Am J Transl Res. 9:683–691.
2017.PubMed/NCBI
|
|
143
|
Patel N, Garikapati KR, Ramaiah MJ,
Polavarapu KK, Bhadra U and Bhadra MP: miR-15a/miR-16 induces
mitochondrial dependent apoptosis in breast cancer cells by
suppressing oncogene BMI1. Life Sci. 164:60–70. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Hamdi K, Blancato J, Goerlitz D, Islam M,
Neili B, Abidi A, Gat A, Ayed FB, Chivi S, Loffredo C, et al:
Circulating Cell-free miRNA expression and its association with
clinicopathologic features in inflammatory and non-inflammatory
breast cancer. Asian Pac J Cancer Prev. 17:1801–1810. 2016.
View Article : Google Scholar
|
|
145
|
Shinden Y, Akiyoshi S, Ueo H, Nambara S,
Saito T, Komatsu H, Ueda M, Hirata H, Sakimura S, Uchi R, et al:
Diminished expression of MiR-15a is an independent prognostic
marker for breast cancer cases. Anticancer Res. 35:123–127.
2015.PubMed/NCBI
|
|
146
|
Li F, Xu Y, Deng S, Li Z, Zou D, Yi S, Sui
W, Hao M and Qiu L: MicroRNA-15a/161 cluster located at chromosome
13q14 is down-regulated but displays different expression pattern
and prognostic significance in multiple myeloma. Oncotarget.
6:38270–38282. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Patel N, Garikapati KR, Makani VKK, Nair
AD, Vangara N, Bhadra U and Pal Bhadra M: Regulating BMI1
expression via miRNAs promote Mesenchymal to Epithelial Transition
(MET) and sensitizes breast cancer cell to chemotherapeutic drug.
PLoS One. 13:e01902452018. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Rodriguez-Aguayo C, Monroig PDC, Redis RS,
Bayraktar E, Almeida MI, Ivan C, Fuentes-Mattei E, Rashed MH,
Chavez-Reyes A, Ozpolat B, et al: Regulation of hnRNPA1 by
microRNAs controls the miR-18a-K-RAS axis in chemotherapy-resistant
ovarian cancer. Cell Discov. 3:170292017. View Article : Google Scholar
|
|
149
|
Wang L, Zhang X, Sheng L, Qiu C and Luo R:
LINC00473 promotes the Taxol resistance via miR-15a in colorectal
cancer. Biosci Rep. 38:pii: BSR20180790. 2018.
|
|
150
|
Chu J, Zhu Y, Liu Y, Sun L, Lv X, Wu Y, Hu
P, Su F, Gong C, Song E, et al: E2F7 overexpression leads to
tamoxifen resistance in breast cancer cells by competing with E2F1
at miR-15a/16 promoter. Oncotarget. 6:31944–31957. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Jurkovicova D, Smolkova B, Magyerkova M,
Sestakova Z, Kajabova VH, Kulcsar L, Zmetakova I, Kalinkova L,
Krivulcik T, Karaba M, et al: Down-regulation of traditional
oncomiRs in plasma of breast cancer patients. Oncotarget.
8:77369–77384. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Zhang N, Zhang H, Liu Y, Su P, Zhang J,
Wang X, Sun M, Chen B, Zhao W, Wang L, et al: SREBP1, targeted by
miR-18a-5p, modulates epithelial-mesenchymal transition in breast
cancer via forming a co-repressor complex with Snail and HDAC1/2.
Cell Death Differ. 26:843–859. 2019. View Article : Google Scholar
|
|
153
|
Yang F, Li Y, Xu L, Zhu Y, Gao H, Zhen L
and Fang L: miR-17 as a diagnostic biomarker regulates cell
proliferation in breast cancer. Onco Targets Ther. 10:543–550.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Wang Y, Li J, Dai L, Zheng J, Yi Z and
Chen L: MiR-17-5p may serve as a novel predictor for breast cancer
recurrence. Cancer Biomark. 22:721–726. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Sueta A, Yamamoto Y, Tomiguchi M,
Takeshita T, Yamamoto-Ibusuki M and Iwase H: Differential
expression of exosomal miRNAs between breast cancer patients with
and without recurrence. Oncotarget. 8:69934–69944. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Hesari A, Azizian M, Darabi H, Nesaei A,
Hosseini SA, Salarinia R, Motaghi AA and Ghasemi F: Expression of
circulating miR-17, miR-25, and miR-133 in breast cancer patients.
J Cell Biochem. Nov 28–2018. View Article : Google Scholar : Epub ahead of
print.
|
|
157
|
Li H, Liu J, Chen J, Wang H, Yang L, Chen
F, Fan S, Wang J, Shao B, Yin D, et al: A serum microRNA signature
predicts trastuzumab benefit in HER2-positive metastatic breast
cancer patients. Nat Commun. 9:16142018. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Braicu C, Raduly L, Morar-Bolba G,
Cojocneanu R, Jurj A, Pop LA, Pileczki V, Ciocan C, Moldovan A,
Irimie A, et al: Aberrant miRNAs expressed in HER-2 negative breast
cancers patient. J Exp Cli. Cancer Res. 37:2572018.
|
|
159
|
Li J, Lai Y, Ma J, Liu Y, Bi J, Zhang L,
Chen L, Yao C, Lv W, Chang G, et al: miR-17-5p suppresses cell
proliferation and invasion by targeting ETV1 in triple-negative
breast cancer. BMC Cancer. 17:7452017. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Li X, Wu B, Chen L, Ju Y, Li C and Meng S:
Urokinase-type plasminogen activator receptor inhibits apoptosis in
triple-negative breast cancer through miR-17/20a suppression of
death receptors 4 and 5. Oncotarget. 8:88645–88657. 2017.PubMed/NCBI
|
|
161
|
Cui YH, Kim H, Lee M, Yi JM, Kim RK, Uddin
N, Yoo KC, Kang JH, Choi MY, Cha HJ, et al: FBXL14 abolishes breast
cancer progression by targeting CDCP1 for proteasomal degradation.
Oncogene. 37:5794–5809. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Wang ST, Liu LB, Li XM, Wang YF, Xie PJ,
Li Q, Wang R, Wei Q, Kang YH, Meng R, et al: Circ-ITCH regulates
triple-negative breast cancer progression through the Wnt/p-catenin
pathway. Neoplasma. 66:232–239. 2019. View Article : Google Scholar
|
|
163
|
Jia J, Zhan D, Li J, Li Z, Li H and Qian
J: The contrary functions of lncRNA HOTAIR/miR-17-5p/PTEN axis and
Shenqifuzheng injection on chemosensitivity of gastric cancer
cells. J Cell Mol Med. 23:656–669. 2019. View Article : Google Scholar
|
|
164
|
Wu DM, Hong XW, Wang LL, Cui XF, Lu J,
Chen GQ and Zheng YL: MicroRNA-17 inhibition overcomes
chemoresistance and suppresses epithelial-mesenchymal transition
through a DEDD-dependent mechanism in gastric cancer. Int J Biochem
Cell Biol. 102:59–70. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Fan B, Shen C, Wu M, Zhao J, Guo Q and Luo
Y: miR-17-92 cluster is connected with disease progression and
oxaliplatin/capecitabine chemotherapy efficacy in advanced gastric
cancer patients: A preliminary study. Medicine (Baltimore).
97:e120072018. View Article : Google Scholar
|
|
166
|
Huang FX, Chen HJ, Zheng FX, Gao ZY, Sun
PF, Peng Q, Liu Y, Deng X, Huang YH, Zhao C and Miao LJ: LncRNA
BLACAT1 is involved in chemoresistance of nonsmall cell lung cancer
cells by regulating autophagy. Int J Oncol. 54:339–347. 2019.
|
|
167
|
Gu J, Wang D, Zhang J, Zhu Y, Li Y, Chen
H, Shi M, Wang X, Shen B, Deng X, et al: GFRa2 prompts cell growth
and chemo- resistance through down-regulating tumor suppressor gene
PTEN via Mir-17-5p in pancreatic cancer. Cancer Lett. 380:434–441.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Cioffi M, Trabulo SM, Sanchez-Ripoll Y,
Miranda-Lorenzo I, Lonardo E, Dorado J, Reis Vieira C, Ramirez JC,
Hidalgo M, Aicher A, et al: The miR-17-92 cluster counteracts
quiescence and chemoresistance in a distinct subpopulation of
pancreatic cancer stem cells. Gut. 64:1936–1948. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Yan HJ, Liu WS, Sun WH, Wu J, Ji M, Wang
Q, Zheng X, Jiang JT and Wu CP: miR-17-5p inhibitor enhances chemo-
sensitivity to gemcitabine via upregulating Bim expression in
pancreatic cancer cells. Dig Dis Sci. 57:3160–3167. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Ao X: Decreased expression of microRNA-17
and microRNA-20b promotes breast cancer resistance to taxol therapy
by upregula- tion of NCOA3. Cell Death Dis. 7:e24632016. View Article : Google Scholar
|
|
171
|
Liao XH, Xiang Y, Yu CX, Li JP, Li H and
Nie Q: STAT3 is required for MiR-17-5p-mediated sensitization to
chemotherapy-induced apoptosis in breast cancer cells. Oncotarget.
8:15763–15774. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Zhu H, Yang SY, Wang J, Wang L and Han SY:
Evidence for miR-s17-92 and miR-134 gene cluster regulation of
ovarian cancer drug resistance. Eur Rev Med Pharmacol Sci.
20:2526–2531. 2016.PubMed/NCBI
|
|
173
|
Fang Y, Xu C and Fu Y: MicroRNA-17-5p
induces drug resistance and invasion of ovarian carcinoma cells by
targeting PTEN signaling. J Biol Res (Thessalon). 22:122015.
View Article : Google Scholar
|
|
174
|
Chatterjee A, Chattopadhyay D and
Chakrabarti G: miR-17-5p downregulation contributes to paclitaxel
resistance of lung cancer cells through altering beclin1
expression. PLoS One. 9:e957162014. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Krutilina R, Sun W, Sethuraman A, Brown M,
Seagroves TN, Pfeffer LM, Ignatova T and Fan M: MicroRNA-18a
inhibits hypoxia-inducible factor 1a activity and lung metastasis
in basal breast cancers. Breast Cancer Res. 16:R782014. View Article : Google Scholar
|
|
176
|
Godfrey AC, Xu Z, Weinberg CR, Getts RC,
Wade PA, DeRoo LA, Sandler DP and Taylor JA: Serum microRNA
expression as an early marker for breast cancer risk in
prospectively collected samples from the Sister Study cohort.
Breast Cancer Res. 15:R422013. View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Shidfar A, Costa FF, Scholtens D, Bischof
JM, Sullivan ME, Ivancic DZ, Vanin EF, Soares MB, Wang J and Khan
SA: Expression of miR-18a and miR-210 in normal breast tissue as
candidate biomarkers of breast cancer risk. Cancer Prev Res
(Phila). 10:89–97. 2017. View Article : Google Scholar
|
|
178
|
Sha LY, Zhang Y, Wang W, Sui X, Liu SK,
Wang T and Zhang H: MiR-18a upregulation decreases Dicer expression
and confers paclitaxel resistance in triple negative breast cancer.
Eur Rev Med Pharmacol Sci. 20:2201–2208. 2016.PubMed/NCBI
|
|
179
|
Xiao H, Liu Y, Liang P, Wang B, Tan H,
Zhang Y, Gao X and Gao J: TP53TG1 enhances cisplatin sensitivity of
non-small cell lung cancer cells through regulating miR-18a/PTEN
axis. Cell Biosci. 8:232018. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Hummel R, Sie C, Watson DI, Wang T, Ansar
A, Michael MZ, Van der Hoek M, Haier J and Hussey DJ: MicroRNA
signatures in chemotherapy resistant esophageal cancer cell lines.
World J Gastroenterol. 20:14904–14912. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Fan YX, Dai YZ, Wang XL, Ren YQ, Han JJ
and Zhang H: MiR-18a upregulation enhances autophagy in triple
negative cancer cells via inhibiting mTOR signaling pathway. Eur
Rev Med Pharmacol Sci. 20:2194–2200. 2016.PubMed/NCBI
|
|
182
|
Zhu HY, Bai WD, Ye XM, Yang AG and Jia LT:
Long non-coding RNA UCA1 desensitizes breast cancer cells to
trastuzumab by impeding miR-18a repression of Yes-associated
protein 1. Biochem Biophys Res Commun. 496:1308–1313. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Li M, Zhou Y, Xia T, Zhou X, Huang Z,
Zhang H, Zhu W, Ding Q and Wang S: Circulating microRNAs from the
miR-106a-363 cluster on chromosome X as novel diagnostic biomarkers
for breast cancer. Breast Cancer Res Treat. 170:257–270. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Li C, Zhang J, Ma Z, Zhang F and Yu W:
miR-19b serves as a prognostic biomarker of breast cancer and
promotes tumor progression through PI3K/AKT signaling pathway. Onco
Targets Ther. 11:4087–4095. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Zhao L, Zhao Y, He Y and Mao Y: miR-19b
promotes breast cancer metastasis through targeting MYLIP and its
related cell adhesion molecules. Oncotarget. 8:64330–64343.
2017.PubMed/NCBI
|
|
186
|
Liu M, Yang R, Urrehman U, Ye C, Yan X,
Cui S, Hong Y, Gu Y, Liu Y, Zhao C, et al: MiR-19b suppresses PTPRG
to promote breast tumorigenesis. Oncotarget. 7:64100–64108.
2016.PubMed/NCBI
|
|
187
|
Maleki E, Ghaedi K, Shahanipoor K and
Karimi Kurdistani Z: Down-regulation of microRNA-19b in hormone
receptor-posi- tive/HER2-negative breast cancer. APMI. S.
126:303–308. 2018.
|
|
188
|
Wu Q, Guo L, Jiang F, Li L, Li Z and Chen
F: Analysis of the miRNA-mRNA-lncRNA networks in ER+ and ER-breast
cancer cell lines. J Cell Mol Med. 19:2874–2887. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Kurokawa K, Tanahashi T, Iima T, Yamamoto
Y, Akaike Y, Nishida K, Masuda K, Kuwano Y, Murakami Y, Fukushima M
and Rokutan K: Role of miR-19b and its target mRNAs in 5-fluo -
rouracil resistance in colon cancer cells. J Gastroenterol.
47:883–895. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Thorne JL, Battaglia S, Baxter DE, Hayes
JL, Hutchinson SA, Jana S, Millican-Slater RA, Smith L, Teske MC,
Wastall LM and Hughes TA: MiR-19b non-canonical binding is directed
by HuR and confers chemosensitivity through regulation of
P-glycoprotein in breast cancer. Biochim Biophys Acta Gene Regul
Mech. 1861:996–1006. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
191
|
Jiang T, Ye L, Han Z, Liu Y, Yang Y, Peng
Z and Fan J: miR-19b-3p promotes colon cancer proliferation and
oxalipl- atin-based chemoresistance by targeting SMAD4: Validation
by bioinformatics and experimental analyses. J Exp Clin Cancer Res.
36:1312017. View Article : Google Scholar
|
|
192
|
Jin J, Sun Z, Yang F, Tang L, Chen W and
Guan X: miR-19b-3p inhibits breast cancer cell proliferation and
reverses saraca- tinib-resistance by regulating PI3K/Akt pathway.
Arch Biochem Biophys. 645:54–60. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
193
|
Tsai HP, Huang SF, Li CF, Chien HT and
Chen SC: Differential microRNA expression in breast cancer with
different onset age. PLoS One. 13:e01911952018. View Article : Google Scholar : PubMed/NCBI
|
|
194
|
Chernyy V, Pustylnyak V, Kozlov V and
Gulyaeva L: Increased expression of miR-155 and miR-222 is
associated with lymph node positive status. J Cancer. 9:135–140.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Xiong DD, Lv J, Wei KL, Feng ZB, Chen JT,
Liu KC, Chen G and Luo DZ: A nine-miRNA signature as a potential
diagnostic marker for breast carcinoma: An integrated study of
1,110 cases. Oncol Rep. 37:3297–3304. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Bahrami A, Aledavood A, Anvari K,
Hassanian SM, Maftouh M, Yaghobzade A, Salarzaee O, ShahidSales S
and Avan A: The prognostic and therapeutic application of microRNAs
in breast cancer: Tissue and circulating microRNAs. J Cell Physiol.
233:774–786. 2018. View Article : Google Scholar
|
|
197
|
Zhu M, Wang X, Gu Y, Wang F, Li L and Qiu
X: MEG3 overexpression inhibits the tumorigenesis of breast cancer
by downregulating miR-21 through the PI3K/Akt pathway. Arch Biochem
Biophys. 661:22–30. 2019. View Article : Google Scholar
|
|
198
|
Wang W, Yuan X, Xu A, Zhu X, Zhan Y, Wang
S and Liu M: Human cancer cells suppress behaviors of endothelial
progenitor cells through miR-21 targeting IL6R. Microvasc Res.
120:21–28. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Han JG, Jiang YD, Zhang CH, Yang YM, Pang
D, Song YN and Zhang GQ: A novel panel of serum
miR-21/miR-155/miR-365 as a potential diagnostic biomarker for
breast cancer. Ann Surg Treat Res. 92:55–66. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Yu X, Liang J, Xu J, Li X, Xing S, Li H,
Liu W, Liu D, Xu J, Huang L and Du H: Identification and validation
of circulating MicroRNA signatures for breast cancer early
detection based on large scale tissue-derived data. J Breast
Cancer. 21:363–370. 2018. View Article : Google Scholar
|
|
201
|
Hu X, Fan J, Duan B, Zhang H, He Y, Duan P
and Li X: Single-molecule catalytic hairpin assembly for rapid and
direct quantification of circulating miRNA biomarkers. Anal Chim.
Acta. 1042:109–115. 2018.
|
|
202
|
Fan T, Mao Y, Sun Q, Liu F, Lin JS, Liu Y,
Cui J and Jiang Y: Branched rolling circle amplification method for
measuring serum circulating microRNA levels for early breast cancer
detection. Cancer Sci. 109:2897–2906. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
203
|
Hannafon BN, Trigoso YD, Calloway CL, Zhao
YD, Lum DH, Welm AL, Zhao ZJ, Blick KE, Dooley WC and Ding WQ:
Plasma exosome microRNAs are indicative of breast cancer. Breast
Cancer Res. 18:902016. View Article : Google Scholar : PubMed/NCBI
|
|
204
|
Wang M, Ji S, Shao G, Zhang J, Zhao K,
Wang Z and Wu A: Effect of exosome biomarkers for diagnosis and
prognosis of breast cancer patients. Clin Transl Oncol. 20:906–911.
2018. View Article : Google Scholar
|
|
205
|
Adhami M, Haghdoost AA, Sadeghi B and
Malekpour Afshar R: Candidate miRNAs in human breast cancer biomark
ers: A systematic review. Breast Cancer. 25:198–205. 2018.
View Article : Google Scholar
|
|
206
|
Petrovic N, Sami A, Martinovic J, Zaric M,
Nakashidze I, Lukic S and Jovanovic-Cupic S: TIMP-3 mRNA expression
levels positively correlates with levels of miR-21 in in situ BC
and negatively in PR positive invasive BC. Pathol Res Pract.
213:1264–1270. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
207
|
Jinling W, Sijing S, Jie Z and Guinian W:
Prognostic value of circulating microRNA-21 for breast cancer: A
systematic review and meta-analysis. Artif Cells Nanomed
Biotechnol. 45:1–6. 2017. View Article : Google Scholar
|
|
208
|
Papadaki C, Stratigos M, Markakis G,
Spiliotaki M, Mastrostamatis G, Nikolaou C, Mavroudis D and Agelaki
S: Circulating microRNAs in the early prediction of disease
recurrence in primary breast cancer. Breast Cancer Res. 20:722018.
View Article : Google Scholar : PubMed/NCBI
|
|
209
|
Huo D, Clayton WM, Yoshimatsu TF, Chen J
and Olopade OI: Identification of a circulating microRNA signature
to distinguish recurrence in breast cancer patients. Oncotarget.
7:55231–55248. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
210
|
Shin VY, Siu JM, Cheuk I, Ng EK and Kwong
A: Circulating cell-free miRNAs as biomarker for triple-negative
breast cancer. Br J Cancer. 112:1751–1759. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
211
|
Fang H, Xie J, Zhang M, Zhao Z, Wan Y and
Yao Y: miRNA-21 promotes proliferation and invasion of
triple-negative breast cancer cells through targeting PTEN. Am J
Transl Res. 9:953–961. 2017.PubMed/NCBI
|
|
212
|
Dong G, Liang X, Wang D, Gao H, Wang L,
Wang L, Liu J and Du Z: High expression of miR-21 in
triple-negative breast cancers was correlated with a poor prognosis
and promoted tumor cell in vitro proliferation. Med Oncol.
31:572014. View Article : Google Scholar : PubMed/NCBI
|
|
213
|
Komatsu S, Ichikawa D, Kawaguchi T,
Miyamae M, Okajima W, Ohashi T, Imamura T, Kiuchi J, Konishi H,
Shiozaki A, et al: Circulating miR-21 as an independent predictive
biomarker for chemoresistance in esophageal squamous cell
carcinoma. Am J Cancer Res. 6:1511–1523. 2016.PubMed/NCBI
|
|
214
|
Campayo M, Navarro A, Benitez JC,
Santasusagna S, Ferrer C, Monzo M and Cirera L: miR-21, miR-99b and
miR-375 combination as predictive response signature for
preoperative chemoradiotherapy in rectal cancer. PLoS One.
13:e02065422018. View Article : Google Scholar : PubMed/NCBI
|
|
215
|
Gaudelot K, Gibier JB, Pottier N, Hemon B,
Van Seuningen I, Glowacki F, Leroy X, Cauffiez C, Gnemmi V, Aubert
S and Perrais M: Targeting miR-21 decreases expression of
multi-drug resistant genes and promotes chemosensitivity of renal
carcinoma. Tumour Biol. 39:10104283177073722017. View Article : Google Scholar : PubMed/NCBI
|
|
216
|
Lin L, Tu HB, Wu L, Liu M and Jiang GN:
MicroRNA-21 regulates non-small cell lung cancer cell invasion and
chemo-sensitivity through SMAD7. Cell Physiol Biochem.
38:2152–2162. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
217
|
Szejniuk WM, Robles AI, McCulloch T,
Falkmer UGI and Roe OD: Epigenetic predictive biomarkers for
response or outcome to platinum-based chemotherapy in non-small
cell lung cancer, current state-of-art. Pharmacogenomics J.
19:5–14. 2019. View Article : Google Scholar
|
|
218
|
Feng Y, Zou W, Hu C, Li G, Zhou S, He Y,
Ma F, Deng C and Sun L: Modulation of CASC2/miR-21/PTEN pathway
sensitizes cervical cancer to cisplatin. Arch Biochem Biophys.
623-624:20–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
219
|
Chan JK, Blansit K, Kiet T, Sherman A,
Wong G, Earle C and Bourguignon LY: The inhibition of miR-21
promotes apoptosis and chemosensitivity in ovarian cancer. Gynecol
Oncol. 132:739–744. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
220
|
Pink RC, Samuel P, Massa D, Caley DP,
Brooks SA and Carter DR: The passenger strand, miR-21-3p, plays a
role in mediating cisplatin resistance in ovarian cancer cells.
Gynecol Oncol. 137:143–151. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
221
|
Dai X, Fang M, Li S, Yan Y, Zhong Y and Du
B: miR-21 is involved in transforming growth factor |31-induced
chemoresis- tance and invasion by targeting PTEN in breast cancer.
Oncol Lett. 14:6929–6936. 2017.PubMed/NCBI
|
|
222
|
Bose RJC, Uday Kumar S, Zeng Y, Afjei R,
Robinson E, Lau K, Bermudez A, Habte F, Pitteri SJ, Sinclair R, et
al: Tumor cell-derived extracellular vesicle-coated nanocarriers:
An efficient theranostic platform for the cancer-specific delivery
of Anti-miR-21 and imaging agents. ACS Nano. 12:10817–10832. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
223
|
Zhou Q, Zeng H, Ye P, Shi Y, Guo J and
Long X: Differential microRNA profiles between
fulvestrant-resistant and tamoxifen-resistant human breast cancer
cells. Anticancer Drugs. 29:539–548. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
224
|
Wu ZH, Tao ZH, Zhang J, Li T, Ni C, Xie J,
Zhang JF and Hu XC: MiRNA-21 induces epithelial to mesenchymal
transition and gemcitabine resistance via the PTEN/AKT pathway in
breast cancer. Tumour Biol. 37:7245–7254. 2016. View Article : Google Scholar
|
|
225
|
Dhayat SA, Mardin WA, Seggewifi J, Strose
AJ, Matuszcak C, Hummel R, Senninger N, Mees ST and Haier J:
MicroRNA profiling implies new markers of gemcitabine
chemoresistance in mutant p53 pancreatic ductal adenocarcinoma.
PLoS One. 10:e01437552015. View Article : Google Scholar : PubMed/NCBI
|
|
226
|
Dong J, Zhao YP, Zhou L, Zhang TP and Chen
G: Bcl-2 upregu- lation induced by miR-21 via a direct interaction
is associated with apoptosis and chemoresistance in MIA PaCa-2
pancreatic cancer cells. Arch Med Res. 42:8–14. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
227
|
Kopczynska E: Role of microRNAs in the
resistance of prostate cancer to docetaxel and paclitaxel. Contemp
Oncol (Pozn). 19:423–427. 2015.
|
|
228
|
Haghnavaz N, Asghari F, Komi Ali Elieh D,
Shanehbandi D, Baradaran B and Kazemi T: HER2 positivity may confer
resistance to therapy with paclitaxel in breast cancer cell lines.
Artif Cells Nanomed Biotechnol. 46:518–523. 2018. View Article : Google Scholar
|
|
229
|
Du G, Cao D and Meng L: miR-21 inhibitor
suppresses cell proliferation and colony formation through
regulating the PTEN/AKT pathway and improves paclitaxel sensitivity
in cervical cancer cells. Mol Med Rep. 15:2713–2719. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
230
|
Jin B, Liu Y and Wang H: Antagonism of
miRNA-21 sensitizes human gastric cancer cells to paclitaxel. Cell
Biochem Biophys. 72:275–282. 2015. View Article : Google Scholar
|
|
231
|
Mei M, Ren Y, Zhou X, Yuan XB, Han L, Wang
GX, Jia Z, Pu PY, Kang CS and Yao Z: Downregulation of miR-21
enhances chemotherapeutic effect of taxol in breast carcinoma
cells. Technol Cancer Res Treat. 9:77–86. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
232
|
Naro Y, Ankenbruck N, Thomas M, Tivon Y,
Connelly CM, Gardner L and Deiters A: Small molecule inhibition of
MicroRNA miR-21 rescues chemosensitivity of renal-cell carcinoma to
topotecan. J Med Chem. 61:5900–5909. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
233
|
Liu Y, Xu J, Choi HH, Han C, Fang Y, Li Y,
Van der Jeught K, Xu H, Zhang L, Frieden M, et al: Targeting 17q23
amplicon to overcome the resistance to anti-HER2 therapy in HER2+
breast cancer. Nat Commun. 9:47182018. View Article : Google Scholar : PubMed/NCBI
|
|
234
|
Decker JT, Hall MS, Blaisdell RB, Schwark
K, Jeruss JS and Shea LD: Dynamic microRNA activity identifies
therapeutic targets in trastuzumab-resistant HER2+
breast cancer. Biotechnol Bioeng. 115:2613–2623. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
235
|
Bahreyni A, Alibolandi M, Ramezani M,
Sarafan Sadeghi A, Abnous K and Taghdisi SM: A novel MUC1
aptamer-modified PLGA-epirubicin-PpAE-antimir-21 nanocomplex
platform for targeted co-delivery of anticancer agents in vitro and
in vivo. Colloids Surf B Biointerfaces. 175:231–238. 2018.
View Article : Google Scholar
|
|
236
|
Vandghanooni S, Eskandani M, Barar J and
Omidi Y: AS1411 aptamer-decorated cisplatin-loaded
poly(lactic-co-glycolic acid) nanoparticles for targeted therapy of
miR-21-inhibited ovarian cancer cells. Nanomedicine (Lond).
13:2729–2758. 2018. View Article : Google Scholar
|
|
237
|
Wang W, Huang S, Yuan J, Xu X, Li H, Lv Z,
Yu W, Duan S and Hu Y: Reverse multidrug resistance in human
HepG2/ADR by Anti-miR-21 combined with hyperthermia mediated by
functionalized gold nanocages. Mol Pharm. 15:3767–3776. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
238
|
Luo J, Zhao Q, Zhang W, Zhang Z, Gao J,
Zhang C, Li Y and Tian Y: A novel panel of microRNAs provides a
sensitive and specific tool for the diagnosis of breast cancer. Mol
Med Rep. 10:785–791. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
239
|
Zhang K, Wang YW, Wang YY, Song Y, Zhu J,
Si PC and Ma R: Identification of microRNA biomarkers in the blood
of breast cancer patients based on microRNA profiling. Gene.
619:10–20. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
240
|
Croset M, Pantano F, Kan CWS, Bonnelye E,
Descotes F, Alix-Panabieres C, Lecellier CH, Bachelier R, Allioli
N, Hong SS, et al: MicroRNA-30 family members inhibit breast cancer
invasion, osteomimicry, and bone destruction by directly targeting
multiple bone metastasis-associated genes. Cancer Res.
78:5259–5273. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
241
|
Ni Q, Stevic I, Pan C, Muller V,
Oliviera-Ferrer L, Pantel K and Schwarzenbach H: Different
signatures of miR-16, miR-30b and miR-93 in exosomes from breast
cancer and DCIS patients. Sci Rep. 8:129742018. View Article : Google Scholar : PubMed/NCBI
|
|
242
|
Xi Z, Si J and Nan J: LncRNA MALAT1
potentiates autophagyassociated cisplatin resistance by regulating
the microRNA30b/autophagyrelated gene 5 axis in gastric cancer. Int
J Oncol. 54:239–248. 2019.
|
|
243
|
Chen JC, Su YH, Chiu CF, Chang YW, Yu YH,
Tseng CF, Chen HA and Su JL: Suppression of Dicer increases
sensitivity to gefitinib in human lung cancer cells. Ann Surg
Oncol. 21(Suppl 4): S555–S563. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
244
|
Tormo E, Adam-Artigues A, Ballester S,
Pineda B, Zazo S, Gonzalez-Alonso P, Albanell J, Rovira A, Rojo F,
Lluch A and Eroles P: The role of miR-26a and miR-30b in HER2+
breast cancer trastuzumab resistance and regulation of the CCNE2
gene. Sci Rep. 7:413092017. View Article : Google Scholar : PubMed/NCBI
|
|
245
|
Gu YF, Zhang H, Su D, Mo ML, Song P, Zhang
F and Zhang SC: miR-30b and miR-30c expression predicted response
to tyrosine kinase inhibitors as first line treatment in non-small
cell lung cancer. Chin Med J (Engl). 126:4435–4439. 2013.
|
|
246
|
Amini S, Abak A, Estiar MA, Montazeri V,
Abhari A and Sakhinia E: Expression Analysis of MicroRNA-222 in
breast cancer. Clin Lab. 64:491–496. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
247
|
Zong Y, Zhang Y, Sun X, Xu T, Cheng X and
Qin Y: miR-221/222 promote tumor growth and suppress apoptosis by
targeting lncRNA GAS5 in breast cancer. Biosci Rep. 39:pii:
BSR20181859. 2019. View Article : Google Scholar :
|
|
248
|
Hoppe R, Fan P, Buttner F, Winter S, Tyagi
AK, Cunliffe H, Jordan VC and Brauch H: Profiles of miRNAs matched
to biology in aromatase inhibitor resistant breast cancer.
Oncotarget. 7:71235–71254. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
249
|
Han SH, Kim HJ, Gwak JM, Kim M, Chung YR
and Park SY: MicroRNA-222 expression as a predictive marker for
tumor progression in hormone receptor-positive breast cancer. J
Breast Cancer. 20:35–44. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
250
|
Kim C, Go EJ and Kim A: Recurrence
prediction using microRNA expression in hormone receptor positive
breast cancer during tamoxifen treatment. Biomarkers. 23:804–811.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
251
|
Zhu W, Liu M, Fan Y, Ma F, Xu N and Xu B:
Dynamics of circulating microRNAs as a novel indicator of clinical
response to neoadjuvant chemotherapy in breast cancer. Cancer Med.
7:4420–4433. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
252
|
Chen X, Lu P, Wang DD, Yang SJ, Wu Y, Shen
HY, Zhong SL, Zhao JH and Tang JH: The role of miRNAs in drug
resistance and prognosis of breast cancer formalin-fixed
paraffin-embedded tissues. Gene. 595:221–226. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
253
|
Chen J, Chen Z, Huang J, Chen F, Ye W,
Ding G and Wang X: Bioinformatics identification of dysregulated
microRNAs in triple negative breast cancer based on microRNA
expression profiling. Oncol Lett. 15:3017–3023. 2018.PubMed/NCBI
|
|
254
|
Li Y, Liang C, Ma H, Zhao Q, Lu Y, Xiang
Z, Li L, Qin J, Chen Y, Cho WC, et al: miR-221/222 promotes S-phase
entry and cellular migration in control of basal-like breast
cancer. Molecules. 19:7122–7137. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
255
|
Zhao L, Ren Y, Tang H, Wang W, He Q, Sun
J, Zhou X and Wang A: Deregulation of the miR-222-ABCG2 regulatory
module in tongue squamous cell carcinoma contributes to
chemoresistance and enhanced migratory/invasive potential.
Oncotarget. 6:44538–44550. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
256
|
Shen H, Wang D, Li L, Yang S, Chen X, Zhou
S, Zhong S, Zhao J and Tang J: MiR-222 promotes drug-resistance of
breast cancer cells to adriamycin via modulation of PTEN/Akt/FOXO1
pathway. Gene. 596:110–118. 2017. View Article : Google Scholar
|
|
257
|
Wang DD, Yang SJ, Chen X, Shen HY, Luo LJ,
Zhang XH, Zhong SL, Zhao JH and Tang JH: miR-222 induces Adriamycin
resistance in breast cancer through PTEN/Akt/p27(kip1) pathway.
Tumour Biol. 37:15315–15324. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
258
|
Rao X, Di Leva G, Li M, Fang F, Devlin C,
Hartman-Frey C, Burow ME, Ivan M, Croce CM and Nephew KP: MicroRNA-
221/222 confers breast cancer fulvestrant resistance by regulating
multiple signaling pathways. Oncogene. 30:1082–1097. 2011.
View Article : Google Scholar
|
|
259
|
Wei F, Ma C, Zhou T, Dong X, Luo Q, Geng
L, Ding L, Zhang Y, Zhang L, Zhang L, et al: Exosomes derived from
gemcitabine-resistant cells transfer malignant phenotypic traits
via delivery of miRNA-222-3p. Mol Cancer. 16:1322017. View Article : Google Scholar : PubMed/NCBI
|
|
260
|
Bliss SA, Sinha G, Sandiford OA, Williams
LM, Engelberth DJ, Guiro K, Isenalumhe LL, Greco SJ, Ayer S, Bryan
M, et al: Mesenchymal stem cell-derived exosomes stimulate cycling
quiescence and early breast cancer dormancy in bone marrow. Cancer
Res. 76:5832–5844. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
261
|
Ding J, Xu Z, Zhang Y, Tan C, Hu W, Wang
M, Xu Y and Tang J: Exosome-mediated miR-222 transferring: An
insight into NF-kappaB-mediated breast cancer metastasis. Exp Cell
Res. 369:129–138. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
262
|
Gan R, Yang Y, Yang X, Zhao L, Lu J and
Meng QH: Downregulation of miR-221/222 enhances sensitivity of
breast cancer cells to tamoxifen through upregulation of TIMP3.
Cancer Gene Ther. 21:290–296. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
263
|
Vishnubalaji R, Hamam R, Yue S, Al-Obeed
O, Kassem M, Liu FF, Aldahmash A and Alajez NM: MicroRNA-320
suppresses colorectal cancer by targeting SOX4, FOXM1, and FOXQ1.
Oncotarget. 7:35789–35802. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
264
|
Iwagami Y, Eguchi H, Nagano H, Akita H,
Hama N, Wada H, Kawamoto K, Kobayashi S, Tomokuni A, Tomimaru Y, et
al: miR-320c regulates gemcitabine-resistance in pancreatic cancer
via SMARCC1. Br J Cancer. 109:502–511. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
265
|
Li M, Zeringer E, Barta T, Schageman J,
Cheng A and Vlassov AV: Analysis of the RNA content of the exosomes
derived from blood serum and urine and its potential as biomarkers.
Philos Trans R Soc Lond B Biol Sci. 369:pii: 201305022014.
View Article : Google Scholar
|