Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most
common aggressive pediatric mature B-cell non-Hodgkin's lymphoma
(NHL), with a high incidence of short- and long-term toxicity after
chemotherapy regimens (1). It is
also common in older adults, comprising 60% of all lymphoid
malignancies in this population (2). DLBCL is characterized by an
infiltration of medium to large cells with large nucleoli and ample
cytoplasm, which disturbs the basic structure of the affected lymph
node (3). The disease is invasive
and patients usually show rapid lymph node enlargement and physical
symptoms, requiring immediate treatment (3). Rituximab, a monoclonal antibody
against CD20, has improved the survival of high-risk patients and
reduced the requirement of total chemotherapy for low-risk patients
(1). Although most patients
receiving rituximab achieve complete remission, approximately 30%
of patients relapse or fail to achieve event-free survival within
24 months after diagnosis, with a median survival time of
approximately 10 months (4). In
such cases, promising approaches are imperative to counter
rituximab resistance.
Tumor necrosis factor necrosis factor alpha-induced
protein 3 (TNFAIP3), also known as A20, is a negative regulator of
nuclear factor kappa B (NF-κB) that has been implicated as a tumor
suppressor in multiple types of B-cell lymphoma (5). In mice, B-cell specific deletion of
TNFAIP3 enhances B-cell proliferation and autoantibody production,
while the germline inactivation of TNFAIP3 results in early
lethality due to inflammation in multiple organs (6). TNFAIP3 genetic alterations are
involved in DLBCL pathogenesis (7). In a previous study, the negative
prognostic significance of TNFAIP3 deletion and somatic mutation
was reported in gastrointestinal DLBCL (8). Notably, TNFAIP3 deletion is
marginally associated with a favorable prognosis in
rituximab-treated populations (9). However, the mechanism of TNFAIP3 in
the sensitivity of DLBCL to rituximab is largely unknown.
Extracellular vesicles (EVs), nanometer-sized,
cell-secreted membrane vesicles, are critical to intercellular
communication between tumor cells and resident cells (10). In particular, EVs obtained from
cancer cells are known to modify the tumor microenvironment and
promote tumor progression (11).
Of note, DLBCL-derived EVs can regulate macrophage polarization and
thus contribute to tumor progression (12). MicroRNAs (miRs), a cargo of EVs,
are also essential for DLBCL development (13). Aberrant expression of miRs has
been shown to have a profound impact on treatment outcomes and
chemoresistance in DLBCL (14).
Therefore, it was hypothesized that TNFAIP3 may be regulated by a
miR shuttled in DLBCL cell-derived EVs, thus modulating the
sensitivity of DLBCL to rituximab. In the present study, the
possible mechanism of TNFAIP3 targeted by the EV-carried miRs in
DLBCL was explored to offer a novel theoretical basis for the
management of DLBCL.
Materials and methods
Ethics statement
The study obtained approval from the Ethics
Committee of The Affiliated Hospital of Southwest Medical
University (China; no. 20180306006). The clinical registration
number is KY2019091. Animal experiments were implemented on the
guide for the care and use of laboratory animals, and approved by
the Ethics Committee of The Affiliated Hospital of Southwest
Medical University (animal ethics no. 20180306006; human ethics no.
KY2019091).
Patients
A total of 34 patients with DLBCL, who underwent
surgical resection at The Affiliated Hospital of Southwest Medical
University from August 2017 to August 2019, were selected as the
DLBCL group, including 20 males and 14 females, aged 38-64 years.
The inclusion criteria were as follows: i) Patients diagnosed with
DLBCL by pathology; ii) patients conforming to surgical
indications; iii) patients who did not receive chemotherapy prior
to surgery; iv) patients with complete clinical and pathological
data and signed informed consent. Patients with other hematological
or solid malignancies, liver and kidney dysfunction, autoimmune
diseases, or infectious diseases were excluded. Furthermore, 30
patients with DLBCL without remission after 6 cycles of rituximab
treatment were selected as the DLBCL-R group, including 18 males
and 12 females, aged 32-64 years. At the same time, another 35
patients with reactive hyperplasia of lymph nodes, confirmed by
pathology at The Affiliated Hospital of Southwest Medical
University, were selected as the control group, including 23 males
and 12 females, aged 34-68 years. There was no significant
difference in the general data among the three groups (Table I, P>0.05).
 | Table IClinical characteristic of DLBCL
patients. |
Table I
Clinical characteristic of DLBCL
patients.
|
Characteristics | Control (N=35) | DLBCL (N=34) | DLBCL+R (N=30) |
χ2/F | P-value |
|---|
| Agea | 34-68
(mean=49.74) | 38-64
(mean=49.03) | 32-64
(mean=45.30) | 2.622 | 0.078 |
| Male | 23/35 | 20/34 | 18/30 | 0.394 | 0.821 |
| Pathology type | | | | | |
| GCB | \ | 15/34 | 13/30 | 0.345 | 0.842 |
| ABC | \ | 14/34 | 11/30 | | |
| Unknown | \ | 5/34 | 6/30 | | |
| Performance status
(ECOG) | | | | | |
| 0-1 | 28/35 | 29/34 | 26/30 | 0.611 | 0.737 |
| 2 | 7/35 | 5/34 | 4/30 | | |
| Lactic
dehydrogenase | | | | | |
| Normal | 23/35 | 20/34 | 19/30 | 0.359 | 0.836 |
| Elevated | 12/35 | 14/34 | 11/30 | | |
| Ann Arbor
stage | | | | 0.779 | 0.446 |
| I-II | \ | 19/34 | 20/30 | | |
| III-IV | \ | 15/34 | 10/30 | | |
Cell culture
DLBCL cell lines SUDHL-4 (SUD, GCB subtype), OCI-LY8
(LY8, GCB subtype), and NU-DUL-1 (DUL, ABC subtype) and 293T cells
were purchased from the American Type Culture Collection (ATCC).
LY8 cells were cultured in Iscove modified Dulbecco's medium
(HyClone) containing 10% fetal bovine serum (FBS; Gibco) and 1%
penicillin/streptomycin. SUD and DUL cells and 293T cells were
cultured in RPMI-1640 medium, containing 10% FBS and 1%
penicillin/streptomycin (HyClone), at 37°C under 5% CO2.
When necessary, 1% antibiotics (penicillin, streptomycin,
amphotericin) (Gibco) were added to the medium to maintain the
culture. Cells were passaged every 2-3 days.
Isolation and identification of EVs
SUD cells were cultured in DMEM or RPMI-1640 medium
with 10% EV-depleted FBS and 1% penicillin/streptomycin for 48 h.
The supernatant was centrifuged at 100 × g for 5 min to remove
cells and cellular debris, and then centrifuged at 3,600 × g and
4°C for 30 min to remove large vesicles. The supernatant was then
filtered through a 0.2 µm filtration membrane (Millipore),
followed by centrifugation at 100,000 × g and 4°C for 2 h. The EVs
were washed with PBS and centrifuged again at 100,000 × g and 4°C
for 2 h. Finally, EVs were suspended in PBS. Similarly, GW4869 (20
µg/ml conditioned medium; Sigma-Aldrich) was added to
EV-depleted FBS, in which SUD cells were cultured for 48 h. The
conditioned medium was then used as a control (GW4896 group). The
concentration of EV protein, detected using the Bicinchoninic Acid
Protein Assay Kit (Thermo Fisher Scientific), was 1.5 mg/ml. The
expression levels of CD9 (ab92726), TSG101 (ab125011), and calnexin
(ab22595) (all from Abcam) were detected using western blot
analysis. EV morphology was observed with transmission electron
microscopy (TEM), and EV particle size was detected using a
nanoparticle tracking analyzer (NTA) (Malvern Panalytical Co.,
Ltd.).
PHK-26 labeling for EV location
EV particles were resuspended in 1 ml diluent C.
PKH-26 (2 µl mixed with 245 µl diluent C;
Sigma-Aldrich) was added to the EV suspension for 5 min, followed
by the addition of 1% bovine serum albumin to terminate the
labeling reaction. Subsequently, LY8 or DUL cells were seeded in
24-well plates and PKH-26-labeled EVs (10 µg) were added to
each well. After fixing the cells with 4% formaldehyde for 24 h,
4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich) was used for
nucleus staining. Finally, the DLBCL cells were observed for EV
internalization, using laser scanning confocal microscopy (LSM710,
Zeiss).
Cell transfection
According to the manufacturer's instructions,
miR-125b-5p inhibitor and its negative control (NC) (50 nM,
GenePharma Co., Ltd.), pcDNA-TNFAIP3, and pcDNA-NC (40 nM, Sangon
Biotech Co., Ltd.) vectors were transfected into LY8 or DUL cells
using Lipofectamine™ 2000 (Thermo Fisher Scientific) according to
the manufacturer's instructions. The DLBCL cells were then
incubated with the conditioned medium from SUD cells treated with
30 µg EVs or GW4869 for 24 h at 37°C.
miR-125b-5p inhibitor and its NC were transfected
into SUD cells using Lipofectamine™ 2000, followed by incubation
for 4 h at 37°C. The cells were cultured in complete medium for 48
h, and then EVs were isolated and incubated with LY8 and DUL
cells.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay
Cells in the logarithmic phase were collected and
treated with rituximab (2.5, 5, 10, 20, and 40 µg/ml)
(15), and the cell suspension
concentration was adjusted to 1×105 cells/ml. The cells
were seeded in 96-well plates (100 µl/well) at 37°C under 5%
CO2 for 48 h and observed using an inverted microscope
(Leica).
Subsequently, 10 µl MTT (5 mg/ml,
Sigma-Aldrich) was added to each well for 4 h. Following
centrifugation at 400 × g and 4°C for 15 min, 200 µl
dimethyl sulfoxide was added to each well and plates were placed in
a decolorizing shaker for 20 min to fully dissolve the crystal.
Optical density (OD) at 570 nm was measured using a microplate
reader (Bio-Rad Laboratories).
Cell apoptosis
Cell apoptosis was detected using the PE Annexin V
Apoptosis Detection Kit with 7-aminoactinomycin D, according to the
manufacturer's instructions (BD Bioscience). The number of
apoptotic cells was quantified using the BD FACSCalibur flow
cytometer system (BD Bioscience) and CellQuest Pro software (BD
Bioscience).
Dual-luciferase reporter gene assay
The TNFAIP3 3′UTR fragment containing the
miR-125b-5p binding site (WT) and the fragment containing the
site-directed mutagenesis modification site (MUT) were cloned into
psiCHECK-2.0 vector (Promega). Then, the 293T cells (ATCC) treated
with miR-125b-5p mimic or miR-125b-5p mimic NC (RiboBio) were
co-transfected with WT or MUT plasmids, with phRL-tk transfection
(Renilla luciferase) as positive control. After 48 h of
transfection, luciferase activity was detected using a luciferase
reporter kit (E1910, Think-Far Technology Co., Ltd, Beijing,
China). Luciferase intensity was expressed in relative luciferase
units (percentage relative to the control group).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
The RNAiso plus kit (Takara, Dalian, China) was used
to extract total RNA from SUD, LY8, and DUL cells, EVs, and
tissues. The total RNA was reverse transcribed into cDNA using
PrimeScript RT reagent kit (Takara). RT-qPCR was performed on a
7500 Real-Time PCR system using the SYBR-Green PCR kit (Takara
Bio). The reaction conditions were pre-denaturation at 95°C for 10
min, denaturation at 95°C for 10 sec, annealing at 60°C for 20 sec,
and extension at 72°C for 34 sec, a total of 40 cycles. U6
or GAPDH served as the internal reference. Quantitative
expression was calculated using the 2−ΔΔCT method
(16). The primers used are shown
in Table II.
 | Table IIPrimer sequences for RT-qPCR. |
Table II
Primer sequences for RT-qPCR.
| Primer | Sequences
(5′-3′) | Accession |
|---|
| TNFAIP3 | F:
ATGGCTGAACAAGTCCTTCCTCAG | NM_001270507 |
| R:
TTAGCCATACATCTGCTTGAACTGA | |
| CD20 | F:
ATGACAACACCCAGAAATTCAGTA | NM_021950 |
| R:
TTAAGGAGAGCTGTCATTTTCTAT | |
|
miR-125b-5p | F:
TCCCTGAGACCCTAACTTGTGA | MIMAT0000423 |
| R:
TCACAAGTTAGGGTCTCAGGGA | |
| U6 | F:
CGCTTCGGCAGCACATATAC | NR_004394 |
| R:
AATATGGAACGCTTCACGA | |
| GAPDH | F:
ATGGTTTACATGTTCCAATATGA | NM_001256799 |
| R:
TTACTCCTTGGAGGCCATGTGG | |
Western blot analysis
Patient focus or mouse tumor tissue homogenate or
cells were lysed in RIPA buffer containing 50 mM Tris-HCl (pH 7.5),
150 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate, 5 mM
EDTA, 0.1 mM PMSF, and 2 mg/ml aprotinin. Protein concentration was
measured using the BCA kit (Beijing Solarbio Science &
Technology Co., Ltd.). Then, the proteins (50 µg/lane) were
separated with 10% sodium dodecyl sulfate polyacrylamide gel
electrophoresis (Beyotime Biotech) and transferred onto
nitrocellulose membranes. The membranes were blocked with 3%
blocking buffer (Beyotime) for 1 h at room temperature and cultured
with the primary antibodies TNFAIP3 (1:1,000, ab92324, Abcam) and
CD20 (1:1,000, Abcam) overnight at 4°C. Subsequently, the membranes
were cultured with the secondary antibody horseradish
peroxidase-conjugated goat anti-rabbit IgG (1:5,000, ab205718,
Abcam) for 2 h. Next, the membranes were visualized using an
enhanced chemiluminescence reagent (Millipore). Protein blotting
was analyzed using the Image Pro Plus 6.0 software (National
Institutes of Health), with β-actin as the internal reference. The
experiment was repeated three times.
Establishment of a mouse model of
DLBCL
Female nude mice (5-6 weeks old) were purchased from
the Shanghai Experimental Animal Center and raised under aseptic
laminar flow conditions. The mice were reared at 50-60% humidity
and 25°C, and maintained in a 12 h light/dark cycle. Food and water
were provided a Extracellular vesicles d libitum.
Each mouse was injected with 2×107 LY8
cells into the right abdomen. When a measurable tumor
(approximately 100 mm3) was observed at the injection
site after 3 weeks, the mice were subjected to drug treatment.
Based on body weight, the mice were randomly allocated into 3
groups, with 10 mice in each group: The LY8 group (injected with
LY8 cells + 10 mg/kg rituximab), GW group (injected with the same
dose of GW4869 medium + 10 mg/kg rituximab), and EV group (injected
with 30 µg EVs + 10 mg/kg rituximab). Five mice in each
group were injected once a week and their tumors were measured
every 4 days to record the long (A) and short (B) diameters of the
tumors. The maximum diameter and volume of the tumor observed in
the mice were 2 cm and 3.4 cm2, respectively. On the
28th day, the mice were sacrificed through cervical dislocation and
tumors were removed. The precise size of the tumor was measured
using a Vernier caliper, and the tumor was weighed.
Statistical analysis
All data were processed using the IBM SPSS
Statistics version 21.0 software (IBM Corp.). Data were first
verified to show normal distribution and homogeneity of variance.
Data were expressed as means ± standard deviation or counts. An
independent samples t-test or χ2 was used to compare two
groups. One-way analysis of variance (ANOVA) was applied for
comparison among multiple groups, followed by Tukey's multiple
comparisons test. P<0.05 indicated a statistically significant
difference.
Results
TNFAIP3 is poorly expressed in DLBCL
DLBCL is the most common histological subtype of NHL
(17-19) and generally responds well to
treatment with rituximab (20,21). TNFAIP3 is an established tumor
suppressor in lymphoma (22-24), but its drug resistance has been
reported in various types of cancer (25,26). It has been hypothesized that
TNFAIP3 may be related to the resistance of DLBCL to rituximab. To
determine the role of TNFAIP3 in DLBCL resistance to treatment,
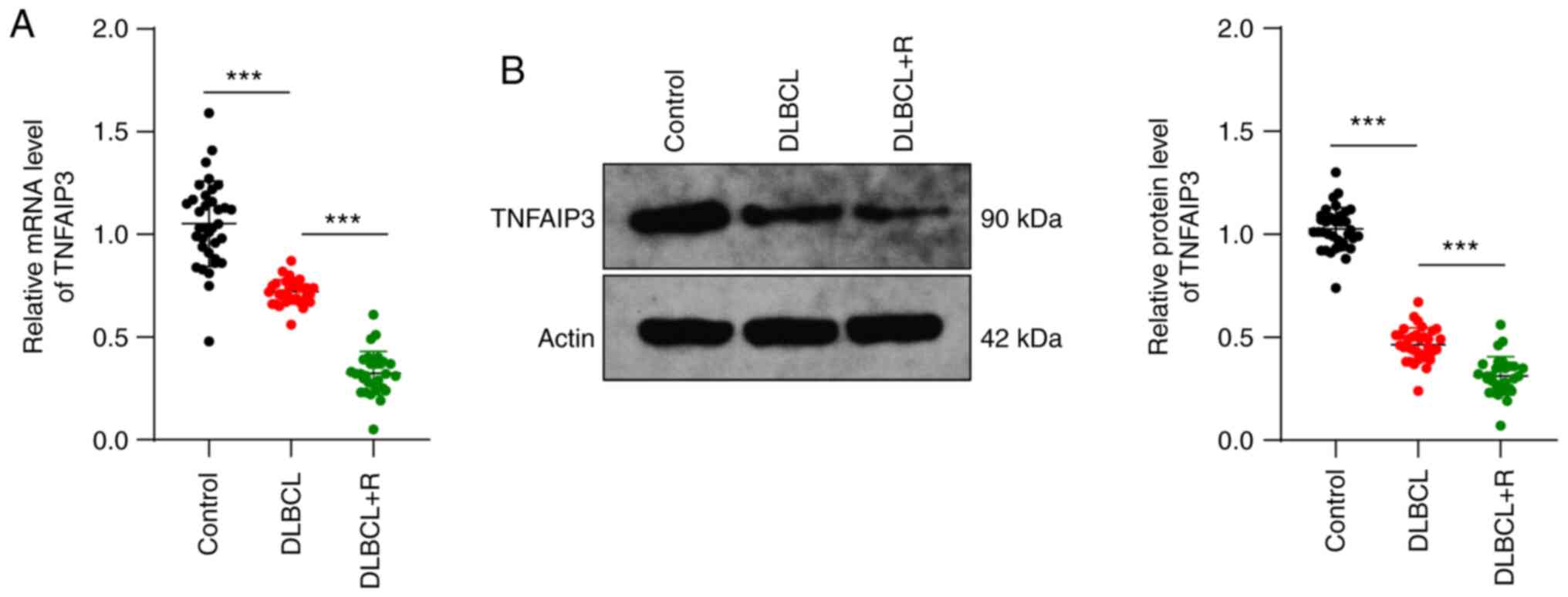
TNFAIP3 levels in patient lesions were first measured. TNFAIP3
levels in the DLBCL group were significantly lower than those in
the control group, and further decreased in the DLBCL-R group (all
P<0.001) (Fig. 1A and B).
Overall, TNFAIP3 expression was low in DLBCL, which may be related
to the development of DLBCL.
Overexpression of TNFAIP3 increases the
sensitivity of DLBCL cells to rituximab
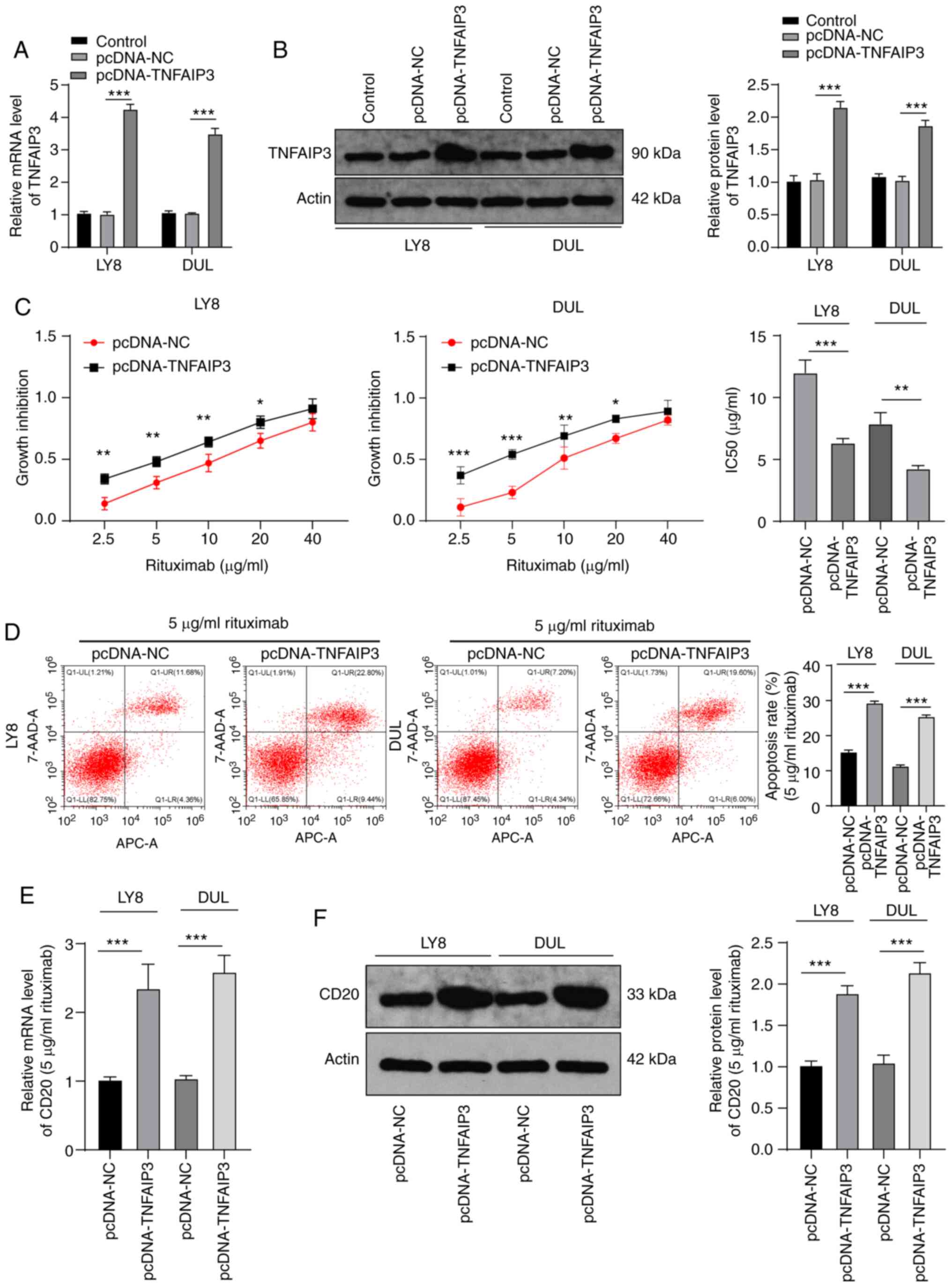
Next, in order to test our hypothesis, the
TNFAIP3-expressing pcDNA vector was transfected into LY8 and DUL
cells, which were then treated with different concentrations of
rituximab. We confirmed the overexpression of TNFAIP3 with RT-qPCR
and western blotting (P<0.001) (Fig. 2A and B). Results of the MTT assay
demonstrated that the IC50 value for cells overexpressing TNFAIP3
was notably reduced compared with that for NC cells (LY8:
12.01±1.02 vs. 6.35±0.35 µg/ml; DUL: 7.89±0.89 vs. 4.25±0.25
µg/ml) (all P<0.001) (Fig.
2C). The cells transfected with pcDNA-TNFAIP3 and treated with
5 µg/ml rituximab had a significantly higher cell apoptosis
rate than the NC group (P<0.001) (Fig. 2D). Rituximab is a chimeric
human-mouse monoclonal antibody targeting CD20 molecules on the
B-cell surface (27). Therefore,
we measured CD20 expression with RT-qPCR and western blotting and
found that CD20 expression in the pcDNA-TNFAIP3 group was
significantly increased compared with that in the NC group
(P<0.001) (Fig. 2E and F).
These results suggested that overexpression of TNFAIP3 increased
DLBCL sensitivity to rituximab.
miR-125b-5p targets TNFAIP3
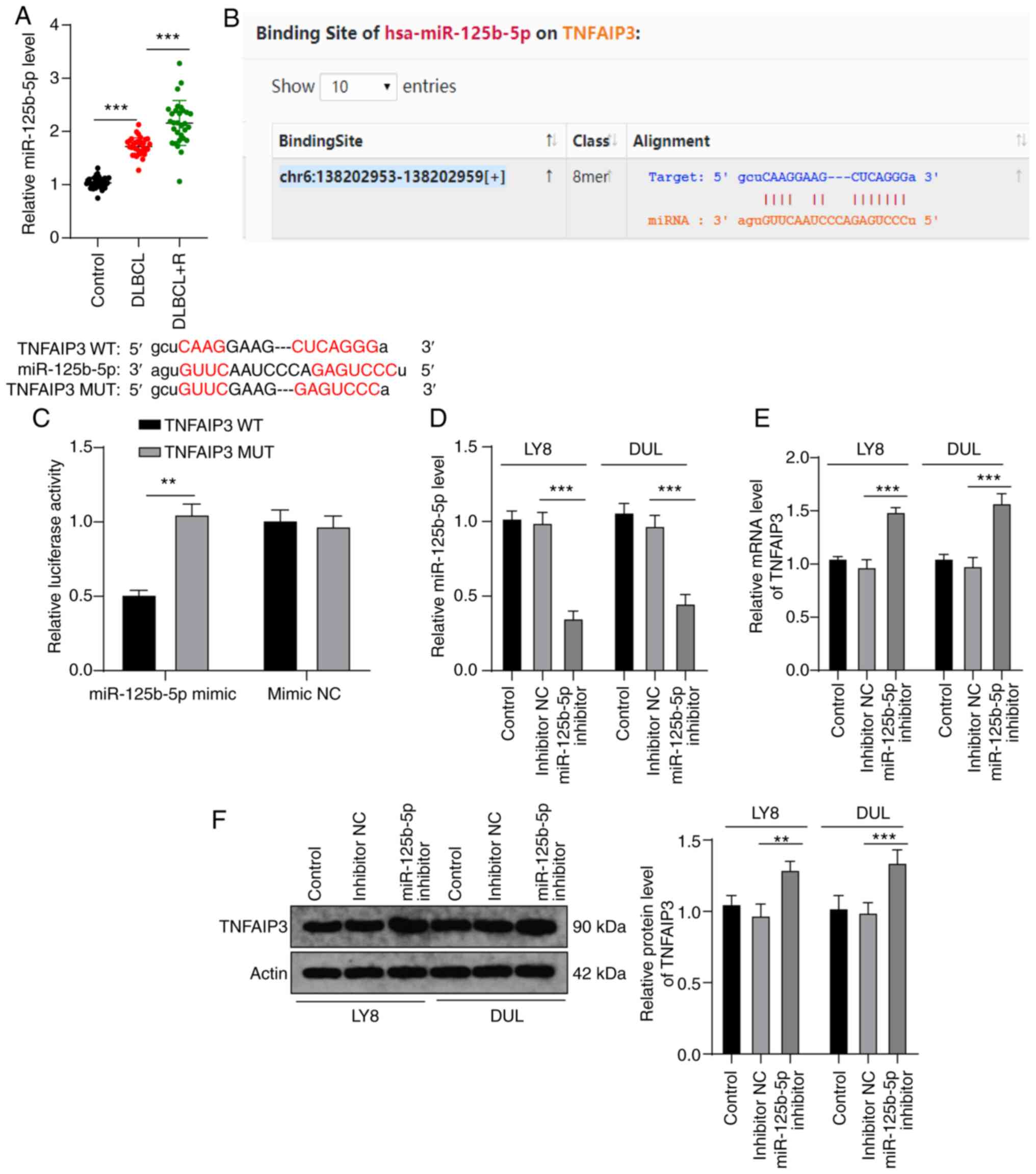
Previous findings have shown that miRs can inhibit
the expression of tumor-specific genes in cancer (28). In order to explore the upstream
molecular mechanism of TNFAIP3 in regulating the resistance of
DLBCL to rituximab, we identified various miRs that target TNFAIP3,
in the starBase database (29);
among these, miR-125b-5p plays an important role in cancer and drug
resistance (30,31). Therefore, it was hypothesized that
miR-125b-5p may regulate rituximab resistance in DLBCL by targeting
TNFAIP3. miR-125b-5p expression was measured in patient lesions and
it was found that miR-125b-5p expression in the DLBCL-R and DLBCL
groups was significantly higher than that in the controls
(P<0.001) (Fig. 3A). Then,
according to TNFAIP3 and miR-125b-5p binding sites in the database,
dual-luciferase experiments (Fig.
3B) verified the targeted binding relationship between TNFAIP3
and miR-125b-5p (P<0.01) (Fig.
3C). In addition, miR-125b-5p expression in LY8 and DUL cells
was significantly reduced using the miR-125b-5p inhibitor (both
P<0.001) (Fig. 3D), which in
turn significantly increased TNFAIP3 expression in LY8 and DUL
cells (all P<0.01) (Fig. 3E and
F). These results indicated that miR-125b-5p targeted TNFAIP3
in DLBCL.
miR-125b-5p downregulation increases the
sensitivity of DLBCL cells to rituximab
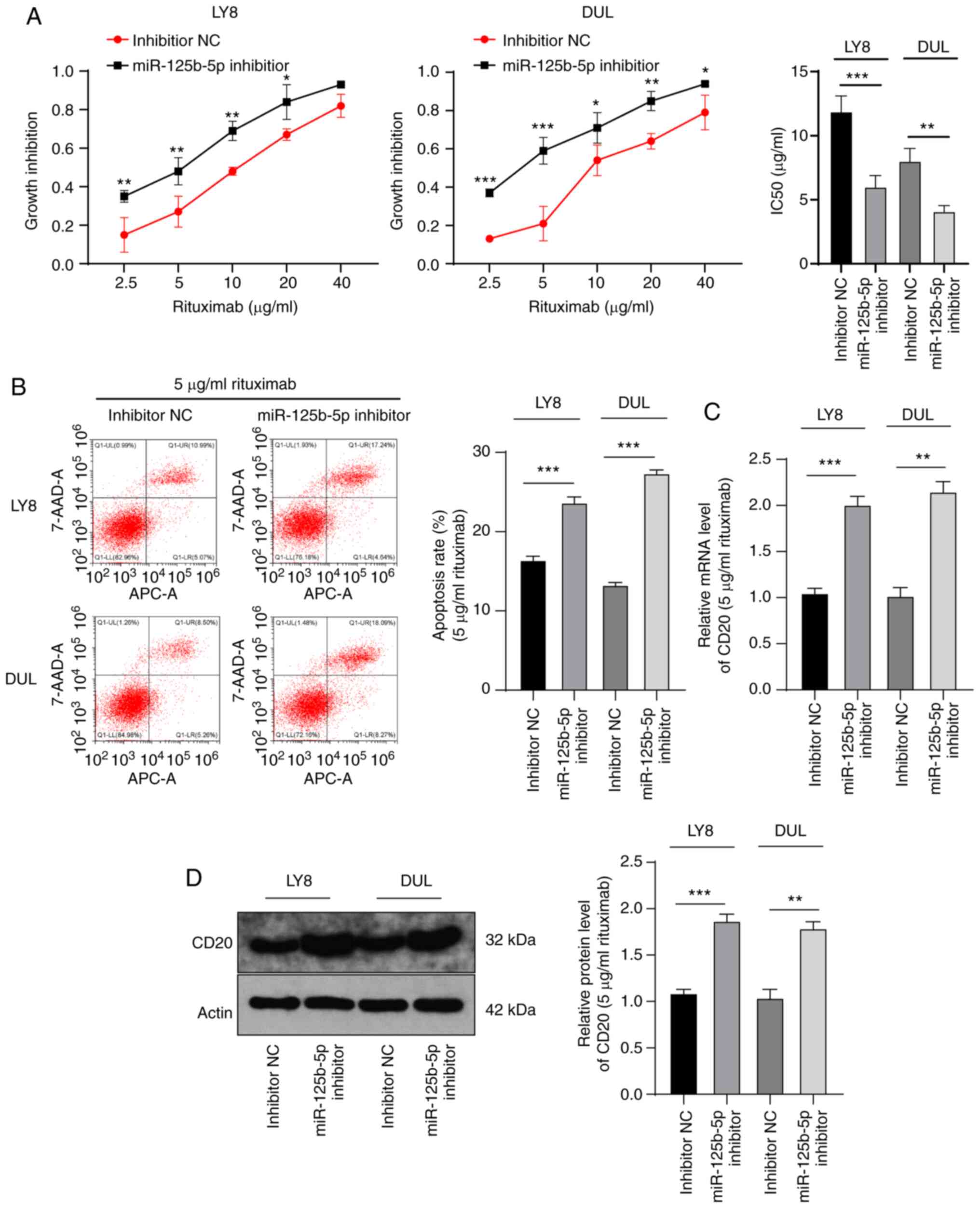
To identify the role of miR-125b-5p in anti-DLBCL
treatment, we used the miR-125b-5p inhibitor to intervene with
miR-125b-5p expression in LY8 and DUL cells, and then treated the
cells with different concentrations of rituximab. The results of
the MTT assay revealed that the IC50 value for cells with low
miR-125b-5p expression was notably reduced compared with that for
NC cells (LY8: 11.89±1.21 vs. 6.01±0.89 µg/ml; DUL:
8.02±0.98 vs. 4.09±0.45 µg/ml) (all P<0.001) (Fig. 4A). The DLBCL cells treated with 5
µg/ml rituximab and miR-125b-5p inhibitor had a
significantly higher apoptosis rate than the NC cells (P<0.001)
(Fig. 4B). Furthermore, CD20
expression in these cells was significantly upregulated
(P<0.001) (Fig. 4C and D).
These results suggested that miR-125b-5p downregulation increased
DLBCL sensitivity to rituximab.
EVs carry miR-125b-5p into DLBCL
cells
Tumor EVs are important mediators for intercellular
communication between tumor cells and other cells located, not only
in the microenvironment, but also at further distances (32,33). Recent findings have shown that the
miRs carried by EVs play an important role in several diseases
(34,35). To validate the role of EVs in
DLBCL resistance to rituximab, we first isolated EVs from DLBCL
cells and observed their morphology. All EVs exhibited the classic
tea tray shape (Fig. 5A). Western
blotting results confirmed that the EV surface markers CD9 and CD63
were expressed in EVs isolated from SUD cells, while the
endoplasmic reticulum marker calnexin was not expressed (Fig. 5B). NTA analysis revealed that EVs
ranged from 45 to 165 nm in size, with an average size of 112 nm
(Fig. 5C). These results
indicated that EVs were successfully isolated from SUD cells.
miR-125b-5p expression in the EV group was significantly higher
than that in the GW4869 group, while there was no difference in
expression between the EV and RNase groups (all P<0.001)
(Fig. 5D), indicating that
miR-125b-5p was carried in SUD cell-derived EVs (SUD-EVs). EVs were
successfully internalized by LY8 and DUL cells after incubation
with the PKH-26-labeled SUD-EVs (Fig.
5E), resulting in significantly increased miR-125b-5p
expression (all P<0.001) (Fig.
5F) and significantly decreased TNFAIP3 expression (P<0.001)
(Fig. 5G and H). These results
suggested that EVs can be internalized by DLBCL cells, carrying
miR-125b-5p into DLBCL cells to upregulate miR-125b-5p expression.
Additionally, miR-125b-5p can target TNFAIP3.
EV-carried miR-125b-5p reduces DLBCL
sensitivity to rituximab
Next, EV-treated LY8 and DUL cells were incubated
with different concentrations of rituximab. Results of the MTT
assay demonstrated that the IC50 value for EV-treated LY8 and DUL
cells was significantly enhanced (LY8: 11.32±1.32 vs. 25.21±1.23
µg/ml; DUL: 8.12±1.39 vs. 18.21±1.21 µg/ml) (all
P<0.001) (Fig. 6A). Next, we
calculated the apoptotic rate of cells treated with 5 µg/ml
rituximab combined with EVs or GW4869. The apoptotic rate and CD20
expression in the EV group were significantly decreased (all
P<0.01) (Fig. 6C-E). To
further verify whether the miR-125b-5p carried by EVs is involved
in DLBCL sensitivity to rituximab, we employed the miR-125b-5p
inhibitor to reduce miR-125b-5p expression in SUD cells, extracted
the EVs, and found that miR-125b-5p expression in the EVs was
reduced (Fig. 6F). Then, the
obtained EVs were incubated with LY8 and DUL cells and treated with
5 µg/ml rituximab. miR-125b-5p expression in the LY8 and DUL
cells incubated with the obtained miR-125b-5p-inhibitor-EVs was
reduced (Fig. 6G), and TNFAIP3
expression was significantly increased (P<0.01) (Fig. 6H and I). Moreover, the activity of
these cells was significantly decreased (Fig. 6B) and the apoptosis rate (Fig. 6C) and CD20 expression level
(Fig. 6D and E) were
significantly increased (all P<0.01). In summary, the
miR-125b-5p carried by EVs reduced the sensitivity of DLBCL to
rituximab.
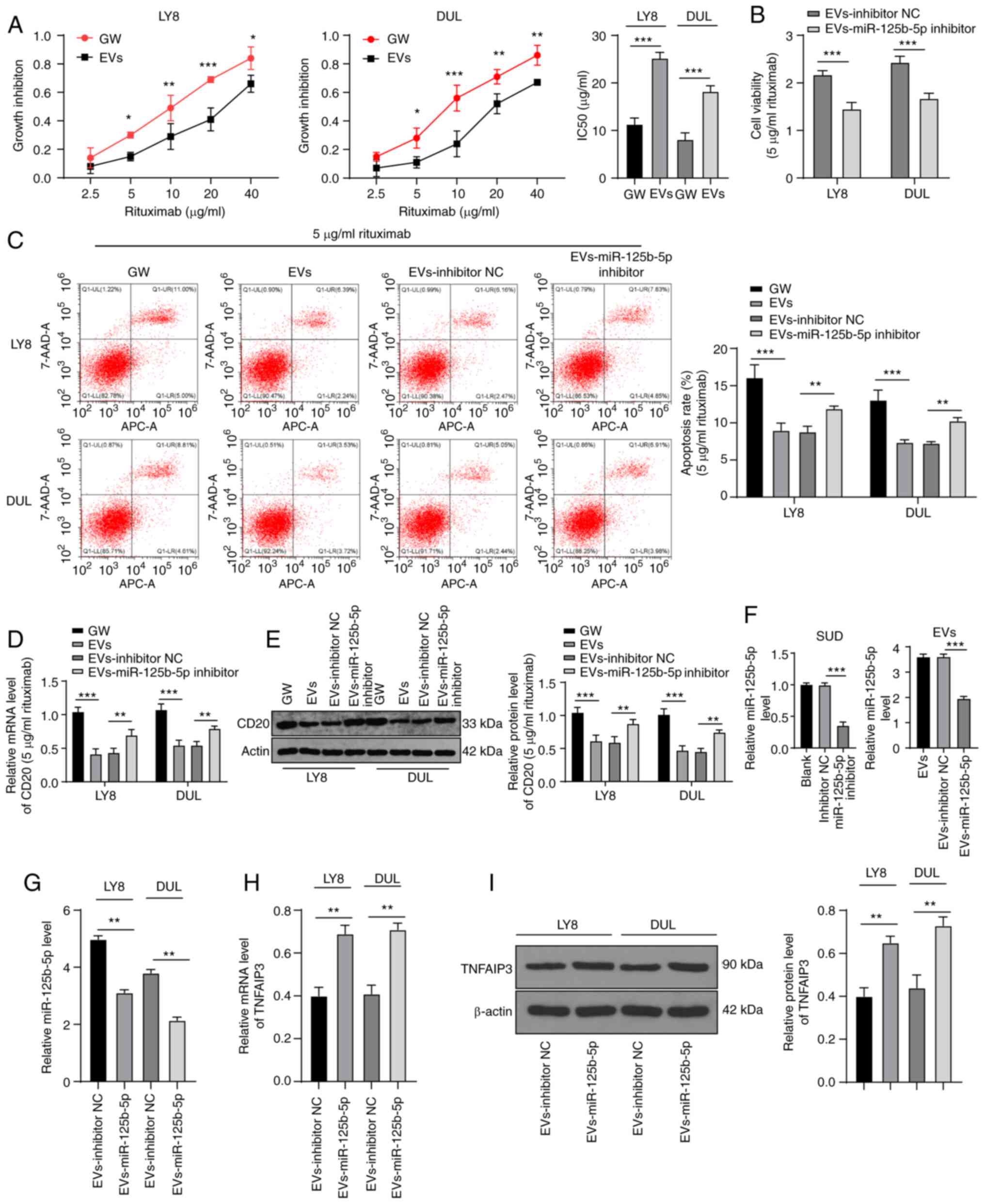 | Figure 6Extracellular vesicles (EVs) carrying
miR-125b-5p reduce the sensitivity of DLBCL to rituximab.
miR-125b-5p inhibitor was transfected into SUD cells, EVs were
extracted, and then used with rituximab to treat LY8 and DUL cells.
(A) Growth inhibition rate and IC50 value of DLBCL cells in each
group, calculated using the MTT assay. (B) DLBCL cell viability
determined using the MTT assay. (C) The apoptosis rate of DLBCL
cells determined with flow cytometry. (D and E) CD20 expression in
DLBCL, analyzed with RT-qPCR and western blotting. (F and G)
miR-125b-5p expression in SUD, EVs, and DLBCL, analyzed with
RT-qPCR. (H and I) TNFAIP3 levels in DLBCL cells, analyzed with
RT-qPCR and western blotting. The experiments were performed three
times. Data are expressed as means ± standard deviations. One-way
ANOVA was used for comparisons among multiple groups, followed by
Tukey's multiple comparisons test; t-test was used for comparisons
between two groups in panels A/B/G/H/I. *P<0.05,
**P<0.01, ***P<0.001. |
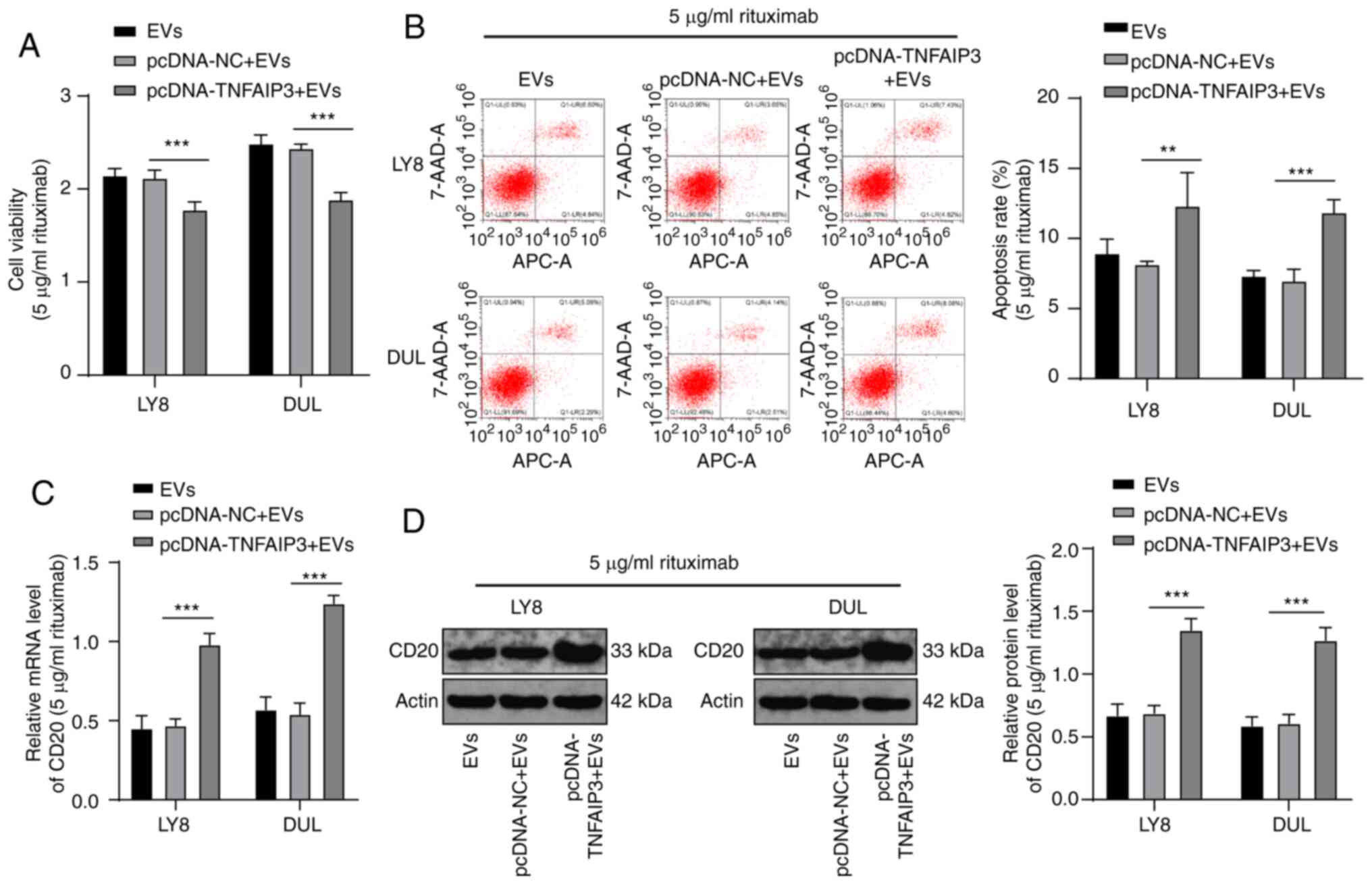
Overexpression of TNFAIP3 increases the
sensitivity of EV-treated DLBCL cells to rituximab
Next, a combined experiment was performed to verify
that the EV-carried miR-125b-5p increased DLBCL resistance to
rituximab by affecting TNFAIP3. The EVs isolated from LY8 cells
were incubated with LY8 and DUL cells transfected with
pcDNA-TNFAIP3 or pcDNA-NC, followed by treatment with 5
µg/ml rituximab. Compared with that of the NC groups, the
activity of cells in the pcDNA-TNFAIP3 + EV group was significantly
decreased (Fig. 7A) and the
apoptosis rate (Fig. 7B) and CD20
expression level were significantly increased (all P<0.001)
(Fig. 7C and D). These results
suggested that overexpression of TNFAIP3 enhanced the sensitivity
of EV-treated DLBCL to rituximab.
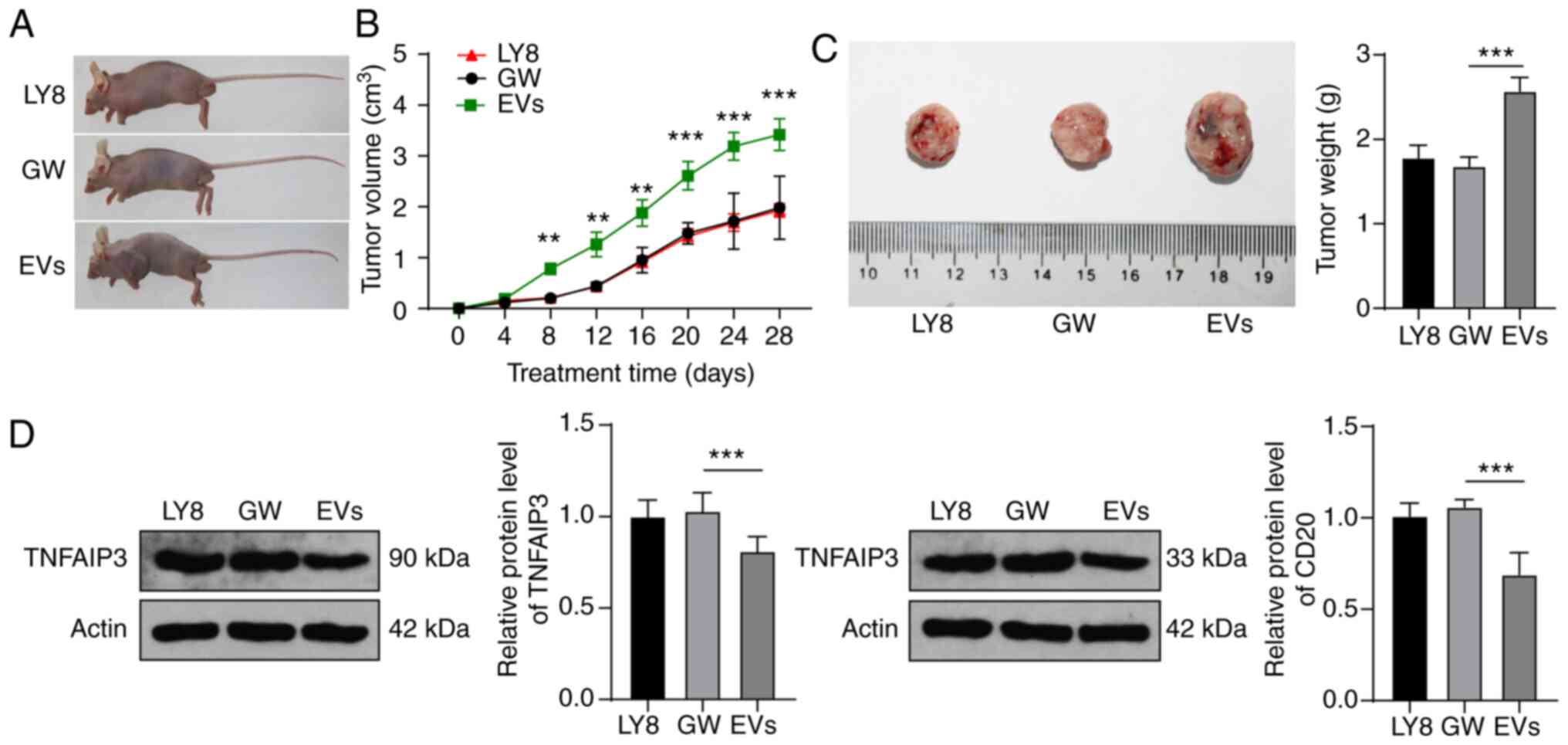
EVs reduce DLBCL sensitivity to rituximab
via the miR-125b-5p/TNFAIP3 axis
Based on the evidence that EVs can increase the
resistance of DLBCL cells to rituximab, we further verified this
effect in a mouse model. Subcutaneous injection of LY8 cells into
mice resulted in the localized formation of tumor tissue masses.
Mice were treated with EVs, GW4869 medium, and rituximab, and their
tumors were monitored and finally extracted for measurement
(Fig. 8A). In the GW group,
rituximab significantly inhibited tumor growth, while the volume
and weight of tumors in the EV group were significantly increased
(all P<0.05) (Fig. 8B and C).
In addition, the expression levels of TNFAIP3 and CD20 in the EV
group were significantly lower than those in the GW group (all
P<0.01) (Fig. 8D).
Collectively, the results indicated that EVs can reduce the
sensitivity of DLBCL mice to rituximab through the
miR-125b-5p/TNFAIP3 axis.
Discussion
TNFAIP3 is frequently inactivated in B-cell
lymphomas (24), and in
particular, is deleted in 18-50% of DLBCL cases (8). miRs transported by EVs have received
great attention due to their roles in intercellular communication
in B-cell lymphomagenesis, possibly by targeting downstream mRNAs
(36). Therefore, we hypothesized
that TNFAIP3 is targeted by an exosomal shuttled miR in DLBCL
cells. As expected, the results of our in vitro and in
vivo experiments supported the finding that tumor-derived EVs
released miR-125b-5p into DLBCL cells, which targeted TNFAIP3, thus
reducing DLBCL sensitivity to rituximab.
Previous findings have emphasized that TNFAIP3
alterations are involved in DLBCL pathogenesis (7,37).
In the present study, TNFAIP3 expression levels in DLBCL patients
were lower than those in control individuals, and were further
decreased in DLBCL-R patients, which concurred with the results of
a previous study in which TNFAIP3 insufficiency was associated with
negative prognosis of DLBCL (8).
Then, TNFAIP3 expression in DLBCL cells was enhanced by
transfection with pcDNA, followed by treatment with different
concentrations of rituximab. The IC50 value for
TNFAIP3-overexpressing cells was notably reduced and the apoptosis
rate was increased. The expression of CD20 in EVs released from
B-cell lymphoma cells has been reported to be negatively correlated
with rituximab treatment (27,38). In the current study, CD20
expression in the TNFAIP3-overexpressing DLBCL cells was
significantly increased. TNFAIP3-deficient cells were previously
shown to stably generate B-cell lymphomas in immunodeficient mice,
whereas tumorigenicity was effectively blocked by TNFAIP3
restoration (39). Of note,
TNFAIP3 deletion has been reported to be marginally associated with
favorable prognosis in rituximab-treated populations (9). Overall, overexpression of TNFAIP3
increased the sensitivity of DLBCL to rituximab.
The risk of drug resistance is reportedly 2.14-fold
higher in DLBCL patients with abnormal miR expression (14). We predicted that miR-125b-5p would
be involved in the upstream mechanism of TNFAIP3 in DLBCL
resistance to rituximab, based on its extensive involvement in
cancer and drug resistance (30,31). miR-125b-5p expression in DLBCL-R
and DLBCL patients was higher than that in the control individuals.
miR-125b and miR-125b-5p are both reportedly upregulated in
rituximab-chemoresistant patients (40,41), and miR-125b reportedly inhibits
TNFAIP3 in DLBCL (42). The
dual-luciferase experiments in the current study verified the
targeted binding relationship between TNFAIP3 and miR-125b-5p.
Subsequently, miR-125b-5p expression in DLBCL cells was suppressed
using the miR-125b-5p inhibitor and the cells were treated with
different concentrations of rituximab. The IC50 value for cells
with low miR-125b-5p expression was notably reduced, while the
apoptosis rate and CD20 expression level were increased. miR-125b
silencing is required for normal B-cell development (43). Indeed, miR-125b was reportedly
upregulated in doxorubicin-resistant Ewing sarcoma, while miR-125b
knockdown enhanced sensitivity to doxorubicin (44). Rituximab-resistant DLBCL in
patients with miR-125b overexpression is more likely to be
refractory to other chemotherapy regimens (41). Briefly, miR-125b-5p downregulation
sensitized DLBCL cells to rituximab.
Tumor-derived exosomal miRs play key roles in tumor
chemoresistance (45). Increasing
evidence supports the significance of EVs in DLBCL progression and
response or resistance to therapies (13). We hypothesized that miR-125b-5p
may be released from SUD cell-derived EVs. Our results demonstrated
that miR-125b-5p expression in the EV group was higher than that in
the GW4869 group, with no difference observed between the EV and
RNase groups, indicating that miR-125b-5p was released by EVs.
Exosomal miR-125b-5p is also described as a potential prognostic
predictor of chemoresistance in the serum of patients with DLBCL
(40). Furthermore, the IC50
value for EV-treated DLBCL cells was significantly enhanced, while
the apoptosis rate and CD20 expression were notably decreased. In
summary, EVs can be internalized by DLBCL cells, carrying
miR-125b-5p that upregulates miR-125b-5p expression, thus reducing
DLBCL sensitivity to rituximab.
Next, a combined experiment was performed to verify
that the miR-125b-5p carried by EVs increased DLBCL resistance to
rituximab by affecting TNFAIP3. pcDNA-transfected DLBCL cells were
treated with LY8-EVs and rituximab, resulting in decreased activity
and enhanced apoptosis rate and CD20 expression. B-cell
lymphoma-derived EVs carry the CD20 target antigen and act as bait,
enabling lymphoma cells to evade immunotherapy (38). These findings suggest that
overexpression of TNFAIP3 can enhance the sensitivity of EV-treated
DLBCL to rituximab. Moreover, rituximab significantly inhibited
tumor growth in vivo, the effects of which were annulled by
EV + rituximab treatment. B-cell lymphoma-derived EVs have been
reported to rescue lymphoma cells from the complement-dependent
cytotoxicity induced by rituximab (46). In addition, enhanced cytolytic
activity against rituximab-treated tumor cells has been observed
after the removal of lymphoma cell EVs (38). In the current study, the
expression levels of TNFAIP3 and CD20 in the EV group were lower
than those in the GW group. Taken together, our results support the
finding that EVs reduced the sensitivity of DLBCL model mice to
rituximab through the miR-125b-5p/TNFAIP3 axis.
Results of the present study demonstrated that the
EVs carrying miR-125b-5p can reduce DLBCL sensitivity to rituximab
by inhibiting TNFAIP3 expression. However, the regulatory mechanism
of the miR-125b-5p/TNFAIP3 axis in the sensitivity of DLBCL to
rituximab requires further investigation. Whether the
miR-125b-5p/TNFAIP3 axis can be used as a therapeutic approach for
DLBCL also requires further study. Future studies aim to focus on
the pathways downstream of the miR-125b-5p/TNFAIP3 axis that
influence the sensitivity of DLBCL to rituximab.
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
LZ conceived and designed the study. SXZ, TJZ, XML
and JLT contributed to research, conducting of experiments,
analysis and review of data, as well as the writing and editing of
the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Study approval was obtained from the Ethics
Committee of The Affiliated Hospital of Southwest Medical
University (no. 20180306006). The clinical registration number is
KY2019091. Patient consent was obtained from the patients. Animal
experiments were implemented on the guide for the care and use of
laboratory animals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
This study was supported by The Key Research Project from
Science and Technology Department of Sichuan Province (no.
2019YFS0301); Doctor Research Foundation of The Affiliated Hospital
of Southwest Medical University (nos. 19032 and 19079); The Key
Research Project from Health and Family Planning Commission of
Sichuan Province (no. 18ZD014); Applied Basic Research Project of
Luzhou Science and Technology Bureau (2019LZXNYDJ54).
References
|
1
|
Harker-Murray PD, Pommert L and Barth MJ:
Novel therapies potentially available for pediatric B-cell
non-hodgkin lymphoma. J Natl Compr Canc Netw. 18:1125–1134. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Allen P: Diffuse large B-cell lymphoma in
the elderly: Current approaches. Curr Oncol Rep. 22:1142020.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu Y and Barta SK: Diffuse large B-cell
lymphoma: 2019 update on diagnosis, risk stratification, and
treatment. Am J Hematol. 94:604–616. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Maurer MJ, Jais JP, Ghesquieres H, Witzig
TE, Hong F, Haioun C, Thompson CA, Thieblemont C, Micallef IN,
Porrata LF, et al: Personalized risk prediction for event-free
survival at 24 months in patients with diffuse large B-cell
lymphoma. Am J Hematol. 91:179–184. 2016. View Article : Google Scholar :
|
|
5
|
Giulino L, Mathew S, Ballon G, Chadburn A,
Barouk S, Antonicelli G, Leoncini L, Liu YF, Gogineni S, Tam W and
Cesarman E: A20 (TNFAIP3) genetic alterations in EBV-associated
AIDS-related lymphoma. Blood. 117:4852–4854. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hovelmeyer N, Reissig S, Xuan NT,
Adams-Quack P, Lukas D, Nikolaev A, Schluter D and Waisman A: A20
deficiency in B cells enhances B-cell proliferation and results in
the development of autoantibodies. Eur J Immunol. 41:595–601. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fujii M, Takata K, Chuang SS,
Miyata-Takata T, Ando M, Sato Y and Yoshino T: A20 (TNFAIP3)
alterations in primary intestinal diffuse large B-cell lymphoma.
Acta Med Okayama. 72:23–30. 2018.PubMed/NCBI
|
|
8
|
Dong G, Chanudet E, Zeng N, Appert A, Chen
YW, Au WY, Hamoudi RA, Watkins AJ, Ye H, Liu H, et al: A20,
ABIN-1/2, and CARD11 mutations and their prognostic value in
gastrointestinal diffuse large B-cell lymphoma. Clin Cancer Res.
17:1440–1451. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Paik JH, Go H, Nam SJ, Kim TM, Heo DS, Kim
CW and Jeon YK: Clinicopathologic implication of A20/TNFAIP3
deletion in diffuse large B-cell lymphoma: An analysis according to
immunohistochemical subgroups and rituximab treatment. Leuk
Lymphoma. 54:1934–1941. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cappariello A and Rucci N: Tumour-derived
extracellular vesicles (EVs): A dangerous 'message in a bottle' for
bone. Int J Mol Sci. 20:48052019. View Article : Google Scholar
|
|
11
|
Urabe F, Kosaka N, Ito K, Kimura T, Egawa
S and Ochiya T: Extracellular vesicles as biomarkers and
therapeutic targets for cancer. Am J Physiol Cell Physiol.
318:C29–C39. 2020. View Article : Google Scholar
|
|
12
|
Liu W, Zhu M, Wang H, Wang W and Lu Y:
Diffuse large B cell lymphoma-derived extracellular vesicles
educate macrophages to promote tumours progression by increasing
PGC-1β. Scand J Immunol. 91:e128412020. View Article : Google Scholar
|
|
13
|
Rutherford SC, Fachel AA, Li S, Sawh S,
Muley A, Ishii J, Saxena A, Dominguez PM, Caldas Lopes E, Agirre X,
et al: Extracellular vesicles in DLBCL provide abundant clues to
aberrant transcriptional programming and genomic alterations.
Blood. 132:e13–e23. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ting CY, Liew SM, Price A, Gan GG, Bee-Lan
Ong D, Tan SY and Bee PC: Clinical significance of aberrant
microRNAs expression in predicting disease relapse/refractoriness
to treatment in diffuse large B-cell lymphoma: A meta-analysis.
Crit Rev Oncol Hematol. 144:1028182019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xu ZZ, Wang WF, Fu WB, Wang AH, Liu ZY,
Chen LY, Guo P and Li JM: Combination of rituximab and mammalian
target of rapamycin inhibitor everolimus (RAD001) in diffuse large
B-cell lymphoma. Leuk Lymphoma. 55:1151–1157. 2014. View Article : Google Scholar
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
17
|
Li S, Young KH and Medeiros LJ: Diffuse
large B-cell lymphoma. Pathology. 50:74–87. 2018. View Article : Google Scholar
|
|
18
|
Sarkozy C and Sehn LH: Management of
relapsed/refractory DLBCL. Best Pract Res Clin Haematol.
31:209–216. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tamma R, Ranieri G, Ingravallo G, Annese
T, Oranger A, Gaudio F, Musto P, Specchia G and Ribatti D:
Inflammatory cells in diffuse large B cell lymphoma. J Clin Med.
9:24182020. View Article : Google Scholar :
|
|
20
|
Habermann TM, Weller EA, Morrison VA,
Gascoyne RD, Cassileth PA, Cohn JB, Dakhil SR, Woda B, Fisher RI,
Peterson BA and Horning SJ: Rituximab-CHOP versus CHOP alone or
with maintenance rituximab in older patients with diffuse large
B-cell lymphoma. J Clin Oncol. 24:3121–3127. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mounier N, Briere J, Gisselbrecht C, Emile
JF, Lederlin P, Sebban C, Berger F, Bosly A, Morel P, Tilly H, et
al: Rituximab plus CHOP (R-CHOP) overcomes bcl-2-associated
resistance to chemotherapy in elderly patients with diffuse large
B-cell lymphoma (DLBCL). Blood. 101:4279–4284. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Honma K, Tsuzuki S, Nakagawa M, Tagawa H,
Nakamura S, Morishima Y and Seto M: TNFAIP3/A20 functions as a
novel tumor suppressor gene in several subtypes of non-Hodgkin
lymphomas. Blood. 114:2467–2475. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Schmitz R, Hansmann ML, Bohle V,
Martin-Subero JI, Hartmann S, Mechtersheimer G, Klapper W, Vater I,
Giefing M, Gesk S, et al: TNFAIP3 (A20) is a tumor suppressor gene
in Hodgkin lymphoma and primary mediastinal B cell lymphoma. J Exp
Med. 206:981–989. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vereecke L, Beyaert R and van Loo G: The
ubiquitin-editing enzyme A20 (TNFAIP3) is a central regulator of
immunopathology. Trends Immunol. 30:383–391. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen S, Xing H, Li S, Yu J, Li H, Liu S,
Tian Z, Tang K, Rao Q, Wang M and Wang J: Up-regulated A20 promotes
proliferation, regulates cell cycle progression and induces
chemotherapy resistance of acute lymphoblastic leukemia cells. Leuk
Res. 39:976–983. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
da Silva CG, Minussi DC, Ferran C and
Bredel M: A20 expressing tumors and anticancer drug resistance. Adv
Exp Med Biol. 809:65–81. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Palanichamy A, Jahn S, Nickles D, Derstine
M, Abounasr A, Hauser SL, Baranzini SE, Leppert D and von Budingen
HC: Rituximab efficiently depletes increased CD20-expressing T
cells in multiple sclerosis patients. J Immunol. 193:580–586. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42:D92–D97. 2014. View Article : Google Scholar
|
|
30
|
Liu S, Chen Q and Wang Y: MiR-125b-5p
suppresses the bladder cancer progression via targeting HK2 and
suppressing PI3K/AKT pathway. Hum Cell. 33:185–194. 2020.
View Article : Google Scholar
|
|
31
|
Yao J, Li Z, Wang X, Xu P, Zhao L and Qian
J: MiR-125a regulates chemo-sensitivity to gemcitabine in human
pancreatic cancer cells through targeting A20. Acta Biochim Biophys
Sin (Shanghai). 48:202–208. 2016. View Article : Google Scholar
|
|
32
|
Kara-Terki L, Treps L, Blanquart C and
Fradin D: Critical roles of tumor extracellular vesicles in the
microenvironment of thoracic cancers. Int J Mol Sci. 21:60242020.
View Article : Google Scholar :
|
|
33
|
Maisano D, Mimmi S, Russo R, Fioravanti A,
Fiume G, Vecchio E, Nistico N, Quinto I and Iaccino E: Uncovering
the exosomes diversity: A window of opportunity for tumor
progression monitoring. Pharmaceuticals (Basel). 13:1802020.
View Article : Google Scholar
|
|
34
|
Gerloff D, Lutzkendorf J, Moritz RKC,
Wersig T, Mader K, Muller LP and Sunderkotter C: Melanoma-derived
exosomal miR-125b-5p educates tumor associated macrophages (TAMs)
by targeting lysosomal acid lipase A (LIPA). Cancers (Basel).
12:4642020. View Article : Google Scholar
|
|
35
|
Wu M, Tan X, Liu P, Yang Y, Huang Y, Liu
X, Meng X, Yu B, Wu Y and Jin H: Role of exosomal microRNA-125b-5p
in conferring the metastatic phenotype among pancreatic cancer
cells with different potential of metastasis. Life Sci.
255:1178572020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Drees EEE and Pegtel DM: Circulating
miRNAs as biomarkers in aggressive B cell lymphomas. Trends Cancer.
6:910–923. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yang W, Li Y, Li P and Wang L: PMA/IONO
affects diffuse large B-cell lymphoma cell growth through
upregulation of A20 expression. Oncol Rep. 36:1069–1075. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Aung T, Chapuy B, Vogel D, Wenzel D,
Oppermann M, Lahmann M, Weinhage T, Menck K, Hupfeld T, Koch R, et
al: Exosomal evasion of humoral immunotherapy in aggressive B-cell
lymphoma modulated by ATP-binding cassette transporter A3. Proc
Natl Acad Sci USA. 108:15336–15341. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kato M, Sanada M, Kato I, Sato Y, Takita
J, Takeuchi K, Niwa A, Chen Y, Nakazaki K, Nomoto J, et al:
Frequent inactivation of A20 in B-cell lymphomas. Nature.
459:712–716. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Feng Y, Zhong M, Zeng S, Wang L, Liu P,
Xiao X and Liu Y: Exosome-derived miRNAs as predictive biomarkers
for diffuse large B-cell lymphoma chemotherapy resistance.
Epigenomics. 11:35–51. 2019. View Article : Google Scholar
|
|
41
|
Yuan WX, Gui YX, Na WN, Chao J and Yang X:
Circulating microRNA-125b and microRNA-130a expression profiles
predict chemoresistance to R-CHOP in diffuse large B-cell lymphoma
patients. Oncol Lett. 11:423–432. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kim SW, Ramasamy K, Bouamar H, Lin AP,
Jiang D and Aguiar RC: MicroRNAs miR-125a and miR-125b
constitutively activate the NF-kB pathway by targeting the tumor
necrosis factor alpha-induced protein 3 (TNFAIP3, A20). Proc Natl
Acad Sci USA. 109:7865–7870. 2012. View Article : Google Scholar
|
|
43
|
Li G, So AY, Sookram R, Wong S, Wang JK,
Ouyang Y, He P, Su Y, Casellas R and Baltimore D: Epigenetic
silencing of miR-125b is required for normal B-cell development.
Blood. 131:1920–1930. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Iida K, Fukushi J, Matsumoto Y, Oda Y,
Takahashi Y, Fujiwara T, Fujiwara-Okada Y, Hatano M, Nabashima A,
Kamura S and Iwamoto Y: miR-125b develops chemoresistance in Ewing
sarcoma/primitive neuroectodermal tumor. Cancer Cell Int.
13:212013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhao L, Liu W, Xiao J and Cao B: The role
of exosomes and 'exosomal shuttle microRNA' in tumorigenesis and
drug resistance. Cancer Lett. 356:339–346. 2015. View Article : Google Scholar
|
|
46
|
Oksvold MP, Kullmann A, Forfang L, Kierulf
B, Li M, Brech A, Vlassov AV, Smeland EB, Neurauter A and Pedersen
KW: Expression of B-cell surface antigens in subpopulations of
exosomes released from B-cell lymphoma cells. Clin Ther. 36:847–862
e841. 2014. View Article : Google Scholar : PubMed/NCBI
|