1. Introduction
The gut microbiota (GM) is composed of >100
trillion of microorganisms (including bacteria, viruses, protozoa
and fungi) resident in the gastro-intestinal lumen, mainly the
large intestine (1). Among all,
bacteria represent the broader category, with thousands of
different species, belonging to the Firmicutes and Bacteroidetes
phyla in particular (2). Gut
microbes have been widely studied given their key role in the
modulation of both the host homeostasis and pathology (3). The main functions of the GM include
the following: i) The maintenance of the host's gut health; ii)
gastrointestinal barrier function; and iii) the neo-synthesis or
transformation of dietary compounds and essential nutrients
(4-6). The fulfillment of all the
aforementioned activities suggests the establishment of a
functional two-way association between GM and the host immune
system (7). This interrelation
guarantees the preservation of a microbial balance or eubiosis
(7). However, the absence of such
an equilibrium (with the concurrent depletion of the gut
microflora) is termed dysbiosis and is associated with a number of
pathologies, including diabetes, inflammation, autoimmune disorders
and cancer (8).
In order to examine the impact of GM on human
health, it is important to characterize what is known as the gut
microbiome, corresponding to the entire genome of the whole GM. The
gut microbiome accounts for 100-fold more genes than the entire
human genome (9). Presently, the
advent of metagenomics allows the transition from merely depicting
the microbiome composition to functionally analyzing the impact of
the microbiome balance vs. imbalance in human health (10-12). The computational analysis of the
16S rRNA amplicons coupled with next-generation sequencing (NGS)
allows the characterization of both the abundance and diversity of
the gut microbiome. Overall, metagenomics, together with
metatranscriptomics, metabolomics and proteomics help to quantify
the impact of specific bacterial species on human health (13-15).
The reconstruction of the whole bacterial genomes
beginning from metagenomic datasets is currently helping to
identify uncharacterized bacterial species, both from the gut and
other body sites, thus expanding the known phylogenetic diversity.
The recent study from Almeida et al (16) established the ultimately most
comprehensive collection of microbial genomes composing the human
gut microbiome, comprised of >200,000 of non-redundant genomes
from 4,644 gut prokaryotes. This will allow their use as a
reference in future metagenomics studies (16). Given the tight interconnection
occurring between the gut microbiome and the human host, it is of
pivotal importance to identify all the host-dependent variables
(e.g., physiology, lifestyle habits and diet), which alter the GM
to further increase both robustness and the reproducibility of
metadata analyses (17). This
will help to identify the members of the GM that are directly
associated with human diseases, including cancer, and ultimately
with the response to anticancer therapy (7).
Cancer is a leading cause of mortality worldwide,
second only to cardiovascular diseases, accounting for almost 10
million deaths in 2020 (18).
Over the past 10 years, anticancer immunotherapy has taken the
central stage in the treatment of a variety of tumors (19). In general, immunotherapy targets
immune cells in order to activate or boost their capacity of
eliminating cancerous cells. Malignant cells are surrounded by a
variety non-cancer cells, including stromal cells and immune cells
[e.g., macrophages, dendritic cells (DCs), natural killer (NK)
cells, T-cells and B-cells], together forming the so-called tumor
microenvironment (20,21). The 'immune contexture' of a given
tumor plays a key role in both the prognosis and treatment of
cancer patients. Despite the presence of cancer immune-evasion
mechanisms, any residual anticancer immune response may suggest a
better prognosis. Moreover, anticancer therapies triggering the
'immune contexture' may produce a more durable anticancer efficacy
(22).
Among the immunotherapies which are currently tested
in clinical practice, immune checkpoint inhibitors (ICIs) have been
shown to efficiently reshape the host immune response against
cancer (23). The discovery of
immune checkpoints led to the designation of the 2018 Nobel Prize
in Physiology or Medicine to the two scientists, James Allison and
Tasuku Honjo (24).
ICIs are monoclonal antibodies designed to inhibit
the immune checkpoint pathway, thereby boosting the host immune
system to efficiently eliminate cancer cells (25). In detail, ICIs trigger cytotoxic
CD8+ T-cells to destroy target malignant cells, thereby
reactivating the cancer immunity cycle (26). In fact, the immune checkpoints are
co-receptors of the T receptor signaling complex, which overall
prevent T-cell overactivation and establish the tolerance against
self-antigens (27). The
suppression of T-cell activation via these co-receptors constitutes
the main immune escape mechanism carried out by neoplastic cells.
In fact, when cancer cells efficiently activate the immune
checkpoint, they may evade the immune system and overgrow (28). However, the administration of ICIs
blocks these inhibitory co-receptors and positively modulates
CD8+ T-cell cytotoxicity, directing it to effectively
eliminating malignant cells (28).
Approved ICIs can target two main co-inhibitory
routes: Either the programmed cell death protein 1 (PD-1) or the
cytotoxic T-lymphocyte antigen-4 (CTLA-4) pathway (29). PD-1 is expressed principally by
T-cells and other immune cells (including NK cells, DCs and
B-cells), whereas its ligand, programmed cell death ligand 1
(PD-L1) is expressed by the antigen-presenting cells (APCs),
including malignant cells (30).
Thus, antibodies directed against PD-1 or PD-L1 block the
inhibitory immune checkpoint interaction between CD8+
T-cells and tumor cells, rehabilitating cytotoxic T-cells to
efficiently eliminate target cancer cells (31). Instead, CTLA4 is a receptor
expressed by T- and B-cells, and inhibits the binding of CD28
receptor with its B7 ligand, expressed by APCs at the earlier
phases of antigen presentation (32). Consequently, ICIs targeting CTLA4
receptor reactivate the cellular-mediated immune response earlier
in the cancer immunity cycle (33).
Since the first ICI was authorized by the US Food
and Drug Administration (FDA) in 2011, a wide range of ICIs has
been further approved (34). ICIs
have been presently employed for the treatment of ~50 cancer types
either as late-line, first-line, or as neoadjuvant therapies. They
are administered either as single agents or in combination (with
chemotherapy or with another ICI) (34). ICIs are currently under study in
>60% of all ongoing oncology clinical trials (for some examples
on the specific use of ICIs in oncology in relation to the gut
microbiome, please see the studies listed in Tables I and II) (35,36). Impressively, for a number of
recalcitrant and otherwise uncurable tumors, such as advanced
melanoma (AM), metastatic melanoma (MM) or advanced non-small cell
lung cancer (NSCLC), the use of ICIs has led to a substantial
long-term remission (37,38).
 | Table ICurrent ongoing clinical trials
registered at clinicaltrials.gov that analyze the intestinal
microbiota through metagenomics during immune-checkpoint
immunotherapy (alone or in combination with other anti-cancer
therapies). |
Table I
Current ongoing clinical trials
registered at clinicaltrials.gov that analyze the intestinal
microbiota through metagenomics during immune-checkpoint
immunotherapy (alone or in combination with other anti-cancer
therapies).
| Tim | Condition(s) | Anticancer
therapy | Enrollment | Start date | (Refs.) |
|---|
| NCT02600143 | Melanoma | ICIs | 123 | 2013 | n.a. |
| NCT01896999 | Hodgkin
lymphoma | Ipilimumab;
nivolumab; brentuximab | 126 | 2014 | (146) |
| NCT02478099 | Advanced solid
tumors | MPDL3280A | 98 | 2016 | (147) |
| NCT02681302 | Multiple myeloma;
lymphoma | Ipilimumab;
nivolumab | 42 | 2016 | n.a. |
| NCT04204434 | Advanced solid
tumors | ICIs | 150 | 2016 | n.a. |
| NCT02858921 | Melanoma | Dabrafenib;
trametinib; pembrolizumab | 60 | 2017 | n.a. |
| NCT03083691 | Non-small cell lung
cancer | Ipilimumab;
nivolumab | 106 | 2017 | n.a. |
| NCT03161756 | Melanoma | Ipilimumab;
nivolumab; denosumab | 72 | 2017 | n.a. |
| NCT03164993 | Breast cancer | Atezolizumab;
doxorubicin; cyclophosphamide | 75 | 2017 | (148) |
| NCT03168464 | Non-small cell lung
cancer | Ipilimumab;
nivolumab; radiotherapy | 45 | 2017 | n.a. |
| NCT03331562 | Pancreatic
cancer | Pembrolizumab | 24 | 2017 | n.a. |
| NCT03289819 | Breast cancer | Pembrolizumab;
paclitaxel; epirubicin; cyclophosphamide | 50 | 2018 | n.a. |
| NCT03688347 | Lung cancer | ICIs | 60 | 2018 | n.a. |
| NCT04054908 | Gastrointestinal
cancer | SOC, ICIs | 60 | 2018 | n.a. |
| NCT04169867 | Melanoma | Nivolumab;
ipilimumab; atezolizumab | 1160 | 2018 | n.a. |
| NCT04579978 | Advanced solid
tumors | ICIs | 60 | 2018 | n.a. |
| NCT03694834 | Endometrial
cancer | Pembrolizumab | 20 | 2019 | n.a. |
| NCT03799744 | Head and neck
cancer | VCN-01;
durvalumab | 20 | 2019 | n.a. |
| NCT03818061 | Head and neck
cancer | Atezolizumab;
bevacizumab | 110 | 2019 | n.a. |
| NCT03894007 | Breast cancer | Docetaxel;
carboplatin; trastuzumab; pertuzumab; epirubicin; cyclophosphamide;
atezolizumab | 190 | 2019 | n.a. |
| NCT04006262 | Oeso-gastric
cancer | Ipilimumab;
nivolumab | 32 | 2019 | (149) |
| NCT04013542 | Lung cancer | Ipilimumab;
nivolumab; radiotherapy | 20 | 2019 | n.a. |
| NCT04133948 | Melanoma | Nivolumab;
ipilimumab; domatinostat | 45 | 2019 | n.a. |
| NCT04136470 | Non-small cell lung
cancer; melanoma | ICIs | 130 | 2019 | n.a. |
| NCT04196465 | Gastric cancer;
esophageal cancer; liver cancer | IMC-001 | 48 | 2019 | n.a. |
| NCT04291755 | Non-small-cell lung
cancer; colorectal cancer | Pembrolizumab | 100 | 2019 | n.a. |
| NCT03977571 | Renal cell
cancer | Ipilimumab;
Nivolumab | 400 | 2020 | n.a. |
| NCT04063501 | Lung cancer | Anti-PD-1
antibodies | 80 | 2020 | n.a. |
| NCT04090710 | Renal cell
cancer | Ipilimumab;
nivolumab; radiotherapy | 78 | 2020 | n.a. |
| NCT04107168 | Melanoma; renal
cancer; lung cancer | Nivolumab;
pembrolizumab; ipilimumab; durvalumab; tremelimumab; atezolizumab;
bevacizumab | 1800 | 2020 | n.a. |
| NCT04189679 | Non-small cell lung
cancer | ICIs | 60 | 2020 | n.a. |
| NCT04207086 | Melanoma | Pembrolizumab;
lenvatinib | 20 | 2020 | n.a. |
| NCT04271384 | Non-small cell lung
cancer | Nivolumab; SOC | 30 | 2020 | n.a. |
| NCT04312308 | Non-small cell lung
cancer | Atezolizumab | 100 | 2020 | n.a. |
| NCT04333004 | Non-small cell lung
cancer (brain metastases) | Pembrolizumab;
chemotherapy | 40 | 2020 | n.a. |
| NCT04392505 | Non-small cell lung
cancer | Durvalumab | 100 | 2020 | n.a. |
| NCT04435964 | Melanoma; lung
cancer; head and neck cancer; urogenital cancer; breast cancer | ICIs | 400 | 2020 | n.a. |
| NCT04566029 | Urothelial
cancer | SOC, ICIs | 40 | 2020 | n.a. |
| NCT04636775 | Non-small cell lung
cancer | ICIs | 46 | 2020 | n.a. |
| NCT04638751 | Non-small cell lung
cancer; colorectal cancer; triple negative breast cancer; pancreas
cancer | ICIs,
chemotherapy | 4000 | 2020 | n.a. |
| NCT04680377 | Non-small cell
lung; advanced lung cancer | Durvalumab | 44 | 2020 | n.a. |
| NCT04169074 | Head and neck
cancer | Nivolumab;
abemaciclib | 20 | 2021 | n.a. |
| NCT04602078 | Urothelial
cancer | Atezolizumab;
gemcitabine; cisplatin | 66 | 2021 | n.a. |
| NCT04698161 | Non-Small cell lung
cancer; melanoma | ICIs | 50 | 2021 | n.a. |
| NCT04711330 | Non-small cell lung
cancer | Durvalumab | 126 | 2021 | n.a. |
| NCT04743752 | Non-small cell lung
cancer | ICIs | 200 | 2021 | n.a. |
| NCT04804137 | Non-small cell lung
cancer; metastatic lung cancer | ICIs | 80 | 2021 | n.a. |
 | Table IICurrent ongoing clinical trials
registered at clinicaltrials.gov using interventions to modulate
intestinal microbiota in association with immune-checkpoint
immunotherapy. |
Table II
Current ongoing clinical trials
registered at clinicaltrials.gov using interventions to modulate
intestinal microbiota in association with immune-checkpoint
immunotherapy.
| NCT Number | Condition(s) | Anticancer
therapy | Gut microbial
modulation | Enrollment | Start date |
|---|
| NCT03353402 | Melanoma | ICIs | Fecal microbiota
transplantation (from patients treated with ICIs in remission from
1 year) | 40 | 2017 |
| NCT03686202 | Advanced ssolid
tumors | ICIs | MET-4 (microbial
ecosystem therapeutics) | 65 | 2018 |
| NCT04056026 | Mesothelioma | Pembrolizumab | Fecal microbiota
transplantation (From healthy family donors) | 1 | 2018 |
| NCT03341143 | Melanoma | Pembrolizumab | Fecal microbiota
transplantation | 20 | 2018 |
| NCT03595683 | Melanoma | Pembrolizumab | EDP1503
(Bifidobacterium animalis) | 70 | 2018 |
| NCT03775850 | Colorectal cancer;
triple negative breast cancer; non-small cell lung cancer; bladder
cancer; oeso-gastric cancer; renal cell cancer | Pembrolizumab | EDP1503
(Bifidobacterium animalis) | 120 | 2018 |
| NCT03817125 | Melanoma | Nivolumab | Vancomycin
pretreatment; SER-401 (adjunctive microbiome therapy) | 14 | 2019 |
| NCT03637803 | Advanced solid
tumors | Pembrolizumab | MRx0518
(Enterococcus gallinarum) | 132 | 2019 |
| NCT03772899 | Melanoma | ICIs | Fecal microbial
transplantation (From single healthy donor) | 20 | 2019 |
| NCT04116775 | Prostate
cancer | Pembrolizumab;
enzalutamide | Fecal microbial
transplantation | 32 | 2019 |
| NCT03829111 | Renal cell
cancer | Ipilimumab;
nivolumab | CBM-588
(Clostridium butyricum) | 30 | 2019 |
| NCT04105270 | Lung cancer | Durvalumab;
cisplatin; carboplatin | Oral restorative
microbiota therapy (microbial ecosystem therapeutics) | 30 | 2020 |
| NCT04114136 | Advanced solid
tumors | Nivolumab;
pembrolizumab | Metformin;
rosiglitazone (metabolism modulatory molecules) | 108 | 2020 |
| NCT04601402 | Non-small cell lung
cancer; head and neck cancer; urothelial cancer | Avelumab | GEN-001 (Single
strain bacteria isolated from gut of healthy human volunteers) | 93 | 2020 |
| NCT04577729 | Melanoma | ICIs | Fecal microbial
transplantation (From patients treated with ICIs in remission from
1 year) | 60 | 2020 |
| NCT04130763 | Gastrointestinal
cancer | Anti-PD-1
antibodies | Fecal microbial
transplantation (From healthy donors) | 10 | 2020 |
| NCT04163289 | Renal cell
cancer | ICIs | Fecal microbial
transplantation (From healthy donors) | 20 | 2020 |
| NCT03819296 | Melanoma | Infliximab;
prednisone; vedolizumab | Fecal microbial
transplantation | 800 | 2020 |
| NCT04038619 | Genitourinary
cancer | ICIs | Fecal microbial
transplantation | 40 | 2020 |
| NCT04521075 | Melanoma | Nivolumab | Fecal microbial
transplantation | 50 | 2020 |
| NCT04758507 | Renal cell
cancer | ICIs | Fecal microbial
transplantation | 50 | 2021 |
Despite this substantial clinical success, the
administration of ICIs is accompanied by some limitations. Amongst
the reported issues, there is the modification of the main clinical
endpoints due to the often-associated delay in the appearance of
positive effects mediated by ICI-based immunotherapy (21,39). Additionally, in the majority of
cases, patients with advanced disease finally develop resistance to
ICIs, mainly due to the development of innate and adaptive
immune-resistance to the checkpoint blockade (40,41). Finally, ICI blockage may be
associated with the occurrence of a broad range of immune-related
adverse events (irAEs), caused by the potential immune and
pro-inflammatory overactivation of the host's immune system. The
reported irAEs are several, and include: Colitis, intestinal
mucositis, diarrhea, thyroiditis, hepatitis, dermatological
manifestations, pneumonitis, myocarditis and others. The outcome of
irAEs can range from mild to severe and in some cases, fatal events
occur (42,43).
The differential response of patients observed with
the use of ICIs, in terms of both efficacy and tolerability, can be
linked to the intrinsic individual diversity of the immune system
and other host-related factors (44). Thus, identifying effective methods
with which to identify the specific features of the individual
immune system and direct it to better respond to ICIs represents
the current challenge of ICI research (44). As it will be largely discussed
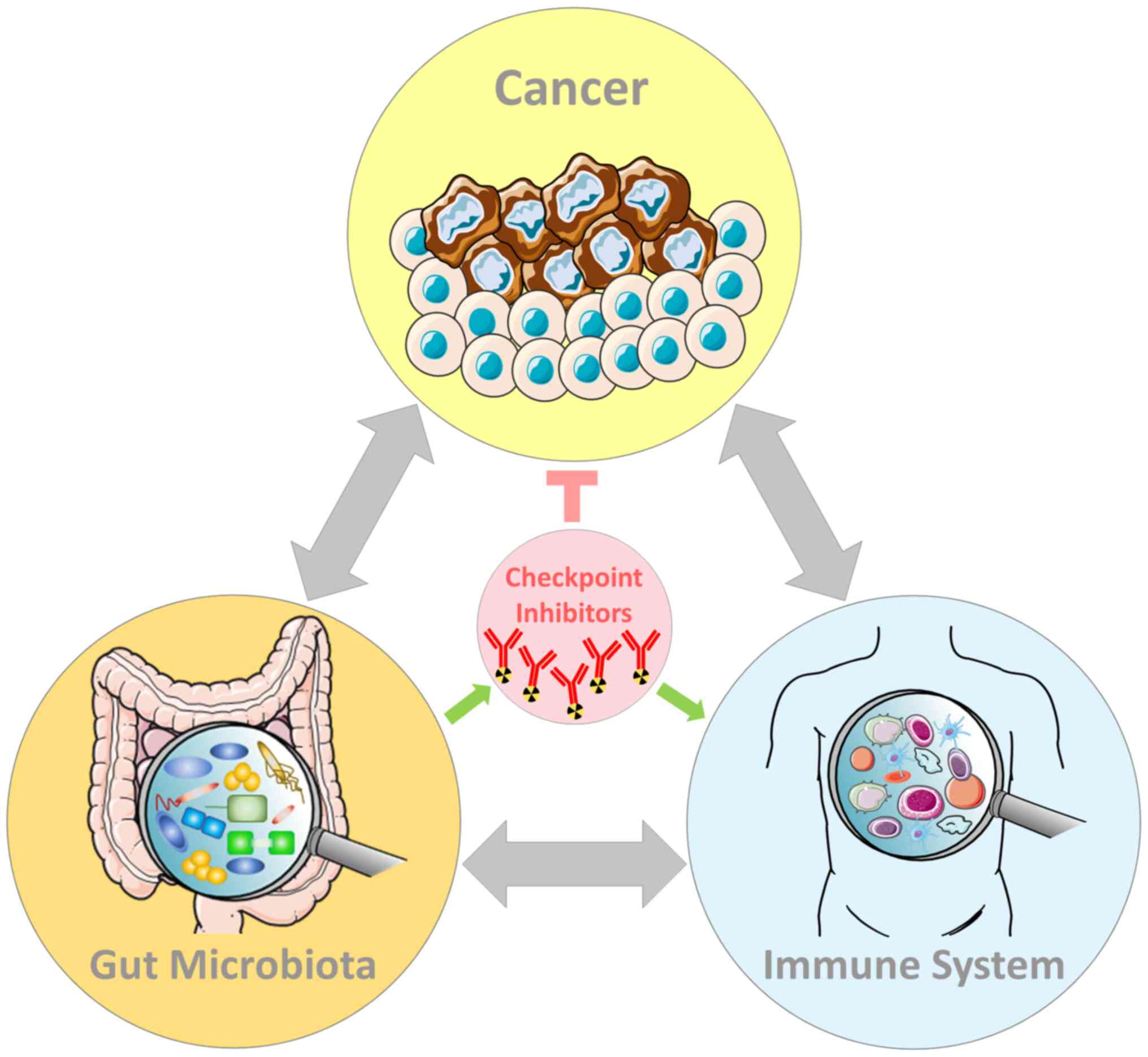
below, the GM is a master regulator of the immune system;
therefore, it directly affects both the efficacy and toxicity of
ICIs (45,46) (Fig.
1).
Consequently, the GM can be used as a powerful
source to identify novel diagnostic and prognostic
microbial-derived biomarkers, as well as innovative therapeutic
targets (47,48). In fact, recent milestone findings
have highlighted the presence of specific gut bacterial species
which are able to improve both the compliance to, as well as the
effectiveness of anticancer therapies, particularly ICIs (49). Notably, in 2021, for the first
time, to the best of our knowledge, two groundbreaking studies
demonstrated that fecal microbiota transplantation (FMT) can
efficiently boost the anticancer efficacy of ICIs in patients with
AM and MM (50,51).
The present review summarizes the up-to-date studies
on the role played by the GM in modulating the host immune system,
thus influencing both the safety and the outcome to ICI-based
anticancer therapy in cancer patients. Current findings indicate
that each individual cancer patient has a specific GM footprint.
Research efforts are presently focusing on developing effective
strategies which can be used to manipulate the GM in a
patient-tailored manner, with the aims of: i) Improving the
efficacy of ICIs; and ii) actively reducing the occurrence of irAEs
linked with ICI administration.
Compared with the existing literature on this topic,
the general aim of the present review was to provide a concise and
complete overview of the milestone studies that have contributed to
deciphering the complicated association between gut microbial
health, the host immune system and ICI activity over the past
decade. Overall, the hidden potential of treating cancer patients
with a more holistic therapeutic approach is strongly emerging,
through the administration of integrated therapies (e.g., ICIs
combined with GM modulators) tailored around the features of each
specific patient (including gut microbial composition and immune
system reactivity).
2. Gut microbiota and the host immune
system
A dynamic two-way association occurs between GM and
the host immune system through the course of a lifetime (52). The GM plays a key role in both
shaping and modulating the immune system. In turn, the immune
system regulates the gut microbial balance and it helps to maintain
a healthy gut homeostasis (53).
Any imbalance in this association could contribute to the
development of several pathological conditions, including
immune-mediated disorders, as well as cancer (54).
As regards innate immunity, since the very early
years of life, the GM composition is actively shaped by the immune
system and, in turn, the GM affects the development of the immune
system (55). It has been
demonstrated that since the maternal acquisition of the gut
microbes during childbirth, critical interactions between the GM
and the immune system may determine the establishment of both a
eubiotic GM and a fully-functional immune activity (56). Any imbalance in the GM composition
due, for example, to antibiotic-mediated depletion, may determine
the instauration of immune-related diseases which can appear later
in the adult life (such as asthma, or inflammatory bowel disease)
(57,58).
The intestinal mucosal barrier represents the
interface between the gut microbiota and the human body (59). Below the mucosal layer resides the
gut lumen which is composed of the following: i) Intestinal
epithelial cells; ii) enteroendocrine cells; and iii)
intraepithelial lymphocytes and other immune cells (60). This conserved structure allows the
interaction of the commensal GM with the host immune system,
together forming the so-called gut-immune axis (54). Although the immune system in the
intestine evolves to fight invading pathogens, there is a delicate
balance which allows the development of tolerance to non-dangerous
commensals, as well as food antigens, although it fights pathogenic
microbes that may otherwise invade the gut lumen and trespass the
gut barrier (61).
At the gut lumen, intestinal goblet cells secrete
high levels of hyperglycosylated mucin able to compartmentalize gut
microbes within the mucosal surface and distant from the epithelial
surface (62). Moreover, the
glycans bound to mucin deliver tolerogenic signals, inducing
intestinal local DCs to switch towards an anti-inflammatory state
(62). Such DCs, once they
internalize commensal microbes and express their antigens at the
cell surface, selectively induce resident plasma cells to secrete
IgA and protect the host from microbial invasion (63). In addition, Paneth cells of the
small intestine secrete a range of antimicrobial peptides (AMPs),
which restrain the expansion of potential microbial pathogens and
hence help to maintain the GM homeostasis (64).
Pathogen-associated molecular patterns (PAMPs)
comprise all the microbial-derived molecules, produced from both
pathogens and commensals (65).
PAMPs are actively recognized by pattern recognition receptors
(PRRs), which are expressed by gut epithelial cells and local
immune myeloid cells, and represent the main innate immune
recognition pathway (65). PRRs
are constantly exposed to PAMPs, also during gut homeostasis, in
the absence of any infection (66).
PAMPs produced by gut commensals usually do not
elicit a pro-inflammatory response. The specific context determines
the outcome upon PRR activation (67). Only in the presence of epithelial
damage, PAMPs enter the cytosolic cellular epithelial compartment
(68). The elicited inflammatory
response determines the activation of NF-κB signaling, further
promoting the local secretion of pro-inflammatory cytokines, such
as interferons (IFNs) (69). This
pro-inflammatory response actively protects the intestine against
microbial infections. On the contrary, in the absence of concurrent
epithelial damage, PRR activation can be beneficial and may promote
immune tolerance (70).
Similar to the innate response, the adaptive immune
response is modulated in a two-way manner by GM, both locally and
systemically (71). For instance,
gut-associated B-cells secrete several grams of IgA per day within
the gut lumen (72). This
secretion can be either T-cell-independent or T-cell-dependent
(72). Recently, it was also
reported that gut mesenchymal cells can induce plasma cells to
secrete IgA (73). In general,
Foxp3+ regulatory T-cells (Tregs) induce a diversified
IgA repertoire, which in turn maintains a heterogeneous and
eubiotic GM. In turn, the healthy GM sustains the homeostatic IgA
responses in a positive feedback loop (74). IgA in the lumen coat pathogenic
bacteria, preventing their potential invasion and a subsequent
pro-inflammatory response (75).
T-cells play a pivotal role in regulating both local
and systemic adaptive immunity related to GM homeostasis, as well
as pathology. GM and GM-derived molecules can induce
CD4+ T-cell differentiation towards the main types: Th1,
Th2, Tregs and Th17 (76). Th1
cells are essential against intracellular pathogens. Th2 cells are
necessary during parasite-mediated infection. Tregs and Th17 are
cellular phenotypes involved in the containment of the immune
response. In particular, Tregs regulate the instauration of the
immune tolerance (77). It has
been observed that polysaccharide A (PSA) produced by
Bacteroides fragilis induces the Th1 phenotype, while
segmented filamentous bacteria (SFB) potently trigger Th17
differentiation (78,79).
Th17 cells play a major role in mucosal immunity as
they are able to prevent pathogen infection within the lamina
propria, by secreting cytokines, including IL-17. IL-17 induces
intestinal epithelial cells to express tight junctions and to
secrete AMPs (80). Moreover,
IL-17 further stimulates the release of other pro-inflammatory
cytokines by neutrophils, which can be recruited from the main
bloodstream and directed towards the gut (81).
Additionally, GM-mediated immune-cell priming can
shape the systemic immune response. When APCs, such as DCs, present
their antigens within the mesenteric lymph nodes to Tregs and Th17
cells, these T-cells can travel through the bloodstream and promote
distal immune responses against cross-reacting antigens located in
other sites of the body (82,83). As a consequence, dysbiosis may
affect systemic immune functions, thus increasing the
susceptibility to certain infections and, for example, altering the
response to vaccines (84).
Commensal gut microbes can actively secrete de
novo produced molecules or transform the host's metabolites,
which all may be sensed by nearby gut epithelial cells (85). Such bioproducts may have profound
effects on the health of a host, including: i) Inducing
immune-mediated protection against microbial pathogens; ii)
maintaining gut barrier integrity; iii) metabolizing xenobiotics;
iv) modulating the host's metabolism; and v) shaping and
activating/inactivating the host immune system (86-89).
The GM is involved in the production of essential
micronutrients, including vitamins K and B (90). In addition, a number of gut
commensals can convert certain amino acids into signaling
molecules, such as glutamate into gamma-aminobutyric acid (GABA) or
histidine into histamine (90).
Importantly, gut bacteria ferment dietary fibers to obtain a class
of hormone-like bioproducts known as short-chain fatty acids
(SCFAs), with multiple known functions in human health (91). For instance, SCFAs are transported
to the liver representing an energy source. Additionally, SCFAs may
control both glucose and lipid metabolism through the modulation of
peptide hormone secretion by gut epithelial cells (92). Moreover, SCFAs (including
butyrate) can enhance immunity, triggering the production of IgA by
plasma cells (93). In turn, IgA
inhibits bacterial adhesion to gut epithelial cells, hence blocking
invasion (94,95). Moreover, SCFAs can interfere with
the balance between anti-inflammatory and pro-inflammatory
cytokines secreted by immune cells, both locally and systemically.
The consequence is the modulation of homeostatic Treg vs. the Th17
cell ratio, resulting in an immune imbalance (96,97).
In summary, the preservation of a fine gut microbial
equilibrium (in terms of the presence and relative abundance of
commensal species) is imperative for sustaining and accomplish all
the vital immune functions of the host. A healthy GM positively
influences the immune system both locally and systemically.
Conversely, once activated, the immune system may alter the GM
balance. Gut eubiosis is thus of utmost importance for the
maintenance of immune health (Fig.
2).
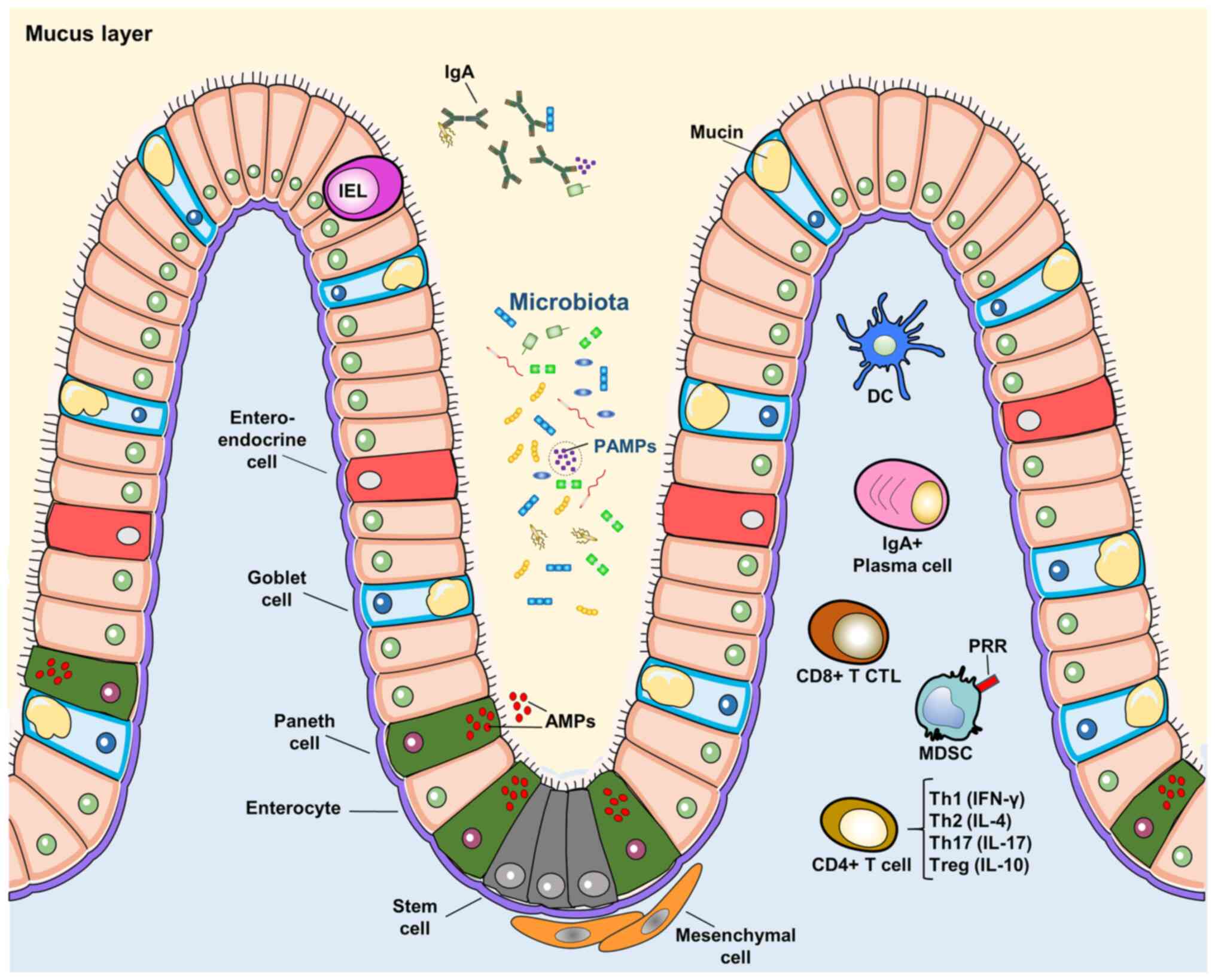 | Figure 2The gut-immune axis. The
gastrointestinal lumen represents the interface between the GM and
immune system. Intestinal cells constitute the villi structure and
include: Enteroendocrine cells, goblet cells, Paneth cells,
enterocytes and stem cells. Paneth cells secrete AMPs. IELs may
reside within the epithelial structure. Goblet cells secrete mucin
which enriches the intraluminal mucus layer. Gut microbiota and
their derived molecules form PAMPs. PAMPs are recognized by PRRs
expressed by immune cells and gut epithelial cells. IgA are
secreted in the lumen and help to bind microbes and
microbial-deriving molecules. Immune cells are pivotal in the
instauration of the immune-tolerance versus commensals and
immune-reactivity against pathogens. Both innate and adaptive
immunity are involved. Immune cells include: DCs, IgA-producing
plasma cells, CD8+ CTLs, MDSCs and CD4+ T-cells. The latter can
differentiate into different phenotypes involved in immune
reactivity or tolerance (i.e., Th1, Th2, Th17 and Tregs). GM, gut
microbiota; AMPs, anti-microbial peptides; IELs, intestinal
intraepithelial lymphocytes; PAMPs, pathogen-associated molecular
patterns; PRRs, pattern recognition receptors; DCs, dendritic
cells; CTLs, cytotoxic T lymphocytes; MDSCs, myeloid-derived
suppressor cells. |
Notably, there is increasing evidence to indicate
that gut dysbiosis may specifically affect local and systemic
anti-tumor immunity. In fact, recurrent antibiotic exposure (which
can impair intestinal eubiosis and favor the expansion of gut
pathogens) is directly associated with an increased risk of cancer
(98). In general, dysbiosis may
influence tumor formation or ICI-based therapy failure (99). As largely depicted in the present
review, a healthy (both enriched and diverse) GM can activate the
immune system to: i) Fight cancer; and ii) efficiently respond to
anticancer immunotherapies, in particular ICIs (100).
3. Gut microbiota and cancer
Cancer is a multifactorial disease resulting from a
combination of intrinsic factors (e.g., the stochastic accumulation
of gene mutations and epigenetic alterations), environmental
exposures (e.g., pollution, sunlight exposure and infections) and
lifestyle habits (cigarette smoking, diet and sport) (101,102). The resulting overall risk of
developing a given malignancy is mainly dependent on the dose,
duration, as well as the combination of exposures, all coupled with
the specific genetic and epigenetic background (103).
Presently, several biological agents are listed as
carcinogens by the International Agency of Research on Cancer
(IARC), including a number of viruses (i.e., Epstein-Barr virus,
hepatitis B virus, hepatitis C virus, Kaposi's sarcoma-associated
herpesvirus, human immunodeficiency virus-1, human papillomaviruses
and human T-cell lymphotropic virus type 1), as well as the
gastrointestinal bacterial pathogen, Helicobacter pylori
(104). Individuals with
Helicobacter pylori infection have a higher risk of
developing stomach cancer as the infection directly causes chronic
inflammation (105).
Generally, the influence of gastrointestinal
microorganisms on cancer development is complex (106). In fact, the GM plays a dual role
in cancer, as gut microbes can either positively or negatively
affect tumorigenesis, depending on their nature (49). Bacteria per se or their
products may directly or indirectly affect tumorigenesis. In
general, beneficial bacteria which are normally part of an eubiotic
GM exert an antitumor effect (107). On the contrary, pathogens
prevailing in a dysbiotic GM are pro-tumorigenic, either directly
or through the production of microbial-derived toxins. These
effects have been documented in both local colorectal cancers
(CRCs), as well as in distant tumors (108). The authors recently reviewed all
the relevant findings regarding the dual role played by the GM in
cancer, focusing on the specific gut microorganisms involved
(49).
Several preclinical findings have demonstrated the
pro-tumorigenic role of a number of gut microbes. For instance, a
number of bacteria, mostly pathogens, can release toxins within the
intestinal lumen which, once internalized by lumen epithelial
cells, can directly promote genotoxic damage or, alternatively,
they can activate pro-proliferative or anti-apoptotic pathways
(49). For example, cytolethal
distending toxins (CDTs), Shiga-like and Shiga toxins secreted
respectively by Escherichia coli, Helicobacter spp.,
Shigella dysente-riae, as well as several others, may
directly induce DNA damage (109). Moreover, the surface molecule
FadA from Fusobacterium nucleatum, CagL from Helicobacter
pylori and SopB from Salmonella typhimurium can trigger
the WNT/B-catenin, MEK-ERK and STAT3 pathways, respectively, all
inducing target cells to over-proliferate and/or not undergo
apoptosis (110-112).
Additionally, the GM may alter the immune response
activated by the host against the neoplasm, as observed in several
tumor-bearing mice models (49,113,114). For example, enterotoxigenic
Bacteroides fragilis infection in a mouse model of CRC
attracts a colonic immune infiltrate. In particular, it has been
demonstrated that Bacteroides-derived enterotoxin BFT
promotes the differentiation of pro-tumoral myeloid-derived
suppressor cells (MDSC), which in turn produce NO and suppress
CD4+ T-cell proliferation (115,116). In addition, the Treg response
upon enterotoxigenic Bacteroides fragilis infection in mice
with CRC triggers IL-17 and induces Th17 development which in turn,
promotes tumorigenesis (117).
Furthermore, Fusobacterium nucleatum has been shown to
potentiate CRC tumorigenesis in a mouse model of CRC, driving MDSC
infiltration within the intestinal tumors and inducing the
activation of a general pro-tumoral immune-milieu (118).
By contrast, some bacteria have been shown to
possess an anticancer function through the stimulation of the
immune system of their host. For example, Bifidobacterium
spp. promotes antitumor immunity in tumor-bearing mice through the
activation of DCs, which enhance cytotoxic CD8+ T-cell
activity directed against tumor cells (119). Akkermansia muciniphila
and Enterococcus hirae orally administered to tumor-bearing
mice have been shown to induce DCs to secrete IL-12, thus
triggering the recruitment of CD8+ cytotoxic T-cells and
inhibiting tumor growth (120)
(Fig. 3).
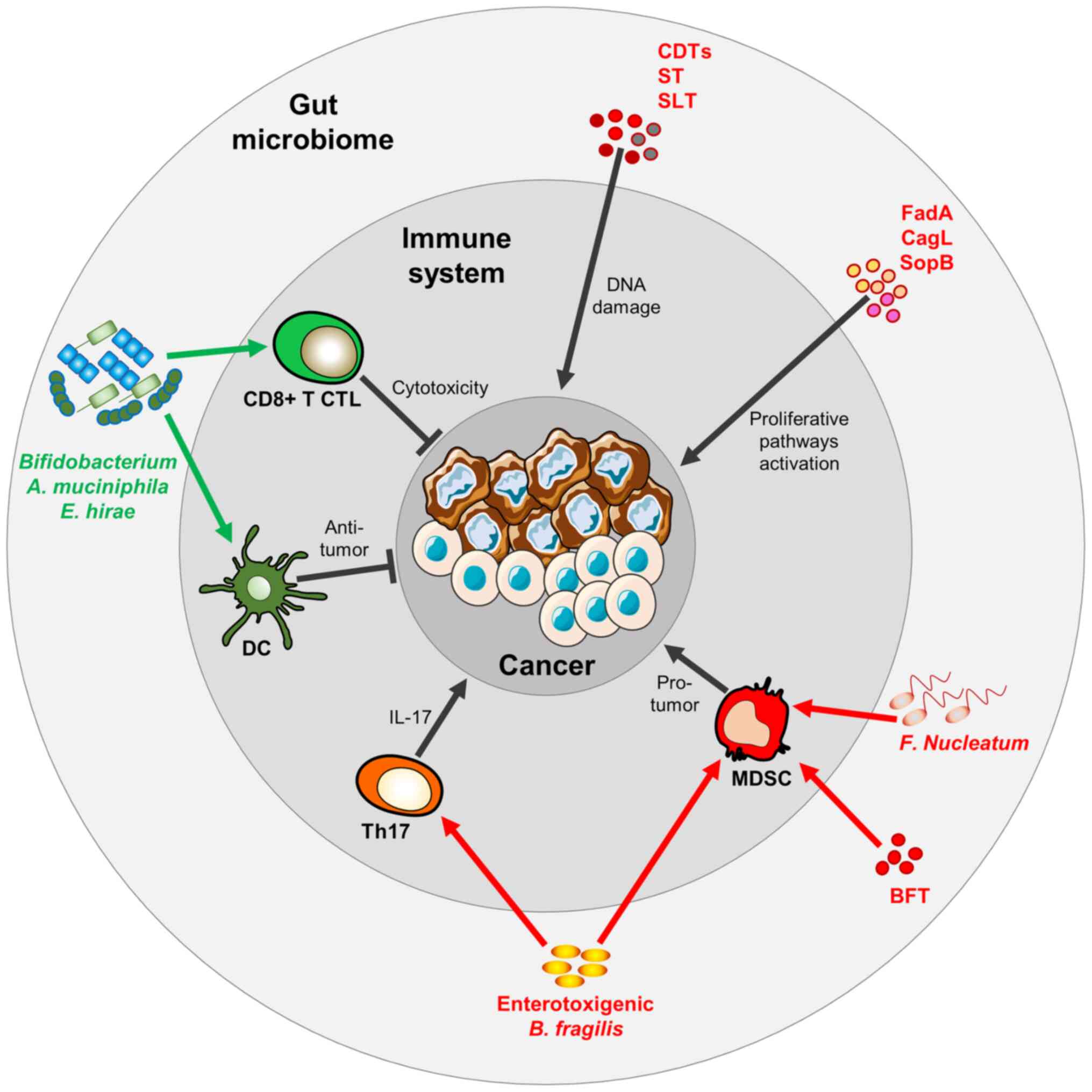 | Figure 3Gut microbiota play a dual role in
cancer. Microbial-derived molecules, including CDTs, SLTs and STs
may directly induce DNA damage and trigger cancer mutation. Other
microbial surface molecules, such as FadA from Fusobacterium
nucleatum, CagL from Helicobacter pylori and SopB from
Salmonella typhimurium induce cancer cell proliferation.
Fusobacterium nucleatum, Bacteroides fragilis or its
derived Bacteroides fragilis toxin (BFT) can boost MDSCs which, in
turn, favor a pro-tumoral milieu. Bacteroides fragilis may
also trigger the Th17 T-cell phenotype, which is immunosuppressive
and hence, pro-tumorigenic. Bifidobacterium spp.,
Akkermansia muciniphila, Enterococcus hirae promote
DC activation and CD8+ cytotoxic T-cell activation, both
triggering an anti-tumor immune response. CDTs, cytolethal
distending toxins; SLT, Shiga-like toxin; ST, Shiga toxin; MDSCs,
myeloid-derived suppressor cells; DC, dendritic cell. |
Given all the aforementioned preclinical examples,
it is clear that the maintenance of a healthy GM through the life
of an individual may represent a good strategy with which to
prevent cancer. A number of groundbreaking studies (described in
detail below) have further expanded this concept, demonstrating
that GM contains diagnostic and prognostic cancer biomarkers,
suitable to identify therapy-responder patients. More importantly,
either the modulation of the GM towards a healthy phenotype or the
selective enrichment of GM in specific beneficial bacterial types
may represent an enforcement to anticancer therapy. In this
context, different studies have demonstrated how the administration
of specific probiotic strains may positively influence the GM with
beneficial effects for cancer patients and human health (121,122). This concept particularly applies
to immunomodulatory treatments and specifically to ICIs.
4. Gut microbiota and immune checkpoint
inhibition
A growing number of studies have shed further light
on the association between the safety and efficacy of ICI-based
immunotherapy and GM features in cancer patients. In this section,
the authors aim to provide a temporal timeline demonstrating the
progress made and milestones accomplished in this field.
In 2015, Vétizou et al (123) observed for the first time that
tumors in antibiotic-treated or in germ-free mice did not respond
to anti-CTLA4 immunotherapy. Pivotally, they demonstrated that the
antitumor effect of anti-CTLA4 was dependent on Bacteroides
spp., and in particular on Bacteroides fragilis. In fact, a
significant antitumor response was observed: i) With oral gavage of
Bacteroides fragilis; or ii) with immunization with
Bacteroides fragilis-derived polysaccharides; or iii) with
the adoptive transfer of Bacteroides fragilis-specific in
vitro activated T-cells (123). The authors of that study
performed FMT in tumor-bearing mice treated with anti-CTLA4
antibody. Specifically, they employed stools from melanoma patient
donors with fecal abundance of Bacteroides spp. Following
FMT, mice-derived feces were found to be selectively enriched in
Bacteroides fragilis. This feature was negatively associated
with tumor size following the CTLA-4 blockade in recipient mice.
Hence, anti-CTLA4 antibody treatment could actively modify the
abundance of immunogenic Bacteroides spp. in the gut, which
in turn affected ICI-anticancer efficacy (123).
In the same year, Sivan et al (119) compared the growth of melanoma in
mice grown in different breading facilities, thus bearing different
GM compositions. They demonstrated significant differences in
melanoma growth, which reflected the different cancer-specific
T-cell response. Pivotally, anti-PD-L1-non-responder mice, when
orally receiving either feces obtained from responder mice or
Bifidobacterium alone, augmented their response to
anti-PD-L1 therapy (119). In
particular, a significant reduction in tumor outgrowth coupled with
an augmented DC activity, leading to increased CD8+
T-cell priming and T-cell accumulation in the tumor
microenvironment was observed (119).
Both studies strongly suggested that manipulating
the GM could enhance the antitumor efficacy of ICIs (119,123). These important observations
paved the way for subsequent translational observations. In 2016,
Dubin et al published a prospective study aimed at analyzing
GM features in patients with MM treated with ICIs and developing
colitis (124). The use of
anti-CTLA4 antibody in cancer patients is often associated with
dysbiosis and with inflammatory colitis as irAEs (124). The authors associated the fecal
microbial composition at the baseline with the one following
colitis manifestation. Notably, they found that an increased
representation of bacteria belonging to the Bacteroidetes
phylum was associated with augmented resistance to the development
of ICI-induced colitis. On the contrary, microbiome analysis
confirmed that patients lacking genetic pathways involved in
polyamine transport and B vitamin biosynthesis had an increased
risk of developing colitis (124).
In 2017, Frankel et al (125), through the use of metagenomic
shotgun sequencing coupled with metabolomic profiling, identified
the specific footprint of GM associated with the efficacy of ICIs
in patients with MM. Of the 39 patients with MM, only a group
exhibited a response to ICIs (corresponding to 67% of ipilimumab
plus nivolumab-, and 23% of pembrolizumab-treated subjects).
Despite the specific ICI used, feces from ICI-responders were
enriched for Bacteroides caccae (125). In particular, responders treated
with a combination of anti-CTLA4 and anti-PD-L1 had a GM enriched
in Faecalibacterium prausnitzii, Bacteroides
thetaiotamicron and Holdemania filiformis. However,
responders treated with anti-PD-1 antibody had a GM enriched in
Dorea formicogenerans. Among all the GM obtained from
responders, the metabolite resulting consistently enriched was
anacardic acid (125).
Also in 2017, Chaput et al (126) performed a prospective analysis
on the fecal microbiota composition in 26 patients with MM. The
analysis revealed that at the baseline, prior to any anti-CTLA4
antibody infusion, patients whose baseline microbiota was driven by
Bacteroides exhibited a longer progression-free survival
(PFS) than patients whose baseline microbiota was driven by
Faecalibacterium genus plus other Firmicutes.
Additionally, baseline colitis-associated phylotypes were
selectively associated with the presence of Firmicutes
(126). Upon ICI treatment,
patients with MM belonging to the Faecalibacterium-driven
cluster who developed anti-CTLA4-induced colitis exhibited a
significant increase in the CD4+ T-cell population and,
in particular, in CD4+ T-cells expressing the T-cell
inducible T-cell COStimulator surface marker. This observation
suggested that the baseline GM composition may represent an
important determinant of the immune response in cancer patients, as
well as of anti-CTLA-4 associated-colitis (126).
Overall, the aforementioned prospective studies have
demonstrated the importance of combining metagenomics and
metabolomics to study the GM in cancer patients. These studies
clearly demonstrate that GM is affected by ICI treatments. Thus, a
specific GM composition and/or metabolite enrichment may be
predictive of a better prognosis.
In 2018, three landmark studies were published in
Science, clearly demonstrating, for the first time, a direct
association between GM composition and efficacy of ICI-based
therapy (120,127,128). In particular, Routy et al
(120) examined the effects of
FMT from NSCLC and renal cell carcinoma (RCC) ICI-responder and
ICI-non-responder donors, in recipient epithelial (melanoma and
sarcoma) tumor-bearing mice (either germ-free or
antibiotic-treated). They found that FMT from responders
significantly enhanced the efficacy of ICIs in reducing tumor
growth in mice, whereas FMT from non-responder patients did not
exert any effect (120).
Metagenomics analyses of fecal samples from responders and
non-responders clearly revealed that the GM composition affected
the primary immune-resistance to ICIs. In particular, a positive
association was observed between the relative abundance of
Akkermansia muciniphila and the clinical response to ICIs.
The oral gavage of Akkermansia muciniphila in mice receiving
FMT from non-responders significantly restored the efficacy of
anti-PD-1 and augmented the recruitment of CD4+ T-cells
at the tumor site, and increased the local secretion of IL-12 by
DCs (120).
Additionally, Matson et al (127) analyzed the baseline GM
composition of stool samples from 42 patients with MM prior to
receiving ICI therapy. Metagenomics analysis revealed that
Bifidobacterium longum, Collinsella aerofaciens and
Enterococcus faecium species were significantly enriched in
ICI-responder patients (127).
FMT from responder donors in recipient tumor-bearing mice improved
the T-cell response and the anti-PD-L1 anticancer efficacy. In
addition, from fecal analyses, 10 bacterial species were found to
be differentially enriched in responder vs. non-responder mice. In
total, eight of these (i.e., Enterococcus faecium,
Collinsella aerofaciens, Bifidobacterium
adolescentis, Klebsiella pneumoniae, Veillonella
parvula, Parabacteroides merdae, Lactobacillus
spp., and Bifidobacterium longum) were more abundant in
responders, whereas two (i.e., Ruminococcus obeum and
Roseburia intestinalis) were more abundant in non-responders
(127).
Finally, Gopalakrishnan et al (128) analyzed the gut microbiome of 112
patients with MM, demonstrating that responders had a significantly
higher alpha diversity and a relative abundance of bacteria
belonging to the Ruminococcaceae family. Moreover, from
metabolomics analyses, the authors of that study observed a
significant enrichment of anabolic pathways. Responder patients
also exhibited an enhanced systemic and antitumor immunity, with
increased cytotoxic CD8+ T-cell tumor infiltrates
(128). Accordingly, germ-free
mice receiving FMT from responders exhibited favorable immune
profiling and an improved response to anti-PD-L1 antibody treatment
in terms of tumor growth, which was significantly reduced.
Importantly, gut microbiome analyses revealed that feces of
responders were enriched in Clostridiales, whereas the feces of
non-responders were rich in Bacteroidales (128).
Since these milestone studies, several others
conducted on cancer patients have further revealed the existence of
a relevant bacterial gut footprint in ICI-responders vs.
non-responders. In 2018, Derosa et al (129) demonstrated how antibiotics can
alter GM health, triggering antibiotic-associated dysbiosis. In
turn, dysbiosis may invalidate the response to ICIs. In the
retrospective analysis, patients with advanced RCC and NSCLC (121
and 239 patients, respectively) treated with anti-PD-L1 antibody
(either as monotherapy or in combination) were considered. Above
all, 13% of patients with RCC and 20% of patients with MM received
antibiotics 30 days prior to the ICI administration. Of note, the
antibiotics significantly reduced the benefits of ICIs in the
cancer patients. In particular, PFS was significantly reduced in
patients with RCC, whereas overall survival (OS) was decreased in
patients with NSCLC (129).
In 2019, Jin et al (130) additionally confirmed that a
favorable GM, as well as a healthy immune profile were associated
with an improved response to anti-PD-1 immunotherapy. The authors
considered 37 patients with advanced NSCLC. They performed the
fecal microbiome analysis: i) At the moment of the anti-PD-1
therapy; ii) at the clinical evaluation; iii) following the
progression of the disease. According to the Response Evaluation
Criteria in Solid Tumor (RECST) scale, patients were divided in
responders and non-responders (130). Responders which exhibited a
significantly prolonged PFS had a high microbiome diversity.
Compared with the baseline, the feces of responders were enriched
in Alistipes putredinis, Bifidobacterium longum and
Prevotella copri. However, non-responders exhibitd a
prevalence in Ruminococcus spp. In addition, responders
manifested an increase in CD8+ T-cell and NK cell
subsets in response to anti-PD-1 therapy (130).
Also in 2019, Zheng et al (131) performed gut microbiome profiling
of a small cohort of patients with hepatocellular carcinoma (HCC),
using metagenomic sequencing. Fecal samples from patients
responding to anti-PD-1 immunotherapy exhibited a higher taxa
richness and more gene counts than those of non-responders.
Furthermore, dynamic sequencing analyses demonstrated that
anti-PD-1 therapy increased the GM dissimilarity between responders
and non-responders, with a prominence in such differences at 6
weeks post-treatment. A total of 20 responder-enriched species
(including Akkermansia muciniphila and
Ruminococcaceae spp.) were further identified as prominent.
Subsequent metabolic pathway analysis demonstrated that
carbohydrate metabolism and methanogenesis were selectively
enriched in the responders (131).
Additionally, in 2020, Salgia et al
(132) characterized the stool
microbiome from 31 patients with metastatic RCC receiving ICIs (as
single agents or combination) to assess treatment-related changes
in GM composition over the course of treatment. They found that a
higher microbial diversity was associated with better treatment
outcomes. Temporal profiling of the microbiome indicated that the
relative abundance of Akkermansia muciniphila significantly
increased in patients obtaining clinical benefits from ICIs. Hence,
dynamic changes in GM composition suggested the potential utility
of modulating GM to reach more successful outcomes with ICIs
(132).
Taken together, the clinical studies evidenced
several microbial candidates, which can be suggested as predictive
biomarkers of the patient population that may truly benefit from
ICIs. Of equal importance is the elucidation of the molecular
mechanisms responsible for the beneficial effect of the GM. For
this purpose, several important preclinical studies have recently
been published.
In 2019, Tanoue et al (114) isolated a consortium of 11
bacterial strains (comprised of seven Bacteroidales and four
non-Bacteroidales species) from healthy human donor feces,
capable of augmenting IFN-γ-secreting CD8 T-cell levels in the gut.
The consortium, when inoculated into syngeneic CRC tumor-bearing
germ-free mice, induced a robust MHCI expression in DCs and
enhanced the therapeutic efficacy of ICIs, with a concurrent
decrease in tumor growth (114).
In 2020, Xu et al (133) evaluated the effects of GM in
MSS-type CRC tumor-bearing mice treated with different antibiotics
prior to anti-PD-1 anticancer treatment. The injection of
antibiotics significantly counteracted the efficacy of anti-PD-1
antibody in inhibiting tumor growth. Furthermore, metabolomics
analysis demonstrated the enrichment in the glycerophospholipid
metabolic pathway, specifically within the antibiotic-treated
group. Changes in GM composition may drive these metabolic changes.
In fact, Xu et al (133)
demonstrated that Prevotella spp. and Akkermansia
spp. were able to support the efficacy of anti-PD-1 by affecting
the metabolism of glycerolipids in MSS-type CRC tumor-bearing
mice.
Of note, in 2020, Mager et al (134) explored the underlying mechanisms
through which the GM enhanced ICI-mediated antitumor immunity, with
particular focus on the T-cell adaptive response. In detail,
Bifidobacterium pseudolongum, Lactobacillus
johnsonii and Olsenella spp. significantly increased the
efficacy of ICIs in four different mouse models of cancer. In
particular, Bifidobacterium pseudolongum modulated the
ICI response through the production of the metabolite, inosine
(134). They further assessed
that a decreased gut barrier functionality induced by
ICI-immunotherapy increased the systemic translocation of inosine,
which in turn activated antitumor CD8+ T-cell activity.
Importantly, the effect of bacterial-derived inosine was strictly
dependent on the T-cell surface expression of the adenosine A2A
receptor, whose stimulation was specifically required (134).
More recently, in 2021, Si et al (135) evaluated the therapeutic efficacy
of administering a single bacterial strain, Lactobacillus
rhamnosus GG (LGG) in combination with ICI immunotherapy.
Pivotally, the oral administration of LGG significantly improved
the efficacy of ICIs and reduced tumor growth in CRC and melanoma
mouse models. The authors further explored the molecular mechanism,
evidencing that the augmented anti-tumor activity of anti-PD-1 was
associated with increased tumor-infiltrating DCs and
CD8+ T-cells (135).
Moreover, treatments with live LGG alone or in combination with
anti-PD-1 triggered type I IFN production by DCs, enhancing the
cross-priming of CD8+ cytotoxic T-cells. In DCs, cyclic
GMP-AMP synthase (cGAS)/stimulator of IFN genes (STING) was
required for IFN-β induction in response to LGG. In fact, LGG
significantly boosted IFN-β production via the cGAS/STING axis in
DCs (135). The role of the
STING pathway in potentiating the efficacy of immunotherapy, was
also proven by Shi et al (136). In agreement with the findings of
Si et al (135), Shi
et al (136) observed
that a specific gut microbe, Bifidobacterium spp.,
potentiated anti-CD47 immunotherapy via the stimulation of STING in
DCs in tumor-bearing mice.
2021 was a breakthrough year in GM research
associated with ICI therapeutic efficacy in human studies. In this
year, two important publications in Science demonstrated,
for the first time, that FMT significantly overcame the resistance
to anti-PD-1 therapy in patients with MM (50,51). Firstly, Davar et al
(50) demonstrated that in
patients with MM, the GM composition was associated with the
anti-PD-1 response. Notably, the resistance to anti-PD-1 was
overcome by directly modulating GM composition. The authors of that
study evaluated both the safety and efficacy of responder-derived
feces transplanted together with anti-PD-1 in recipient patients
with PD-1-refractory melanoma (50). FMT with anti-PD-1 was
well-tolerated and provided clinical benefit in 6 out of 15
patients. The combined treatment induced a rapid and robust gut
microbiota perturbation. The six responders exhibited an increased
abundance of taxa, such as Ruminococcaceae and Bifidobacteriaceae,
that were previously shown to be associated with a response to
anti-PD-1 associated with an increased CD8+ T-cell
activation, as well as with a decreased frequency of myeloid cells
secreting IL-8 (119,128). In addition, the responders
exhibited distinct proteomic and metabolomic signatures. The
trans-kingdom network analysis confirmed that the GM directly
regulated these changes (50).
In parallel, Baruch et al (51) performed a pilot phase I clinical
study to assess both the safety and feasibility of FMT and
anti-PD-1 immunotherapy re-induction 10 patients with
anti-PD-1-refractory MM. Notably, they observed a positive clinical
response in 3 out of 10 patients (two partial responses and one
complete response). Treatment with FMT was associated with
favorable changes in immune cell infiltrates and immune-related
gene expression profiles locally (at the level of the gut lamina
propria), as well as distally (within the tumor microenvironment)
(51).
Taken together, the reported findings provide
strong evidence on the key role that the GM plays in modulating the
ICI therapeutic response and potential-associated toxicity.
Pivotally, Davar et al (50) and Baruch et al (51) demonstrated that gut microbial
modulation can reverse ICI resistance in patients with MM through
the specific modulation of the individual immune system, both
locally and systemically (Fig.
4).
Since 2015, a number of clinical trials have been
performed with two main goals. One important aim is to characterize
the gut microbiome signature, as well as the associated immune
system changes upon a specific ICI immunotherapy. The outcomes of
these studies may reveal the existence of specific diagnostic and
prognostic gut microbial biomarkers (Table I).
Additionally, a growing number of clinical trials
is currently evaluating both the safety and efficacy of actively
modulating GM composition in association with ICI immunotherapy.
Currently, a number of companies are investing in this direction
and the number of registered trials is increasing exponentially
(100). As presented in Table II, in order to 'turn bugs into
drugs' a number of strategies are currently being tested, including
the administration of: i) Single probiotics; ii) microbial
communities; iii) synbiotics; iv) FMT; and v) metabolic
modulators.
Of note, for certain malignancies, such as prostate
cancer, researchers have shown the limited efficacy of ICIs
(137). This may be partially
explained by the fact that prostate cancer is immunologically
'cold' compared to both melanoma and lung cancer, thus with a lower
tumor mutational burden (TMB) (138,139). Previous clinical studies have
tested ICIs administered in combination with miscellaneous agents
that may improve the overall efficacy of ICIs (140,141). For instance, the concurrent
administration of androgen inhibitors has been shown to
significantly improve the OS of patients with metastatic
castration-sensitive prostate cancer (142). Coherently, one ongoing trial
reported (presented in Table II)
is evaluating both the safety and efficacy of combining anti-PD-1
antibody with the anti-androgen enzalutamide and the FMT in
prostate cancer patients not responding to initial ICI and androgen
deprivation therapy. Prostate cancer patients with A significant
initial response to anti-PD-1 and anti-androgens, will be used as
fecal donors (NCT04116775).
5. Conclusions
Since birth, each individual inherits a specific GM
footprint. Moreover, the GM is constantly shaped with age, diet and
lifetime exposures. Current research has demonstrated the dual role
played by the GM in cancer (49).
In particular, the GM deeply affects the host immune system and
vice versa. During dysbiosis, several microbial species can
overpopulate the gut. Consequently, they may enhance inflammation
and promote the formation of a pro-cancerogenic environment
(143).
On the contrary, the re-establishment of a healthy
GM may be beneficial for cancer patients, inducing a healthy immune
system. In line with this observation, it has been further
demonstrated that a specific ICI-responder gut microbial footprint
is associated with a reduction in irAEs and an improvement of
ICI-efficacy (120,127,128). Furthermore, specific gut
microbial modulatory therapy (based on FMT from healthy/responders
donors) may revert ICI-resistance in patients with advanced cancer
(50,51). This latter groundbreaking
observation is paving the way towards a growing number of ongoing
clinical trials in cancer patients to test the administration of
'good' gut microbes as adjuvants in association with ICI-based
therapy (Table II).
In spite of the tremendous progress made, some
questions still remain unanswered. In fact, since cancer patients
are often immunocompromised, special care needs to be taken to
select the correct therapeutic strategy to modulate the GM, in
order to maximize the positive outcomes of microbial-modulators,
and reduce the potential harmful side-effects due to the possibly
fragile immune system. For instance, a number of factors may affect
the overall clinical outcome, including the use of concurrent
medications such as antibiotics, which have been associated with a
reduced response to ICIs (144).
In addition, specific dietary associations are crucial and they can
be efficiently used as predictors of FMT success, as demonstrated
for patients with a higher intake of dietary fiber (145). The consumption of specific
nutrients or bioactive food-derivatives may favor the FMT
engraftment, particularly in cancer patients, who are often
affected by nutritional and metabolic issues (e.g., vomiting,
swallowing difficulties, reduced food adsorption and inadequate
food consumption).
In conclusion, anticancer therapy is increasingly
becoming holistic. In the near future, anticancer treatments will
be tailored to the specific cancer patient, based on the GM if the
individual, as well as the immune signature. Henceforward, larger
clinical longitudinal studies will help to increase the current
knowledge on the long-term safety and robustness of FMT as
adjuvants of ICIs, in order to expand and standardize their use in
a number of types of cancer.
Availability of data and materials
Not applicable.
Authors' contributions
All authors (SV, LF, GCL, MS and ML) participated
in the writing of the manuscript, and preparing the figures and
tables, as well as in the revisions. SV and LF confirm the
authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
ML is the principal investigator of a research
grant founded by Dicofarm Spa to his University Department. In
light of the fact that this manuscript is a Review of the existing
literature, as well as of the ongoing clinical studies, the
Dicofarm Spa will not benefit specifically from any of the findings
described. The other authors declare that the research was
conducted in the absence of any commercial or financial
relationships that could be construed as a potential competing
interest.
Acknowledgments
The authors would like to thank the Italian League
Against Cancer (LILT) for its support.
References
|
1
|
Leviatan S and Segal E: Identifying gut
microbes that affect human health. Nature. 587:373–374. 2020.
View Article : Google Scholar
|
|
2
|
Greenhalgh K, Meyer KM, Aagaard KM and
Wilmes P: The human gut microbiome in health: Establishment and
resilience of microbiota over a lifetime. Environ Microbiol.
18:2103–2116. 2016. View Article : Google Scholar
|
|
3
|
Feng Q, Chen WD and Wang YD: Gut
microbiota: An integral moderator in health and disease. Front
Microbiol. 9:1512018. View Article : Google Scholar
|
|
4
|
Vaishnava S, Behrendt CL, Ismail AS,
Eckmann L and Hooper LV: Paneth cells directly sense gut commensals
and maintain homeostasis at the intestinal host-microbial
interface. Proc Natl Acad Sci USA. 105:20858–20863. 2008.
View Article : Google Scholar
|
|
5
|
Belkaid Y and Naik S: Compartmentalized
and systemic control of tissue immunity by commensals. Nat Immunol.
14:646–653. 2013. View Article : Google Scholar
|
|
6
|
Magnúsdóttir S, Ravcheev D, de
Crécy-Lagard V and Thiele I: Systematic genome assessment of
B-vitamin biosynthesis suggests co-operation among gut microbes.
Front Genet. 6:1482015. View Article : Google Scholar
|
|
7
|
Zheng D, Liwinski T and Elinav E:
Interaction between microbiota and immunity in health and disease.
Cell Res. 30:492–506. 2020. View Article : Google Scholar
|
|
8
|
Sharma VR, Singh M, Kumar V, Yadav M,
Sehrawat N, Sharma DK and Sharma AK: Microbiome dysbiosis in
cancer: Exploring therapeutic strategies to counter the disease.
Semin Cancer Biol. 70:61–70. 2021. View Article : Google Scholar
|
|
9
|
Sepich-Poore GD, Zitvogel L, Straussman R,
Hasty J, Wargo JA and Knight R: The microbiome and human cancer.
Science. 371:eabc45522021. View Article : Google Scholar
|
|
10
|
The integrative human microbiome project.
Nature. 569:641–648. 2019. View Article : Google Scholar
|
|
11
|
Rothschild D, Weissbrod O, Barkan E,
Kurilshikov A, Korem T, Zeevi D, Costea PI, Godneva A, Kalka IN,
Bar N, et al: Environment dominates over host genetics in shaping
human gut microbiota. Nature. 555:210–215. 2018. View Article : Google Scholar
|
|
12
|
Korem T, Zeevi D, Suez J, Weinberger A,
Avnit-Sagi T, Pompan-Lotan M, Matot E, Jona G, Harmelin A, Cohen N,
et al: Growth dynamics of gut microbiota in health and disease
inferred from single metagenomic samples. Science. 349:1101–1106.
2015. View Article : Google Scholar
|
|
13
|
Whon TW, Shin NR, Kim JY and Roh SW: Omics
in gut microbiome analysis. J Microbiol. 59:292–297. 2021.
View Article : Google Scholar
|
|
14
|
Geva-Zatorsky N, Sefik E, Kua L, Pasman L,
Tan TG, Ortiz-Lopez A, Yanortsang TB, Yang L, Jupp R, Mathis D, et
al: Mining the human gut microbiota for immunomodulatory organisms.
Cell. 168:928–943.e11. 2017. View Article : Google Scholar
|
|
15
|
Haber AL, Biton M, Rogel N, Herbst RH,
Shekhar K, Smillie C, Burgin G, Delorey TM, Howitt MR, Katz Y, et
al: A single-cell survey of the small intestinal epithelium.
Nature. 551:333–339. 2017. View Article : Google Scholar
|
|
16
|
Almeida A, Nayfach S, Boland M, Strozzi F,
Beracochea M, Shi ZJ, Pollard KS, Sakharova E, Parks DH, Hugenholtz
P, et al: A unified catalog of 204,938 reference genomes from the
human gut microbiome. Nat Biotechnol. 39:105–114. 2021. View Article : Google Scholar
|
|
17
|
Vujkovic-Cvijin I, Sklar J, Jiang L,
Natarajan L, Knight R and Belkaid Y: Host variables confound gut
microbiota studies of human disease. Nature. 587:448–454. 2020.
View Article : Google Scholar
|
|
18
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar
|
|
19
|
Esfahani K, Roudaia L, Buhlaiga N, Del
Rincon SV, Papneja N and Miller WH Jr: A review of cancer
immunotherapy: From the past, to the present, to the future. Curr
Oncol. 27(Suppl 2): S87–S97. 2020. View Article : Google Scholar
|
|
20
|
Baghban R, Roshangar L, Jahanban-Esfahlan
R, Seidi K, Ebrahimi-Kalan A, Jaymand M, Kolahian S, Javaheri T and
Zare P: Tumor microenvironment complexity and therapeutic
implications at a glance. Cell Commun Signal. 18:592020. View Article : Google Scholar
|
|
21
|
Robert C: A decade of immune-checkpoint
inhibitors in cancer therapy. Nat Commun. 11:38012020. View Article : Google Scholar
|
|
22
|
Fridman WH, Zitvogel L, Sautès-Fridman C
and Kroemer G: The immune contexture in cancer prognosis and
treatment. Nat Rev Clin Oncol. 14:717–734. 2017. View Article : Google Scholar
|
|
23
|
Hiam-Galvez KJ, Allen BM and Spitzer MH:
Systemic immunity in cancer. Nat Rev Cancer. 21:345–359. 2021.
View Article : Google Scholar
|
|
24
|
Ledford H, Else H and Warren M: Cancer
immunologists scoop medicine nobel prize. Nature. 562:20–21. 2018.
View Article : Google Scholar
|
|
25
|
Shin EC: Cancer immunotherapy: Special
issue of BMB Reports in 2021. BMB Rep. 54:12021. View Article : Google Scholar
|
|
26
|
Chen DS and Mellman I: Oncology meets
immunology: The cancer-immunity cycle. Immunity. 39:1–10. 2013.
View Article : Google Scholar
|
|
27
|
Huang PW and Chang JWC: Immune checkpoint
inhibitors win the 2018 nobel prize. Biomed J. 42:299–306. 2019.
View Article : Google Scholar
|
|
28
|
Waldman AD, Fritz JM and Lenardo MJ: A
guide to cancer immunotherapy: From T cell basic science to
clinical practice. Nat Rev Immunol. 20:651–668. 2020. View Article : Google Scholar
|
|
29
|
Shen CR and Chen YS: Immune checkpoint
blockade therapy: The 2014 tang prize in biopharmaceutical science.
Biomed J. 38:5–8. 2015. View Article : Google Scholar
|
|
30
|
Topalian SL, Hodi FS, Brahmer JR,
Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD,
Sosman JA, Atkins MB, et al: Safety, activity, and immune
correlates of anti-PD-1 antibody in cancer. N Engl J Med.
366:2443–2454. 2012. View Article : Google Scholar
|
|
31
|
Long J, Qi Z and Rongxin Z: PD-1/PD-L1
pathway blockade works as an effective and practical therapy for
cancer immunotherapy. Cancer Biol Med. 15:116–123. 2018. View Article : Google Scholar
|
|
32
|
Seidel JA, Otsuka A and Kabashima K:
Anti-PD-1 and anti-CTLA-4 therapies in cancer: Mechanisms of
action, efficacy, and limitations. Front Oncol. 8:862018.
View Article : Google Scholar
|
|
33
|
Wei SC, Duffy CR and Allison JP:
Fundamental mechanisms of immune checkpoint blockade therapy.
Cancer Discov. 8:1069–1086. 2018. View Article : Google Scholar
|
|
34
|
Twomey JD and Zhang B: Cancer
immunotherapy update: FDA-approved checkpoint inhibitors and
companion diagnostics. AAPS J. 23:392021. View Article : Google Scholar
|
|
35
|
Xin Yu J, Hubbard-Lucey VM and Tang J:
Immuno-oncology drug development goes global. Nat Rev Drug Discov.
18:899–900. 2019. View Article : Google Scholar
|
|
36
|
Alsaab HO, Sau S, Alzhrani R, Tatiparti K,
Bhise K, Kashaw SK and Iyer AK: PD-1 and PD-L1 checkpoint signaling
inhibition for cancer immunotherapy: Mechanism, combinations, and
clinical outcome. Front Pharmacol. 8:5612017. View Article : Google Scholar
|
|
37
|
Weber J, Mandala M, Del Vecchio M, Gogas
HJ, Arance AM, Cowey CL, Dalle S, Schenker M, Chiarion-Sileni V,
Marquez-Rodas I, et al: Adjuvant nivolumab versus ipilimumab in
resected stage III or IV melanoma. N Engl J Med. 377:1824–1835.
2017. View Article : Google Scholar
|
|
38
|
Eggermont AMM, Blank CU, Mandala M, Long
GV, Atkinson V, Dalle S, Haydon A, Lichinitser M, Khattak A,
Carlino MS, et al: Adjuvant pembrolizumab versus placebo in
resected stage III melanoma. N Engl J Med. 378:1789–1801. 2018.
View Article : Google Scholar
|
|
39
|
Kennedy LB and Salama AKS: A review of
cancer immunotherapy toxicity. CA Cancer J Clin. 70:86–104. 2020.
View Article : Google Scholar
|
|
40
|
Barrueto L, Caminero F, Cash L, Makris C,
Lamichhane P and Deshmukh RR: Resistance to checkpoint inhibition
in cancer immunotherapy. Transl Oncol. 13:1007382020. View Article : Google Scholar
|
|
41
|
Chen PL, Roh W, Reuben A, Cooper ZA,
Spencer CN, Prieto PA, Miller JP, Bassett RL, Gopalakrishnan V,
Wani K, et al: Analysis of immune signatures in longitudinal tumor
samples yields insight into biomarkers of response and mechanisms
of resistance to immune checkpoint blockade. Cancer Discov.
6:827–837. 2016. View Article : Google Scholar
|
|
42
|
Moslehi JJ, Salem JE, Sosman JA,
Lebrun-Vignes B and Johnson DB: Increased reporting of fatal immune
checkpoint inhibitor-associated myocarditis. Lancet. 391:9332018.
View Article : Google Scholar
|
|
43
|
Teixidor E and Bosch-Barrera J: The dark
side of immunotherapy: Challenges facing the new hope in cancer
treatment. Ann Transl Med. 7(Suppl 6): S1832019. View Article : Google Scholar
|
|
44
|
Havel JJ, Chowell D and Chan TA: The
evolving landscape of biomarkers for checkpoint inhibitor
immunotherapy. Nat Rev Cancer. 19:133–150. 2019. View Article : Google Scholar
|
|
45
|
Pezo RC, Wong M and Martin A: Impact of
the gut microbiota on immune checkpoint inhibitor-associated
toxicities. Therap Adv Gastroenterol. 12:17562848198709112019.
View Article : Google Scholar
|
|
46
|
Chang AE, Golob JL, Schmidt TM, Peltier
DC, Lao CD and Tewari M: Targeting the gut microbiome to mitigate
immunotherapy-induced colitis in cancer. Trends Cancer. 7:583–593.
2021. View Article : Google Scholar
|
|
47
|
Sarshar M, Scribano D, Ambrosi C, Palamara
AT and Masotti A: Fecal microRNAs as innovative biomarkers of
intestinal diseases and effective players in host-microbiome
interactions. Cancers (Basel). 12:21742020. View Article : Google Scholar
|
|
48
|
Murciano-Goroff YR, Warner AB and Wolchok
JD: The future of cancer immunotherapy: Microenvironment-targeting
combinations. Cell Res. 30:507–519. 2020. View Article : Google Scholar
|
|
49
|
Vivarelli S, Salemi R, Candido S, Falzone
L, Santagati M, Stefani S, Torino F, Banna GL, Tonini G and Libra
M: Gut microbiota and cancer: From pathogenesis to therapy. Cancers
(Basel). 11:382019. View Article : Google Scholar
|
|
50
|
Davar D, Dzutsev AK, McCulloch JA,
Rodrigues RR, Chauvin JM, Morrison RM, Deblasio RN, Menna C, Ding
Q, Pagliano O, et al: Fecal microbiota transplant overcomes
resistance to anti-PD-1 therapy in melanoma patients. Science.
371:595–602. 2021. View Article : Google Scholar
|
|
51
|
Baruch EN, Youngster I, Ben-Betzalel G,
Ortenberg R, Lahat A, Katz L, Adler K, Dick-Necula D, Raskin S,
Bloch N, et al: Fecal microbiota transplant promotes response in
immunotherapy-refractory melanoma patients. Science. 371:602–609.
2021. View Article : Google Scholar
|
|
52
|
Maynard CL, Elson CO, Hatton RD and Weaver
CT: Reciprocal interactions of the intestinal microbiota and immune
system. Nature. 489:231–241. 2012. View Article : Google Scholar
|
|
53
|
Belkaid Y and Hand TW: Role of the
microbiota in immunity and Inflammation. Cell. 157:121–141. 2014.
View Article : Google Scholar
|
|
54
|
Jain T, Sharma P, Are AC, Vickers SM and
Dudeja V: New insights into the cancer-microbiome-immune axis:
Decrypting a decade of discoveries. Front Immunol. 12:6220642021.
View Article : Google Scholar
|
|
55
|
Koenig JE, Spor A, Scalfone N, Fricker AD,
Stombaugh J, Knight R, Angenent LT and Ley RE: Succession of
microbial consortia in the developing infant gut microbiome. Proc
Natl Acad Sci USA. 108(Suppl 1): S4578–S4585. 2011. View Article : Google Scholar
|
|
56
|
Gomez de Agüero M, Ganal-Vonarburg SC,
Fuhrer T, Rupp S, Uchimura Y, Li H, Steinert A, Heikenwalder M,
Hapfelmeier S, Sauer U, et al: The maternal microbiota drives early
postnatal innate immune development. Science. 351:1296–1302. 2016.
View Article : Google Scholar
|
|
57
|
Russell SL, Gold MJ, Hartmann M, Willing
BP, Thorson L, Wlodarska M, Gill N, Blanchet MR, Mohn WW, McNagny
KM and Finlay BB: Early life antibiotic-driven changes in
microbiota enhance susceptibility to allergic asthma. EMBO Rep.
13:440–447. 2012. View Article : Google Scholar
|
|
58
|
Cahenzli J, Köller Y, Wyss M, Geuking MB
and McCoy KD: Intestinal microbial diversity during early-life
colonization shapes long-term IgE levels. Cell Host Microbe.
14:559–570. 2013. View Article : Google Scholar
|
|
59
|
Ge Y, Wang X, Guo Y, Yan J, Abuduwaili A,
Aximujiang K, Yan J and Wu M: Gut microbiota influence tumor
development and alter interactions with the human immune system. J
Exp Clin Cancer Res. 40:422021. View Article : Google Scholar
|
|
60
|
Shi N, Li N, Duan X and Niu H: Interaction
between the gut microbiome and mucosal immune system. Mil Med Res.
4:142017.
|
|
61
|
Lazar V, Ditu LM, Pircalabioru GG,
Gheorghe I, Curutiu C, Holban AM, Picu A, Petcu L and Chifiriuc MC:
Aspects of gut microbiota and immune system interactions in
infectious diseases, immunopathology, and cancer. Front Immunol.
9:18302018. View Article : Google Scholar
|
|
62
|
Shan M, Gentile M, Yeiser JR, Walland AC,
Bornstein VU, Chen K, He B, Cassis L, Bigas A, Cols M, et al: Mucus
enhances gut homeostasis and oral tolerance by delivering
immunoregulatory signals. Science. 342:447–453. 2013. View Article : Google Scholar
|
|
63
|
Macpherson AJ and Uhr T: Induction of
protective IgA by intestinal dendritic cells carrying commensal
bacteria. Science. 303:1662–1665. 2004. View Article : Google Scholar
|
|
64
|
Bevins CL and Salzman NH: Paneth cells,
antimicrobial peptides and maintenance of intestinal homeostasis.
Nat Rev Microbiol. 9:356–368. 2011. View Article : Google Scholar
|
|
65
|
Chu H and Mazmanian SK: Innate immune
recognition of the microbiota promotes host-microbial symbiosis.
Nat Immunol. 14:668–675. 2013. View Article : Google Scholar
|
|
66
|
Vance RE, Isberg RR and Portnoy DA:
Patterns of pathogenesis: Discrimination of pathogenic and
nonpathogenic microbes by the innate immune system. Cell Host
Microbe. 6:10–21. 2009. View Article : Google Scholar
|
|
67
|
Mogensen TH: Pathogen recognition and
inflammatory signaling in innate immune defenses. Clin Microbiol
Rev. 22:240–273. 2009. View Article : Google Scholar
|
|
68
|
Suresh R and Mosser DM: Pattern
recognition receptors in innate immunity, host defense, and
immunopathology. Adv Physiol Educ. 37:284–291. 2013. View Article : Google Scholar
|
|
69
|
Gaudet RG, Guo CX, Molinaro R, Kottwitz H,
Rohde JR, Dangeard AS, Arrieumerlou C, Girardin SE and Gray-Owen
SD: Innate recognition of intracellular bacterial growth is driven
by the TIFA-dependent cytosolic surveillance pathway. Cell Rep.
19:1418–1430. 2017. View Article : Google Scholar
|
|
70
|
Bai L, Li W, Zheng W, Xu D, Chen N and Cui
J: Promising targets based on pattern recognition receptors for
cancer immunotherapy. Pharmacol Res. 159:1050172020. View Article : Google Scholar
|
|
71
|
Luis Muñoz-Carrillo J, Francisco
Contreras-Cordero J, Gutiérrez-Coronado O, Trinidad
Villalobos-Gutiérrez P, Guillermo Ramos-Gracia L and Elizabeth
Hernández-Reyes V: Cytokine profiling plays a crucial role in
activating immune system to clear infectious pathogens. Immune
Response Activation and Immunomodulation. IntechOpen; 2019,
View Article : Google Scholar
|
|
72
|
Kim M and Kim CH: Regulation of humoral
immunity by gut microbial products. Gut Microbes. 8:392–399. 2017.
View Article : Google Scholar
|
|
73
|
Nagashima K, Sawa S, Nitta T, Tsutsumi M,
Okamura T, Penninger JM, Nakashima T and Takayanagi H:
Identification of subepithelial mesenchymal cells that induce IgA
and diversify gut microbiota. Nat Immunol. 18:675–682. 2017.
View Article : Google Scholar
|
|
74
|
Cong Y, Feng T, Fujihashi K, Schoeb TR and
Elson CO: A dominant, coordinated T regulatory cell-IgA response to
the intestinal microbiota. Proc Natl Acad Sci USA. 106:19256–19261.
2009. View Article : Google Scholar
|
|
75
|
Tezuka H and Ohteki T: Regulation of IgA
production by intestinal dendritic cells and related cells. Front
Immunol. 10:18912019. View Article : Google Scholar
|
|
76
|
Wu HJ and Wu E: The role of gut microbiota
in immune homeostasis and autoimmunity. Gut Microbes. 3:4–14. 2012.
View Article : Google Scholar
|
|
77
|
Lee GR: The Balance of Th17 versus Treg
cells in autoimmunity. Int J Mol Sci. 19:7302018. View Article : Google Scholar
|
|
78
|
Mazmanian SK, Liu CH, Tzianabos AO and
Kasper DL: An immunomodulatory molecule of symbiotic bacteria
directs maturation of the host immune system. Cell. 122:107–118.
2005. View Article : Google Scholar
|
|
79
|
Gaboriau-Routhiau V, Rakotobe S, Lécuyer
E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M,
Brandi G, et al: The key role of segmented filamentous bacteria in
the coordinated maturation of gut helper T cell responses.
Immunity. 31:677–689. 2009. View Article : Google Scholar
|
|
80
|
Valeri M and Raffatellu M: Cytokines IL-17
and IL-22 in the host response to infection. Pathog Dis.
74:ftw1112016. View Article : Google Scholar
|
|
81
|
Liu R, Lauridsen HM, Amezquita RA, Pierce
RW, Jane-Wit D, Fang C, Pellowe AS, Kirkiles-Smith NC, Gonzalez AL
and Pober JS: IL-17 promotes neutrophil-mediated immunity by
activating microvascular pericytes and not endothelium. J Immunol.
197:2400–2408. 2016. View Article : Google Scholar
|
|
82
|
Stary G, Olive A, Radovic-Moreno AF,
Gondek D, Alvarez D, Basto PA, Perro M, Vrbanac VD, Tager AM, Shi
J, et al: VACCINES. A mucosal vaccine against chlamydia trachomatis
generates two waves of protective memory T cells. Science.
348:aaa82052015. View Article : Google Scholar
|
|
83
|
Hirota K, Duarte JH, Veldhoen M, Hornsby
E, Li Y, Cua DJ, Ahlfors H, Wilhelm C, Tolaini M, Menzel U, et al:
Fate mapping of IL-17-producing T cells in inflammatory responses.
Nat Immunol. 12:255–263. 2011. View Article : Google Scholar
|
|
84
|
Oh JZ, Ravindran R, Chassaing B, Carvalho
FA, Maddur MS, Bower M, Hakimpour P, Gill KP, Nakaya HI, Yarovinsky
F, et al: TLR5-mediated sensing of gut microbiota is necessary for
antibody responses to seasonal influenza vaccination. Immunity.
41:478–492. 2014. View Article : Google Scholar
|
|
85
|
Sandrini S, Aldriwesh M, Alruways M and
Freestone P: Microbial endocrinology: Host-bacteria communication
within the gut microbiome. J Endocrinol. 225:R21–R34. 2015.
View Article : Google Scholar
|
|
86
|
Gensollen T, Iyer SS, Kasper DL and
Blumberg RS: How colonization by microbiota in early life shapes
the immune system. Science. 352:539–544. 2016. View Article : Google Scholar
|
|
87
|
Schmidt TSB, Raes J and Bork P: The human
gut microbiome: From association to modulation. Cell.
172:1198–1215. 2018. View Article : Google Scholar
|
|
88
|
Bultman SJ: Emerging roles of the
microbiome in cancer. Carcinogenesis. 35:249–255. 2014. View Article : Google Scholar
|
|
89
|
Cani PD: Human gut microbiome: Hopes,
threats and promises. Gut. 67:1716–1725. 2018. View Article : Google Scholar
|
|
90
|
LeBlanc JG, Chain F, Martín R,
Bermúdez-Humarán LG, Courau S and Langella P: Beneficial effects on
host energy metabolism of short-chain fatty acids and vitamins
produced by commensal and probiotic bacteria. Microb Cell Fact.
16:792017. View Article : Google Scholar
|
|
91
|
Nakkarach A, Foo HL, Song AA, Mutalib NEA,
Nitisinprasert S and Withayagiat U: Anti-cancer and
anti-inflammatory effects elicited by short chain fatty acids
produced by Escherichia coli isolated from healthy human gut
microbiota. Microb Cell Fact. 20:362021. View Article : Google Scholar
|
|
92
|
Clarke G, Stilling RM, Kennedy PJ, Stanton
C, Cryan JF and Dinan TG: Minireview: Gut microbiota: The neglected
endocrine organ. Mol Endocrinol. 28:1221–1238. 2014. View Article : Google Scholar
|
|
93
|
Pabst O: New concepts in the generation
and functions of IgA. Nat Rev Immunol. 12:821–832. 2012. View Article : Google Scholar
|
|
94
|
Mantis NJ, Rol N and Corthésy B: Secretory
IgA's complex roles in immunity and mucosal homeostasis in the gut.
Mucosal Immunol. 4:603–611. 2011. View Article : Google Scholar
|
|
95
|
Mathias A, Pais B, Favre L, Benyacoub J
and Corthésy B: Role of secretory IgA in the mucosal sensing of
commensal bacteria. Gut Microbes. 5:688–695. 2014. View Article : Google Scholar
|
|
96
|
Levy M, Kolodziejczyk AA, Thaiss CA and
Elinav E: Dysbiosis and the immune system. Nat Rev Immunol.
17:219–232. 2017. View Article : Google Scholar
|
|
97
|
Round JL and Mazmanian SK: Inducible
Foxp3+ regulatory T-cell development by a commensal
bacterium of the intestinal microbiota. Proc Natl Acad Sci USA.
107:12204–12209. 2010. View Article : Google Scholar
|
|
98
|
Cianci R, Franza L, Schinzari G, Rossi E,
Ianiro G, Tortora G, Gasbarrini A, Gambassi G and Cammarota G: The
interplay between immunity and microbiota at intestinal
immunological niche: The case of cancer. Int J Mol Sci. 20:5012019.
View Article : Google Scholar
|
|
99
|
Shui L, Yang X, Li J, Yi C, Sun Q and Zhu
H: Gut microbiome as a potential factor for modulating resistance
to cancer immunotherapy. Front Immunol. 10:29892020. View Article : Google Scholar
|
|
100
|
Zipkin M: Fecal microbiota potentiate
checkpoint inhibitors, unleash microbiome startups. Nat Biotechnol.
39:529–532. 2021. View Article : Google Scholar
|
|
101
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar
|
|
102
|
Niedzwiecki MM, Walker DI, Vermeulen R,
Chadeau-Hyam M, Jones DP and Miller GW: The exposome: Molecules to
populations. Annu Rev Pharmacol Toxicol. 59:107–127. 2019.
View Article : Google Scholar
|
|
103
|
Arem H and Loftfield E: Cancer
epidemiology: A survey of modifiable risk factors for prevention
and survivorship. Am J Lifestyle Med. 12:200–210. 2017. View Article : Google Scholar
|
|
104
|
IARC Working Group on the Evaluation of
Carcinogenic Risks to Humans: Biological agents. Volume 100 B. A
review of human carcinogens. IARC Monogr Eval Carcinog Risks Hum.
100:1–441. 2012.
|
|
105
|
Wroblewski LE, Peek RM Jr and Wilson KT:
Helicobacter pylori and gastric cancer: Factors that modulate
disease risk. Clin Microbiol Rev. 23:713–739. 2010. View Article : Google Scholar
|
|
106
|
Whisner CM and Athena Aktipis C: The role
of the microbiome in cancer initiation and progression: How
microbes and cancer cells utilize excess energy and promote one
another's growth. Curr Nutr Rep. 8:42–51. 2019. View Article : Google Scholar
|
|
107
|
Villéger R, Lopès A, Carrier G, Veziant J,
Billard E, Barnich N, Gagnière J, Vazeille E and Bonnet M:
Intestinal microbiota: A novel target to improve anti-tumor
treatment? Int J Mol Sci. 20:45842019. View Article : Google Scholar
|
|
108
|
Li W, Deng X and Chen T: Exploring the
modulatory effects of gut microbiota in anti-cancer therapy. Front
Oncol. 11:6444542021. View Article : Google Scholar
|
|
109
|
Guerra L, Cortes-Bratti X, Guidi R and
Frisan T: The biology of the cytolethal distending toxins. Toxins
(Basel). 3:172–190. 2011. View Article : Google Scholar
|
|
110
|
Sun J, Hobert ME, Duan Y, Rao AS, He TC,
Chang EB and Madara JL: Crosstalk between NF-kappaB and
beta-catenin pathways in bacterial-colonized intestinal epithelial
cells. Am J Physiol Gastrointest Liver Physiol. 289:G129–G137.
2005. View Article : Google Scholar
|
|
111
|
Liu X, Lu R, Wu S and Sun J: Salmonella
regulation of intestinal stem cells through the Wnt/beta-catenin
pathway. FEBS Lett. 584:911–916. 2010. View Article : Google Scholar
|
|
112
|
Scanu T, Spaapen RM, Bakker JM, Pratap CB,
Wu LE, Hofland I, Broeks A, Shukla VK, Kumar M, Janssen H, et al:
Salmonella manipulation of host signaling pathways provokes
cellular transformation associated with gallbladder carcinoma. Cell
Host Microbe. 17:763–774. 2015. View Article : Google Scholar
|
|
113
|
Li Y, Tinoco R, Elmé L, Segota I, Xian Y,
Fujita Y, Sahu A, Zarecki R, Marie K, Feng Y, et al: Gut microbiota
dependent anti-tumor immunity restricts melanoma growth in
Rnf5−/− mice. Nat Commun. 10:14922019. View Article : Google Scholar
|
|
114
|
Tanoue T, Morita S, Plichta DR, Skelly AN,
Suda W, Sugiura Y, Narushima S, Vlamakis H, Motoo I, Sugita K, et
al: A defined commensal consortium elicits CD8 T cells and
anti-cancer immunity. Nature. 565:600–605. 2019. View Article : Google Scholar
|
|
115
|
Valguarnera E and Wardenburg JB: Good gone
bad: One toxin away from disease for Bacteroides fragilis. J Mol
Biol. 432:765–785. 2020. View Article : Google Scholar
|
|
116
|
Thiele Orberg E, Fan H, Tam AJ, Dejea CM,
Destefano Shields CE, Wu S, Chung L, Finard BB, Wu X, Fathi P, et
al: The myeloid immune signature of enterotoxigenic Bacteroides
fragilis-induced murine colon tumorigenesis. Mucosal Immunol.
10:421–433. 2017. View Article : Google Scholar
|
|
117
|
Geis AL, Fan H, Wu X, Wu S, Huso DL, Wolfe
JL, Sears CL, Pardoll DM and Housseau F: Regulatory T-cell response
to enterotoxigenic Bacteroides fragilis colonization triggers
IL17-dependent colon carcinogenesis. Cancer Discov. 5:1098–1109.
2015. View Article : Google Scholar
|
|
118
|
Kostic AD, Chun E, Robertson L, Glickman
JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold
GL, et al: Fusobacterium nucleatum potentiates intestinal
tumorigenesis and modulates the tumor-immune microenvironment. Cell
Host Microbe. 14:207–215. 2013. View Article : Google Scholar
|
|
119
|
Sivan A, Corrales L, Hubert N, Williams
JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B,
Alegre ML, et al: Commensal Bifidobacterium promotes antitumor
immunity and facilitates anti-PD-L1 efficacy. Science.
350:1084–1089. 2015. View Article : Google Scholar
|
|
120
|
Routy B, Le Chatelier E, Derosa L, Duong
CPM, Alou MT, Daillère R, Fluckiger A, Messaoudene M, Rauber C,
Roberti MP, et al: Gut microbiome influences efficacy of PD-1-based
immunotherapy against epithelial tumors. Science. 359:91–97. 2018.
View Article : Google Scholar
|
|
121
|
Sharifi-Rad J, Rodrigues CF,
Stojanović-Radić Z, Dimitrijević M, Aleksić A, Neffe-Skocińska K,
Zielińska D, Kołożyn-Krajewska D, Salehi B, Milton Prabu S, et al:
Probiotics: Versatile bioactive components in promoting human
health. Medicina (Kaunas). 56:4332020. View Article : Google Scholar
|
|
122
|
Banna GL, Torino F, Marletta F, Santagati
M, Salemi R, Cannarozzo E, Falzone L, Ferraù F and Libra M:
Lactobacillus rhamnosus GG: An overview to explore the rationale of
its use in cancer. Front Pharmacol. 8:6032017. View Article : Google Scholar
|
|
123
|
Vétizou M, Pitt JM, Daillère R, Lepage P,
Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong
CP, et al: Anticancer immunotherapy by CTLA-4 blockade relies on
the gut microbiota. Science. 350:1079–1084. 2015. View Article : Google Scholar
|
|
124
|
Dubin K, Callahan MK, Ren B, Khanin R,
Viale A, Ling L, No D, Gobourne A, Littmann E, Huttenhower C, et
al: Intestinal microbiome analyses identify melanoma patients at
risk for checkpoint-blockade-induced colitis. Nat Commun.
7:103912016. View Article : Google Scholar
|
|
125
|
Frankel AE, Coughlin LA, Kim J, Froehlich
TW, Xie Y, Frenkel EP and Koh AY: Metagenomic shotgun sequencing
and unbiased metabolomic profiling identify specific human gut
microbiota and metabolites associated with immune checkpoint
therapy efficacy in melanoma patients. Neoplasia. 19:848–855. 2017.
View Article : Google Scholar
|
|
126
|
Chaput N, Lepage P, Coutzac C, Soularue E,
Le Roux K, Monot C, Boselli L, Routier E, Cassard L, Collins M, et
al: Baseline gut microbiota predicts clinical response and colitis
in metastatic melanoma patients treated with ipilimumab. Ann Oncol.
28:1368–1379. 2017. View Article : Google Scholar
|
|
127
|
Matson V, Fessler J, Bao R, Chongsuwat T,
Zha Y, Alegre ML, Luke JJ and Gajewski TF: The commensal microbiome
is associated with anti-PD-1 efficacy in metastatic melanoma
patients. Science. 359:104–108. 2018. View Article : Google Scholar
|
|
128
|
Gopalakrishnan V, Spencer CN, Nezi L,
Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman
K, Wei SC, et al: Gut microbiome modulates response to anti-PD-1
immunotherapy in melanoma patients. Science. 359:97–103. 2018.
View Article : Google Scholar
|
|
129
|
Derosa L, Hellmann MD, Spaziano M,
Halpenny D, Fidelle M, Rizvi H, Long N, Plodkowski AJ, Arbour KC,
Chaft JE, et al: Negative association of antibiotics on clinical
activity of immune checkpoint inhibitors in patients with advanced
renal cell and non-small-cell lung cancer. Ann Oncol. 29:1437–1444.
2018. View Article : Google Scholar
|
|
130
|
Jin Y, Dong H, Xia L, Yang Y, Zhu Y, Shen
Y, Zheng H, Yao C, Wang Y and Lu S: The diversity of gut microbiome
is associated with favorable responses to anti-programmed death 1
immunotherapy in chinese patients with NSCLC. J Thorac Oncol.
14:1378–1389. 2019. View Article : Google Scholar
|
|
131
|
Zheng Y, Wang T, Tu X, Huang Y, Zhang H,
Tan D, Jiang W, Cai S, Zhao P, Song R, et al: Gut microbiome
affects the response to anti-PD-1 immunotherapy in patients with
hepatocellular carcinoma. J Immunother Cancer. 7:1932019.
View Article : Google Scholar
|
|
132
|
Salgia NJ, Bergerot PG, Maia MC, Dizman N,
Hsu J, Gillece JD, Folkerts M, Reining L, Trent J, Highlander SK
and Pal SK: Stool microbiome profiling of patients with metastatic
renal cell carcinoma receiving anti-PD-1 immune checkpoint
inhibitors. Eur Urol. 78:498–502. 2020. View Article : Google Scholar
|
|
133
|
Xu X, Lv J, Guo F, Li J, Jia Y, Jiang D,
Wang N, Zhang C, Kong L, Liu Y, et al: Gut Microbiome influences
the efficacy of PD-1 antibody immunotherapy on MSS-type colorectal
cancer via metabolic pathway. Front Microbiol. 11:8142020.
View Article : Google Scholar
|
|
134
|
Mager LF, Burkhard R, Pett N, Cooke NCA,
Brown K, Ramay H, Paik S, Stagg J, Groves RA, Gallo M, et al:
Microbiome-derived inosine modulates response to checkpoint
inhibitor immunotherapy. Science. 369:1481–1489. 2020. View Article : Google Scholar
|
|
135
|
Si W, Liang H, Bugno J, Xu Q, Ding X, Yang
K, Fu Y, Weichselbaum RR, Zhao X and Wang L: Lactobacillus
rhamnosus GG induces cGAS/STING-dependent type I interferon and
improves response to immune checkpoint blockade. Gut.
gutjnl-2020-323426. 2021.Epub ahead of print. View Article : Google Scholar
|
|
136
|
Shi Y, Zheng W, Yang K, Harris KG, Ni K,
Xue L, Lin W, Chang EB, Weichselbaum RR and Fu YX: Intratumoral
accumulation of gut microbiota facilitates CD47-based immunotherapy
via STING signaling. J Exp Med. 217:e201922822020. View Article : Google Scholar
|
|
137
|
Beer TM, Kwon ED, Drake CG, Fizazi K,
Logothetis C, Gravis G, Ganju V, Polikoff J, Saad F, Humanski P, et
al: Randomized, double-blind, phase III Trial of ipilimumab versus
placebo in asymptomatic or minimally symptomatic patients with
metastatic chemotherapy-naive castration-resistant prostate cancer.
J Clin Oncol. 35:40–47. 2017. View Article : Google Scholar
|
|
138
|
Berger MF, Lawrence MS, Demichelis F,
Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger
D, Sougnez C, et al: The genomic complexity of primary human
prostate cancer. Nature. 470:214–220. 2011. View Article : Google Scholar
|
|
139
|
Nicholson LT and Fong L: Immune checkpoint
inhibition in prostate cancer. Trends Cancer. 6:174–177. 2020.
View Article : Google Scholar
|
|
140
|
Venkatachalam S, McFarland TR, Agarwal N
and Swami U: Immune checkpoint inhibitors in prostate cancer.
Cancers (Basel). 13. pp. 21872021, View Article : Google Scholar
|
|
141
|
Crocetto F, Boccellino M, Barone B, Di
Zazzo E, Sciarra A, Galasso G, Settembre G, Quagliuolo L, Imbimbo
C, Boffo S, et al: The crosstalk between prostate cancer and
microbiota inflammation: Nutraceutical products are useful to
balance this interplay? Nutrients. 12:26482020. View Article : Google Scholar
|
|
142
|
Ferro M, Lucarelli G, Crocetto F, Dolce P,
Verde A, La Civita E, Zappavigna S, de Cobelli O, Di Lorenzo G,
Facchini BA, et al: First-line systemic therapy for metastatic
castration-sensitive prostate cancer: An updated systematic review
with novel findings. Crit Rev Oncol Hematol. 157:1031982021.
View Article : Google Scholar
|
|
143
|
Barbosa AM, Gomes-Gonçalves A, Castro AG
and Torrado E: Immune system efficiency in cancer and the
microbiota influence. Pathobiology. 88:170–186. 2021. View Article : Google Scholar
|
|
144
|
Chen D, Wu J, Jin D, Wang B and Cao H:
Fecal microbiota transplantation in cancer management: Current
status and perspectives. Int J Cancer. 145:2021–2031. 2019.
View Article : Google Scholar
|
|
145
|
Thompson S, Guetterman H, Taylor A, Bogner
A, Martin D, Farrell JJ, Swanson KS and Holscher H: Dietary
predictors of fecal microbiota transplantation success. J Acad Nutr
Diet. 116(Suppl 9): A762016. View Article : Google Scholar
|
|
146
|
Diefenbach CS, Hong F, Ambinder RF, Cohen
JB, Robertson MJ, David KA, Advani RH, Fenske TS, Barta SK,
Palmisiano ND, et al: Ipilimumab, nivolumab, and brentuximab
vedotin combination therapies in patients with relapsed or
refractory Hodgkin lymphoma: Phase 1 results of an open-label,
multicentre, phase 1/2 trial. Lancet Haematol. 7:e660–e670. 2020.
View Article : Google Scholar
|
|
147
|
Bensch F, van der Veen EL, Lub-de Hooge
MN, Jorritsma-Smit A, Boellaard R, Kok IC, Oosting SF, Schröder CP,
Hiltermann TJN, van der Wekken AJ, et al:
89Zr-atezolizumab imaging as a non-invasive approach to
assess clinical response to PD-L1 blockade in cancer. Nat Med.
24:1852–1858. 2018. View Article : Google Scholar
|
|
148
|
Kyte JA, Røssevold A, Falk RS and Naume B:
ALICE: A randomized placebo-controlled phase II study evaluating
atezolizumab combined with immunogenic chemotherapy in patients
with metastatic triple-negative breast cancer. J Transl Med.
18:2522020. View Article : Google Scholar
|
|
149
|
Cohen R, Pudlarz T, Garcia-Larnicol ML,
Vernerey D, Dray X, Clavel L, Jary M, Piessen G, Zaanan A, Aparicio
T, et al: Localized MSI/dMMR gastric cancer patients, perioperative
immunotherapy instead of chemotherapy: The GERCOR NEONIPIGA phase
II study is opened to recruitment. Bull Cancer. 107:438–446.
2020.In French. View Article : Google Scholar
|