Introduction
Pancreatic cancer (PC) is one of the most aggressive
and devastating types of cancer owing to its poor prognosis and
deadly characteristics. It is the 4th leading cause of cancer-led
mortality in the USA, with only 8% chance of surviving longer than
5 years (1), and it will be
ranked second due to its high mortality by 2030 (2). PC has the most dismal diagnosis
among all digestive malignancies due to its late clinical
appearance, lack of an effective cure and a constantly increasing
global incidence ratio (3,4).
Therefore, it is particularly important to reveal the molecular
mechanisms that lead to PC.
Human Williams-Beuren syndrome chromosomal region 22
(WBSCR22) was initially identified as one of the 26 contiguous
genes that are deleted in Williams-Beuren syndrome (WBS). WBS is a
neurological disorder that displays clinically unique features,
such as mental retardation, dysmorphic facial appearances, sunken
chest, vascular and congenital heart disease, hypertension and
infantile hypocalcemia. The etiology of this disease is associated
with the induction of oxidative stress. The main contributing
genes, including superoxide dismutase-1 and neutrophil cytosolic
factor-1, are primarily involved in the regulation of the
redox-state (5-7). WBSCR22 is localized on chromosome 7
(7q11.23) and constitutes an S-adenosyl-L-methionine (SAM) motif
and nuclear localization signal with well-preserved features of
methyltransferase family proteins (8,9).
Subsequently, the human WBSCR22 protein was identified as a
methyltransferase for 18S rRNA m7G, which was involved
in pre-rRNA processing and 40S ribosome subunit biogenesis
(10-12). In addition, WBSCR22 has been
revealed to modulate histone methylation (13).
Previous studies have revealed that WBSCR22
expression is upregulated in several types of cancer, including
invasive breast cancer (13),
multiple myeloma, plasma cells (14), colorectal cancer (15), lung cancer (16) and hepatocellular carcinoma
(17). In addition, upregulation
of WBSCR22 expression promotes invasion, proliferation and
migration, while knockdown of this gene exhibits the opposite
effects in glioma cells (18).
WBSCR22 exhibits tumor-promoting potential in several types of
cancer, whereas WBSCR22 loss has also been reported in specific
inflammatory-type human lung pathologies (16). However, the function of WBSCR22 in
PC remains unknown. In the present study, it was unexpectedly
revealed that WBSCR22 played a tumor suppressor role in PC.
Interferon-stimulated gene 15 (ISG15) is a 17-kDa
protein consisting of 165 amino acids, which is induced by
interferon (INF-Type I) treatment and is extensively recognized as
an antiviral protein (19-21).
ISG15 is a ubiquitin-like protein that covalently binds to several
nuclear and cytoplasmic proteins, commonly referred to as the
ISGylation process (22,23). Similar to ubiquitination,
ISGylation requires a three-step enzymatic system, including
activating E1 enzyme (UBE1L), conjugating E2 enzyme (UBCH8) and
ligating E3 (24-26). It has been widely accepted that
the function of ISG15 or the effects of ISGylation are tissue
specific (27). Certain studies
have suggested its role in multiple cellular functions, including
protein turnover (28), protein
stability (27),
interferon-induced immune responses and in the production of type
II IFN, which enhances the proliferation and activity of natural
killer cells, particularly neutrophils (29-31). Elevated expression levels of ISG15
have been detected in various human cancer cells, including
pancreatic adenocarcinoma, and breast, colorectal, ovarian and
prostate carcinomas (32).
Additionally, the oncogenic capacities of ISG15, including the
promotion of migration, proliferation and tumorigenesis, have been
well established in multiple types of cancer (33-35). Conversely, a tumor suppressive
function of ISG15 has also been reported in hepatocellular,
cervical and breast cancer (36-39). ISG15 exhibits inconsistent
biological functions that are explained by its ability to act both
as a tumor promoter and suppressor, depending on the cancer type.
To the best of our knowledge, the role of ISG15 in PC remains
unclear and requires further investigation.
Several methyltransferases require accessory
proteins for their activity and stability. The human protein tRNA
methyltransferase activator subunit 11-2 (TRMT112) shares homology
with the TRMT112 protein of Saccharomyces cerevisiae. The
TRMT112 protein is highly conserved, and is expressed at high
levels in multiple tissues and organs, particularly during early
mouse development (40,41). TRMT112 acts as a cofactor for
various methyltransferases, including methylated rRNA, tRNA and
proteins (42-44). It has been reported that TRMT112
is the interaction partner of WBSCR22, and enhances the stability
of WBSCR22 (45). The interaction
between WBSCR22 and TRMT112 was also reported to be involved in the
processing of pre-rRNA, leading to the generation of 18S-rRNA
(10). However, the biological
function of the WBSCR22-TRMT112 interaction in pancreatic tumor
development remains unclear.
In the present study, the tumor suppressor role of
WBSCR22 was reported for the first time in PC. Overexpression of
WBSCR22 significantly suppressed the proliferation, migration,
invasion and tumorigenesis of PANC-1 cells in vivo and in
vitro, while knockdown of its expression exhibited the opposite
effects. In addition, RNA-sequencing (RNA-seq) analysis revealed
that ISG15 is a downstream target of WBSCR22, as its mRNA
expression and protein levels were markedly reduced in
WBSCR22-overexpressing cells. Additionally, ISG15 plays an
oncogenic role in PC, as upregulated expression of ISG15 promoted
the proliferation, migration, invasion and tumorigenesis of PC.
Furthermore, it was suggested that TRMT112 and WBSCR22 may function
synergistically in PC and that ectopic OE of WBSCR22 and TRMT112
may simultaneously promote the tumor suppressive potential of
WBSCR22 in PC. WBSCR22 is a clinically important gene in PC and the
newly identified WBSCR22/ISG15 axis may represent an innovative
approach for therapeutic purposes.
Materials and methods
Analysis of the cancer genome atlas
(TCGA) and human protein atlas
The genes of interest, which were differentially
expressed in PC cells were investigated in comparison with adjacent
normal non-neoplastic tissues using TCGA (http://gepia.cancer-pku.cn/ and http://tcga-data.nci.nih.gov/tcga/). The WBSCR22
prognostic value, clinical significance, overall survival and
expression profiles were examined in the TCGA cohort.
Cell culture
The human PC cell lines PANC-1 (resource no.
1101HUM-PUMC000023), BXPC3 (resource no. 1101HUM-PUMC000274) and
ASPC1 (resource no. 1101HUM-PUMC000214), a normal human pancreatic
ductal epithelial cell line, HPDE6-C7 (resource no. C1248;
https://www.whelab.com/pro/580.html),
and the human embryonic kidney cell line, 293T (resource no.
4201HUM-CCTCC00187) were purchased from The Cell Bank of Type
Culture Collection of Chinese Academy of Sciences. All cell lines
were authenticated at the beginning of the present study by
species-specific PCR evaluation (Chinese Academy of Sciences). The
cell line authentication profile is provided in Table SI. All the cell lines were
cultured in Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher
Scientific, Inc.) high glucose, supplemented with 10% fetal bovine
serum (FBS; cat. no. 04-007-1A; Biological Industries). In order to
inhibit residual bacterial activity two potential antibiotics were
supplemented with a concentration of 100 U/ml penicillin and
streptomycin (100 ng/ml) reagent (both from Hyclone; Cytiva). The
cell culture was performed at 37°C in a humidified incubator with
5% CO2.
Generation of stable cell lines
The PC cell line PANC-1 was cultured and incubated
in a humidified incubator at 37°C with 5% CO2.
WBSCR22-OE and ISG15-OE stably transfected cells were generated and
the CDS portion of WBSCR22 was subcloned into the lentiviral vector
pCDH-EF1α-MCS-T2A-Puro (cat. no. CD527A-1; System Biosciences).
Small hairpin (sh)WBSCR22 stable cell line generation was achieved
by subcloning the shRNA sequence of WBSCR22 into the
pLVX-shRNA2-BSD lentiviral vector upgraded from pLVX-shRNA2 (cat.
no. 632179; Clonetech laboratories Inc.). The 293T cells were
transfected with the aforementioned lentiviral plasmids (0.5
µg/µl) and the second-generation lentivirus packing
system (0.5 µg/µl pMD2.0 and 0.5 µg/µl
pspAX2; mixed ratio was pMD2.0: pspAX2: lentivirus, 1:3:4) using
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) for
72 h at 37°C. Subsequently, the supernatant containing the
infectious lentivirus was collected and transduced to PANC-1 cells
for 72 h, at a multiplicity of infection (MOI) of 10. Following 36
h of incubation after transfection, puromycin (2 µg/ml) or
Blasticidin S (5 µg/ml) was added to the cultured cells to
select for overexpressing and knockdown cells, respectively. The
selection was performed for 14 days, and overexpression/knockdown
in all positive viable cells was further confirmed by reverse
transcription-quantitative PCR (RT-qPCR) and western blotting.
Wound healing assay
The cells cultured previously were harvested during
the logarithmic growth phase and seeded in a six-well plate to
create a confluent monolayer. When the confluency reached 70-80%, a
horizontal scratch was made using a sterile 200-µl
microliter pipette tip. The medium was removed and the cells were
washed twice with cold PBS to remove the cells detached during
wound scratching. Fresh DMEM supplemented with 2% FBS was added to
the cells. Images of the selected areas were obtained at 0 h and
subsequently the culture was incubated overnight at 37°C with 5%
CO2. The selected wound area was imaged following 24 and
48 h of culture using a fluorescence microscope (Leica
Microsystems, Inc.), and the wound area was assessed using ImageJ
software (version 1.53e). Wound healing was quantified by
calculating the percent of wound closure: The % of wound
closure=1-(wound surface area at the indicated time-point/initial
wound surface area). The assay was performed in triplicate.
Colony formation assay
To evaluate the reproductive potential of the cells,
the colony formation assay was performed. The cells that were
cultured previously were harvested at 70-90% confluency and
resuspended in 1-2 ml DMEM. Cells were counted, and ~200 cells/well
were seeded into a 6-well plate. Fresh DMEM with 10% FBS was added
and incubated in a humidified incubator for 14 days at 37°C
supplied with 5% CO2. Following the incubation period,
the media was removed and the cells were washed with cold PBS twice
to remove excess media. Cells were fixed with 1% formaldehyde for
15 min followed by staining with 0.5% crystal violet in 25%
methanol for 30 min at room temperature. Cell differentiation was
performed by washing the cells with tap water and subsequent
drying. The purple dots (representing a colony grown during the
incubation period, containing >50 cells) were counted manually.
The assay was performed in triplicate.
Transwell invasion assay
The cell lines cultured previously were harvested
and resuspended in serum-free DMEM. The Transwell apparatus
consisted of two chambers. The upper chamber of the insert was a
Matrigel-coated Millicell chamber (BD Biosciences) with an
8-µm pore size membrane, to which cells were added at a
density of 6×103 cells/well for all Transwell assays in
this study. Except for one set of experiments involving three cell
lines (Ctrl, WBSCR22-OE and WBSCR22-OE + TRMT112OE cells), the cell
seeding density in this set was 1×104 cells/well. A
total of ~200 µl serum-free DMEM was added to the upper
chamber. The lower chamber contained ~500 µl DMEM
supplemented with 10% FBS. The 24-well plate was incubated for 24 h
in a humidified incubator at 37°C with 5% CO2. Following
incubation, the media were removed and washed with cold PBS twice.
The upper chamber was fixed with 1% formaldehyde for 15 min and
subsequently stained with 0.5% crystal violet for 15 min at room
temperature. The inner side of the upper insert was cleaned with a
cotton swab to remove the excess stain and cells. The chamber was
air-dried and the cells were counted in three different fields of
view using a light microscopy at a magnification of ×100 (Leica
Microsystems, Inc.).
Cell proliferation assay
The cell proliferative potential was analyzed using
a Cell Counting Kit-8 (CCK-8) assay. The cells were seeded in a
96-well plate at a concentration of 2×103 cells/well.
The plate was incubated in a humidified incubator at 37°C with 5%
CO2 and 10 ml CCK-8 (Beijing Solarbio Science &
Technology Co., Ltd.) was added to each well. Following incubation
for 2 h, the optical density was measured at 450 nm using a
Microplate reader (Bio-Rad Laboratories, Inc.). The experiment was
performed in triplicate for a single cell line and the optical
density was expressed as a percentage of viable cells.
Promoter luciferase reporter assay
A promoter sequence 2001 bp in length, ranging from
the-2000 base site to the +77 base site of the ISG15 gene, was
copied into the modified pGL4.11[luc2P] (Promega Corporation)
vector. The Renilla luciferase gene sequence was added to
the vector for the normalization of transfection efficiency. Cells
were cotransfected with the pcDNA3.1-WBSCR22 plasmid or pcDNA3.1
empty vector and 50 ng of the pGL-ISG15-promoter vector using
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. Firefly luciferase
activities and Renilla signals were measured 48 h after
transfection using a Dual-luciferase reporter assay kit (Promega
Corporation) according to the manufacturer's instructions.
RT-qPCR
Total RNA was isolated using an RNA purification kit
(cat. no. DP430; Tiangen Biotech Co., Ltd.) according to the
manufacturer's protocol. The isolated RNA was reverse transcribed
to cDNA using a HiScript III 1st Strand cDNA Synthesis kit (Vazyme
Biotech Co., Ltd.) according to the manufacturer's protocol. qPCR
was performed using ChamQ SYBR Color qPCR MasterMix (Vazyme Biotech
Co., Ltd.) on an ABI StepOne system (Applied Biosystems; Thermo
Fisher Scientific, Inc.) with the following thermocycling
conditions: Initial denaturation step at 95°C for 3 min, followed
by 40 cycles of denaturation at 95°C for 15 sec, annealing at 60°C
for 15 sec and extension at 72°C for 30 sec. The 2−ΔΔCq
method was calculated to analyze the relative changes in gene
expression according to the previous reported method (46). Normalization of the expression
levels was performed using β-actin as the internal control. The
sequences for the corresponding forward and reverse primers used
for WBSCR22 and β-actin were as follows: human WBSCR22 forward,
5′-ATG AGA GGG AAG GTG GAG CA-3′ and reverse, 5′-AGA ACC GCG TGG
TGA CTT AG-3′; and human β-actin forward, 5′-TGA CGT GGA CAT CCG
CAA AG-3′ and reverse, 5′-CTG GAA GGT GGA CAG CGA GG-3′.
Western blot analysis
Total protein was extracted using cell lysis buffer
(Beyotime Institute of Biotechnology) supplemented with a protease
and phosphatase inhibitor cocktail (Beyotime Institute of
Biotechnology). The protein concentration was quantified using a
bicinchoninic acid protein estimation kit (Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. A total
of 20 µg protein/per lane was loaded on 4-20% SDS-gels and
resolved using SDS-PAGE, and subsequently transferred to a PVDF
membrane (MilliporeSigma). The membranes were rinsed with
Trisbuffered saline with 0.1% Tween-20 (TBST) for 5 min and blocked
by 5% skimmed milk powder at room temperature for 2 h. The
membranes were then subjected to immunoblotting with the
appropriate primary (WBSCR22; cat. no. A7317; 1:1,000; ABclonal
Biotech Co. and ISG15; cat. no. P05161; 1:1,000; Cusabio technology
LLC.) at 4°C overnight and goat anti-rabbit (HRP) secondary
antibodies (cat. no. orb43514; 1:5,000; Biorbyt) for 2 h at room
temperature. The GAPDH antibody (cat. no. orb555879; 1:5,000;
Biorbyt) was used as a control. Signals were visualized using the
BeyoECL Moon super sensitivity detection kit (Beyotime Institute of
Biotechnology) according to the manufacturer's protocol, and the
blots were quantified using Image Pro Plus v6.0 software (Media
Cybernetics, Inc.).
RNA isolation, cDNA library preparation
and sequencing
Cells were harvested and total RNA was extracted
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). Assessment of RNA quality was performed using a
Qubit Flourometer (Agilent 2100 Bioanalyzer; Agilent Technologies,
Inc.). Total RNA samples, which exhibited optimal quality, were
used in subsequent experiments. The following criteria were used:
RNA integrity number >7.0 and 28S to 18S ratio >1.8. RNA-seq
libraries were generated and sequenced by CapitalBio Technology,
Inc. All assays were run in triplicate and an independent library
was constructed in order to perform sequencing. The construction of
the library for sequencing was performed using the NEB Next Ultra
RNA Library Prep for Illumina, Inc. (New England Biolabs, Inc.). A
total of 1 µg total RNA was used. The mRNA Poly (A) tailed
enrichment was performed using the NEB Next Poly (A) mRNA Magnetic
Isolation Module kit (New England Biolabs, Inc.), and fragments of
~200 base pairs were generated. The first-strand and second-strand
cDNAs were generated from mRNA fragments using random hexamer
primers, reverse transcriptase and DNA polymerase-I and RNase H,
respectively. A single adenine base was added to the ends of the
cDNA fragments followed by adapter ligation. The end products were
first purified and subsequently enriched via PCR to amplify the DNA
library. The quantification of the final product of the DNA
libraries was performed using Agilent 2100 Bioanalyzer and the KAPA
Library quantification kit (Kapa Biosystems; Roche Diagnostics
GmbH) according to the manufacturer's protocol. All the RT-qPCR
validated libraries were processed by paired-end sequencing with a
pair end of 150-base pair length reading on the Illumina Novaseq
sequencer (Illumina, Inc.).
RNA-seq and data analysis
The hg38 human genome version was used as a
reference (Homo_sapiens.GRCh38.92.gtf). The assessment of the
quality of the sequencing was performed using FastQC (v0.11.5;
https://www.bioinformatics.babraham.ac.uk/projects/fastqc/)
and the filtration of the low quality data was achieved through
NGSQC (v2.3.3; http://hpc.ilri.cgiar.org/_export/xhtml/ngsqctoolkit-software#installation).
The alignment of the clear and clean reads was performed using
HISAT2 (v2.2.0; https://daehwankimlab.github.io/hisat2/). Each sample
was aligned from the processed reads against the reference genome
using HISAT2. The differentially expressed genes (DEGs) in the
samples were analyzed using DESeq9 (v1.28.0) (https://www.huber.embl.de/users/anders/DESeq/).
Multiple independent statistical hypotheses were separately
performed on the DEGs. The obtained P-value was corrected using the
FDR method. The BH method was used to calculate the corrected
P-value (q-value). Significance analysis was performed using the
P-value. The different parameters used for categorizing the
significant DEGs were based on ≥2-fold differences (|log2FC|≥1;
where FC is the fold change in expression) in the transcript
abundance and with P≤0.05. Annotation of DEGs was performed on the
basis of the information obtained from Uniprot, Gene Ontology (GO),
NCBI, ENSEMBL and Kyoto Encyclopedia of Genes and Genomes (KEGG).
The RNA-seq data were submitted to the Gene Expression Omnibus
(GEO; GSE186154).
In vivo tumorigenicity assays
A total of 12 female nude mice (6 to 8 weeks old;
20-25 g) were purchased from Weitong Lihua Experimental Animal
Technology Co., Ltd. The mice were allowed to grow in a specific
pathogen-free facility (12-h light/dark cycle; 18-23°C; and
humidity 50-60%) and were provided with ad libitum access to
all nutritional supplements. A total of 2×106 WBSCR22-OE
cells or the corresponding control cells in 200 µl PBS were
subcutaneously injected into the left and right flanks of the mice.
Following 4-5 weeks of injection, the mice were anesthetized with
1% pentobarbital sodium (45 mg/kg, intraperitoneal) and
subsequently euthanized by cervical dislocation. The tumor volume
and weight were assessed. The experiments were executed in
triplicate. All animal experiments performed in the present study
were approved (approval no. IRB-1507) by the Ethics Committee of
the Beijing University of Technology (Beijing, China). The maximum
tumor volume allowed by the Ethics Committee of the Beijing
University of Technology was 300 mm3 per tumor (or 1.5
cm in diameter).
Statistical analysis
Data are presented as the mean ± standard deviation
of at least three independent experiments. Quantitative results
were compared using GraphPad Prism version 5.0 (GraphPad Software,
Inc.). Data normality tests were performed using the Shapiro-Wilk
and Kolmogorov-Smirnov tests. A two-tailed paired Student's t-test
was used to assess the significance between two groups. The
differences between multiple groups were compared using a one-way
or two-way ANOVA with a Tukey's post-hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
WBSCR22 gene expression is downregulated
in PC cells
According to data obtained from TCGA, the expression
levels of WBSCR22 are associated with the overall survival rate of
patients with PC. High expression levels of WBSCR22 were
significantly associated with an improved 5-year survival rate
(40%) (PDAC, n=103), while low expression levels were associated
with a reduced 5-year survival rate (9%) (PDAC, n=73; Fig. 1A). In addition, analysis of
results from the Human Protein Atlas indicated WBSCR22 protein
levels were downregulated in PC tissues compared with those of the
surrounding normal non-neoplastic healthy tissues. Reduced staining
was observed for WBSCR22 in PC tissues, whereas high WBSCR22
staining was observed in normal pancreatic tissues (Fig. 1B). Furthermore, western blot
analysis of the three PC cell lines (PANC-1, BXPC3 and ASPC1) and
one pancreatic ductal epithelial cell line (HPDE6-C7) confirmed the
decreased WBSCR22 expression levels in PDAC cells (Fig. 1C). These analyses suggested that
high WBSCR22 expression levels may have a direct tumor suppressor
role in PC.
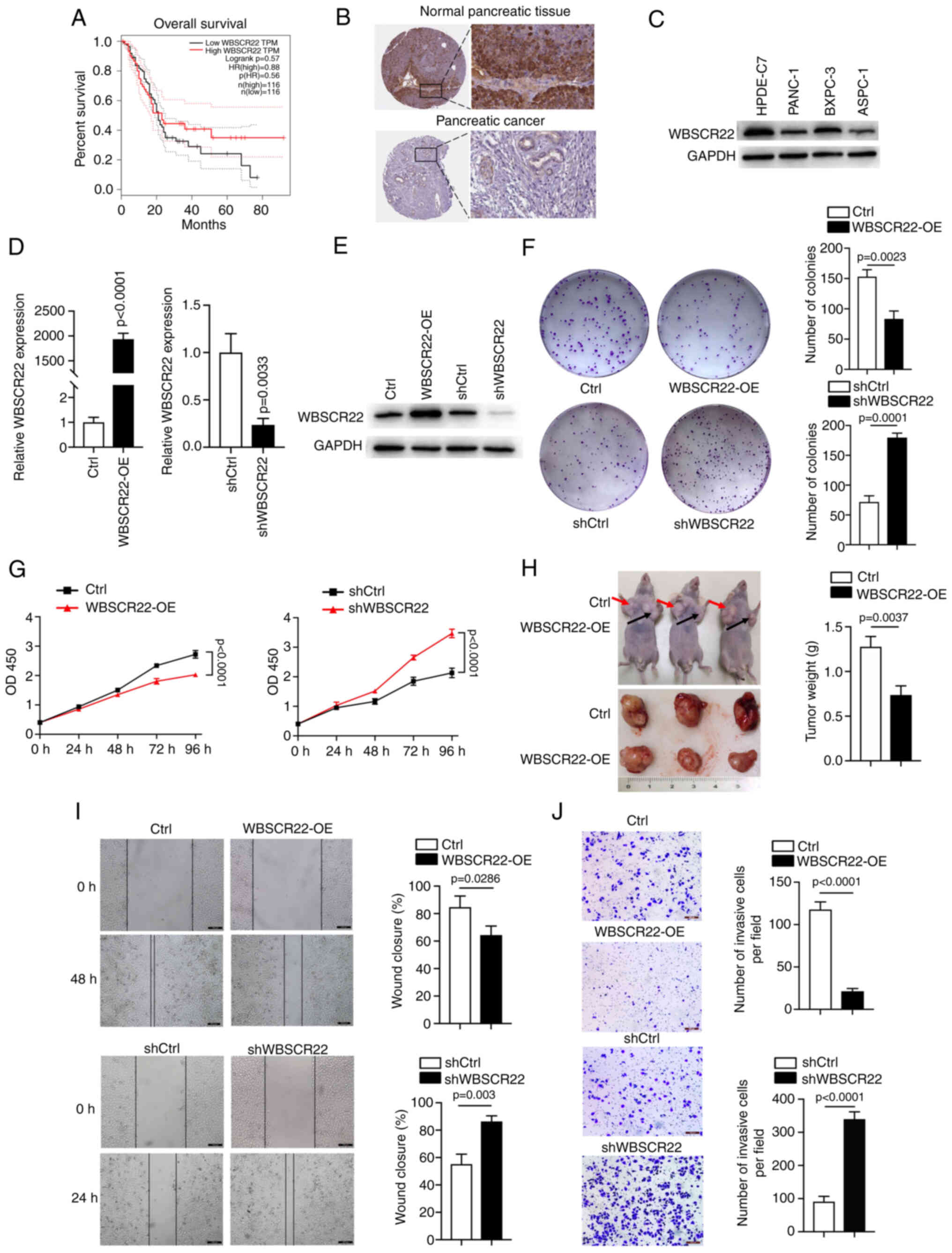 | Figure 1Upregulation of WBSCR22 expression
suppresses PC progression. (A) Survival plot indicating that
upregulation of WBSCR22 is associated with an increased overall
survival rate. (B) Data analysis results from the Human Protein
Atlas indicating downregulation of WBSCR22 protein in PC compared
with the corresponding expression noted in normal pancreatic
tissues. Decreased staining was observed for WBSCR22 in PC tissues,
whereas increased WBSCR22 staining was observed in normal
pancreatic tissues. (C) The protein levels of WBSCR22 were assessed
by western blot analysis. Consistent data were obtained from three
independent experiments. (D) mRNA expression levels of WBSCR22 in
WBSCR22-OE, WBSCR22-KD (shWBSCR22) and the corresponding control
cell lines. (E) WBSCR22 protein levels were confirmed in
WBSCR22-OE, WBSCR22-KD (shWBSCR22) and in the corresponding control
cell lines. Consistent data were obtained from three independent
experiments. (F) Colony formation assay of the WBSCR22-OE,
WBSCR22-KD (shWBSCR22) and corresponding control cells. (G) Cell
proliferation assays were performed in WBSCR22-OE, WBSCR22-KD
(shWBSCR22) and in the corresponding control cells. (H) A tumor
xenograft model was used to investigate the in vivo effect
of WBSCR22 (n=3 independent samples for each group). (I) Wound
healing and (J) Transwell assays were performed to evaluate the
migratory and invasive capacities of WBSCR22-overexpressing and
WBSCR22-KD (shWBSCR22) cells relative to their corresponding
control cell lines. The data are presented as the mean value ±
standard deviation; n=3 biologically independent repeats. The data
in D, F, H-J were analyzed using a two-tailed, unpaired t-test. The
data in G were analyzed using a two-way ANOVA, followed by
Bonferroni corrections. P-values are indicated. WBSCR22,
Williams-Beuren syndrome chromosomal region 22; PC, pancreatic
cancer; OE, overexpression; KD, knockdown; ANOVA, analysis of
variance. |
WBSCR22 functions as a tumor suppressor
in PC
To investigate the oncogenic or tumor suppressive
behavior of WBSCR22, stably transfected PANC-1 cells were
established, using WBSCR22 knockdown (KD) (PANC-1-shWBSCR22) and
WBSCR22-OE (PANC-1-WBSCR22-OE) models and their corresponding
controls. The transfection efficiency was confirmed at the mRNA and
protein level (Fig. 1D and E). OE
of WBSCR22 significantly suppressed the proliferation and colony
formation capacity of PANC-1 cells, while knockdown of WBSCR22
expression promoted these processes in vitro (Fig. 1F and G). In addition, a
subcutaneous xenograft mouse model (n=3/group) was established to
further verify the role of WBSCR22 in tumorigenesis in vivo.
WBSCR22-OE significantly suppressed the tumor weight and volume
compared with the corresponding control xenografts (Fig. 1H). In addition, the Transwell
assays indicated that WBSCR22-OE significantly suppressed the
migratory and invasive capacities of PANC-1 cells, whereas WBSCR22
knockdown exhibited the opposite effects (Fig. 1I and J). Furthermore, the tumor
suppressor function of WBSCR22 was confirmed in BXPC-3 cells
(Fig. S1). Collectively, these
data demonstrated that WBSCR22-OE suppressed proliferation,
migration, invasion and tumorigenesis in vivo and in
vitro. The results revealed for the first time the tumor
suppressor function of the WBSCR22 gene in PC.
ISG15 is a downstream target of WBSCR22
in PC
To explore the downstream pathways of WBSCR22 in PC,
RNA-seq was performed using shWBSCR22 and the corresponding control
cells. A total of 329 upregulated and 264 downregulated genes were
identified in response to WBSCR22 knockdown (FC≥2, P<0.05;
Fig. 2A). ISG15 was one of the
most significantly upregulated differentially expressed genes
(DEGs) in response to WBSCR22 knockdown, and its role in PC has not
been studied, to the best of our knowledge (Fig. 2B). ISG15 exerts a vital role in
protein turnover, protein stability and most importantly,
ubiquitin-like modification of several nuclear and cytoplasmic
proteins. ISG15 targets >300 proteins, including p53, by
altering the cellular metabolic pathway and ISGylation as well as
the ubiquitin proteasome degradation pathway (32). Therefore, it was hypothesized that
the tumor suppressor function of WBSCR22 in PC may be connected to
cellular metabolic, ubiquitin proteasome or RNA degradation
pathways via the regulation of ISG15-mediated ISGylation.
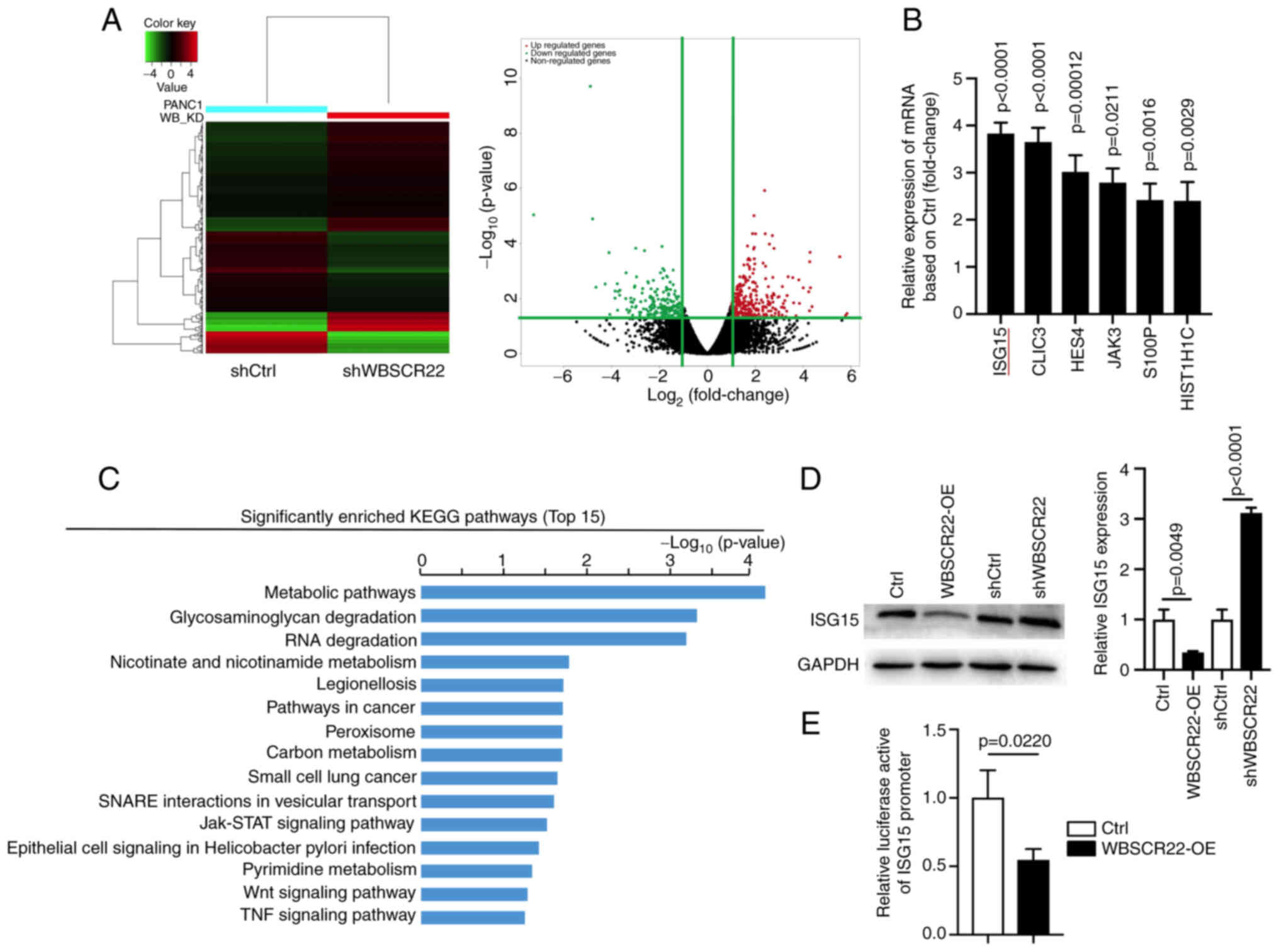 | Figure 2WBSCR22 regulates the downstream
expression of ISG15. (A) Hierarchical clustering plots and volcano
plots were used to identify the DEGs (fold change >2, P<0.05)
between wild-type and WBSCR22-knockdown PANC-1 cells. (B) Relative
mRNA expression levels of genes in WBSCR22-knockdown PANC-1 cells
as determined by RNA-seq. (C) KEGG pathway enrichment analysis of
DEGs in WBSCR22-knockdown PANC-1 cells. The graphs indicating the
top 15 signaling pathways were listed based on the log10 (P-value).
(D) Protein and mRNA levels of ISG15 in WBSCR22-overexpressing,
shWBSCR22 and control cells, respectively. (E) A luciferase
reporter assay was performed to investigate the possible effect of
WBSCR22-OE on the transcriptional activity of ISG15 in PANC-1
cells. The data indicated the relative ratio of firefly luciferase
activity and Renilla luciferase activity. The data are
presented as the mean value ± standard deviation; n=3 biologically
independent repeats. The data in B, D and E were analyzed using a
two-tailed, unpaired t-test. P-values are indicated. WBSCR22,
Williams-Beuren syndrome chromosomal region 22; ISG15,
interferon-stimulated gene 15; DEGs, differentially expressed
genes; RNA-seq, RNA sequencing; KEGG, Kyoto Encyclopedia of Genes
and Genomes; OE, overexpression. |
Subsequently, KEGG enrichment pathway analysis
revealed that the 'metabolic' and 'RNA degradation pathways' were
among the top 5 most enriched pathways in which ISG15 was involved
(Fig. 2C). Additionally, several
pathways closely related to tumor development, such as the
'JAK-STAT', 'Wnt' and 'TNF signaling pathways', were among the top
15 most enriched pathways, suggesting that WBSCR22 may play a
critical role in cancer development/progression. RT-qPCR and
western blot assays confirmed the reduced levels of ISG15 in
WBSCR22-OE cells (Fig. 2D). The
transcriptional regulatory role of WBSCR22 on ISG15 was evaluated
by a luciferase promoter reporter assay using the ISG15 promoter.
WBSCR22-OE significantly reduced the activity of the ISG15 promoter
(Fig. 2E). Collectively, these
results suggested that WBSCR22 is an upstream regulator of ISG15 in
PC.
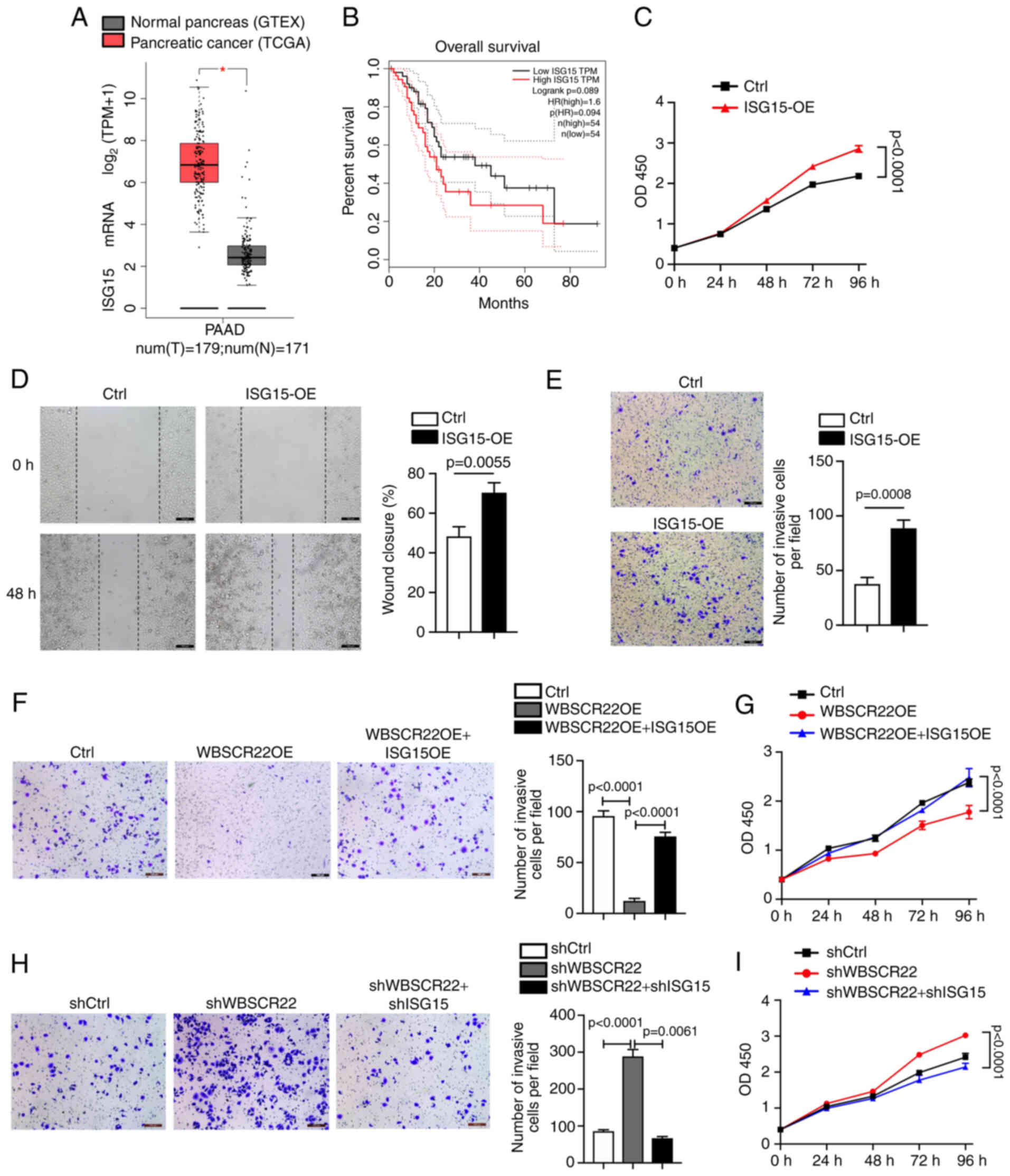
ISG15 promotes tumorigenesis in PC
Elevated expression levels of ISG15 have been
previously reported in multiple types of cancer, including PC
(32-35). According to TCGA database
analysis, the ISG15 gene is preferentially upregulated in PC
specimens (PDAC; n=179) compared with adjacent normal tissues
(n=171; Fig. 3A). In addition,
high expression levels of ISG15 in PC were associated with poor
survival (Fig. 3B). To further
determine the possible oncogenic function of ISG15 in PC, ISG15 was
overexpressed in PANC-1 cells. OE of ISG15 significantly promoted
the proliferation and migration of PANC-1 cells compared with those
of the corresponding controls (Fig.
3C and D). Furthermore, the Transwell assay confirmed that
ISG15 OE promoted the invasion of PANC-1 cells compared with that
of the corresponding controls (Fig.
3E).
In addition, rescue experiments were performed to
further confirm the oncogenic role of ISG15 and its functional
association with WBSCR22. Ectopic OE and knockdown ISG15 models
were established in WBSCR22-OE and shWBSCR22 PANC-1 cells,
respectively. The data indicated that the tumor-inhibitory capacity
of WBSCR22-OE could be significantly rescued by ISG15 OE in PC
(Fig. 3F and G). In contrast to
these observations, the tumor-promoting function caused by WBSCR22
knockdown (shWBSCR22) was significantly abolished by shISG15
(Fig. 3H and I). Collectively,
the data indicated that ISG15 functioned as an oncogene in PC
cells, and that the antitumor function of WBSCR22 in PC was closely
associated with its negative regulatory effect on ISG15.
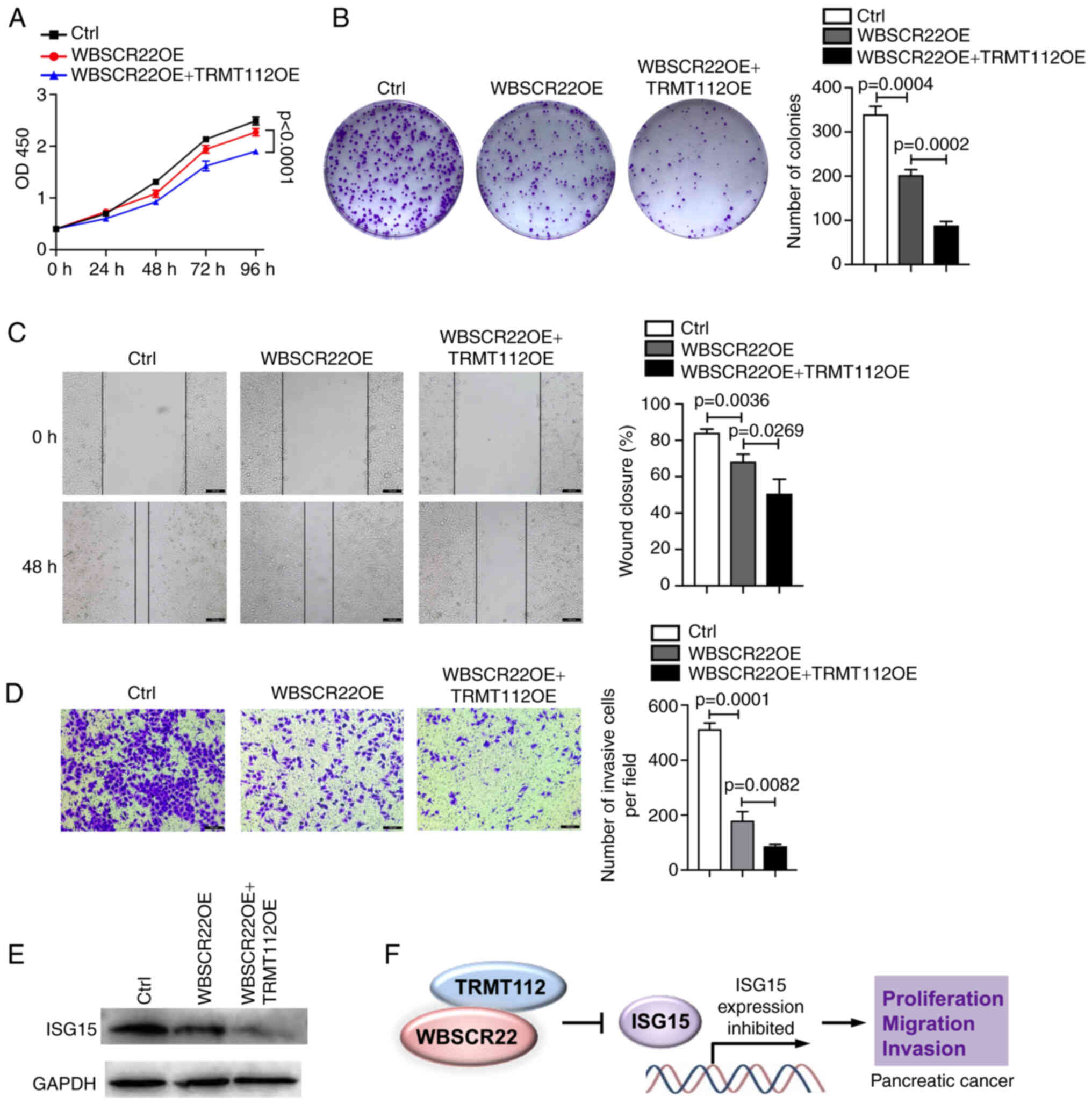
TRMT112 promotes the tumor suppressive
function of WBSCR22
The interaction of WBSCR22 with TRMT112 was
verified; the stability of WBSCR22 affected its expression. The
present study examined further whether this interaction could
influence the function of WBSCR22 in PC. To test this hypothesis,
the ability of WBSCR22 to suppress PC progression was investigated
using PANC-1 cells transfected with WBSCR22-OE alone or
co-transfected with WBSCR22-OE and TRMT112-OE. As expected, the
tumor suppressive capacity of WBSCR22 in PC was significantly
enhanced when TRMT112 was concurrently overexpressed with WBSCR22.
The synergistic ectopic OE of WBSCR22-OE + TRMT112-OE significantly
decreased cellular proliferation and colony formation of PANC-1
cells compared with that noted in WBSCR22-OE alone and in the
corresponding control cells (Fig. 4A
and B). Wound healing and Transwell assays further confirmed
that WBSCR22-OE + TRMT112-OE suppressed the cellular migration and
invasive capacities of PANC-1 cells compared with those of
WBSCR22-OE alone and the corresponding controls (Fig. 4C and D). Furthermore, as
aforementioned, the tumor suppressive role of WBSCR22 in PC was
revealed to be associated with its negative regulatory effect on
ISG15. It was hypothesized that TRMT112 may also be involved in
regulating the expression of ISG15. As expected, TRMT112 further
enhanced the inhibitory effect of WBSCR22 on ISG15 expression
(Fig. 4E). It was also revealed
that the tumor-inhibitory capacity of WBSCR22-OE + TRMT112-OE could
be partially rescued by ISG15-OE in PC (Fig. S2). Collectively, these results
demonstrated that WBSCR22 and TRMT112 may function together in PC.
Simultaneous targeting of WBSCR22 and TRMT112 may represent a novel
therapeutic strategy for the treatment of PC (Fig. 4F).
Discussion
The human WBSCR22 protein has been identified as a
methyltransferase for 18S rRNA m7G and is involved in
pre-rRNA processing and 40S ribosome subunit biogenesis. The
elevated expression levels and the tumor-promoting potential of
WBSCR22 have been observed in several types of cancer (10-18). However, the role of WBSCR22 in PC
remains unknown. In the present study, WBSCR22 was revealed for the
first time to the best of our knowledge, to act as a tumor
suppressor by attenuating cellular proliferation, migration,
invasion and tumorigenesis of PC. The effects mediated by WBSCR22
in PC may involve the downstream regulation of ISG15. In addition,
ectopic expression of TRMT112 further promoted the tumor
suppressive potential of WBSCR22 in PC. These results propose a
tumor suppressive role of the TRMT112/WBSCR22/ISG15 axis in PC that
may represent a novel therapeutic strategy for the treatment of
this disease.
According to RNA-seq using shWBSCR22 and control
cells, the metabolic pathway was among the top five enriched KEGG
pathways. In addition, the carbon and pyrimidine metabolic pathways
and Wnt and Janus kinase-STAT signaling pathways were among the top
15 enriched KEGG pathways. ISG15, a crucial member of the metabolic
pathway, plays a vital role in protein turnover, protein stability
and ISGylation (27,28). The present study demonstrated that
ISG15 was transcriptionally regulated by WBSCR22. Therefore, it was
hypothesized that WBSCR22 suppressed the progression and metastasis
of PC by regulating ISG15. The oncogenic role of ISG15 and its
functional association with WBSCR22 were also confirmed. ISG15
functions as an oncogene in PC cells and its antitumor function in
PC is closely related to its negative regulatory effect on ISG15.
However, the mechanism by which WBSCR22 regulates ISG15 is not
fully known. In addition, the WBSCR22 protein has been identified
as a methyltransferase for 18S rRNA m7G involved in
pre-rRNA processing and ribosome 40S subunit biogenesis. It would
be interesting to investigate whether the regulation of ISG15 by
WBSCR22 is associated with its catalytic activity in future
studies. In addition, the Human Protein Atlas database indicated
that the WBSCR22 protein was localized in the nucleoli and
nucleoplasm of cells. Therefore, it is worth investigating whether
WBSCR22 exerts a direct transcriptional regulatory role on ISG15
via an interaction with transcription-related proteins.
Numerous studies have established the oncogenic role
of WBSCR22 in multiple malignancies. WBSCR22 has been reported as
an oncogene in several carcinomas, including invasive breast
cancer, multiple myeloma, plasma cell carcinoma, colorectal cancer,
lung cancer and hepatocellular carcinoma (13-17). In glioma cells, upregulation of
WBSCR22 promotes proliferation, invasion, tumorigenesis and
migration, while its knockdown exerts the opposite effects
(18). In contrast to these
findings, WBSCR22 loss has also been reported in certain neoplastic
and inflammatory types of human lung pathologies, which indicates
that the role of WBSCR22 in different cancer types is tissue
specific (16). It will be
interesting to investigate the mechanism and the diverse roles of
WBSCR22 in other cancer types.
Aberrant cellular metabolism is the primary effect
by which WBSCR22 promotes tumor initiation, progression and cancer
cell metastatic dissemination. Cancer cells undergo substantial
metabolic rewiring to attain metastatic traits and survive in
varying cellular environmental conditions, including oxygen
concentration, nutrient availability and extracellular signals
(47,48). ISG15 functions as a ubiquitin-like
modifier of various nuclear and cytoplasmic proteins by targeting
>300 proteins (including p53) through ubiquitination of a
cellular metabolic pathway (32).
ISG15 primarily targets proteins that play a role in altering
cellular metabolic processes (49). However, the biological function of
ISG15 is not consistent across different types of cancer. In the
present study, the oncogenic role of ISG15 was demonstrated in PC,
and that the upregulated ISG15 expression promoted the
proliferation, migration, invasion and tumorigenesis of PC. A
recent study indicated that ISG15 and ISGylation were essential for
maintaining PC stem cell metabolic plasticity and mitophagy
(23). An additional study
reported that ISG15 was secreted into the tumor microenvironment
and that extracellular-free ISG15 played an important role in the
maintenance of cancer stem cell-like features of PDACs (50). These data are consistent with our
investigation indicating that ISG15 is preferentially upregulated
in PC cells in order to promote PC progression. However, the
mechanism of ISG15 in PC is still not fully understood. Whether the
role of ISG15 in PC depends on the ISGylation of key proteins
involved in cellular metabolism and cancer progression or involves
completely different regulatory mechanisms is worthy of further
investigation.
TRMT112 has been validated as the interaction
partner of WBSCR22, and enhances the stability of WBSCR22. The
present study examined further whether this interaction could
influence the function of WBSCR22 in PC. To test this hypothesis,
the ability of WBSCR22 to suppress PC progression was investigated
using PANC-1 cells transfected with WBSCR22-OE alone or
co-transfected with WBSCR22-OE and TRMT112-OE. As expected, the
tumor suppressive capacity of WBSCR22 in PC was significantly
enhanced when TRMT112 was concurrently overexpressed with WBSCR22.
A previous study has demonstrated that the stability of WBSCR22 is
regulated by interaction with TRMT112 through the
ubiquitin-proteasome degradation pathway, resulting in the tight
control of WBSCR22 in cells (45). The present study investigated the
biological function and significance of the WBSCR22-TRMT112
interaction in the development of PC.
In conclusion, the present study described a novel
regulatory network for WBSCR22 in PC. The data verified for the
first time that WBSCR22 functions as a tumor suppressor in PC by
significantly suppressing cellular proliferation, migration,
invasion and tumorigenesis in vivo and in vitro. In
addition, it was confirmed that WBSCR22 regulated the downstream
transcriptional activity of ISG15, which acted as an oncogene in PC
by promoting the proliferation, migration, invasion and
tumorigenesis of PC. Furthermore, the data confirmed that TRMT112
and WBSCR22 functioned cooperatively in PC. Simultaneous ectopic OE
of WBSCR22 and TRMT112 further promoted the tumor suppressive
potential of WBSCR22 in PC. WBSCR22 is a clinically important gene
in PC and the newly identified WBSCR22/ISG15 axis may represent an
innovative approach for therapeutic purposes.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
AAK, HH and XL conceived the idea and designed the
experiments of the present study. SW, HL, YZ and RP performed the
experiments. AAK and XL confirm the authenticity of all the raw
data. HH, AAK and XL were involved in writing the manuscript. XL
and HH supervised the overall project. All authors read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved (approval no.
IRB-1507) by the Ethics Committee of Beijing University of
Technology (Beijing, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors are thankful and highly acknowledge the
cooperation of the Chinese Scholarship Council for providing a
platform for conducting quality research.
Funding
The present study was supported by the National Natural Science
Foundation China (grant no. 81702802), the programs of the Beijing
Municipal Education Commission (grant no. KM201910005005) and
Beijing Municipal Science and Technology Commission (grant no.
K2015311201501).
Abbreviations:
|
PC
|
pancreatic cancer
|
|
WBSCR22
|
Williams-Beuren syndrome chromosomal
region 22
|
|
TRMT112
|
tRNA methyltransferase subunit
11-2
|
|
PDAC
|
pancreatic ductal adenocarcinoma
|
|
SAM
|
S-adenosyl-L-methionine
|
|
DEGs
|
differentially expressed genes
|
|
WBS
|
Williams-Beuren syndrome
|
|
ISG15
|
interferon-stimulated gene 15
|
References
|
1
|
Khan AA, Liu X, Yan X, Tahir M, Ali S and
Huang H: An overview of genetic mutations and epigenetic signatures
in the course of pancreatic cancer progression. Cancer Metastasis
Rev. 40:245–272. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cao W, Chen HD, Yu YW, Li N and Chen WQ:
Changing profiles of cancer burden worldwide and in China: A
secondary analysis of the global cancer statistics 2020. Chin Med J
(Engl). 134:783–791. 2021. View Article : Google Scholar
|
|
3
|
Aier I, Semwal R, Sharma A and Varadwaj
PK: A systematic assessment of statistics, risk factors, and
underlying features involved in pancreatic cancer. Cancer
Epidemiol. 58:104–110. 2019. View Article : Google Scholar
|
|
4
|
Hung YH, Hsu MC, Chen LT, Hung WC and Pan
MR: Alteration of epigenetic modifiers in pancreatic cancer and its
clinical implication. J Clin Med. 8:9032019. View Article : Google Scholar :
|
|
5
|
Ferrari M and Stagi S: Oxidative stress in
down and Williams-Beuren syndromes: An overview. Molecules.
26:31392021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pangallo E, Cianci P, Favuzza F, Milani D,
Vimercati C, Moretti A, Picchi R, De Paoli A, Agosti M and
Selicorni A: Pulmonary function in Williams-Beuren syndrome:
Spirometric data of 22 Italian patients. Am J Med Genet A.
185:390–396. 2021. View Article : Google Scholar
|
|
7
|
Wang LX, Leng J, Li ZH, Yan L, Gou P, Tang
F, Su N, Gong CZ and Cheng XR: Clinical and genetic characteristics
of two cases with Williams-Beuren syndrome. Transl Pediatr.
10:1743–1747. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Merla G, Ucla C, Guipponi M and Reymond A:
Identification of additional transcripts in the Williams-Beuren
syndrome critical region. Hum Genet. 110:429–438. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Alesi V, Loddo S, Orlando V, Genovese S,
Di Tommaso S, Liambo MT, Pompili D, Ferretti D, Calacci C, Catino
G, et al: Atypical 7q11.23 deletions excluding ELN gene result in
Williams-Beuren syndrome craniofacial features and neurocognitive
profile. Am J Med Genet A. 185:242–249. 2021. View Article : Google Scholar
|
|
10
|
Zorbas C, Nicolas E, Wacheul L, Huvelle E,
Heurgue-Hamard V and Lafontaine DL: The human 18S rRNA base
methyltransferases DIMT1L and WBSCR22-TRMT112 but not rRNA
modification are required for ribosome biogenesis. Mol Biol Cell.
26:2080–2095. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Õunap K, Käsper L, Kurg A and Kurg R: The
human WBSCR22 protein is involved in the biogenesis of the 40S
ribosomal subunits in mammalian cells. PLoS One. 8:e756862013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Garcia BCB, Horie M, Kojima S, Makino A
and Tomonaga K: BUD23-TRMT112 interacts with the L protein of Borna
disease virus and mediates the chromosomal tethering of viral
ribonucleoproteins. Microbiol Immunol. 65:492–504. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nakazawa Y, Arai H and Fujita N: The novel
metastasis promoter Merm1/Wbscr22 enhances tumor cell survival in
the vasculature by suppressing Zac1/p53-dependent apoptosis. Cancer
Res. 71:1146–1155. 2011. View Article : Google Scholar
|
|
14
|
Tiedemann RE, Zhu YX, Schmidt J, Shi CX,
Sereduk C, Yin H, Mousses S and Stewart AK: Identification of
molecular vulnerabilities in human multiple myeloma cells by RNA
interference lethality screening of the druggable genome. Cancer
Res. 72:757–768. 2012. View Article : Google Scholar
|
|
15
|
Yan D, Tu L, Yuan H, Fang J, Cheng L,
Zheng X and Wang X: WBSCR22 confers oxaliplatin resistance in human
colorectal cancer. Sci Rep. 7:154432017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jangani M, Poolman TM, Matthews L, Yang N,
Farrow SN, Berry A, Hanley N, Williamson AJ, Whetton AD, Donn R and
Ray DW: The methyltransferase WBSCR22/Merm1 enhances glucocorticoid
receptor function and is regulated in lung inflammation and cancer.
J Biol Chem. 289:8931–8946. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Stefanska B, Cheishvili D, Suderman M,
Arakelian A, Huang J, Hallett M, Han ZG, Al-Mahtab M, Akbar SM,
Khan WA, et al: Genome-wide study of hypomethylated and induced
genes in patients with liver cancer unravels novel anticancer
targets. Clin Cancer Res. 20:3118–3132. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chi YJ, Liang Z, Guo YW, Chen D, Lu L, Lin
J, Qiu S, Wang X, Qiu E, Lin F, et al: WBSCR22 confers cell
survival and predicts poor prognosis in glioma. Brain Res Bull.
161:1–12. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Reich N, Evans B, Levy D, Fahey D, Knight
E Jr and Darnell JE Jr: Interferon-induced transcription of a gene
encoding a 15-kDa protein depends on an upstream enhancer element.
Proc Natl Acad Sci USA. 84:6394–6398. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen RH, Xiao ZW, Yan XQ, Han P, Liang FY,
Wang JY, Yu ST, Zhang TZ, Chen SQ, Zhong Q and Huang XM: Tumor
cell-secreted ISG15 promotes tumor cell migration and immune
suppression by inducing the macrophage M2-like phenotype. Front
Immunol. 11:5947752020. View Article : Google Scholar
|
|
21
|
Freitas BT, Scholte FEM, Bergeron É and
Pegan SD: How ISG15 combats viral infection. Virus Res.
286:1980362020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Loeb KR and Haas AL: The
interferon-inducible 15-kDa ubiquitin homolog conjugates to
intracellular proteins. J Biol Chem. 267:7806–7813. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Alcalá S, Sancho P, Martinelli P, Navarro
D, Pedrero C, Martín-Hijano L, Valle S, Earl J, Rodríguez-Serrano
M, Ruiz-Cañas L, et al: ISG15 and ISGylation is required for
pancreatic cancer stem cell mitophagy and metabolic plasticity. Nat
Commun. 11:26822020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yuan WM and Krug RM: Influenza B virus NS1
protein inhibits conjugation of the interferon (IFN)-induced
ubiquitin-like ISG15 protein. EMBO J. 20:362–371. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Villarroya-Beltri C, Guerra S and
Sánchez-Madrid F: ISGylation-a key to lock the cell gates for
preventing the spread of threats. J Cell Sci. 130:2961–2969.
2017.PubMed/NCBI
|
|
26
|
Dang F, Nie L and Wei W: Ubiquitin
signaling in cell cycle control and tumorigenesis. Cell Death
Differ. 28:427–438. 2021. View Article : Google Scholar :
|
|
27
|
Zhang D and Zhang DE:
Interferon-stimulated gene 15 and the protein ISGylation system. J
Interferon Cytokine Res. 31:119–130. 2011. View Article : Google Scholar :
|
|
28
|
Liu MJ, Li XL and Hassel BA: Proteasomes
modulate conjugation to the ubiquitin-like protein, ISG15. J Biol
Chem. 278:1594–1602. 2003. View Article : Google Scholar
|
|
29
|
Bogunovic D, Boisson-Dupuis S and Casanova
JL: ISG15: Leading a double life as a secreted molecule. Exp Mol
Med. 45:e182013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
D'Cunha J, Knight E Jr, Haas AL, Truitt RL
and Borden EC: Immunoregulatory properties of ISG15, an
interferon-induced cytokine. Proc Natl Acad Sci USA. 93:211–215.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Held T, Basler M, Knobeloch KP and
Groettrup M: Evidence for an involvement of the ubiquitin-like
modifier ISG15 in MHC class I antigen presentation. Eur J Immunol.
51:138–150. 2021. View Article : Google Scholar
|
|
32
|
Desai SD, Haas AL, Wood LM, Tsai YC,
Pestka S, Rubin EH, Saleem A, Nur-E-Kamal A and Liu LF: Elevated
expression of ISG15 in tumor cells interferes with the
ubiquitin/26S proteasome pathway. Cancer Res. 66:921–928. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li C, Wang J, Zhang H, Zhu M, Chen F, Hu
Y, Liu H and Zhu H: Interferon-stimulated gene 15 (ISG15) is a
trigger for tumorigenesis and metastasis of hepatocellular
carcinoma. Oncotarget. 5:8429–8441. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Burks J, Reed RE and Desai SD: ISGylation
governs the oncogenic function of Ki-Ras in breast cancer.
Oncogene. 33:794–803. 2014. View Article : Google Scholar
|
|
35
|
Burks J, Fleury A, Livingston S and Smith
JP: ISG15 pathway knockdown reverses pancreatic cancer cell
transformation and decreases murine pancreatic tumor growth via
downregulation of PDL-1 expression. Cancer Immunol Immunother.
68:2029–2039. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wan XX, Chen HC, Khan MA, Xu AH, Yang FL,
Zhang YY and Zhang DZ: ISG15 inhibits IFN-α-resistant liver cancer
cell growth. Biomed Res Int. 2013:5709092013. View Article : Google Scholar
|
|
37
|
Zhou MJ, Chen FZ, Chen HC, Wan XX, Zhou X,
Fang Q and Zhang DZ: ISG15 inhibits cancer cell growth and promotes
apoptosis. Int J Mol Med. 39:446–452. 2017. View Article : Google Scholar
|
|
38
|
Jeon YJ, Jo MG, Yoo HM, Hong SH, Park JM,
Ka SH, Oh KH, Seol JH, Jung YK and Chung CH: Chemosensitivity is
controlled by p63 modification with ubiquitin-like protein ISG15. J
Clin Invest. 122:2622–2636. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Burks J, Reed RE and Desai SD: Free ISG15
triggers an antitumor immune response against breast cancer: A new
perspective. Oncotarget. 6:7221–7231. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liger D, Mora L, Lazar N, Figaro S, Henri
J, Scrima N, Buckingham RH, van Tilbeurgh H, Heurgué-Hamard V and
Graille M: Mechanism of activation of methyltransferases involved
in translation by the Trm112 'hub' protein. Nucleic Acids Res.
39:6249–6259. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gu T, He H, Zhang Y, Han Z, Hou G, Zeng T,
Liu Q and Wu Q: Trmt112 gene expression in mouse embryonic
development. Acta Histochem Cytochem. 45:113–119. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Figaro S, Wacheul L, Schillewaert S,
Graille M, Huvelle E, Mongeard R, Zorbas C, Lafontaine DL and
Heurgué-Hamard V: Trm112 is required for Bud23-mediated methylation
of the 18S rRNA at position G1575. Mol Cell Biol. 32:2254–2267.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sardana R and Johnson AW: The
methyltransferase adaptor protein Trm112 is involved in biogenesis
of both ribosomal subunits. Mol Biol Cell. 23:4313–4322. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Leetsi L, Õunap K, Abroi A and Kurg R: The
common partner of several methyltransferases TRMT112 regulates the
expression of N6AMT1 isoforms in mammalian cells. Biomolecules.
9:4222019. View Article : Google Scholar :
|
|
45
|
Õunap K, Leetsi L, Matsoo M and Kurg R:
The stability of ribosome biogenesis factor WBSCR22 is regulated by
interaction with TRMT112 via ubiquitin-proteasome pathway. PLoS
One. 10:e01338412015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
47
|
Teoh ST and Lunt SY: Metabolism in cancer
metastasis: Bioenergetics, biosynthesis, and beyond: Metabolism in
cancer metastasis. Wires Syst Biol Med. 10:e14062018. View Article : Google Scholar
|
|
48
|
Bergers G and Fendt SM: The metabolism of
cancer cells during metastasis. Nat Rev Cancer. 21:162–180. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhang YF, Thery F, Wu NC, Luhmann EK,
Dussurget O, Foecke M, Bredow C, Jiménez-Fernández D, Leandro K,
Beling A, et al: The in vivo ISGylome links ISG15 to metabolic
pathways and autophagy upon Listeria monocytogenes infection. Nat
Commun. 10:53832019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Sun J, Yan J, Qiao HY, Zhao FY, Li C,
Jiang JY, Liu BQ, Meng XN and Wang HQ: Loss of TRIM29 suppresses
cancer stem cell-like characteristics of PDACs via accelerating
ISG15 degradation. Oncogene. 39:546–559. 2020. View Article : Google Scholar
|