Breast and ovarian cancer are two of the most common
malignancies affecting the female population both in industrialized
and developing countries. According to GLOBOCAN 2020, breast cancer
is the most frequently diagnosed tumor among females with 2,261,419
new cases (24.5%) and the first leading cause of cancer-related
death with 684,996 deaths (15.5%). On the other hand, ovarian
cancer represents the eighth most common cancer for incidence and
mortality in females, accounting for 313,959 newly diagnosed cases
(3,4%) and 207,252 deaths (4.7%) (1).
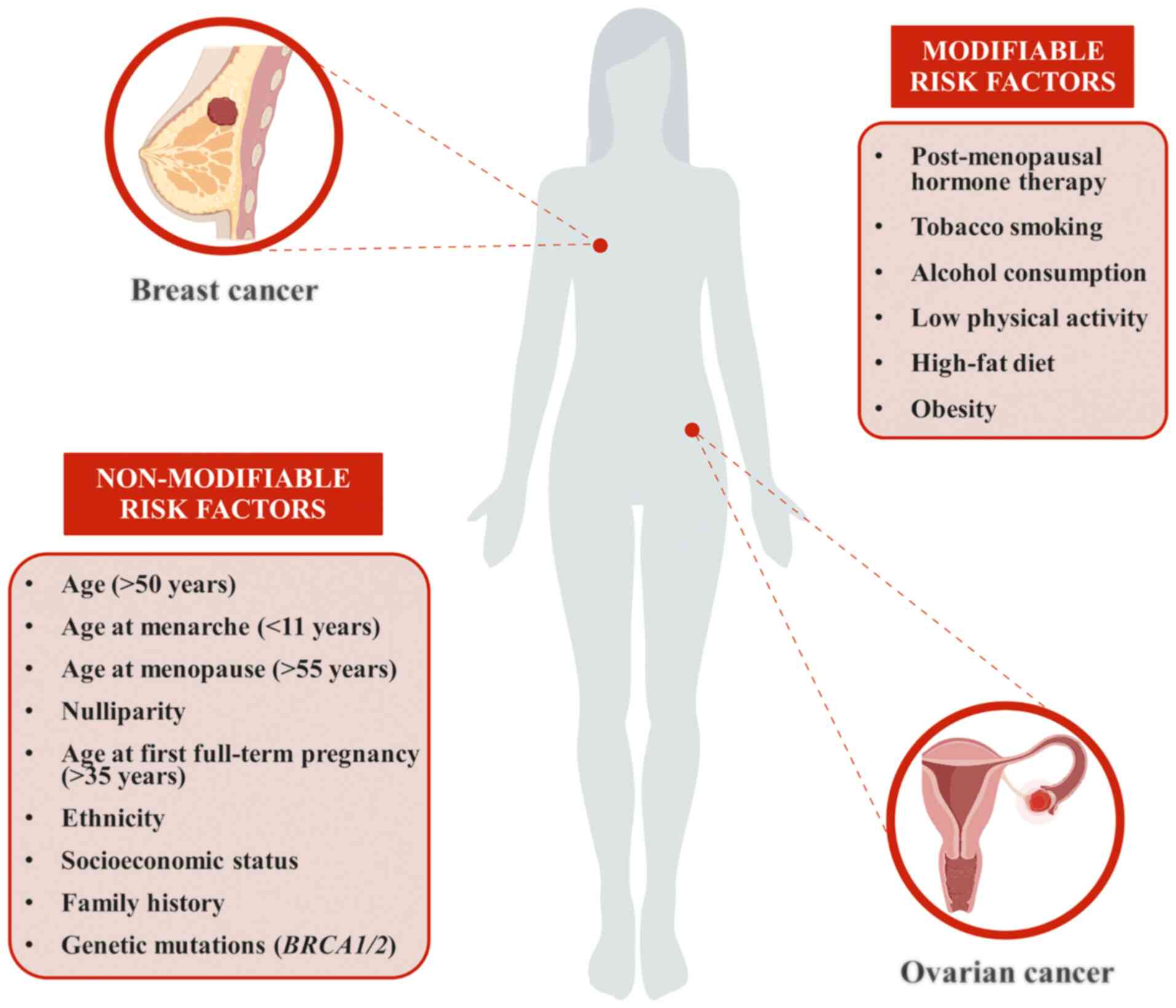
Over the years, several risk factors have been
associated with the onset and progression of both breast and
ovarian cancer (Fig. 1). As
widely reported in the literature, more than half of all cases
diagnosed are females aged >50 years, indicating that age is one
of the major non-modifiable risk factors (2,3).
Similarly, reproductive factors, such as age at menarche and
menopause (before 11 and after 55 years of age, respectively),
nulliparity and age at first full-term pregnancy (>35 years),
are well-established risk factors for both breast and ovarian
cancer (4,5). Furthermore, post-menopausal hormone
therapies, based on the administration of estrogens plus progestin,
significantly increase the risk of cancer development (6,7).
Of note, the use of oral contraceptives for birth control has only
been described as a risk factor for breast cancer, while oral
contraceptive pills represent a protective factor for ovarian
cancer, reducing the risk by at least 50% when used for 10 years or
more (8-10). Other risk factors for both of
these female cancer types include ethnicity, tobacco smoking,
alcohol consumption, low physical activity, high-fat diet, obesity
and socioeconomic status (11-16). Several studies have demonstrated
that environmental, lifestyle and epigenetic factors are associated
with the development of both breast and ovarian cancer (17-20).
Besides the aforementioned modifiable and
non-modifiable risk factors, a family history of breast and/or
ovarian cancer, as well as genetic mutations, may have a key role
in increasing cancer susceptibility. Of note, a growing body of
evidence suggests that the risk of developing these female cancers
is significantly increased in females having a first-degree
relative affected by breast cancer or ovarian cancer or when the
affected relative was under 50 years of age (21-24). Of note, over the years, several
genetic mutations have been reported to be highly associated with
an increased risk of both breast and ovarian cancer. Among these,
BReast CAncer 1 (BRCA1) and BRCA2 are two tumor suppressor genes
with high penetrance, which are involved in the activation of DNA
repair processes and cell-cycle checkpoints in response to DNA
damage (25,26). Functional deficiencies due to
BRCA1/2 mutations induce genome instability, cell-cycle
dysregulation and accumulation of other mutations (27,28). In this field, it has been widely
demonstrated that BRCA1/2 mutations increase the lifetime risk to
develop breast or ovarian cancer. Specifically, 5-10% of patients
with breast cancer and 25% of ovarian cancer cases are due to an
inherited genetic mutation affecting BRCA1 or BRCA2 (29,30). Furthermore, healthy individuals
harboring germline mutations of BRCA1/2 had a 60-70% increased risk
to develop breast cancer and a 15-40% increased risk for ovarian
cancer (31,32). Therefore, the assessment of
cancer-related risk factors, particularly the identification of
BRCA1 and BRCA2 gene mutations, is crucial for the clinical
management of females at a high risk of developing breast or
ovarian cancer, which includes annual screening, chemoprevention
and preventive surgery (33,34).
Although the impairment of DNA repair mechanisms due
to BRCA mutations is associated with an increased risk of breast
and ovarian cancer, patients developing these tumors benefit from
therapies further affecting the DNA repair machinery aimed at
killing cancer cells through the accumulation of several DNA
alterations resulting in tumor cell death (35,36). Among these therapies, the use of
poly [ADP-ribose] polymerase (PARP) inhibitors directed at PARP
proteins involved in DNA repair mechanisms proved to be highly
efficient in both ovarian and breast cancer (37,38).
The precise molecular characterization of both
breast and ovarian tumors is essential to correctly classify cancer
lesions and predict the prognosis of patients. With regard to
breast cancer, the presence of hormone receptors (estrogen receptor
and progesterone receptor), membrane receptors (epidermal growth
factor receptor) and other molecular markers, including BRCA1/2
mutations, are used to classify breast cancer into different
molecular subtypes (39). Besides
its molecular classification, breast cancer may be divided
according to histological features, e.g. ductal carcinoma, lobular
carcinoma, mucinous carcinoma or spindle cell carcinoma (40,41).
As mentioned above, the precise identification of
molecular markers such as BRCA1/2 mutations is essential to predict
the prognosis of patients. In this context, both BRCA1 and BRCA2
mutations are associated with a higher aggressiveness of both
breast and ovarian cancer (42).
However, due to the development of novel targeted therapies using
high-effective PARP inhibitors (olaparib, niraparib, recuparib and
veliparib) for the treatment of tumors with BRCA mutations, the
presence of these mutations in ovarian cancer is associated with a
favorable prognosis (43).
Despite the prognostic importance of BRCA1/2 mutations, the
identification of other proteins or genetic and epigenetic factors
is essential to predict the efficacy of treatments and the survival
of patients (44).
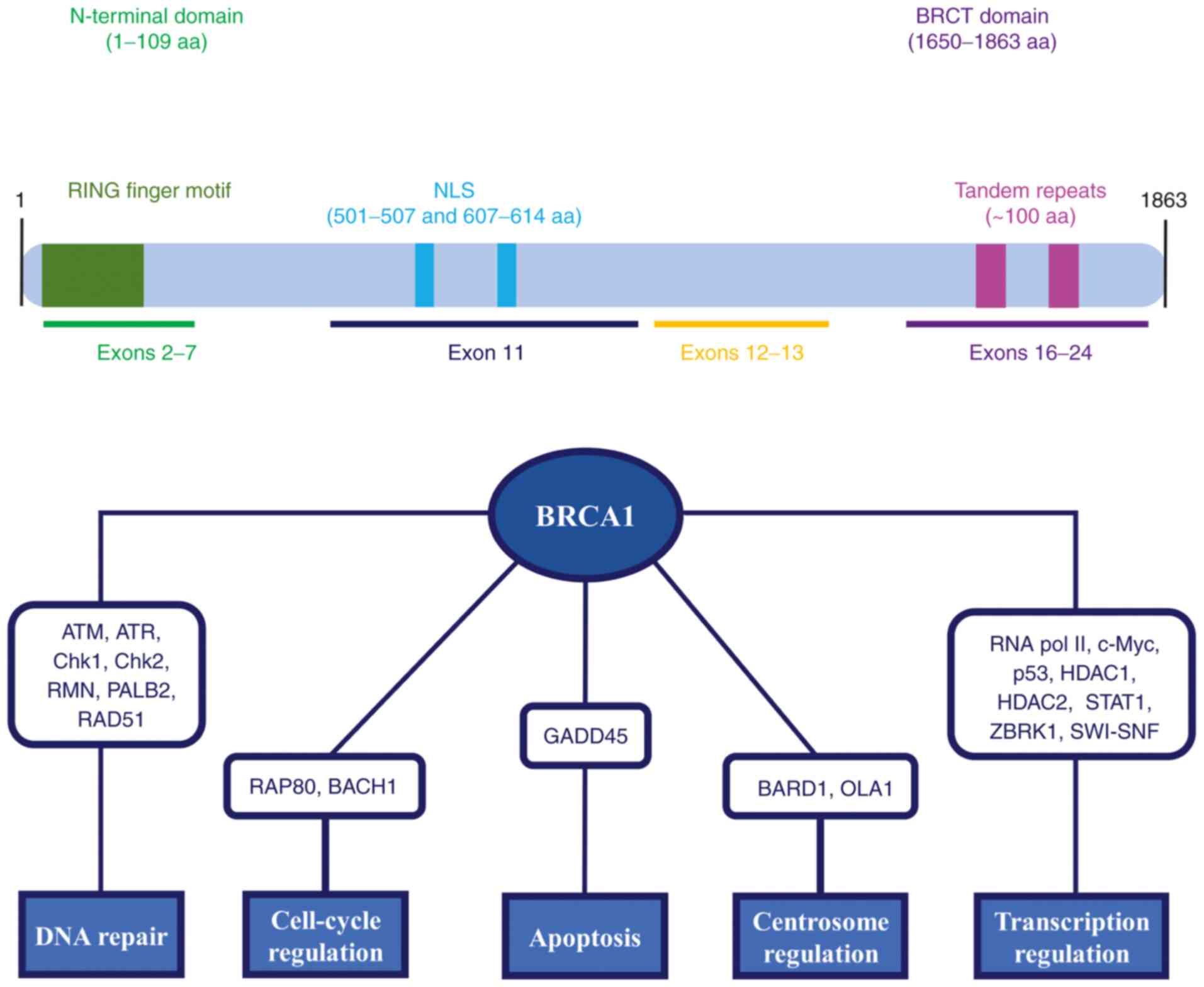
The BRCA1 gene is located on the long arm of
chromosome 17 (17q21) and it is composed of 24 exons (45). BRCA1 encodes for a
multi-functional protein of 1,863 amino acids, which consists of an
amino (N)-terminal RING domain, a carboxyl (C)-terminus, also known
as the BRCT domain, and coding regions of exons 11-13 (Fig. 2) (46,47). These domains have a crucial role
in the interaction between BRCA1 and several partner proteins. The
RING domain (amino acids 1-109) is a highly conserved domain
encoded by exons 2-7, which is characterized by a RING finger motif
involved in the ubiquitination pathway. Of note, it heterodimerizes
with BRCA1 associated RING domain 1 (BARD1) to form a dimeric RING
ubiquitin-ligase (E3) (48). The
BRCT domain is encoded by exons 16-24 and spans from amino acids
1,650-1,863, including two tandem repeats (~100 amino acids) linked
by 22 amino acids. This domain binds to the phosphorylated
serine-proline-x-phenylalanine motifs of different partner
proteins, such as BTB domain and CNC homolog 1 (BACH1), BRCA1
interacting helicase 1, BRCA1 A complex subunit and C-terminal
binding protein 1, to form functional macromolecular complexes that
allow selecting the substrate for BRCA1-BARD1 activity (49-52). Compared to other domains, exons
11-13 cover a large part of the BRCA1 protein. Of note, exon 11
comprises two nuclear localization sequences (NLS) (amino acids
501-507 and 607-614), which facilitate the nuclear import process
of BRCA1 interacting with importin α (53).
As widely described in the literature, BRCA1 may be
considered a tumor suppressor gene whose derived protein is
involved in several molecular pathways both in the nucleus and
cytoplasm, including DNA double-strand break (DSB) repair,
cell-cycle checkpoints, genome stability, transcription regulation,
apoptosis, chromosomal segregation, mitochondrial genome repair,
cytoskeletal rearrangements and centrosome regulation (Fig. 2) (54-57).
It has been reported that DNA DSBs activate several
kinases, such as ATM, ATM-related kinase, checkpoint kinase 1
(Chk1) and Chk2, which phosphorylate BRCA1 (58,59). The hyperphosphorylated BRCA1 then
interacts with several protein complexes that repair DSBs via
homologous recombination repair (HRR) and the activation of
cell-cycle checkpoints. Of note, BRCA1 is involved in HRR through
the interaction with the RAD50-MRE11-NBS1 complex, as well as
partner and localizer of BRCA2 (PALB2) and RAD51 DNA repair
proteins (60-62). Regarding cell-cycle checkpoints,
other BRCA1 complexes have been described. Specifically, G2/M
checkpoint signaling is activated by BRCA1-receptor-associated
protein 80, while the BRCA1-BACH1 complex is required during the
S-phase (63,64). BRCA1 also regulates gene
expression at the transcriptional level, interacting with RNA
polymerase II and several transcription factors, including c-Myc,
p53, histone deacetylase 1 and 2, signal transducer and activator
of transcription 1 and zinc finger and BRCA1-interacting protein
with KRAB domain-1, as well as the SWItch/sucrose non-fermentable
complex (65-70). Of note, the ubiquitin-ligase
activity of the BRCA1-BARD1 complex has a critical role in
centrosome regulation. In particular, BRCA1 and BARD1 bind to
Obg-Like ATPase 1, favoring the maintenance of centrosome numbers
at S- and G2/M-phases (71).
Furthermore, BRCA1 has apoptotic properties due to its nuclear
export and the activation of the p53-independent growth arrest and
DNA damage-inducible 45 regulatory sequences (72).
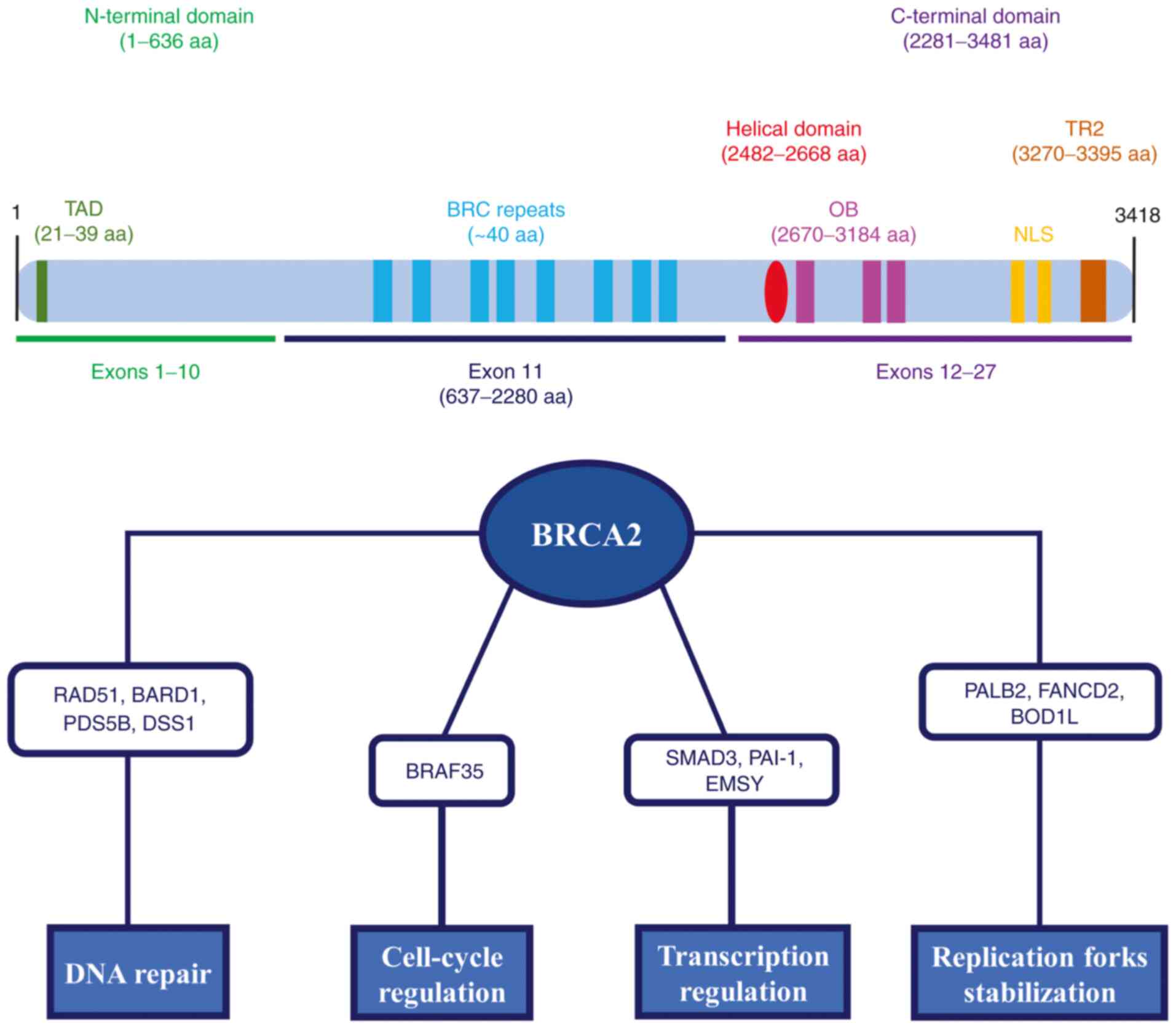
Similar to BRCA1, the BRCA2 gene is considered a
caretaker of genome stability that has a key role in several
biological pathways. Of note, it has been reported that BRCA2
interacts with different partner proteins for the formation of
macromolecular complexes performing distinct cellular functions,
such as DNA DSBs repair by HRR, DNA replication fork stabilization,
transcription regulation and cell cycle checkpoint regulation
(Fig. 3) (79-82).
For instance, the interaction between BRCA2 and
RAD51 is implicated in the repair of DNA damage by the HRR pathway.
Of note, BRC repeats, as well as the C-terminal domain of BRCA2,
regulate the assembly of RAD51 into a nucleoprotein filament,
promoting strand invasion and the search for homologous DNA
(83,84). Other partner proteins interact
with BRCA2 forming macromolecular complexes that have a crucial
role in DNA DSB repair and maintenance of genome stability,
including BARD1, PDS5 cohesin-associated factor B and SEM1 26S
proteasome subunit (85-87). BRCA2 also promotes the
stabilization of stalled DNA replication forks. Specifically, BRCA2
directly interacts with PALB2 through TAD of the N-terminal domain
to sustain the recruitment of polymerase η at blocked replication
forks (88). Furthermore, Fanconi
anemia complementation group D2 and biorientation of chromosomes in
cell division 1-like proteins have been described to interact with
BRCA2 and promote stalled fork protection (89,90). Of note, BRCA2 may act as a
transcriptional co-regulator forming a functional complex with
mothers against decapentaplegic homolog 3 (SMAD3). BRCA2 induces
SMAD3-dependent transcriptional activation of plasminogen activator
inhibitor 1, while SMAD3 increases the transcriptional activity of
BRCA2, indicating a synergistic activity of these two proteins
(91). Furthermore, it has been
reported that the binding between BRCA2 exon 3 and the nuclear
protein EMSY (BRCA2-interacting transcriptional repressor) is
involved in chromatin remodeling and transcription regulation
(92). Finally, although the
direct role of BRCA2 in cell-cycle checkpoints remains to be fully
clarified, the interaction between BRCA2 and BRCA2-associated
factor 35 may be responsible for G2/M checkpoint modulation
(93).
On these bases, the present study aimed to provide
an update of the current knowledge on BRCA1 and BRCA2 mutations and
cancer susceptibility, focusing on their physiological functions,
mutation frequency and clinical impact, as well as the available
genetic tests and new potential technologies for the detection of
BRCA1/2 mutations.
The analysis of BRCA1 and BRCA2 mutations was
performed using the Breast Cancer Information Core (BIC)
(https://research.nhgri.nih.gov/bic/,
accessed on 3 February 2022), BRCA Exchange (https://brcaexchange.org/, accessed on 15 February
2022) and ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/, accessed on 7
February 2022) public databases. Specifically, a total of 15,311
BRCA1 and 14,914 BRCA2 mutations were registered in the BIC
database. Among these, the top 10 most common BRCA1 and BRCA2
genetic variants were selected for evaluation of their clinical
significance. In this regard, the BIC designation of BRCA1/2
mutations was matched with data reported in the BRCA Exchange and
ClinVar databases in order to classify them into variants that are
benign, likely benign, pathogenetic or of uncertain
significance.
A literature search of studies published from 1995
until November 2021 was conducted using the PubMed public database
(https://pubmed.ncbi.nlm.nih.gov/) in
order to investigate interethnic mutation frequencies. The key
words 'breast cancer', 'ovarian cancer', 'BRCA1 mutations', 'BRCA2
mutations', 'BRCA founder mutations', 'BRCA germline mutations' or
'BRCA somatic mutations' were used to identify potentially relevant
studies. In addition, the references contained in the most relevant
studies were manually retrieved to find relevant articles not
retrieved by PubMed exploration. Of note, the mutation frequency
analysis was performed focusing on specific regions and ethnic
groups, including Ashkenazi Jews, as well as populations from
China, Denmark, Finland, France, Germany, Italy, Japan, Korea,
Norway, Philippines, Poland, Russia and Sweden. In cases of
overlapping data with the other published articles, the latest
published and/or the larger sample size study was selected.
Articles with ambiguous annotation of data, published in
non-English or in Chinese language and duplicate publications were
not included.
Both endogenous and external DNA-damaging agents,
including mutagens and radiation, as well as spontaneously
occurring mutations, constantly threaten the integrity of the
genome. Of note, DSBs represent the most damaging lesions of DNA,
which may lead to chromosomal aberrations and mutations, increasing
the risk of developing genetic disorders strictly related to cancer
susceptibility (94). In this
field, it has been widely reported that BRCA1 and BRCA2 tumor
suppressor genes are involved in the repair of DNA DSBs by HRR.
Specifically, BRCA1 promotes end resection and recruits PALB2,
inducing chromatin localization of BRCA2. On the other hand, BRCA2
facilitates the recruitment of RAD51 recombinase, which inhibits
the annealing of complementary ssDNA into the deleterious single
strand (95). However, BRCA1 and
BRCA2 mutations may increase the susceptibility to several tumor
types, particularly breast and ovarian cancer (96,97). Indeed, BRCA1/2 loss of functions
leads to genomic instability, which may result in the oncogenic
transformation of normal cells into tumor-initiating cells
(42).
BRCA1/2 mutations may be classified into germline
and somatic mutations. Germline mutations are inherited in an
autosomal dominant manner, while somatic mutations may arise de
novo in tumor tissues due to a combination of genetic and
environmental factors (98).
Regarding BRCA1/2 germline mutations, loss of heterozygosity
results in a non-functional protein that leads to Hereditary Breast
and Ovarian Cancer (HBOC) syndrome, which is associated with
increased susceptibility for these female tumors (99). Furthermore, inherited bi-allelic
mutations of both BRCA1 and BRCA2 may cause congenital syndromes
that are strictly associated with developmental anomalies,
chromosomal fragility and increased cancer risk (100). In particular, the risk of breast
cancer for females with a pathogenic BRCA1 or BRCA2 germline
variant is 55-72% and 45-69%, respectively. Similarly, the risk for
ovarian cancer is 39-44% for females with a BRCA1 germline variant
and 11-17% for those with a BRCA2 germline variant (101). On the other hand, it has been
reported that BRCA somatic mutations account for 15-30% of all
BRCA1 and BRCA2 mutations. In addition, these non-inherited
mutations are only present in 3% of all breast cancer cases
(102).
Over the years, a large number of BRCA1/2 mutations
have been described, and several of them are reliably known to
increase cancer susceptibility (103). As reported in ClinVar, thousands
of pathogenic or likely pathogenic BRCA1 and BRCA2 variants have
been identified (>2,900 and >3,500, respectively) (https://www.ncbi.nlm.nih.gov/clinvar/,
accessed 7 February 2022). The pathogenic or likely pathogenic
mutations account for 80% of all mutations and result in a
premature termination codon and truncated protein. Furthermore,
missense mutations encoding a stable mutant protein account for 10%
of all missense variants. Of note, frameshift mutations are more
common in BRCA1, whereas missense mutations are more frequent in
BRCA2 (104).
In the BIC database, a total of 15,311 BRCA1
mutations were registered, of which 6,133 are frameshift mutations,
4,577 are missense mutations and 1,421 are nonsense mutations.
Regarding BRCA2, 14,914 mutations were registered (3,567
frameshift, 7,156 missense and 1,040 nonsense mutations). Of note,
the mutation with the highest number of entries for BRCA1 was
185delAG, followed by 5382insC, whereas the most frequent mutation
for BRCA2 was 6174delT (https://research.nhgri.nih.gov/bic/, accessed on 3
February 2022). Table I
summarizes the most common BRCA1 and BRCA2 mutations registered in
the BIC database.
Furthermore, according to the BRCA Exchange and
ClinVar database, numerous BRCA1/2 mutations among the 10 most
frequent in the BIC database are classified as benign or likely
benign variants. Specifically, 4427T>C, S1613G, 2430T>C,
2201C>T, IVS18+66G>A, IVS16°68A>G and IVS16°92A>G are
benign or likely benign BRCA1 variants, while 185delAG, 5382insC
and C61G are certainly pathogenetic. Regarding BRCA2, 10°90A>C,
IVS16-14T>C, IVS21-66T>C, K3326X, I2490T, 3°24A>G and
IVS11+80delTTAA are registered as benign variants, H372N and F599S
are variants with uncertain significance, and 6174delT is a
pathogenic variant (https://brcaexchange.org/, accessed on 15 February
2022; https://www.ncbi.nlm.nih.gov/clinvar/, accessed on 7
February 2022).
In the last decades, an increasing number of studies
have reported that certain BRCA1/2 mutations, also known as founder
mutations, were more frequent in specific regions and ethnic
groups. For instance, 185delAG, 5382insC and 6174delT mutations
have been detected in Ashkenazi Jews, which accounted for 99% of
the pathogenic variants identified in this population. Of note, the
5382insC founder mutation has also been identified in other
countries, such as Poland and Russia, accounting for 94 and 60% of
BRCA1 mutations, respectively. Other BRCA1 founder mutations have
been observed in these countries, including C61G and 4154delA among
the Polish population and G1706A in Russians (105-108).
Regarding the Northern European countries, the most
common BRCA1 founder mutations detected among the Norwegian
population were represented by 1675delA, 816delGT, 3347delAG and
1135insA (109-111). Furthermore, several BRCA1/2
mutations have been exclusively detected in individuals born in
Finland, including IVS1°+3A>G and R1443X for BRCA1 and
IVS23+1G>A, 7708C>T and T8555G for BRCA2 (112). Of note, 2594delC, E1107X and
G1706A represented the most common BRCA1 mutations in the Danish
population, while 3171ins5 and 6601delA were detected among the
Swedish (113,114).
As for Central European regions, it has been
reported that 3600del11 and G1710X were the most frequently
detected BRCA1 mutations in the French population (115). On the other hand, numerous
founder mutations were detected among Germans, such as G1706A,
C61G, 2804delAA and IVS12-1643del3835 for BRCA1 and 5579insA and
6503delTT for BRCA2 (116). Of
note, the 5083del19 mutation exhibited a high rate in Italian
families from Calabria, while 8765delAG was observed in certain
Sardinian families (117-119).
Finally, different BRCA1/2 founder mutations have
been also discovered in Asian populations. Of note, L63X, Q934X and
5802delAATT have been identified in Japanese, while the most common
mutations among Koreans were 1041del3insT for BRCA1 and 7708C>T
for BRCA2 (120-123). In addition, 1100delAT,
3337C>T and 9325insA were more frequently detected among
Chinese, whereas 5454delC and 4859delA were the most common BRCA1/2
founder mutations in the Philippine population (124,125). The distribution of BRCA1/2
founder mutations is summarized in Tables II and III.
To date, thousands of mutations have been identified
in the BRCA1 and BRCA2 genes. Most of the described mutations are
caused by small insertions or deletions, large genomic
rearrangements (LGRs), as well as nonsense mutations and splice
variants (126). As previously
described, certain mutations have been more frequently detected in
specific geographical areas and ethnicities (127). Furthermore, it has been reported
that each family group may carry a specific mutation that may be
considered unique (128).
Functional deficiencies due to these pathogenic mutations increase
the lifetime risk to develop both breast and ovarian cancer. Of
note, the most common cause of these female tumors is HBOC
syndrome. Females harboring BRCA1/2 germline mutations have a
higher risk of cancer development compared to other subjects
(129,130). Therefore, the usage of reliable
genetic tests for the assessment of BRCA1/2 mutations in high-risk
females is crucial for the clinical management of patients.
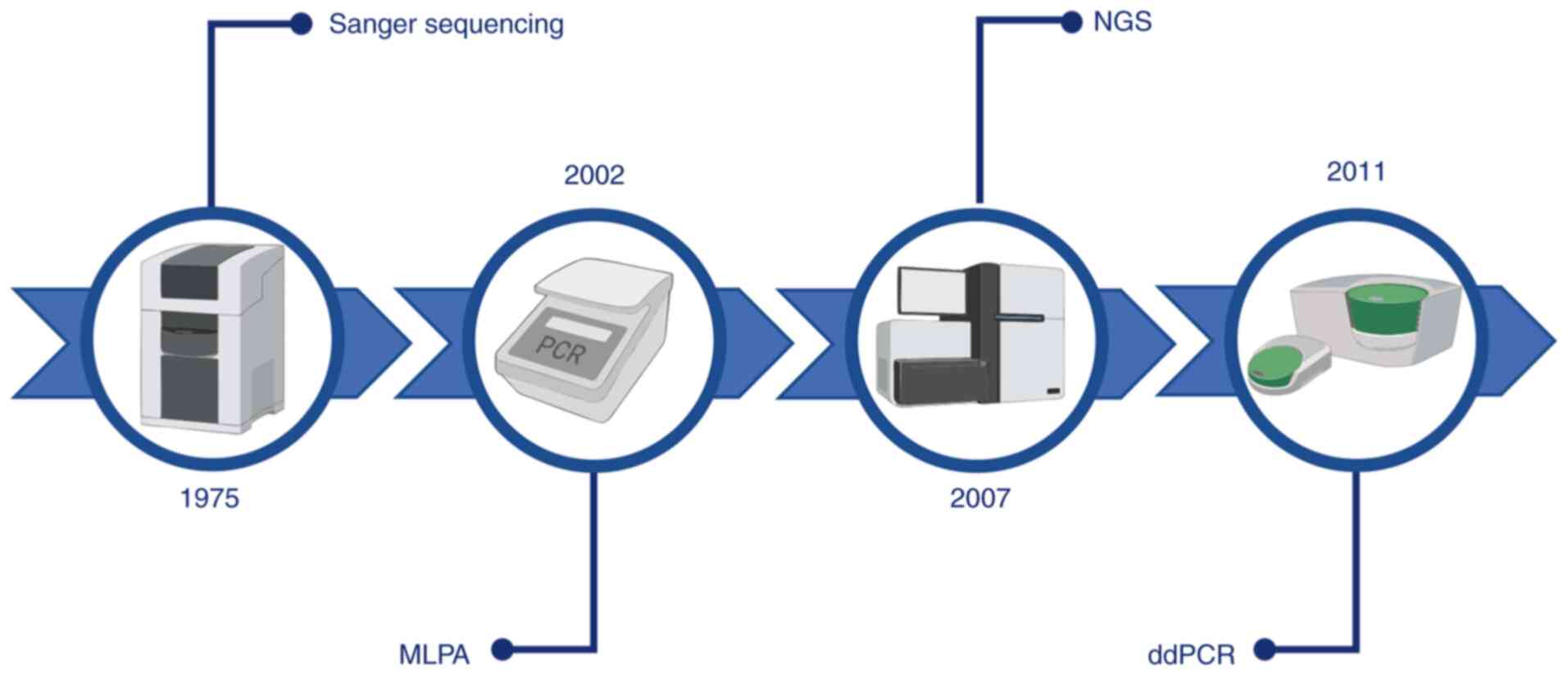
Over the years, different platforms have been
developed for the detection of BRCA mutations (Fig. 4). Among these, the Sanger method
has been widely employed for genomic DNA sequencing. In brief, this
method allows division of large genomic DNA into small fragments
that are sequenced separately (131,132). Although the Sanger sequencing
method is considered a reliable technology with a relatively simple
workflow, it has several limits, such as sequencing throughput (a
single DNA fragment at a time), time-consuming analysis and low
cost-effectiveness (133).
Currently, the gold standard genetic test for the
identification of BRCA1/2 mutations is next-generation sequencing
(NGS). NGS is a high-throughput technology based on synthesis by
sequencing millions of DNA fragments at once. Compared to Sanger
sequencing, NGS has numerous other advantages, including automated
analysis, faster turnaround time and higher sensitivity to detect
LGRs and low-frequency variants with deep sequencing (134,135). However, NGS technology may also
have certain disadvantages, such as complex workflow, high rates of
variants with uncertain significance, higher cost and the reduction
of sensitivity for large insertions/deletions (>20 bp) (136,137). The currently used standard
protocol for the identification of BRCA1 and BRCA2 mutations
includes comprehensive sequencing and the assessment of LGRs.
Furthermore, a single-site target may also be analyzed for patients
with a first-degree relative affected by BRCA1/2 mutation (138,139).
The high cost of these technologies has prompted
researchers and clinicians to develop novel high-sensitivity and
low-cost strategies for the detection of BRCA1/2 mutations. Among
these, reliable results were obtained by using the droplet digital
PCR (ddPCR) platform.
ddPCR has recently emerged as a reliable tool with
high sensitivity and specificity. In brief, the ddPCR system is
based on a water-oil emulsion of the reaction mixture, which
consists of a DNA sample, ddPCR Mastermix, primers and probe in a
final volume of 20 µl (140). This procedure allows division of
the sample into ~20,000 droplets that are transferred into a
96-well PCR plate for amplification. Since the sample is fractioned
into thousands of droplets, PCR amplification takes place in each
droplet. A droplet reader then detects the positive/negative signal
of droplets depending on their amplified target and fluorescence
amplitude (141).
Of note, ddPCR represents a valuable alternative to
standard methods for the analysis of different clinical samples,
such as fresh tumor biopsies and formalin-fixed paraffin-embedded
(FFPE) tissues, as well as liquid biopsy samples (peripheral blood,
sputum, urine, cerebrospinal fluid, stool, pleural effusions and
ascites fluid), overcoming the limits due to poor DNA quality
(142). Over the years, the
potential clinical application of ddPCR has been demonstrated for
absolute allele quantification, viral load quantification, DNA
methylation, DNA copy number variation, germline/somatic mutation
detection, circulating mutation detection, analysis of microRNAs,
long non-coding RNAs and gene rearrangements (143-147). In this field, a growing number
of recent studies have focused on the ddPCR system as a promising
tool for the identification of specific BRCA1/2 mutations.
Although no studies have been performed on the use
of ddPCR for the detection of specific BRCA mutations in relatives
of BRCA-positive cancer patients, the use of ddPCR for the
detection of already known mutations in familial clusters would
reduce the higher costs related to the whole sequencing of both
BRCA1 and BRCA2 genes. Indeed, at present, the cost of a single
test for the detection of BRCA mutations is 172$, while Garcia and
colleagues described that the analysis of gene mutations performed
by ddPCR cost half as much as NGS or other methods (152,153).
Besides the cost-effectiveness, ddPCR may also be
used for the detection of circulating BRCA1/2 mutations in liquid
biopsy samples. Indeed, ddPCR was effectively used for the
detection of circulating mutations in patients with ovarian cancer,
demonstrating how the detection of both germline and somatic ctDNA
mutations is a valuable complementary tool for the diagnosis of
BRCA-positive tumors and to establish the effectiveness of PARP
inhibitors and the prognosis of patients with ovarian cancer
(154).
All of these data suggest that ddPCR may be used for
the detection of already known BRCA1/2 mutations in familial
clusters of patients with a diagnosis of hereditary breast or
ovarian cancer.
In conclusion, BRCA1 and BRCA2 germline mutations
are well-established risk factors for the onset of breast or
ovarian cancer. Overall, the computational investigations performed
in the present study suggested that the BRCA1/2 mutations with the
highest number of entries are classified as certainly pathogenic
variants, such as 185delAG, 5382insC and C61G for BRCA1 and
6174delT for BRCA2. Furthermore, several of these mutations, also
known as founder mutations, appear to be more frequently detected
in specific geographical regions and ethnic groups. In this
context, the development of low-cost and reliable genetic tests is
fundamental to improve the screening program for the identification
of females with germline BRCA1/2 mutations and the management of
relatives of patients with a known diagnosis of hereditary breast
or ovarian cancer. Currently, NGS is the gold standard for the
assessment of BRCA1/2 mutations. In this field, ddPCR has recently
emerged as a highly sensitive and specific technique. According to
the studies described in the present article, ddPCR may be
considered a promising alternative strategy for the detection of
BRCA1/2 pathogenetic mutations. However, further studies should be
performed to validate the application of ddPCR in routine clinical
practice and surveillance strategies for hereditary female
tumors.
ML and AS conceptualized the study. AL, RR, GG and
SC wrote the original draft of the manuscript. ML, AS, GNZ, GC and
LF provided critical revisions. AL and RR prepared the tables and
figures, conducted the formal analysis and critically analyzed the
literature. All authors contributed to manuscript revision and read
and approved the final version of the manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
This work was supported by the Italian League Against Cancer
(LILT) - Grant Ricerca Sanitaria 2018 LILT.
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
Cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Łukasiewicz S, Czeczelewski M, Forma A,
Baj J, Sitarz R and Stanisławek A: Breast cancer-epidemiology, risk
factors, classification, prognostic markers, and current treatment
strategies-an updated review. Cancers (Basel). 13:42872021.
View Article : Google Scholar
|
|
3
|
Falzone L, Scandurra G, Lombardo V,
Gattuso G, Lavoro A, Distefano AB, Scibilia G and Scollo P: A
multidisciplinary approach remains the best strategy to improve and
strengthen the management of ovarian cancer (Review). Int J Oncol.
59:532021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Winters S, Martin C, Murphy D and Shokar
NK: Breast cancer epidemiology, prevention, and screening. Prog Mol
Biol Transl Sci. 151:1–32. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
La Vecchia C: Ovarian cancer: Epidemiology
and risk factors. Eur J Cancer Prev. 26:55–62. 2017. View Article : Google Scholar
|
|
6
|
D'Alonzo M, Bounous VE, Villa M and Biglia
N: Current evidence of the oncological benefit-risk profile of
hormone replacement therapy. Medicina (Kaunas). 55:5732019.
View Article : Google Scholar
|
|
7
|
Prentice RL, Aragaki AK, Chlebowski RT,
Rossouw JE, Anderson GL, Stefanick ML, Wactawski-Wende J, Kuller
LH, Wallace R, Johnson KC, et al: Randomized trial evaluation of
the benefits and risks of menopausal hormone therapy among women
50-59 years of age. Am J Epidemiol. 190:365–375. 2021. View Article : Google Scholar
|
|
8
|
Beaber EF, Malone KE, Tang MT, Barlow WE,
Porter PL, Daling JR and Li CI: Oral contraceptives and breast
cancer risk overall and by molecular subtype among young women.
Cancer Epidemiol Biomarkers Prev. 23:755–764. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Havrilesky LJ, Moorman PG, Lowery WJ,
Gierisch JM, Coeytaux RR, Urrutia RP, Dinan M, McBroom AJ,
Hasselblad V, Sanders GD and Myers ER: Oral contraceptive pills as
primary prevention for ovarian cancer: A systematic review and
meta-analysis. Obstet Gynecol. 112:139–147. 2013. View Article : Google Scholar
|
|
10
|
Benfatto G, Zanghì G, Catalano F, Di
Stefano G, Fancello R, Mugavero F and Giovanetto A: Day surgery for
breast cancer in the elderly. G Chir. 27:49–52. 2006.In Italian.
PubMed/NCBI
|
|
11
|
Shoemaker ML, White MC, Wu M, Weir HK and
Romieu I: Differences in breast cancer incidence among young women
aged 20-49 years by stage and tumor characteristics, age, race, and
ethnicity, 2004-2013. Breast Cancer Res Treat. 169:595–606. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sarink D, Wu AH, Le Marchand L, White KK,
Park SY, Setiawan VW, Hernandez BY, Wilkens LR and Merritt MA:
Racial/ethnic differences in ovarian cancer risk: Results from the
multiethnic cohort study. Cancer Epidemiol Biomarkers Prev.
29:2019–2025. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zeinomar N, Knight JA, Genkinger JM,
Phillips KA, Daly MB, Milne RL, Dite GS, Kehm RD, Liao Y, Southey
MC, et al: Alcohol consumption, cigarette smoking, and familial
breast cancer risk: Findings from the prospective family study
cohort (ProF-SC). Breast Cancer Res. 21:1282019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Friedenreich CM, Ryder-Burbidge C and
McNeil J: Physical activity, obesity and sedentary behavior in
cancer etiology: Epidemiologic evidence and biologic mechanisms.
Mol Oncol. 15:790–800. 2021. View Article : Google Scholar :
|
|
15
|
Dunneram Y, Greenwood DC and Cade JE:
Diet, menopause and the risk of ovarian, endometrial and breast
cancer. Proc Nutr Soc. 78:438–448. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bryere J, Dejardin O, Launay L, Colonna M,
Grosclaude P and Launoy G; French Network of Cancer Registries
(FRANCIM): Socioeconomic status and site-specific cancer incidence,
a Bayesian approach in a French cancer registries network study.
Eur J Cancer Prev. 27:391–398. 2018. View Article : Google Scholar
|
|
17
|
Falzone L, Grimaldi M, Celentano E,
Augustin LSA and Libra M: Identification of modulated MicroRNAs
associated with breast cancer, diet, and physical activity. Cancers
(Basel). 12:25552020. View Article : Google Scholar
|
|
18
|
Park HL: Epigenetic biomarkers for
environmental exposures and personalized breast cancer prevention.
Int J Environ Res Public Health. 17:11812020. View Article : Google Scholar :
|
|
19
|
Singh A, Gupta S and Sachan M: Epigenetic
biomarkers in the management of ovarian cancer: Current
prospectives. Front Cell Dev Biol. 7:1822019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang M, Xiao J, Nasca PC, Liu C, Lu Y,
Lawrence WR, Wang L, Chen Q and Lin S: Do multiple environmental
factors impact four cancers in women in the contiguous United
States? Environ Res. 179(PtA): 1087822019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Brewer HR, Jones ME, Schoemaker MJ,
Ashworth A and Swerdlow AJ: Family history and risk of breast
cancer: An analysis accounting for family structure. Breast Cancer
Res Treat. 165:193–200. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Flaum N, Crosbie EJ, Edmondson RJ, Smith
MJ and Evans DG: Epithelial ovarian cancer risk: A review of the
current genetic landscape. Clin Genet. 97:54–63. 2020. View Article : Google Scholar :
|
|
23
|
Bethea TN, Ochs-Balcom HM, Bandera EV,
Beeghly-Fadiel A, Camacho F, Chyn D, Cloyd EK, Harris HR, Joslin
CE, Myers E, et al: First- and second-degree family history of
ovarian and breast cancer in relation to risk of invasive ovarian
cancer in African American and white women. Int J Cancer.
148:2964–2973. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu L, Hao X, Song Z, Zhi X, Zhang S and
Zhang J: Correlation between family history and characteristics of
breast cancer. Sci Rep. 11:63602021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Welcsh PL and King MC: BRCA1 and BRCA2 and
the genetics of breast and ovarian cancer. Hum Mol Genet.
10:705–713. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yoshida K and Miki Y: Role of BRCA1 and
BRCA2 as regulators of DNA repair, transcription, and cell cycle in
response to DNA damage. Cancer Sci. 95:866–871. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Paul A and Paul S: The breast cancer
susceptibility genes (BRCA) in breast and ovarian cancers. Front
Biosci (Landmark Ed). 19:605–618. 2014. View Article : Google Scholar
|
|
28
|
Venkitaraman AR: How do mutations
affecting the breast cancer genes BRCA1 and BRCA2 cause cancer
susceptibility? DNA Repair (Amst). 81:1026682019. View Article : Google Scholar
|
|
29
|
Paalosalo-Harris K and Skirton H: Mixed
method systematic review: The relationship between breast cancer
risk perception and health-protective behaviour in women with
family history of breast cancer. J Adv Nurs. 73:760–764. 2017.
View Article : Google Scholar
|
|
30
|
Hanley GE, McAlpine JN, Miller D, Huntsman
D, Schrader KA, Gilks CB and Mitchell G: A population-based
analysis of germline BRCA1 and BRCA2 testing among ovarian cancer
patients in an era of histotype-specific approaches to ovarian
cancer prevention. BMC Cancer. 18:2542018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Moschetta M, George A, Kaye SB and
Banerjee S: BRCA somatic mutations and epigenetic BRCA
modifications in serous ovarian cancer. Ann Oncol. 27:1449–1455.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kuchenbaecker KB, Hopper JL, Barnes DR,
Phillips KA, Mooij TM, Roos-Blom MJ, Jervis S, van Leeuwen FE,
Milne RL, Andrieu N, et al: Risks of breast, ovarian, and
contralateral breast cancer for BRCA1 and BRCA2 mutation carriers.
JAMA. 317:2402–2416. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Neff RT, Senter L and Salani R: BRCA
mutation in ovarian cancer: Testing, implications and treatment
considerations. Ther Adv Med Oncol. 9:519–531. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kotsopoulos J: BRCA mutations and breast
cancer prevention. Cancers (Basel). 10:5242018. View Article : Google Scholar
|
|
35
|
Cortesi L, Piombino C and Toss A: Germline
mutations in other homologous recombination repair-related genes
than BRCA1/2: Predictive or prognostic factors? J Pers Med.
11:2452021. View Article : Google Scholar :
|
|
36
|
Zhao W, Hu H, Mo Q, Guan Y, Li Y, Du Y and
Li L: Function and mechanism of combined PARP-1 and BRCA genes in
regulating the radiosensitivity of breast cancer cells. Int J Clin
Exp Pathol. 12:3915–3920. 2019.
|
|
37
|
Liu X, Wu K, Zheng D, Luo C, Fan Y, Zhong
X and Zheng H: Efficacy and safety of PARP inhibitors in advanced
or metastatic triple-negative breast cancer: A systematic review
and meta-analysis. Front Oncol. 11:7421392021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dickson KA, Xie T, Evenhuis C, Ma Y and
Marsh DJ: PARP inhibitors display differential efficacy in models
of BRCA mutant high-grade serous ovarian cancer. Int J Mol Sci.
22:85062021. View Article : Google Scholar :
|
|
39
|
Al-Thoubaity FK: Molecular classification
of breast cancer: A retrospective cohort study. Ann Med Surg
(Lond). 49:44–48. 2019. View Article : Google Scholar
|
|
40
|
Makki J: Diversity of breast carcinoma:
Histological subtypes and clinical relevance. Clin Med Insights
Pathol. 8:23–31. 2015. View Article : Google Scholar
|
|
41
|
Magro G, Salvatorelli L, Puzzo L, Piombino
E, Bartoloni G, Broggi G and Vecchio GM: Practical approach to
diagnosis of bland-looking spindle cell lesions of the breast.
Pathologica. 111:344–360. 2019. View Article : Google Scholar
|
|
42
|
Gorodetska I, Kozeretska I and Dubrovska
A: BRCA genes: The role in genome stability, cancer stemness and
therapy resistance. J Cancer. 10:2109–2127. 2019. View Article : Google Scholar :
|
|
43
|
Tung NM and Garber JE: BRCA1/2 testing:
Therapeutic implications for breast cancer management. Br J Cancer.
119:141–152. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Broggi G, Filetti V, Ieni A, Rapisarda V,
Ledda C, Vitale E, Varricchio S, Russo D, Lombardo C, Tuccari G, et
al: MacroH2A1 immunoexpression in breast cancer. Front Oncol.
10:15192020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Miki Y, Swensen J, Shattuck-Eidens D,
Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM
and Ding W: A strong candidate for the breast and ovarian cancer
susceptibility gene BRCA1. Science. 266:66–71. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Nelson AC and Holt JT: Impact of RING and
BRCT domain mutations on BRCA1 protein stability, localization and
recruitment to DNA damage. Radiat Res. 174:1–13. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Christou CM and Kyriacou K: BRCA1 and its
network of interacting partners. Biology (Basel). 2:40–63.
2013.
|
|
48
|
Xia Y, Pao GM, Chen HW, Verma IM and
Hunter T: Enhancement of BRCA1 E3 ubiquitin ligase activity through
direct interaction with the BARD1 protein. J Biol Chem.
278:5255–5263. 2003. View Article : Google Scholar
|
|
49
|
Manke IA, Lowery DM, Nguyen A and Yaffe
MB: BRCT repeats as phosphopeptide-binding modules involved in
protein targeting. Science. 302:636–639. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Clapperton JA, Manke IA, Lowery DM, Ho T,
Haire LF, Yaffe MB and Smerdon SJ: Structure and mechanism of BRCA1
BRCT domain recognition of phosphorylated BACH1 with implications
for cancer. Nat Struct Mol Biol. 11:512–518. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Thanassoulas A, Nomikos M, Theodoridou M,
Yannoukakos D, Mastellos D and Nounesis G: Thermodynamic study of
the BRCT domain of BARD1 and its interaction with the
-pSER-X-X-Phemotif-containing BRIP1 peptide. Biochim Biophys Acta.
1804:1908–1916. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wang B, Matsuoka S, Ballif BA, Zhang D,
Smogorzewska A, Gygi SP and Elledge SJ: Abraxas and RAP80 form a
BRCA1 protein complex required for the DNA damage response.
Science. 316:1194–1198. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chen CF, Li S, Chen Y, Chen PL, Sharp ZD
and Lee WH: The nuclear localization sequences of the BRCA1 protein
interact with the importin-alpha subunit of the nuclear transport
signal receptor. J Biol Chem. 271:32863–32868. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Caestecker KW and Van de Walle GR: The
role of BRCA1 in DNA double-strand repair: past and present. Exp
Cell Res. 319:575–587. 2013. View Article : Google Scholar
|
|
55
|
Savage KI and Harkin DP: BRCA1, a
'complex' protein involved in the maintenance of genomic stability.
FEBS J. 282:630–646. 2015. View Article : Google Scholar
|
|
56
|
Sharma B, Kaur RP, Raut S and Munshi A:
BRCA1 mutation spectrum, functions, and therapeutic strategies: The
story so far. Curr Probl Cancer. 42:189–207. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Takaoka M and Miki Y: BRCA1 gene: Function
and deficiency. Int J Clin Oncol. 23:36–44. 2018. View Article : Google Scholar
|
|
58
|
Matsuoka S, Ballif BA, Smogorzewska A,
McDonald ER III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini
N, Lerenthal Y, et al: ATM and ATR substrate analysis reveals
extensive protein networks responsive to DNA damage. Science.
316:1160–1166. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Smith J, Tho LM, Xu N and Gillespie DA:
The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and
cancer. Adv Cancer Res. 108:73–112. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Syed A and Tainer JA: The MRE11-RAD50-NBS1
complex conducts the orchestration of damage signaling and outcomes
to stress in DNA replication and repair. Annu Rev Biochem.
87:263–294. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Simhadri S, Vincelli G, Huo Y, Misenko S,
Foo TK, Ahlskog J, Sørensen CS, Oakley GG, Ganesan S, Bunting SF
and Xia B: PALB2 connects BRCA1 and BRCA2 in the G2/M checkpoint
response. Oncogene. 38:1585–1596. 2019. View Article : Google Scholar :
|
|
62
|
Zhao W, Steinfeld JB, Liang F, Chen X,
Maranon DG, Ma CJ, Kwon Y, Rao T, Wang W, Sheng C, et al:
BRCA1-BARD1 promotes RAD51-mediated homologous DNA pairing. Nature.
550:360–365. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Coleman KA and Greenberg RA: The
BRCA1-RAP80 complex regulates DNA repair mechanism utilization by
restricting end resection. J Biol Chem. 286:13669–13680. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Kumaraswamy E and Shiekhattar R:
Activation of BRCA1/BRCA2-associated helicase BACH1 is required for
timely progression through S phase. Mol Cell Biol. 27:6733–6741.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wang Q, Zhang H, Kajino K and Greene MI:
BRCA1 binds c-Myc and inhibits its transcriptional and transforming
activity in cells. Oncogene. 17:1939–1948. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Chai YL, Cui J, Shao N, Shyam E, Reddy P
and Rao VN: The second BRCT domain of BRCA1 proteins interacts with
p53 and stimulates transcription from the p21WAF1/CIP1 promoter.
Oncogene. 18:263–268. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Thurn KT, Thomas S, Raha P, Qureshi I and
Munster PN: Histone deacetylase regulation of ATM-mediated DNA
damage signaling. Mol Cancer Ther. 12:2078–2087. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Buckley NE, Hosey AM, Gorski JJ, Purcell
JW, Mulligan JM, Harkin DP and Mullan PB: BRCA1 regulates IFN-gamma
signaling through a mechanism involving the type I IFNs. Mol Cancer
Res. 5:261–270. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Tan W, Zheng L, Lee WH and Boyer TG:
Functional dissection of transcription factor ZBRK1 reveals zinc
fingers with dual roles in DNA-binding and BRCA1-dependent
transcriptional repression. J Biol Chem. 279:6576–6587. 2004.
View Article : Google Scholar
|
|
70
|
Harte MT, O'Brien GJ, Ryan NM, Gorski JJ,
Savage KI, Crawford NT, Mullan PB and Harkin DP: BRD7, a subunit of
SWI/SNF complexes, binds directly to BRCA1 and regulates
BRCA1-dependent transcription. Cancer Res. 70:2538–2547. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Yoshino Y, Qi H, Fujita H, Shirota M, Abe
S, Komiyama Y, Shindo K, Nakayama M, Matsuzawa A, Kobayashi A, et
al: BRCA1-interacting protein OLA1 requires interaction with BARD1
to regulate centrosome number. Mol Cancer Res. 16:1499–1511. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Harkin DP, Bean JM, Miklos D, Song YH,
Truong VB, Englert C, Christians FC, Ellisen LW, Maheswaran S,
Oliner JD and Haber DA: Induction of GADD45 and JNK/SAPK-dependent
apoptosis following inducible expression of BRCA1. Cell.
97:575–586. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Wooster R, Bignell G, Lancaster J, Swift
S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C and Micklem G:
Identification of the breast cancer susceptibility gene BRCA2.
Nature. 378:789–792. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Chen J, Silver DP, Walpita D, Cantor SB,
Gazdar AF, Tomlinson G, Couch FJ, Weber BL, Ashley T, Livingston DM
and Scully R: Stable interaction between the products of the BRCA1
and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol
Cell. 2:317–328. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Sharan SK and Bradley A: Functional
characterization of BRCA1 and BRCA2: Clues from their interacting
proteins. J Mammary Gland Biol Neoplasia. 3:413–421. 1998.
View Article : Google Scholar
|
|
76
|
Oliver AW, Swift S, Lord CJ, Ashworth A
and Pearl LH: Structural basis for recruitment of BRCA2 by PALB2.
EMBO Rep. 10:990–996. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Carreira A, Hilario J, Amitani I, Baskin
RJ, Shivji MK, Venkitaraman AR and Kowalczykowski SC: The BRC
repeats of BRCA2 modulate the DNA-binding selectivity of RAD51.
Cell. 136:1032–1043. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Davies OR and Pellegrini L: Interaction
with the BRCA2 C terminus protects RAD51-DNA filaments from
disassembly by BRC repeats. Nat Struct Mol Biol. 14:475–483. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Moynahan ME, Pierce AJ and Jasin M: BRCA2
is required for homology-directed repair of chromosomal breaks. Mol
Cell. 7:263–272. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Roy R, Chun J and Powell SN: BRCA1 and
BRCA2: Different roles in a common pathway of genome protection.
Nat Rev Cancer. 12:68–78. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Yuan SS, Lee SY, Chen G, Song M, Tomlinson
GE and Lee EY: BRCA2 is required for ionizing radiation-induced
assembly of Rad51 complex in vivo. Cancer Res. 59:3547–3551.
1999.PubMed/NCBI
|
|
82
|
Milner J, Ponder B, Hughes-Davies L,
Seltmann M and Kouzarides T: Transcriptional activation functions
in BRCA2. Nature. 386:772–773. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Davies AA, Masson JY, McIlwraith MJ,
Stasiak AZ, Stasiak A, Venkitaraman AR and West SC: Role of BRCA2
in control of the RAD51 recombination and DNA repair protein. Mol
Cell. 7:273–282. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Esashi F, Galkin VE, Yu X, Egelman EH and
West SC: Stabilization of RAD51 nucleoprotein filaments by the
C-terminal region of BRCA2. Nat Struct Mol Biol. 14:468–474. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Henderson BR: Regulation of BRCA1, BRCA2
and BARD1 intracellular trafficking. Bioessays. 27:884–893. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Couturier AM, Fleury H, Patenaude AM,
Bentley VL, Rodrigue A, Coulombe Y, Niraj J, Pauty J, Berman JN,
Dellaire G, et al: Roles for APRIN (PDS5B) in homologous
recombination and in ovarian cancer prediction. Nucleic Acids Res.
44:10879–10897. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Yang H, Jeffrey PD, Miller J, Kinnucan E,
Sun Y, Thoma NH, Zheng N, Chen PL, Lee WH and Pavletich NP: BRCA2
function in DNA binding and recombination from a BRCA2-DSS1-ssDNA
structure. Science. 297:1837–1848. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Buisson R, Niraj J, Pauty J, Maity R, Zhao
W, Coulombe Y, Sung P and Masson JY: Breast cancer proteins PALB2
and BRCA2 stimulate polymerase η in recombination-associated DNA
synthesis at blocked replication forks. Cell Rep. 6:553–564. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Hussain S, Wilson JB, Medhurst AL, Hejna
J, Witt E, Ananth S, Davies A, Masson JY, Moses R, West SC, et al:
Direct interaction of FANCD2 with BRCA2 in DNA damage response
pathways. Hum Mol Genet. 13:1241–1248. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Higgs MR and Stewart GS: Protection or
resection: BOD1L as a novel replication fork protection factor.
Nucleus. 7:34–40. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Preobrazhenska O, Yakymovych M, Kanamoto
T, Yakymovych I, Stoika R, Heldin CH and Souchelnytskyi S: BRCA2
and Smad3 synergize in regulation of gene transcription. Oncogene.
21:5660–5664. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Hughes-Davies L, Huntsman D, Ruas M, Fuks
F, Bye J, Chin SF, Milner J, Brown LA, Hsu F, Gilks B, et al: EMSY
links the BRCA2 pathway to sporadic breast and ovarian cancer.
Cell. 115:523–535. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Marmorstein LY, Kinev AV, Chan GK, Bochar
DA, Beniya H, Epstein JA, Yen TJ and Shiekhattar R: A human BRCA2
complex containing a structural DNA binding component influences
cell cycle progression. Cell. 104:247–257. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Singh JK, Smith R, Rother MB, de Groot
AJL, Wiegant WW, Vreeken K, D'Augustin O, Kim RQ, Qian H, Krawczyk
PM, et al: Zinc finger protein ZNF384 is an adaptor of Ku to DNA
during classical non-homologous end-joining. Nat Commun.
12:65602021. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Prakash R, Zhang Y, Feng W and Jasin M:
Homologous recombination and human health: The roles of BRCA1,
BRCA2, and associated proteins. Cold Spring Harb Perspect Biol.
7:a0166002015. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Huang KL, Mashl RJ, Wu Y, Ritter DI, Wang
J, Oh C, Paczkowska M, Reynolds S, Wyczalkowski MA, Oak N, et al:
Pathogenic Germline variants in 10,389 adult cancers. Cell.
173:355–370. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Van Hout CV, Tachmazidou I, Backman JD,
Hoffman JD, Liu D, Pandey AK, Gonzaga-Jauregui C, Khalid S, Ye B,
Banerjee N, et al: Exome sequencing and characterization of 49,960
individuals in the UK Biobank. Nature. 586:749–756. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Engel C and Fischer C: Breast cancer risks
and risk prediction models. Breast Care (Basel). 10:7–12. 2015.
View Article : Google Scholar
|
|
99
|
Wu H, Wu X and Liang Z: Impact of germline
and somatic BRCA1/2 mutations: Tumor spectrum and detection
platforms. Gene Ther. 24:601–609. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Seo A, Steinberg-Shemer O, Unal S, Casadei
S, Walsh T, Gumruk F, Shalev S, Shimamura A, Akarsu NA, Tamary H
and King MC: Mechanism for survival of homozygous nonsense
mutations in the tumor suppressor gene BRCA1. Proc Natl Acad Sci
USA. 115:5241–5246. 2018. View Article : Google Scholar :
|
|
101
|
Petrucelli N, Daly MB and Pal T: BRCA1-
and BRCA2-Associated Hereditary Breast and Ovarian Cancer.
GeneReviews®. Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH,
Gripp KW, Mirzaa GM and Amemiya A: University of Washington;
Seattle: pp. 1993–2022. 2022
|
|
102
|
Winter C, Nilsson MP, Olsson E, George AM,
Chen Y, Kvist A, Törngren T, Vallon-Christersson J, Hegardt C,
Häkkinen J, et al: Targeted sequencing of BRCA1 and BRCA2 across a
large unselected breast cancer cohort suggests that one-third of
mutations are somatic. Ann Oncol. 27:1532–1538. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Cline MS, Liao RG, Parsons MT, Paten B,
Alquaddoomi F, Antoniou A, Baxter S, Brody L, Cook-Deegan R, Coffin
A, et al: BRCA challenge: BRCA exchange as a global resource for
variants in BRCA1 and BRCA2. PLoS Genet. 26:e10077522018.
View Article : Google Scholar
|
|
104
|
Anczuków O, Ware MD, Buisson M, Zetoune
AB, Stoppa-Lyonnet D, Sinilnikova OM and Mazoyer S: Does the
nonsense-mediated mRNA decay mechanism prevent the synthesis of
truncated BRCA1, CHK2, and p53 proteins? Hum Mutat. 29:65–73. 2008.
View Article : Google Scholar
|
|
105
|
Roa BB, Boyd AA, Volcik K and Richards CS:
Ashkenazi Jewish population frequencies for common mutations in
BRCA1 and BRCA2. Nat Genet. 14:185–187. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Struewing JP, Abeliovich D, Peretz T,
Avishai N, Kaback MM, Collins FS and Brody LC: The carrier
frequency of the BRCA1 185delAG mutation is approximately 1 percent
in Ashkenazi Jewish individuals. Nat Genet. 11:198–200. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Ozolina S, Sinicka O, Jankevics E,
Inashkina I, Lubinski J, Gorski B, Gronwald J, Nasedkina T,
Fedorova O, Lyubchenko L and Tihomirova L: The 4154delA mutation
carriers in the BRCA1 gene share a common ancestry. Fam Cancer.
8:1–4. 2009. View Article : Google Scholar
|
|
108
|
Kaufman B, Laitman Y, Gronwald J, Lubinski
J and Friedman E: Haplotype of the C61G BRCA1 mutation in Polish
and Jewish individuals. Genet Test Mol Biomarkers. 13:465–469.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Borg A, Dørum A, Heimdal K, Maehle L,
Hovig E and Møller P: BRCA1 1675delA and 1135insA account for one
third of Norwegian familial breast-ovarian cancer and are
associated with later disease onset than less frequent mutations.
Dis Markers. 15:79–84. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Møller P, Heimdal K, Apold J, Fredriksen
A, Borg A, Hovig E, Hagen A, Hagen B, Pedersen JC, Maehle L, et al:
Genetic epidemiology of BRCA1 mutations in Norway. Eur J Cancer.
37:2428–2434. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Heimdal K, Maehle L, Apold J, Pedersen JC
and Møller P: The Norwegian founder mutations in BRCA1: High
penetrance confirmed in an incident cancer series and differences
observed in the risk of ovarian cancer. Eur J Cancer. 39:2205–2213.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Sarantaus L, Huusko P, Eerola H, Launonen
V, Vehmanen P, Rapakko K, Gillanders E, Syrjäkoski K, Kainu T,
Vahteristo P, et al: Multiple founder effects and geographical
clustering of BRCA1 and BRCA2 families in Finland. Eur J Hum Genet.
8:757–763. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Thomassen M, Hansen TV, Borg A, Lianee HT,
Wikman F, Pedersen IS, Bisgaard ML, Nielsen FC, Kruse TA and Gerdes
AM: BRCA1 and BRCA2 mutations in Danish families with hereditary
breast and/or ovarian cancer. Acta Oncol. 47:772–777. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Einbeigi Z, Bergman A, Kindblom LG,
Martinsson T, Meis-Kindblom JM, Nordling M, Suurküla M, Wahlström
J, Wallgren A and Karlsson P: A founder mutation of the BRCA1 gene
in Western Sweden associated with a high incidence of breast and
ovarian cancer. Eur J Cancer. 37:1904–1909. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Muller D, Bonaiti-Pellié C, Abecassis J,
Stoppa-Lyonnet D and Fricker JP: BRCA1 testing in breast and/or
ovarian cancer families from northeastern France identifies two
common mutations with a founder effect. Fam Cancer. 3:15–20. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Hartmann C, John AL, Klaes R, Hofmann W,
Bielen R, Koehler R, Janssen B, Bartram CR, Arnold N and Zschocke
J: Large BRCA1 gene deletions are found in 3% of German high-risk
breast cancer families. Hum Mutat. 24:5342004. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Pisano M, Cossu A, Persico I, Palmieri G,
Angius A, Casu G, Palomba G, Sarobba MG, Rocca PC, Dedola MF, et
al: Identification of a founder BRCA2 mutation in Sardinia. Br J
Cancer. 82:553–559. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Baudi F, Quaresima B, Grandinetti C, Cuda
G, Faniello C, Tassone P, Barbieri V, Bisegna R, Ricevuto E,
Conforti S, et al: Evidence of a founder mutation of BRCA1 in a
highly homogeneous population from southern Italy with
breast/ovarian cancer. Hum Mutat. 18:163–164. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Cipollini G, Tommasi S, Paradiso A,
Aretini P, Bonatti F, Brunetti I, Bruno M, Lombardi G, Schittulli
F, Sensi E, et al: Genetic alterations in hereditary breast cancer.
Ann Oncol. 15(Supp 1): SI7–SI13. 2004. View Article : Google Scholar
|
|
120
|
Ikeda N, Miyoshi Y, Yoneda K, Shiba E,
Sekihara Y, Kinoshita M and Noguchi S: Frequency of BRCA1 and BRCA2
germline mutations in Japanese breast cancer families. Int J
Cancer. 91:83–88. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Sekine M, Nagata H, Tsuji S, Hirai Y,
Fujimoto S, Hatae M, Kobayashi I, Fujii T, Nagata I, Ushijima K, et
al: Japanese Familial Ovarian Cancer Study Group. Mutational
analysis of BRCA1 and BRCA2 and clinicopathologic analysis of
ovarian cancer in 82 ovarian cancer families: Two common founder
mutations of BRCA1 in Japanese population. Clin Cancer Res.
7:3144–3150. 2001.PubMed/NCBI
|
|
122
|
Kang E and Kim SW: The korean hereditary
breast cancer study: Review and future perspectives. J Breast
Cancer. 16:245–253. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Kang E, Seong MW, Park SK, Lee JW, Lee J,
Kim LS, Lee JE, Kim SY, Jeong J, Han SA, et al: Korean hereditary
breast cancer study group. The prevalence and spectrum of BRCA1 and
BRCA2 mutations in Korean population: Recent update of the Korean
hereditary breast cancer (KOHBRA) study. Breast Cancer Res Treat.
151:157–168. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Kwong A, Ng EK, Wong CL, Law FB, Au T,
Wong HN, Kurian AW, West DW, Ford JM and Ma ES: Identification of
BRCA1/2 founder mutations in Southern Chinese breast cancer
patients using gene sequencing and high resolution DNA melting
analysis. PLoS One. 7:e439942012. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
De Leon Matsuda ML, Liede A, Kwan E, Mapua
CA, Cutiongco EM, Tan A, Borg A and Narod SA: BRCA1 and BRCA2
mutations among breast cancer patients from the Philippines. Int J
Cancer. 98:596–603. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Concolino P and Capoluongo E: Detection of
BRCA1/2 large genomic rearrangements in breast and ovarian cancer
patients: An overview of the current methods. Expert Rev Mol Diagn.
19:795–802. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Bhaskaran SP, Chandratre K, Gupta H, Zhang
L, Wang X, Cui J, Kim YC, Sinha S, Jiang L, Lu B, et al: Germline
variation in BRCA1/2 is highly ethnic-specific: Evidence from over
30,000 Chinese hereditary breast and ovarian cancer patients. Int J
Cancer. 145:962–973. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Hoogerbrugge N and Jongmans MC: Finding
all BRCA pathogenic mutation carriers: Best practice models. Eur J
Hum Genet. 24(Suppl 1): S19–S26. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Hawsawi YM, Al-Numair NS, Sobahy TM,
Al-Ajmi AM, Al-Harbi RM, Baghdadi MA, Oyouni AA and Alamer OM: The
role of BRCA1/2 in hereditary and familial breast and ovarian
cancers. Mol Genet Genomic Med. 7:e8792019. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Hodgson A and Turashvili G: Pathology of
hereditary breast and ovarian cancer. Front Oncol. 10:5317902020.
View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Venter JC, Adams MD, Sutton GG, Kerlavage
AR, Smith HO and Hunkapiller M: Shotgun sequencing of the human
genome. Science. 280:1540–1542. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Sanger F and Coulson AR: A rapid method
for determining sequences in DNA by primed synthesis with DNA
polymerase. J Mol Biol. 94:441–448. 1975. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Wallace AJ: New challenges for BRCA
testing: A view from the diagnostic laboratory. Eur J Hum Genet.
24(Suppl 1): S10–S18. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Serratì S, De Summa S, Pilato B, Petriella
D, Lacalamita R, Tommasi S and Pinto R: Next-generation sequencing:
Advances and applications in cancer diagnosis. Onco Targets Ther.
9:7355–7365. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Kumar KR, Cowley MJ and Davis RL:
Next-generation sequencing and emerging technologies. Semin Thromb
Hemost. 45:661–673. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Idris SF, Ahmad SS, Scott MA, Vassiliou GS
and Hadfield J: The role of high-throughput technologies in
clinical cancer genomics. Expert Rev Mol Diagn. 13:167–181. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Cheon JY, Mozersky J and Cook-Deegan R:
Variants of uncertain significance in BRCA: A harbinger of ethical
and policy issues to come? Genome Med. 6:1212014. View Article : Google Scholar
|
|
138
|
Wong RSJ and Lee SC: BRCA sequencing of
tumors: Understanding its implications in the oncology community.
Chin Clin Oncol. 9:662020. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Pujol P, Barberis M, Beer P, Friedman E,
Piulats JM, Capoluongo ED, Foncillas JG, Ray-Coquard I,
Penault-Llorca F, Foulkes WD, et al: Clinical practice guidelines
for BRCA1 and BRCA2 genetic testing. Eur J Cancer. 146:30–47. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Hindson BJ, Ness KD, Masquelier DA,
Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY,
Hiddessen AL, Legler TC, et al: High-throughput droplet digital PCR
system for absolute quantitation of DNA copy number. Anal Chem.
83:8604–8610. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Pinheiro LB, Coleman VA, Hindson CM,
Herrmann J, Hindson BJ, Bhat S and Emslie KR: Evaluation of a
droplet digital polymerase chain reaction format for DNA copy
number quantification. Anal Chem. 84:1003–1011. 2012. View Article : Google Scholar :
|
|
142
|
Olmedillas-López S, García-Arranz M and
García-Olmo D: Current and emerging applications of droplet digital
PCR in Oncology. Mol Diagn Ther. 21:493–510. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Postel M, Roosen A, Laurent-Puig P, Taly V
and Wang-Renault SF: Droplet-based digital PCR and next generation
sequencing for monitoring circulating tumor DNA: A cancer
diagnostic perspective. Expert Rev Mol Diagn. 18:7–17. 2018.
View Article : Google Scholar
|
|
144
|
Stella M, Falzone L, Caponnetto A, Gattuso
G, Barbagallo C, Battaglia R, Mirabella F, Broggi G, Altieri R,
Certo F, et al: Serum extracellular vesicle-derived circHIPK3 and
circS-MARCA5 are two novel diagnostic biomarkers for glioblastoma
multiforme. Pharmaceuticals (Basel). 14:6182021. View Article : Google Scholar
|
|
145
|
Crimi S, Falzone L, Gattuso G, Grillo CM,
Candido S, Bianchi A and Libra M: droplet digital PCR analysis of
liquid biopsy samples unveils the diagnostic role of
hsa-miR-133a-3p and hsa-miR-375-3p in oral cancer. Biology (Basel).
9:3792020.
|
|
146
|
Falzone L, Gattuso G, Tsatsakis A,
Spandidos D and Libra M: Current and innovative methods for the
diagnosis of COVID-19 infection (Review). Int J Mol Med.
47:1002021. View Article : Google Scholar :
|
|
147
|
Falzone L, Musso N, Gattuso G, Bongiorno
D, Palermo CI, Scalia G, Libra M and Stefani S: Sensitivity
assessment of droplet digital PCR for SARS-CoV-2 detection. Int J
Mol Med. 46:957–964. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Preobrazhenskaya EV, Bizin IV, Kuligina
ES, Shleykina AY, Suspitsin EN, Zaytseva OA, Anisimova EI, Laptiev
SA, Gorodnova TV, Belyaev AM, et al: Detection of BRCA1 gross
rearrangements by droplet digital PCR. Breast Cancer Res Treat.
165:765–770. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Oscorbin I, Kechin A, Boyarskikh U and
Filipenko M: Multiplex ddPCR assay for screening copy number
variations in BRCA1 gene. Breast Cancer Res Treat. 178:545–555.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Khalique S, Pettitt SJ, Kelly G, Tunariu
N, Natrajan R, Banerjee S and Lord CJ: Longitudinal analysis of a
secondary BRCA2 mutation using digital droplet PCR. J Pathol Clin
Res. 6:3–11. 2020. View Article : Google Scholar :
|
|
151
|
De Paolis E, De Bonis M, Concolino P,
Piermattei A, Fagotti A, Urbani A, Scambia G, Minucci A and
Capoluongo E: Droplet digital PCR for large genomic rearrangements
detection: A promising strategy in tissue BRCA1 testing. Clin Chim
Acta. 513:17–24. 2021. View Article : Google Scholar
|
|
152
|
Manchanda R, Sun L, Patel S, Evans O,
Wilschut J, De Freitas Lopes AC, Gaba F, Brentnall A, Duffy S, Cui
B, et al: Economic evaluation of population-based BRCA1/BRCA2
mutation testing across multiple countries and health systems.
Cancers (Basel). 12:19292020. View Article : Google Scholar
|
|
153
|
Garcia J, Forestier J, Dusserre E, Wozny
AS, Geiguer F, Merle P, Tissot C, Ferraro-Peyret C, Jones FS,
Edelstein DL, et al: Cross-platform comparison for the detection of
RAS mutations in cfDNA (ddPCR Biorad detection assay, BEAMing
assay, and NGS strategy). Oncotarget. 9:21122–21131. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Ratajska M, Koczkowska M, Żuk M,
Gorczyński A, Kuźniacka A, Stukan M, Biernat W, Limon J and Wasąg
B: Detection of BRCA1/2 mutations in circulating tumor DNA from
patients with ovarian cancer. Oncotarget. 8:101325–101332. 2017.
View Article : Google Scholar : PubMed/NCBI
|