Introduction
Tongue squamous cell carcinoma (TSCC) is one of the
most common and lethal types of oral cancer (1) and is characterized by a poor
prognosis, frequent lymphatic metastasis and a high rate of
regional recurrence. Currently, the preferred treatment for TSCC is
surgery combined with postoperative radiotherapy and chemotherapy.
However, the 5-year overall survival rate of TSCC has not improved
significantly over the past decades (2). Diagnosing TSCC in the early stage is
critical. Therefore, elucidating the molecular mechanisms
underlying TSCC and exploring effective molecular therapeutic
targets are essential for improving the survival rate of patients
with TSCC.
Studies have confirmed numerous protein-coding genes
and epigenetic factors that may regulate the microenvironment of
cancer cells and tumors, such as long noncoding RNAs (lncRNAs),
microRNAs (miRNAs/miRs) and histone modifications (3,4).
Less than 2% of the human genome contains protein-coding genes and
most transcripts are nonprotein-coding RNAs (5). Among these RNA transcripts are
lncRNAs, which consist of >200 nucleotides and have been found
to widely interact with biomacromolecules, such as DNA, RNA and
proteins, to regulate fundamental cellular processes at the
transcriptional or translational level (6). LncRNAs have diverse regulatory
mechanisms in various cancers according to their subcellular
localization. LncRNAs in the cytoplasm can function as competitive
endogenous RNAs (ceRNAs). Since the structure of most lncRNAs is
similar to that of mRNAs, miRNAs may negatively regulate the
expression of lncRNAs through a mechanism similar to that of mRNAs.
MiRNAs play an important role in the posttranscriptional regulation
of gene expression by degrading target gene mRNA or inhibiting the
translation of mRNA and are widely involved in cell proliferation,
differentiation and apoptosis and cell cycle regulation (7,8). In
other words, lncRNAs can alleviate the repressive effect of miRNAs
on mRNA expression by adsorbing miRNAs as sponges (9). In addition, lncRNAs can also interact
with RNA-binding proteins (10).
lncRNAs in the nucleus may be involved in the transcriptional
regulation of genes (11).
The expression patterns of cancer-specific lncRNAs
seem to be more tissue- and stage-specific than those of mRNAs,
indicating that lncRNAs are promising alternative biomarkers and
therapeutic targets (12).
Dysregulated expression of lncRNAs has been reported in diverse
pathologies, including TSCC and bladder, prostate, lung, breast and
gastric cancers (13,14). For example, studies have shown that
melatonin protects against oral cancer cell migration through the
epigenetic regulation of prune homolog 2 with BCH domain by
inhibiting the expression and function of the lncRNA MROS-1
(15). The lncRNA colorectal
neoplasia differentially expressed (CRNDE) is associated with the
chemosensitivity of gastric cancer in an analysis of clinical
samples and CRNDE directly binds the splicing protein SRSF6 to
reduce its stability and thus regulate alternative splicing events
(16). In addition, intricate
interactions occur between coding and noncoding RNAs. These two
types of RNA transcripts can serve as ceRNAs, i.e., miRNA
sponges, to precisely regulate signaling networks by competing for
shared miRNAs and thus significantly effect tumorigenesis and
development (17). For example,
the lncRNA ESCCAL-1 promotes the invasion and migration of
esophageal squamous cell carcinoma cells by reducing the inhibitory
effect of miR-590-3p on APOBEC3G expression (18). A study has identified that FAM225A,
an oncogenic lncRNA, enhances ITGB3 expression and activation of
the focal adhesion kinase/PI3K/Akt pathway by sponging miR-590-3p
and miR-1275, thereby promoting the malignant features of
nasopharyngeal carcinoma (19).
The lncRNA Cardiac Mesoderm Enhancer-associated Noncoding is a
predictive biomarker and the host gene of miR143-3p, which
downregulates MCM5, inhibiting DNA replication in breast cancer
(20). Studies have also
identified several lncRNAs that can modulate various aspects of
diverse biological processes in TSCC, including cell proliferation,
tumorigenesis, cell survival, apoptosis and cell migration
(21-23). However, although thousands of
lncRNAs have been identified, the biological functions and
underlying regulatory mechanisms of lncRNAs in tongue cancer
progression remain largely elusive. Thus, further insight into
lncRNA-dependent gene-regulatory mechanisms will provide predictive
biomarkers and individual treatments for TSCC.
The present study, based on our previous research
results (24), performed an
in-depth analysis of lncRNA expression profiles and identified a
novel lncRNA, keratin 16 pseudogene 6 (lncKRT16P6), which was
upregulated in TSCC tissues and cell lines and associated with TSCC
tumor stage and differentiation grade. The aim of the present study
was to explore the specific regulatory mechanism by which
lncKRT16P6 promoted the occurrence and development of TSCC. To this
end, after in vitro and in vivo functional
experiments such as CCK-8, Transwell, clone formation, flow
cytometry, fluorescence in situ hybridization (FISH), dual
luciferase report experiments, it was found that lncKRT16P6
promoted TSCC cell proliferation and metastasis. In addition, as a
ceRNA, lncKRT16P6 sponged miR-3180 to increase GATA zinc finger
domain containing 2A (GATAD2A) expression. The present study
elucidated the clinical significance and regulatory mechanism of
lncKRT16P6 in TSCC and provides a prognostic indicator for TSCC
patients as well as a promising therapeutic target for TSCC.
Materials and methods
Clinical samples and ethical
approval
A total of 50 pairs of tongue cancer specimens were
collected from patients who underwent surgery at the First
Affiliated Hospital of Fujian Medical University (Fuzhou, Fujian,
China) between January 2017 and January 2020. The School and
Hospital of Stomatology and the First Affiliated Hospital are
cooperative units affiliated with Fujian Medical University. All
samples were immediately snap frozen in liquid nitrogen and stored
at −80°C until needed. Two pathologists pathologically confirmed
each sample by hematoxylin-eosin (HE) staining. Written informed
consent was obtained from all participants in accordance with the
Declaration of Helsinki and the study protocol was approved by the
Ethics Committee of the First Affiliated Hospital of Fujian Medical
University [approval number: No. 159 (2020) of the First Affiliated
Hospital of Fujian Medical University] The clinicopathological
characteristics of the patients are summarized in Table I.
 | Table ICorrelations between the expression
level of lncKRT16P6 and the clinicopathological characteristics of
TSCC patients. |
Table I
Correlations between the expression
level of lncKRT16P6 and the clinicopathological characteristics of
TSCC patients.
|
Characteristics | Cases | High expression
level (n=25) | Low expression
level (n=25) | P-value
(χ2) |
|---|
| Age (years) | | | | 0.155969
(2.012882) |
| ≥60 | 23 | 14 | 9 | |
| >60 | 27 | 11 | 16 | |
| Sex | | | | 0.122823
(2.380952) |
| Male | 35 | 15 | 20 | |
| Female | 15 | 10 | 5 | |
| Tumor stage | | | | 0.020921a (5.333333) |
| I-II | 20 | 14 | 6 | |
| III-IV | 30 | 11 | 19 | |
| T stage | | | | 0.575027
(1.987395) |
| T1 | 4 | 4 | 0 | |
| T2 | 17 | 14 | 3 | |
| T3 | 8 | 6 | 2 | |
| T4 | 21 | 18 | 3 | |
| N stage | | | | 0.275065
(3.876923) |
| N0 | 30 | 25 | 5 | |
| N1 | 6 | 5 | 1 | |
| N2 | 13 | 11 | 2 | |
| N3 | 1 | 1 | 0 | |
| Differentiation
grade | | | | 0.041399a (4.159593) |
| High | 19 | 13 | 6 | |
| Moderate-poor | 31 | 12 | 19 | |
Cell lines and cell culture
TSCC cell lines CAL-27 and SCC-9 were purchased from
the American Type Culture Collection and normal human oral
keratinocyte (HOK) cells were generously donated by the research
group of Professor Cheng Hui, School of Stomatology, Fujian Medical
University. The primary HOK cells were purchased from ScienCell
Research Laboratories Inc. In order to acquire adequate cells for
the subsequent study, the immortalization of HOK was conducted. HOK
were maintained in oral keratinocyte medium (OKM; ScienCell
Research Laboratories Inc.) before proceeding with immortalization.
The primary HOK were infected by hTERT retrovirus expressing simian
virus 40 large T antigen (SV40-T). Once the infection of HOK was
performed, the immortalized cells were subsequently selected and
passaged with OKM changed every 3 days (25). CAL-27 cells and HOK cells were
grown in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented
with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.). SCC-9 cells
were cultured in DMEM/F-12 (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS and 0.4 µg/ml hydrocortisone. The
pH of the cell culture medium was 7.0 to 7.4. All cells were
maintained at 37°C in an incubator supplied with 5%
CO2.
RNA preparation and reverse
transcription-quantitative (RT-q) PCR
Total RNA was extracted from tissues or cells using
TRIzol® reagent (Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. The cell density for
RNA extraction was 1×106. The concentration and purity
of RNA were evaluated using a Quawell spectrophotometer (Q5000;
Quawell Technology, Inc.). When the 260/280 ratio of RNA was
>1.8, total RNA was reverse transcribed into cDNA using a
PrimeScript RT reagent kit (Takara Biotechnology Co., Ltd.)
according to the manufacturer's instructions. RT-qPCR analyses were
performed using SYBR Green Master Mix (Takara Biotechnology Co.,
Ltd.) in an Applied Biosystems 7500 Real-Time PCR System (Applied
Biosystems; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. PCR cycling conditions were:
Pre-denaturation: 95°C, 1 min; denaturation: 95°C, 15 sec;
annealing: 60°C, 30 sec; extension: 72°C, 1 min; 72°C, 10 min for a
total of 40 cycles. Target mRNA expression levels were normalized
to that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Relative expression levels were calculated by the 2−∆∆
Cq method (26). All
experiments were repeated three times. MiR-mimics NC, hsa-miR-3180
mimics, miR-inhibitor NC and hsa-miR-3180 inhibitor were designed
and synthesized by Shanghai GenePharma Co., Ltd. and the primers
were provided by Fuzhou Sunya Biotechnology, Co., Ltd. and are
listed in Table SI.
Small interfering RNAs (siRNAs) and
lentiviral transduction
siRNAs targeting lncKRT16P6 and GATAD2A were
designed and synthesized by Shanghai GenePharma Co., Ltd.
(sequences listed in Table SII).
The two siRNAs with the best knockdown efficiency were used in
subsequent functional studies. The cell density of CAL-27 and SCC-9
cells for transfection was 2×105. The mass of mimic,
vector and si-NC used for transfection were 50 pmol. The
transfection reagent was Lipofectamine® 3000 (Thermo
Fisher Scientific, Inc.) and the dosage for each well of a 6-well
plate was 5 µl. Cell transfection steps were performed
according to the product manual. Diluted Lipofectamine®
3000 (Thermo Fisher Scientific, Inc.) in Opti-MEM Medium (Gibco;
Thermo Fisher Scientific, Inc.) and mixed well. The master mix of
DNA was prepared by diluting DNA in Opti-MEM Medium and mixing
well. Diluted DNA was added to each tube of diluted
Lipofectamine® 3000 (Thermo Fisher Scientific, Inc.; 1:1
ratio). After incubation for 20 min at room temperature DNA-lipid
complex was added to the cells. Cells were incubated for 48 h at
37°C before subsequent experiments. For lentiviral transduction,
virus-containing supernatant was collected 48 h after
cotransfection of the packaging plasmids pGag/Pol and pRev, the
envelope plasmid pVSV-G (Shanghai GenePharma Co., Ltd.) and the
short hairpin RNA (shRNA; shlncKRT16P6) vector or empty vector as
control into 293(T) cells and the lentiviral vectors were then
transduced into the target cells. The lentiviral vector
pLV3/H1/GFP&Puro was constructed by Shanghai GenePharma Co.,
Ltd. To establish stable cell lines, tongue cancer cells were
transduced with the above lentiviral vectors in the presence of
polybrene (5 µg/ml, Shanghai GenePharma Co., Ltd.). After
incubation for 72 h, cells were selected with 2 µg/ml
puromycin (GeneChem) for 3 days. Information on the viral vectors
and verification of the transduction efficiency are shown in
Fig. S1.
Cell counting kit-8 (CCK-8) assay
The different groups of treated CAL-27 and SCC-9
cells (1,000) were seeded into each well of a 96-well plate. Cell
viability was determined every 24 h using a CCK-8 assay (Dojindo
Laboratories, Inc.) by measuring the absorbance of the different
cell cultures at 450 nm (BioTek Instruments, Inc.), in accordance
with the manufacturer's instructions. The proliferation of CAL-27
and SCC-9 cells expressing control or lncKRT16P6 siRNA was detected
by CCK-8 assays, CCK-8 assays were performed in CAL-27 and SCC-9
cells stably transfected with sh-lncKRT16P6 with or without the
miR-3180 inhibitor and CCK-8 assays were performed in CAL-27 and
SCC-9 cells transfected with the miR-3180 inhibitor with or without
GATAD2A siRNAs.
Transwell assays
The invasive and migratory capacities of cells were
evaluated using Transwell chambers with 8-µm pore filters.
After 48 h of transfection, cells in serum-free medium
(5×104 cells/100 µl) were plated in the upper
chambers, which contained membranes coated with or without 50
µl of Matrigel (BD Biosciences) at 37°C for 2 h. DMEM
containing 10% FBS was added to the lower chambers as a
chemoattractant. After incubation for 12 h at 37°C, cells that had
migrated to the bottom surface of the membrane were fixed with 4%
paraformaldehyde at room temperature for 30 min and stained with a
crystal violet solution at room temperature for 10 min. Cells in
five random fields were counted and the numbers were averaged.
Colony formation assays
Transfected cells (1×103) were uniformly
seeded in 6-well plates and cultured for 2 weeks at 37°C in an
incubator supplied with 5% CO2, then the colony
formation of the cells was observed under a low magnification (×10)
microscope. The number of cells >10 were counted as a colony,
and then the number of colonies in different treatments counted.
The cells were then washed with phosphate-buffered saline (PBS),
fixed with 4% paraformaldehyde at room temperature for 30 min,
stained with 0.5% crystal violet at room temperature for 10 min and
images captured.
Flow cytometry
Cells were routinely transfected and cultured for 48
h and were then digested with EDTA-free trypsin. An Annexin
V-fluorescein isothiocyanate (FITC) apoptosis assay kit (BD
Biosciences) was used to estimate the apoptosis rate according to
the manufacturer's instructions. Cells were suspended in 1X Annexin
binding buffer and 5 µl of Annexin V reagent and 3 µl
of propidium iodide (PI) reagent were then added to 100 µl
of the cell suspension and mixed. The mixture was incubated in the
dark for 30 min at room temperature and 400 µl of 1X Annexin
binding buffer was then added to each sample to terminate
staining.
For cell cycle analysis, a PI/RNase staining kit (BD
Biosciences) was used according to the manufacturer's instructions.
After treatment, tongue cancer cells were harvested, washed with
ice-cold PBS and fixed with 70% ethanol for 24 h at 4°C. After
staining with PI in the dark for 30 min at 4°C, the apoptosis rate
(the percentage of late apoptotic cells) and cell cycle
distribution were evaluated using a FACSVerse flow cytometer (BD
Biosciences).
RNA FISH
Cy3-labeled probes specific for lncKRT16P6 were
designed and synthesized by Shanghai GenePharma Co., Ltd. Probe
signals were detected with a FISH kit (Shanghai GenePharma Co.,
Ltd.) according to the manufacturer's instructions. In brief, cells
were fixed, permeabilized (1X PBS/0.5% Triton X-100) and
prehybridized. Then, the cells were hybridized in hybridization
buffer with Cy3-labeled probes specific for lncKRT16P6 at 37°C
overnight. Cells were sequentially rinsed with 4X SSC (containing
0.1% Tween 20), 2X SSC and 1X SSC at 42°C and nuclei were stained
with 4′,6-diamidino-2-phenylindole (DAPI) (Beyotime Institute of
Biotechnology). Probe signals were observed via fluorescence
microscopy, the (magnification, ×630). The probe sequences are
listed in Table SIII.
Cell nucleocytoplasmic fractionation
Cell nucleocytoplasmic fractionation was performed
with a Nuclear and Cytoplasmic Protein Extraction kit and RNase
Inhibitor (Beyotime Institute of Biotechnology,) according to the
manufacturer's instructions. Cytoplasmic and nuclear RNA was
separated from tongue cancer cells and reverse transcribed. The
lncKRT16P6 level was evaluated by RT-qPCR as aforementioned and its
percentages in the nuclear and cytoplasmic fractions were
determined.
Xenograft tumor formation and
staining
The Animal Experimentation Ethics Committee of
Fujian Medical University approved the present study and ensured
that all experiments conformed to all relevant regulatory standards
(approval number: FJMU IACUC NO. 2020-0039 of Fujian Medical
University). In the present study, 4-week-old BALB/c athymic male
nude mice (Shanghai SLAC Laboratory Animal Co., Ltd.) were used.
The weight of nude mice was ~20±5 g. All mice were maintained under
controlled temperature (22±1°C) and humidity (50±5%) in a 12 h
light/dark cycle with food and water available ad libitum. CAL-27
cells (5×106 cells/100 µl) transfected with
different constructs were subcutaneously injected into the nude
mice. A total of eight mice were randomly divided into two groups:
i) sh-NC group (control group, mice were injected subcutaneously
with CAL-27 cells transfected with empty vector, n=4); ii)
sh-lncKRT16P6 group (mice were injected subcutaneously with CAL-27
cells transfected with sh-lncKRT16P6, n=4). Tumors were measured
weekly and the tumor volume (V) was calculated using the equation
V=(LxW2)/2, where L and W were the length and width of
the tumor, respectively. After 24 days, the mice were sacrificed;
1% pentobarbital sodium 40 mg/kg was injected intraperitoneally and
the animals were sacrificed by acute exsanguination after they
became unconscious. The tumors were excised for hematoxylin and
eosin (HE) and immunohistochemical (IHC) staining.
For HE staining, in brief, paraffin-embedded tumor
tissues were sliced into 4-µm-thick sections. Deparaffinized
and rehydrated sections were stained with HE (Beyotime Institute of
Biotechnology,) at room temperature for 5 min and the stained
tissues were evaluated under a light microscope (magnification,
×100 and ×200). For IHC staining, the Ready-to-use
Immunohistochemical UltraSensitive SP Detection kit (Mouse/Rabbit;
Fuzhou Maixin Biotech Co., Ltd.) and Enhanced DAB Plus kit (Fuzhou
Maixin Biotech Co., Ltd.) were used according to the manufacturer's
instructions. Paraffin-embedded tumor sections (4-µm-thick)
were deparaffinized and blocked with 5% goat non-immune serum
(UltraSensitive SP kit; Fuzhou Maixin Biotech Co., Ltd.) and
incubated at room temperature for 10 min. Primary antibodies were
diluted in bovine serum albumin and the sections were then
incubated with the anti-Ki67 antibody (1:500; Fuzhou Maixin Biotech
Co., Ltd.) overnight at 4°C in a wet chamber. Then the tumor
sections were incubated with secondary antibodies (UltraSensitive
SP kit; Fuzhou Maixin Biotech Co., Ltd.) at room temperature for 10
min, diaminobenzidine reagent was added and hematoxylin
counterstaining was performed to visualize nuclei.
Bioinformatic analysis
Interaction probabilities between lncRNAs and miRNAs
were predicted using the online in silico tool MicroRNA
Target Prediction Database (miRDB 6.0; http://mirdb.org/). Interaction probabilities between
miRNAs and mRNAs were analyzed using online in silico tools,
including miRDB, the Encyclopedia of RNA Interactomes (ENCORI 3.0,
also known as StarBase; https://starbase.sysu.edu.cn/index.php), Prediction of
MicroRNA Targets (TargetScan 3.1; http://www.targetscan.org/mamm_31/) and
MicroRNA-Target Interactions (miRTarBase 9.0 beta; https://mirtarbase.cuhk.edu.cn/~miRTarBase/miRTar-Base_2022/php/index.php).
Ensembl database (Ensembl 104; http://asia.ensembl.org/index.html) was used to
identify the location of lncKRT16P6 and its sequence was predicted
to exhibit lower protein-coding potential by Coding Potential
Calculator 2 (CPC 2.0; http://cpc2.gao-lab.org/).
Dual-luciferase reporter assay
This experiment used the Dual-Luciferase reporter
gene detection system kit (Promega Corporation), which was used
according to the manufacturer's instructions. GP-transfect-Mate
transfection reagent (Shanghai GenePharma Co., Ltd.) was used for
transfection. Cells were seeded in 12-well plates at a density of
5×105 cells per well for 24 h before transfection.
Reporter plasmids [pGP-miRGLO-Firefly luciferase-Renilla
luciferase, containing lncKRT16P6, the GATAD2A 3′ untranslated
region (UTR), or the corresponding mutant (MUT) sequences] were
designed by Shanghai GenePharma Co., Ltd. 293(T) cells were
cotransfected with a reporter plasmid and either miR-3180 mimic or
the NC mimic (Shanghai GenePharma Co., Ltd.). After 48 h,
luciferase activity was measured using a dual-luciferase reporter
assay system (Promega Corporation) according to the manufacturer's
protocol. Firefly luciferase activity was normalized to Renilla
luciferase activity.
Western blot analysis
Cells were prepared at 4°C in
radio-immunoprecipitation assay (RIPA) buffer containing
phenylmethylsulfonyl fluoride (PMSF; Beijing Solarbio Science &
Technology Co., Ltd.). Then, the protein concentration was
determined using the BCA kit (Beyotime Institute of
Biotechnology,). Proteins (25 µg per lane) were
electrophoretically separated on 10% SDS-PAGE gels and transferred
onto polyvinylidene difluoride membranes. The membrane was placed
in 5% non-fat milk (Hangzhou Fude Biological Technology Co., Ltd.)
and blocked for 3 h at room temperature to remove nonspecific
binding sites. Membranes were then incubated with the appropriate
primary antibodies overnight at 4°C, and incubated with the
appropriate secondary antibodies at room temperature for 30 min.
Primary antibodies specific for the following proteins were used:
E-cadherin (1:1,000; cat. no. 3195; CST Biological Reagents Co.,
Ltd.), N-cadherin (1:1,000; cat. no. 13116; CST Biological Reagents
Co., Ltd.), Vimentin (1:1,000; #5741; CST Biological Reagents Co.,
Ltd.), GATAD2A (1:2,000; cat. no. ab188472, Abcam), Cyclin A
(1:1,000; cat. no. WL01841, Wanleibio Co., Ltd.), Cyclin D1
(1:1,000; cat. no. WL01435a, Wanleibio Co., Ltd.), Cyclin E (1:500;
cat. no. WL01072, Wanleibio Co., Ltd.), Cyclin-dependent kinase 2
(CDK2; 1:1,000; cat. no. WL01543, Wanleibio Co., Ltd.), GAPDH
(1:10,000; cat. no. 60004-1-Ig, ProteinTech Group, Inc.), β-Actin
(1:1,000; cat. no. 3700; CST Biological Reagents Co., Ltd.) and
β-Tubulin (1:10,000; cat. no. BS1482M, Bioworld Technology). Since
each target protein had a different molecular weight distribution,
in order not to overlap with the experimental protein in each case,
three different loading controls were used (GAPDH, β-tubulin,
β-actin) in this experiment. Horseradish peroxidase-conjugated
secondary antibodies, including goat anti-mouse (1:5,000; cat. no.
SA00001-1) and goat anti-rabbit (1:5,000; cat. no. SA00001-2)
antibodies were purchased from ProteinTech Group, Inc.
Immunoreactions were visualized by electrochemiluminescence using a
BeyoECL Moon kit (Beyotime Institute of Biotechnology,). Then
western blot analysis was performed with the ImageJ software
(v1.44p; National Institutes of Health.).
Statistical analyses
Statistical analyses were performed using SPSS 22
software (IBM Corp.). For all continuous variables, Shapiro Wilk
tests were used to test the normality. Experimental data are
presented as the mean ± standard error of the mean of a minimum of
three biological replicates or are representative of three
independent experiments with similar results. Unpaired Student's t
test was used for two-group comparisons. To test for statistical
significance between > two groups, one-way ANOVA coupled with
Dunnett T3′s post-hoc test was used. The chi-square test was used
to determine the statistical significance of differences in
clinicopathological data. Paired Student's t test was used to
determine the statistical significance of differences in the
expression levels in clinical tissue samples. All statistical tests
were two-sided and P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression profile of lncKRT16P6 and
correlation of lncKRT16P6 expression with clinicopathological
characteristics in TSCC
Previously, we screened for differentially expressed
lncRNAs by analyzing six pairs of TSCC and healthy adjacent normal
tissues through high-throughput RNA sequencing (Kangchen BioTech
Co., Ltd.) (24) and identified
the significantly overexpressed lncRNA lncKRT16P6. RT-qPCR was
applied to detect the expression of differentially expressed
lncRNAs. The expression level of lncKRT16P6 was determined in 50
paired human TSCC and normal tissues and lncKRT16P6 expression was
consistently upregulated in the cancerous samples compared with
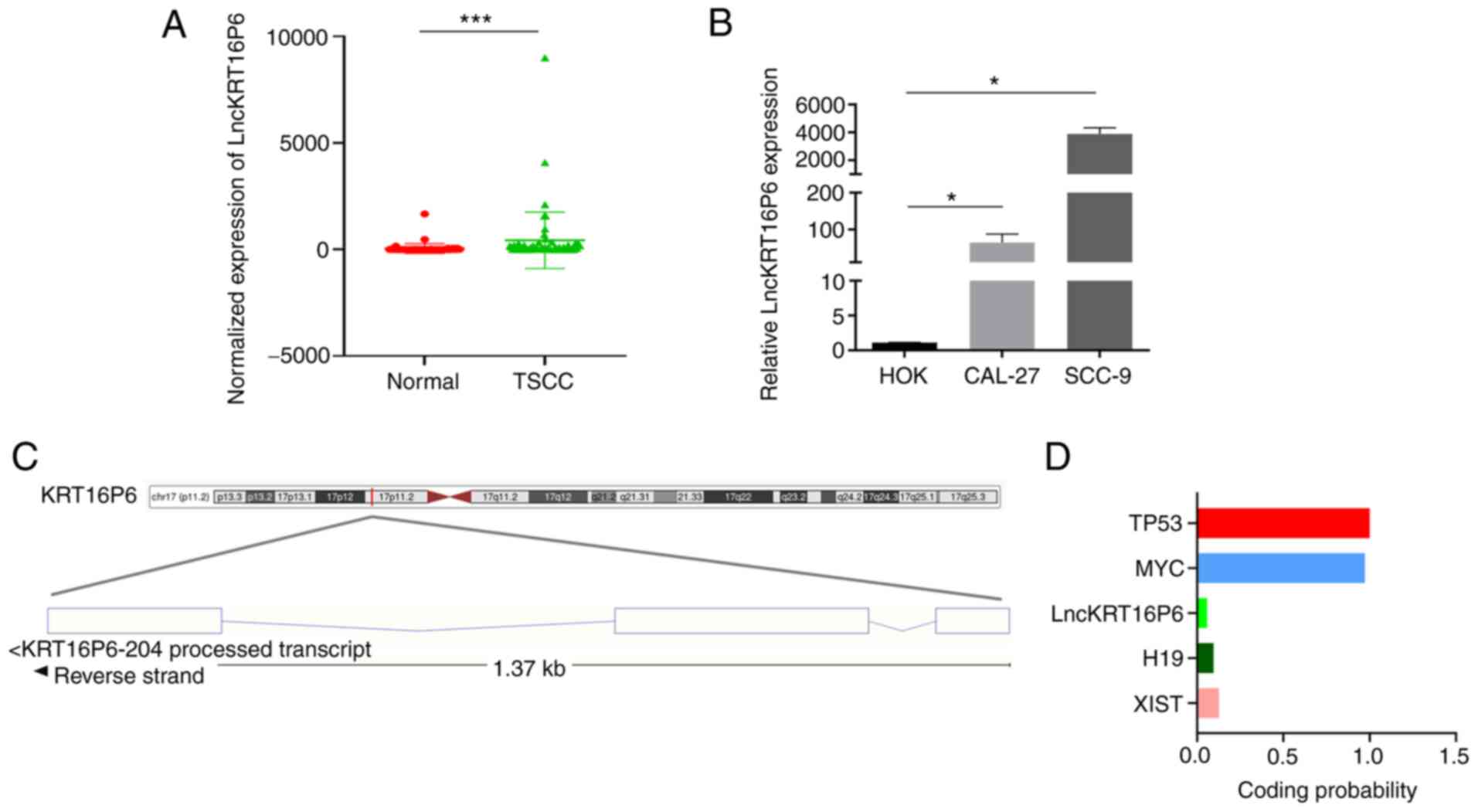
their corresponding normal tissues (P<0.001; Fig. 1A). Notably, high lncKRT16P6
expression was significantly correlated with stage I-II disease
(P=0.0209) and advanced tumor grade (P=0.0414; Table I). Consistent with the above
results, the expression of lncKRT16P6 was dramatically increased in
the human tongue cancer cell lines CAL-27 and SCC-9 compared with
the normal human oral keratinocyte cell line HOK (Fig. 1B). In addition, Ensembl database
analysis indicated that lncKRT16P6 is located on chromosome 17 with
a total length of 715 bp and its sequence was predicted to exhibit
lower protein-coding potential (coding probability, 0.04 by Coding
Potential Calculator 2; http://cpc2.gao-lab.org/) compared with those of the
classical lncRNA H19, XIST and coding genes (TP53 and MYC; Fig. 1).
Depleting lncKRT16P6 inhibits TSCC
progression in vitro and in vivo
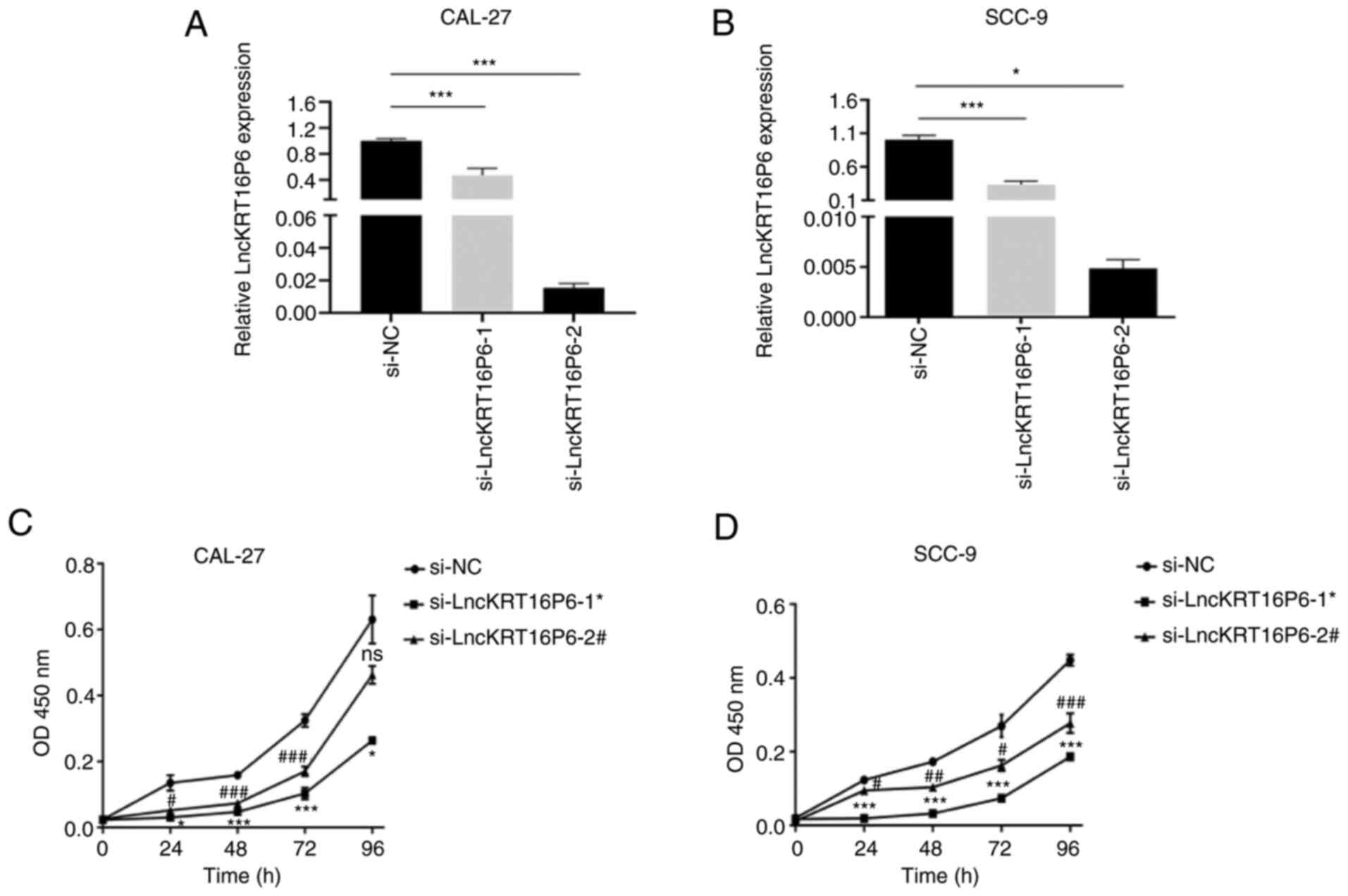
To explore the role of lncKRT16P6 in TSCC cells, two
specific siRNA oligonucleotides to target lncKRT16P6 in CAL-27 and
SCC-9 cells were designed (Fig.
2A-B). The CCK-8 assay showed that knockdown of lncKRT16P6
significantly reduced the proliferation ability of CAL-27 and SCC-9
cells (Fig. 2C-D) and the
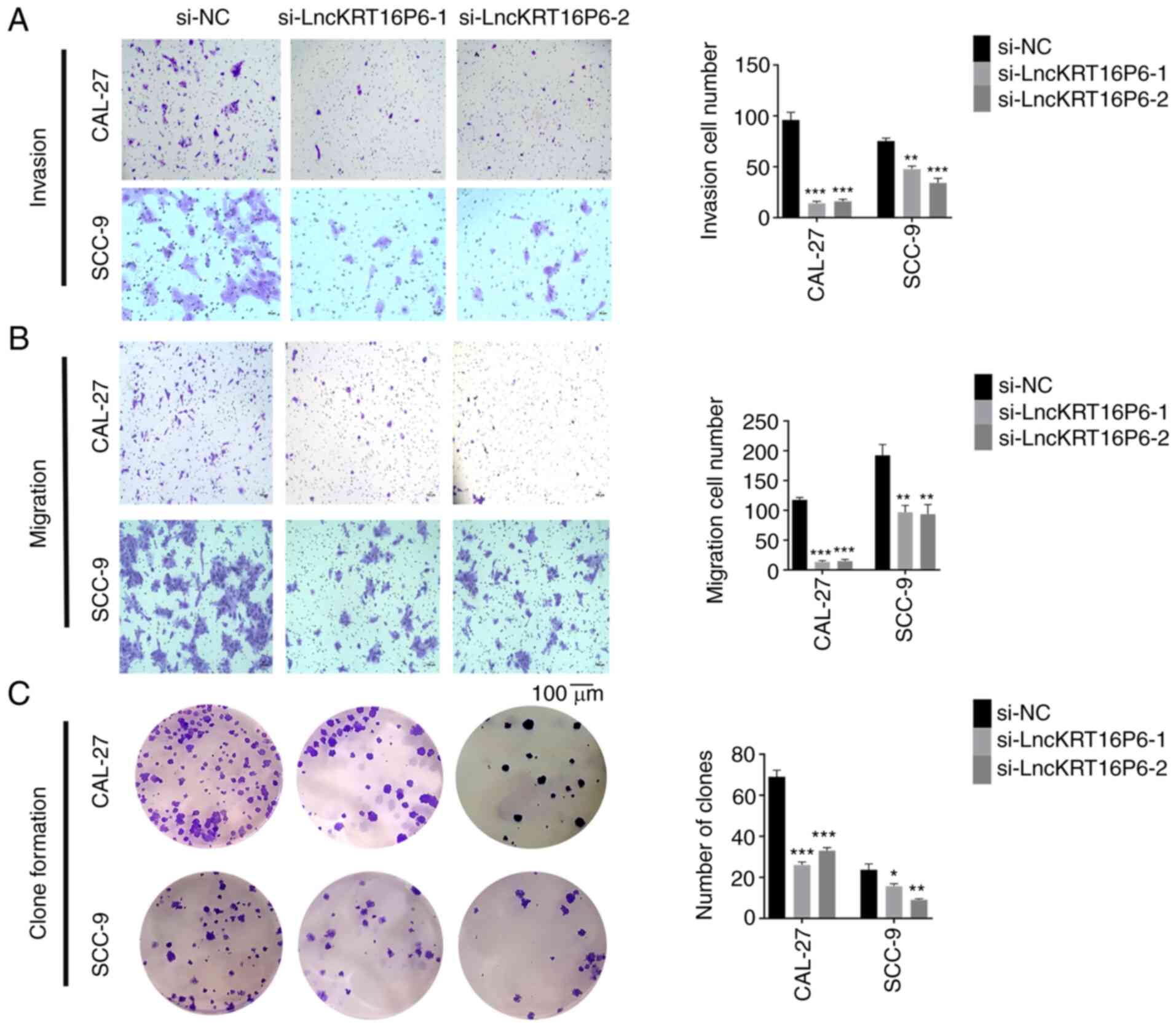
Transwell and colony formation assay results showed that knockdown
of lncKRT16P6 significantly reduced the invasion, migration and
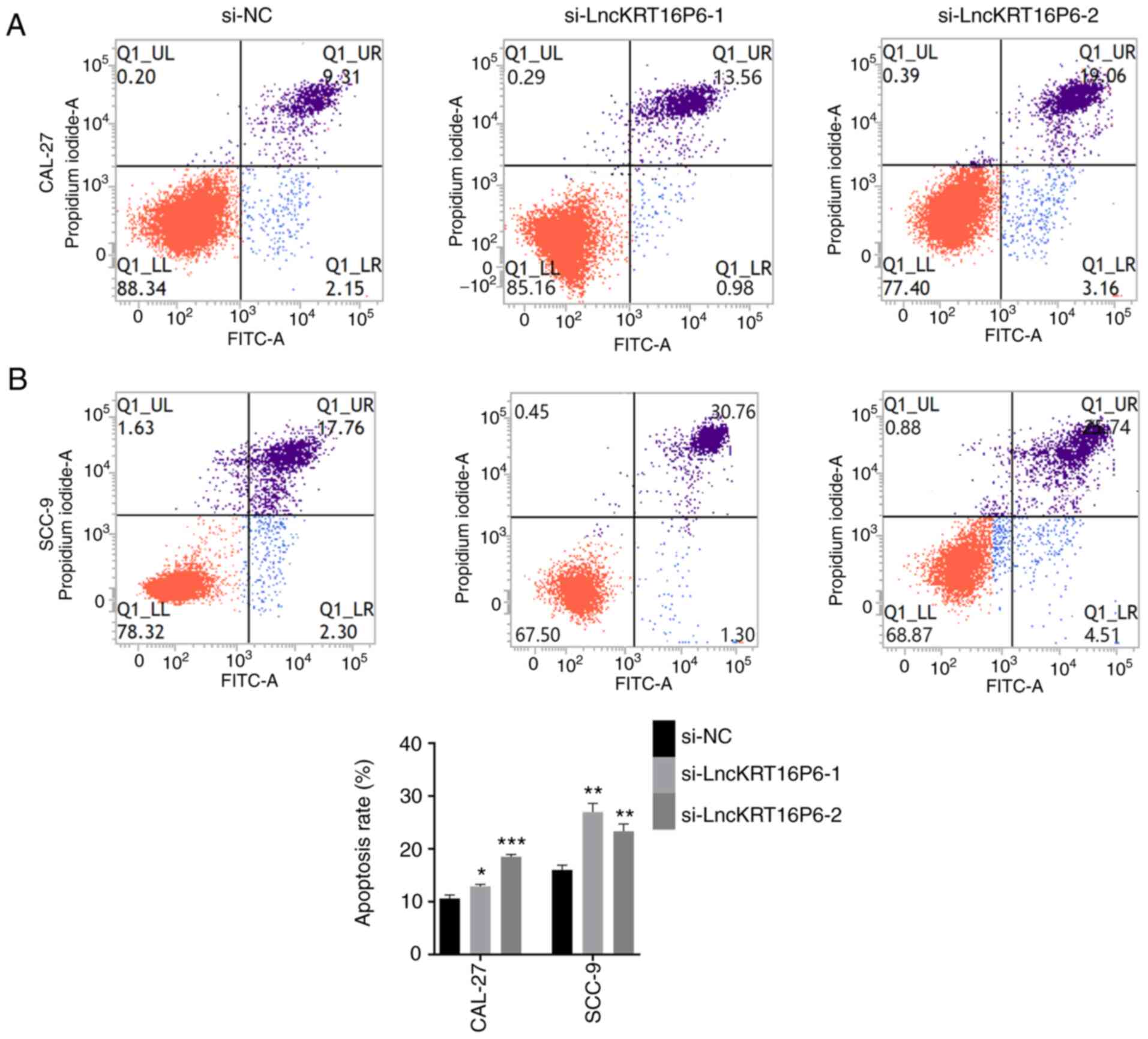
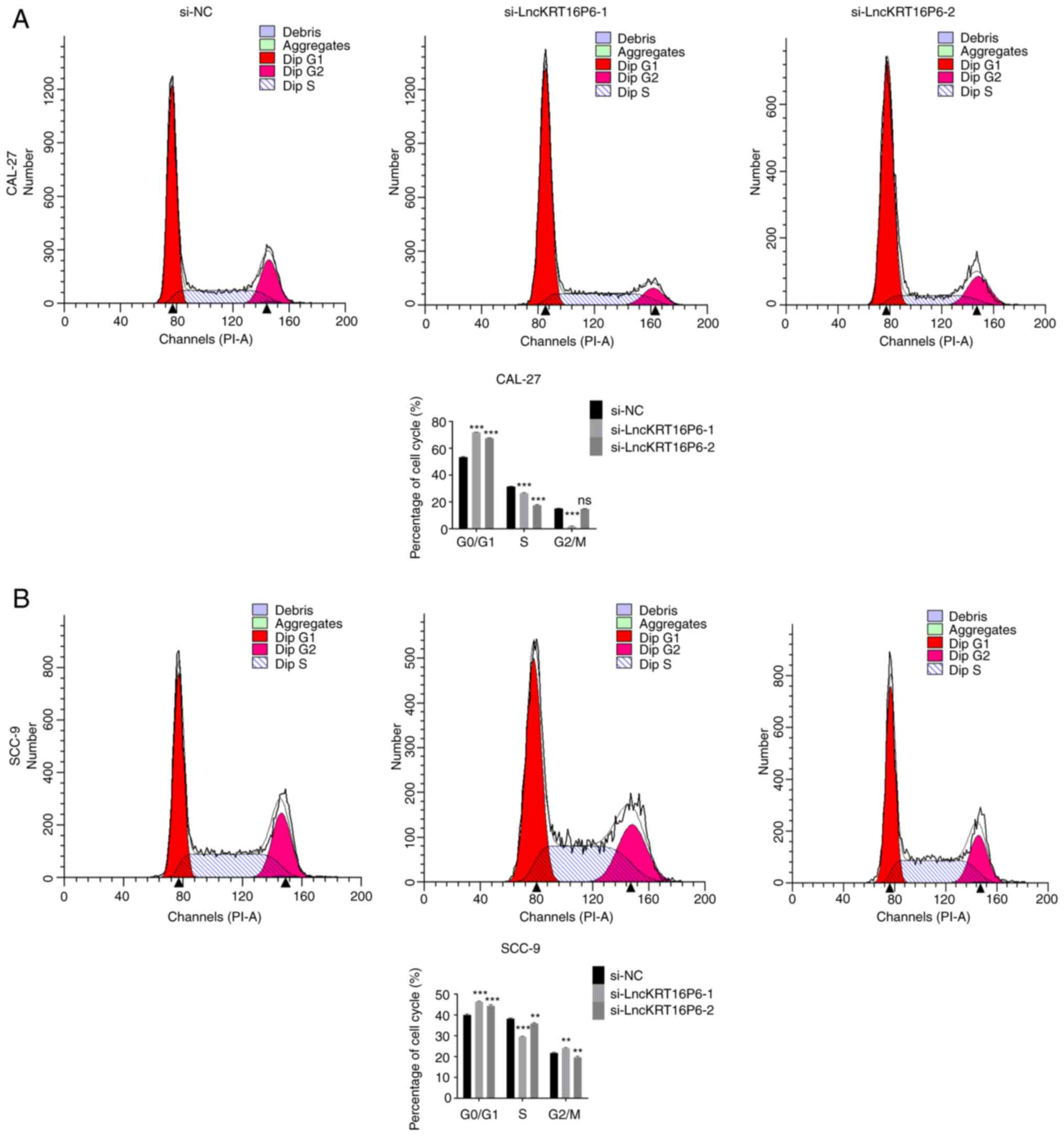
colony-forming abilities of CAL-27 and SCC-9 cells (Fig. 3). Moreover, the flow cytometry
results indicated that knockdown of lncKRT16P6 induced apoptosis
(Fig. 4) and affected cell cycle
progression in CAL-27 and SCC-9 cells; most of the cells arrested
in G0/G1 phase (Fig. 5).
To further investigate the role of lncKRT16P6,
CAL-27 cells with stable knockdown of lncKRT16P6 were generated
(Supplementary Fig. 1B-C).
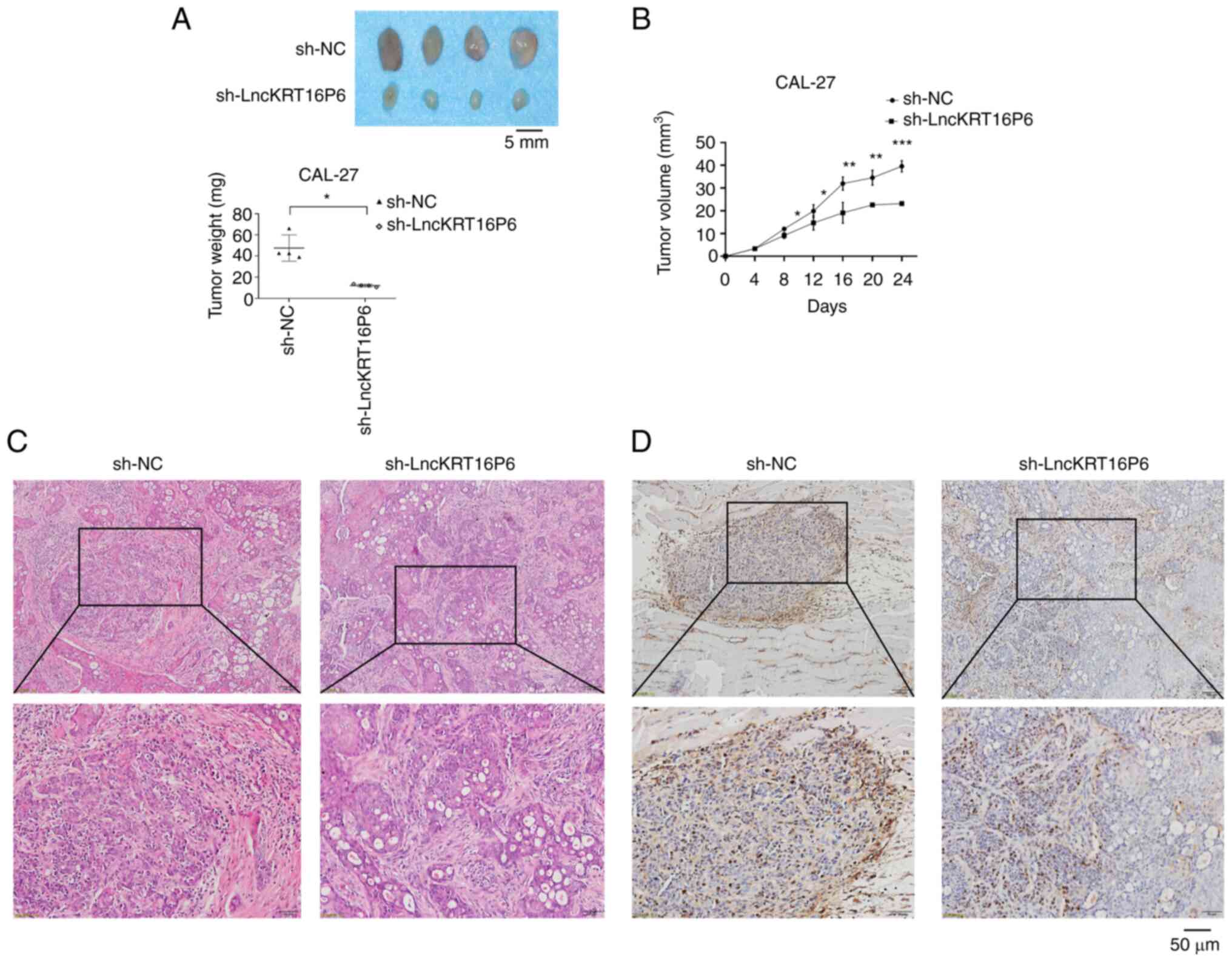
Moreover, to further determine the potential role of lncKRT16P6 in
the progression of TSCC, an in vivo model of subcutaneous
tumorigenesis was established in nude mice. CAL-27 cells with
stable lncKRT16P6 knockdown or CAL-27 cells transfected with the
control vector were inoculated into the right flanks of nude mice
(n=4 mice per group). Markedly, 24 days after injection, lncKRT16P6
knockdown was found to suppress the tumorigenic and proliferation
abilities of tongue cancer cells (Fig.
6A). The volumes of the subcutaneous tumors were significantly
smaller in the lncKRT16P6 knockdown group compared with the control
group (Fig. 6B). HE and IHC
staining indicated that the model of subcutaneous tumorigenesis had
been successfully established in nude mice and that the tumor
tissue was Ki-67-positive (Fig.
6). Collectively, the findings suggested that lncKRT16P6 may
promote the progression of tongue cancer.
lncKRT16P6 functions as a ceRNA to sponge
hsa-miR-3180
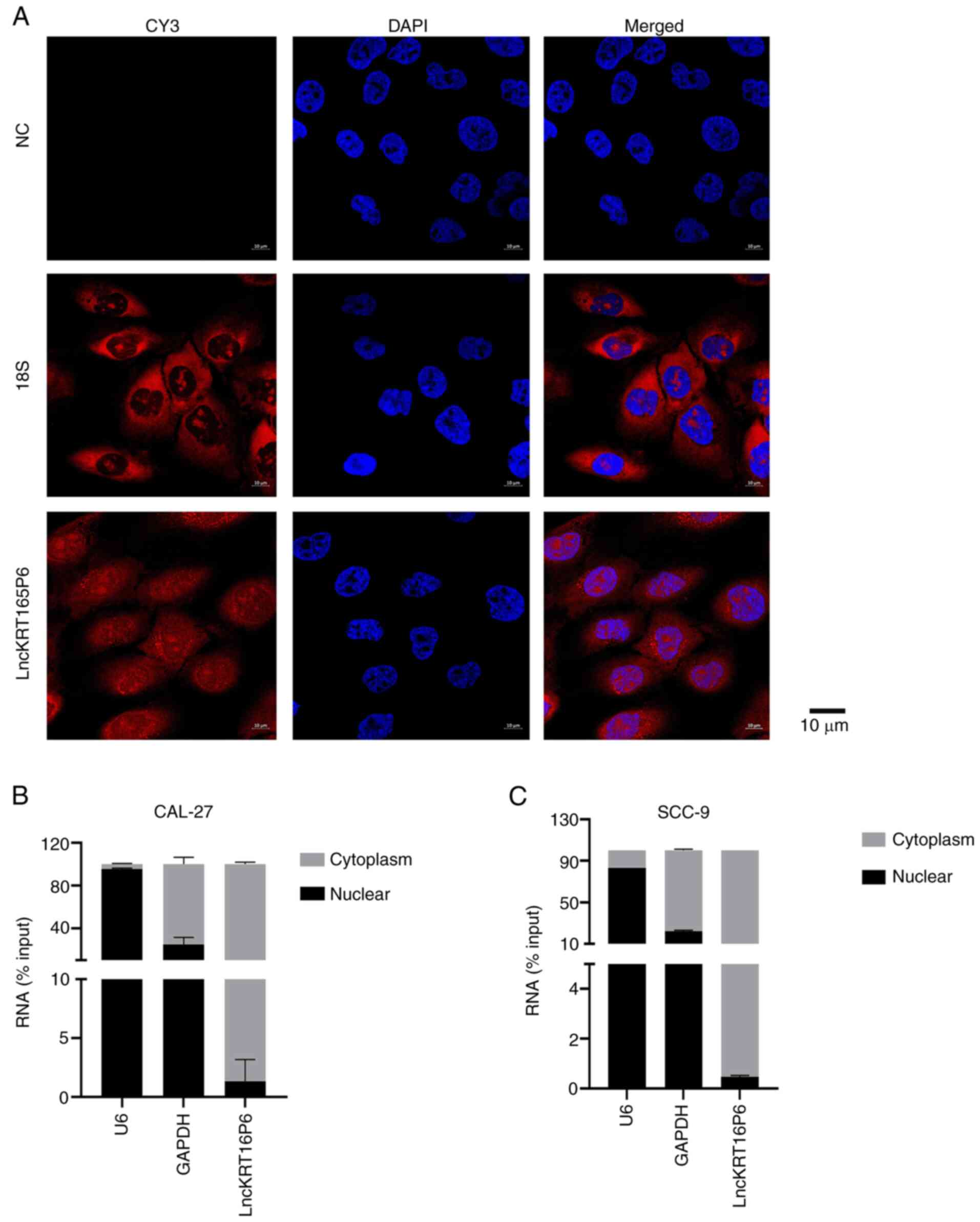
Next, the molecular mechanism of lncKRT16P6 in TSCC
was investigated and FISH and cell nucleocytoplasmic fractionation
used to determine its localization. lncKRT16P6 was localized mainly
in the cytoplasm (Fig. 7),
suggesting that it may function by acting as a sponge for various
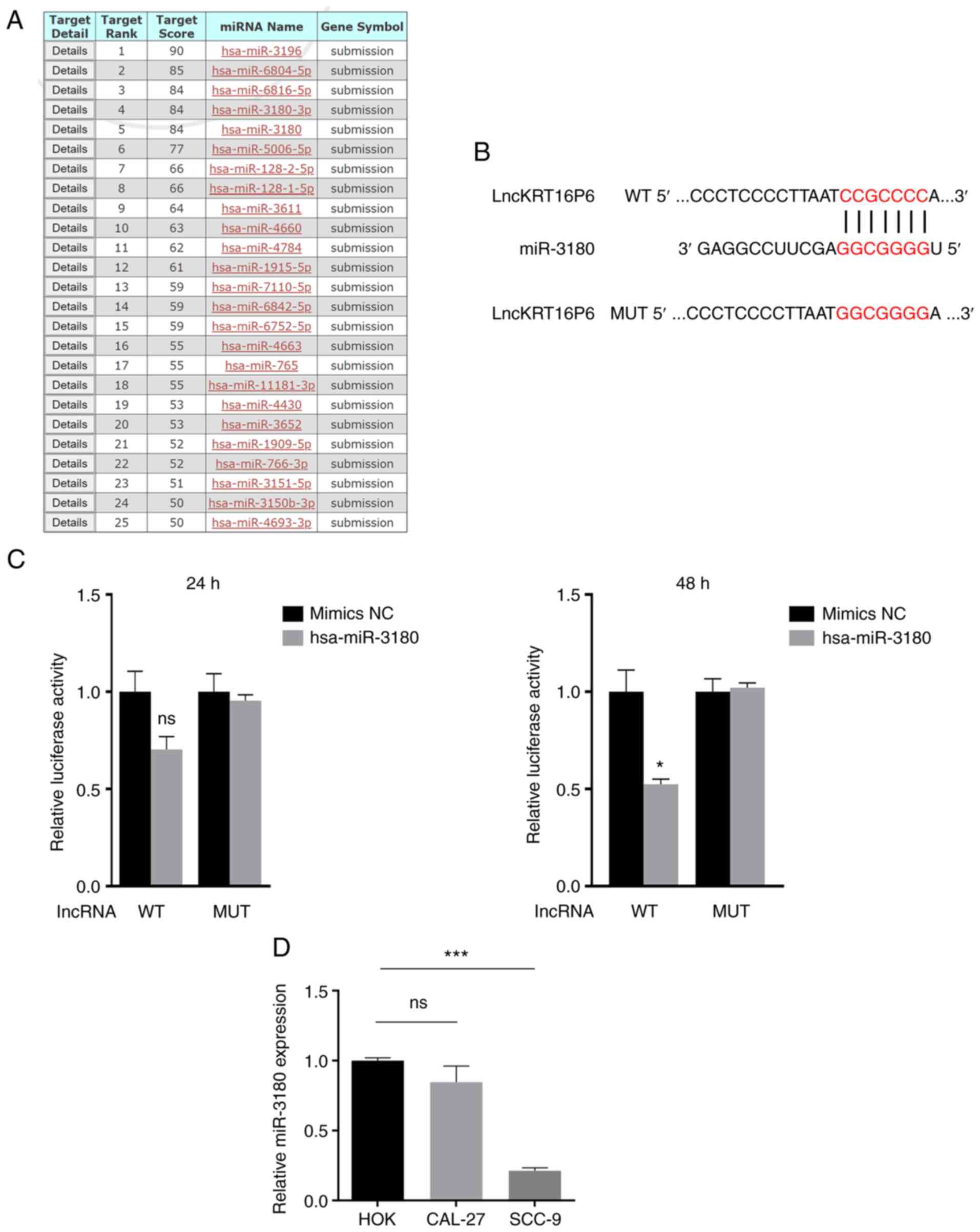
miRNAs. MiRDB was used to predict potential target miRNAs (Fig. 8A) and miR-3180 was selected for
further investigation. To confirm the predicted interactions,
dual-luciferase reporter assays were performed. Bioinformatic
analysis revealed the shared miRNA response elements (MREs) between
lncKRT16P6 and hsa-miR-3180. Therefore, these MREs were mutated
(MUT) and cloned into the luciferase reporter gene in place of the
wild-type (WT) lncKRT16P6 3′ UTR (Fig.
8B). miR-3180 directly bound lncKRT16P6 (Fig. 8C). RT-qPCR was performed to detect
miR-3180 expression in tongue cancer cell lines. The expression of
miR-3180 was significantly reduced in tongue cancer cells compared
with HOK cells (Fig. 8D).
Silencing hsa-miR-3180 reverses the
antitumor effects of lncKRT16P6 depletion in TSCC cells
Rescue experiments were next designed to explore
whether lncKRT16P6 enhanced the malignant behavior of TSCC cells by
interacting with miR-3180. CAL-27 and SCC-9 cells were stably
cotransduced with the sh-lncKRT16P6 lentiviral vector and miR-3180
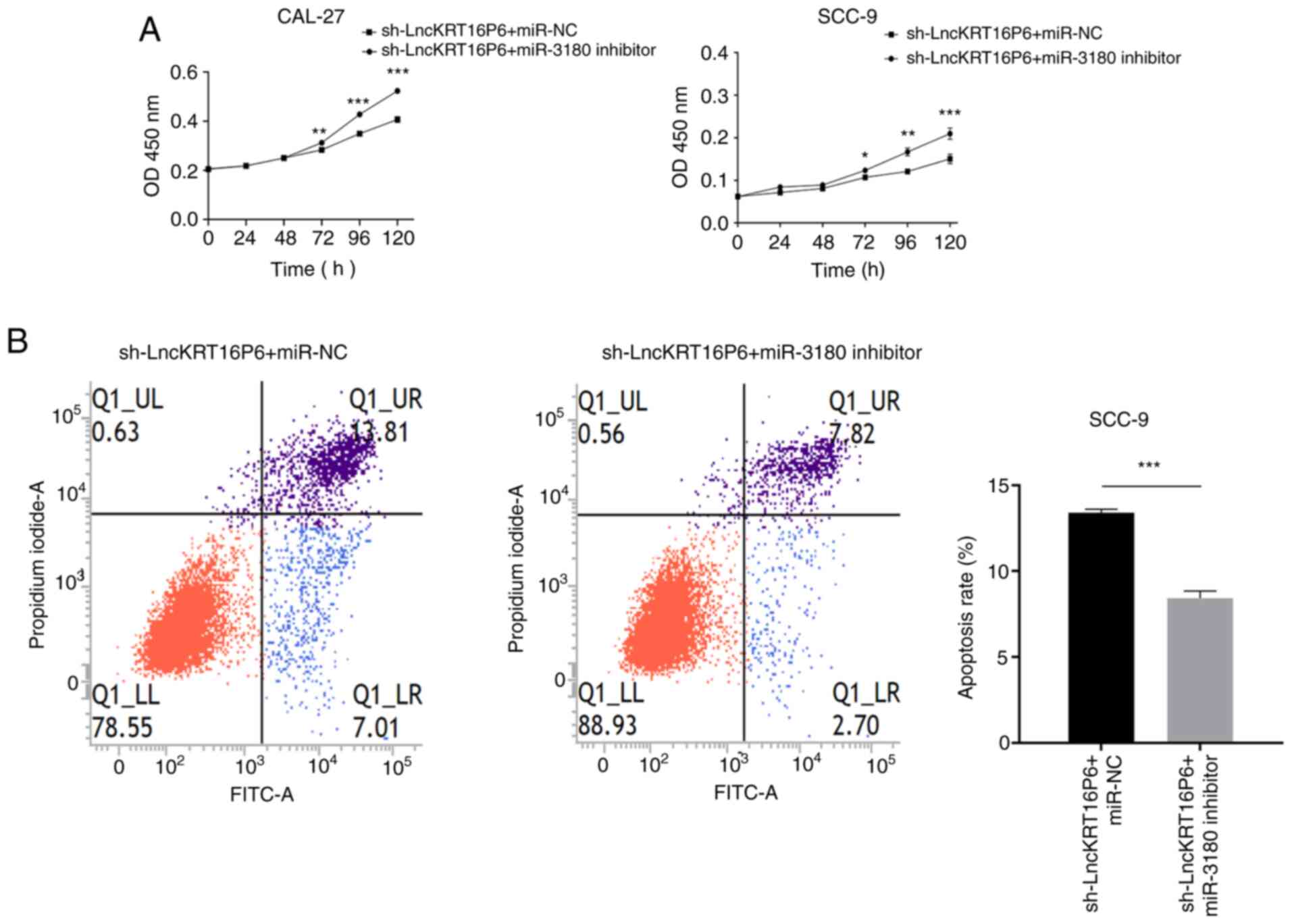
inhibitor. The inhibitory effect on miR-3180 partially attenuated
the reduction in viability caused by sh-lncKRT16P6, as demonstrated
by CCK-8 assays (Fig. 9A). Flow
cytometry showed that the proportion of apoptotic cells was
significantly decreased following sh-lncKRT16P6/miR-3180 inhibitor
cotreatment (Fig. 9B). In
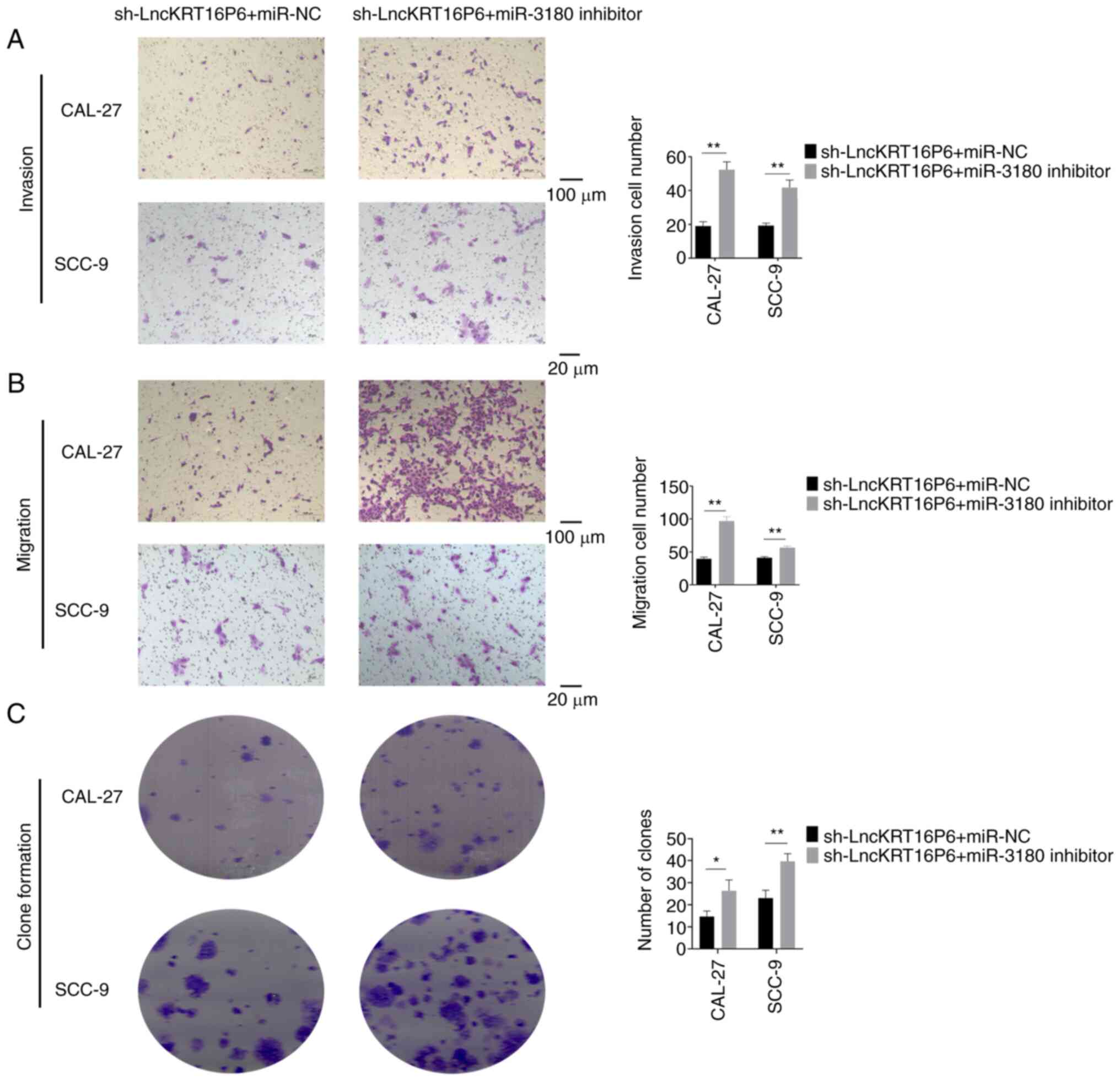
addition, Transwell invasion and migration assays and colony
formation assays demonstrated that miR-3180 partially reversed the
decreases in the invasion, migration and colony formation abilities
caused by lncKRT16P6 depletion (Fig.
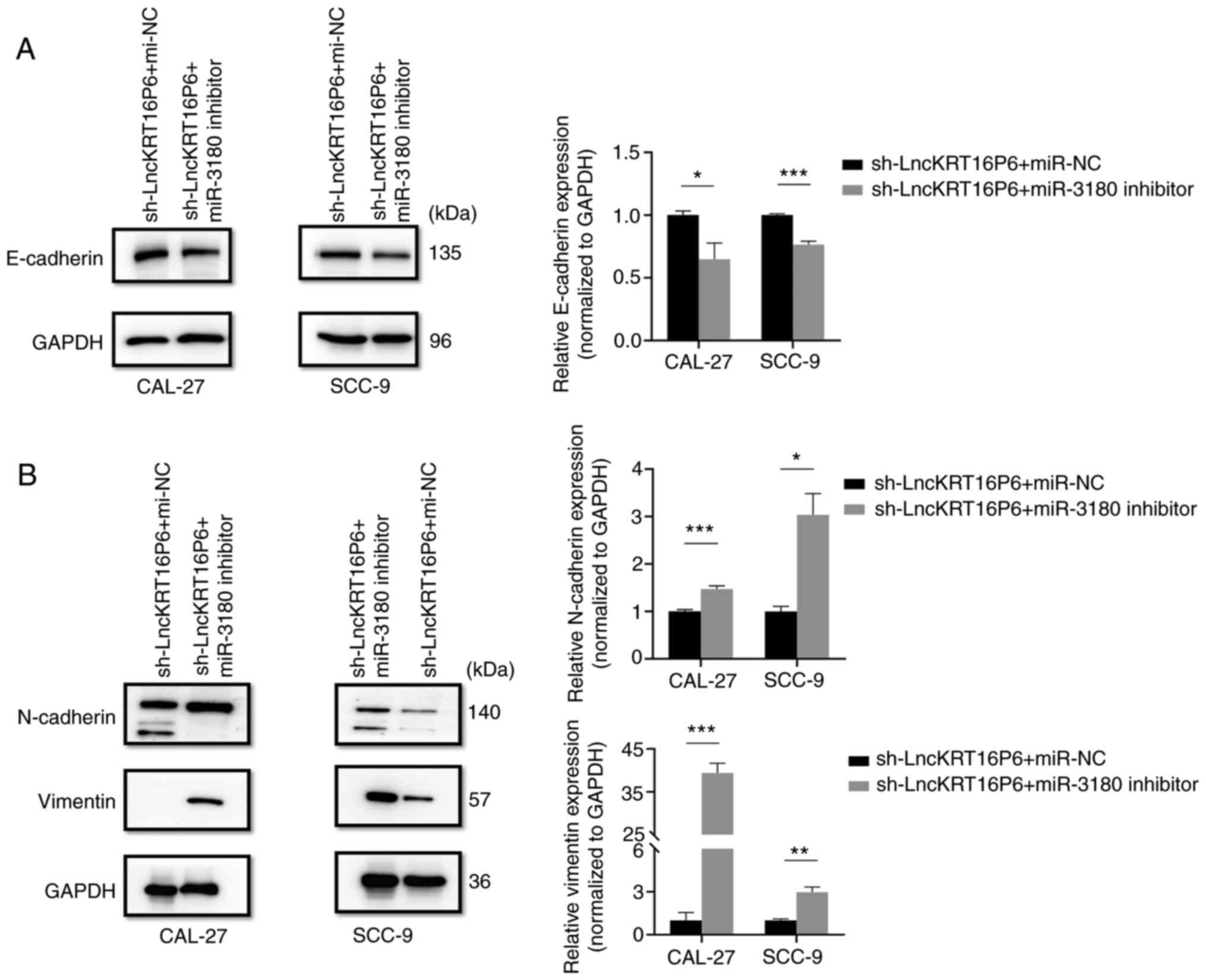
10). Western blot analysis was used to detect the expression of
invasion- and migration-related marker proteins, such as
E-cadherin, N-cadherin and Vimentin and the results were consistent
with the Transwell results (Fig.
11). These findings indicate that lncKRT16P6 promotes TSCC
progression by eliminating the antitumor effects of miR-3180.
GATAD2A is a direct target of
hsa-miR-3180 and acts as an oncogene in TSCC
Then, the target genes of miR-3180 in TSCC were
investigated. Four databases (ENCORI, miRDB, TargetScan and
miRTarBase) were used to predict potential target genes (Fig. 12A) and GATAD2A was selected for
further investigation. To confirm whether GATAD2A is a potential
target gene of miR-3180, 3′ UTR sensors downstream of the
luciferase reporter sequence were generated and cotransfected into
293(T) cells with miR-3180 mimics. When miR-3180 was overexpressed,
GATAD2A showed reduced luciferase activity (Fig. 12B and C). The expression of
GATAD2A in TSCC and head and neck squamous cell carcinoma (HNSC)
tissues and cell lines was higher than that in healthy normal
tissues and cells (Fig.
12D-F).
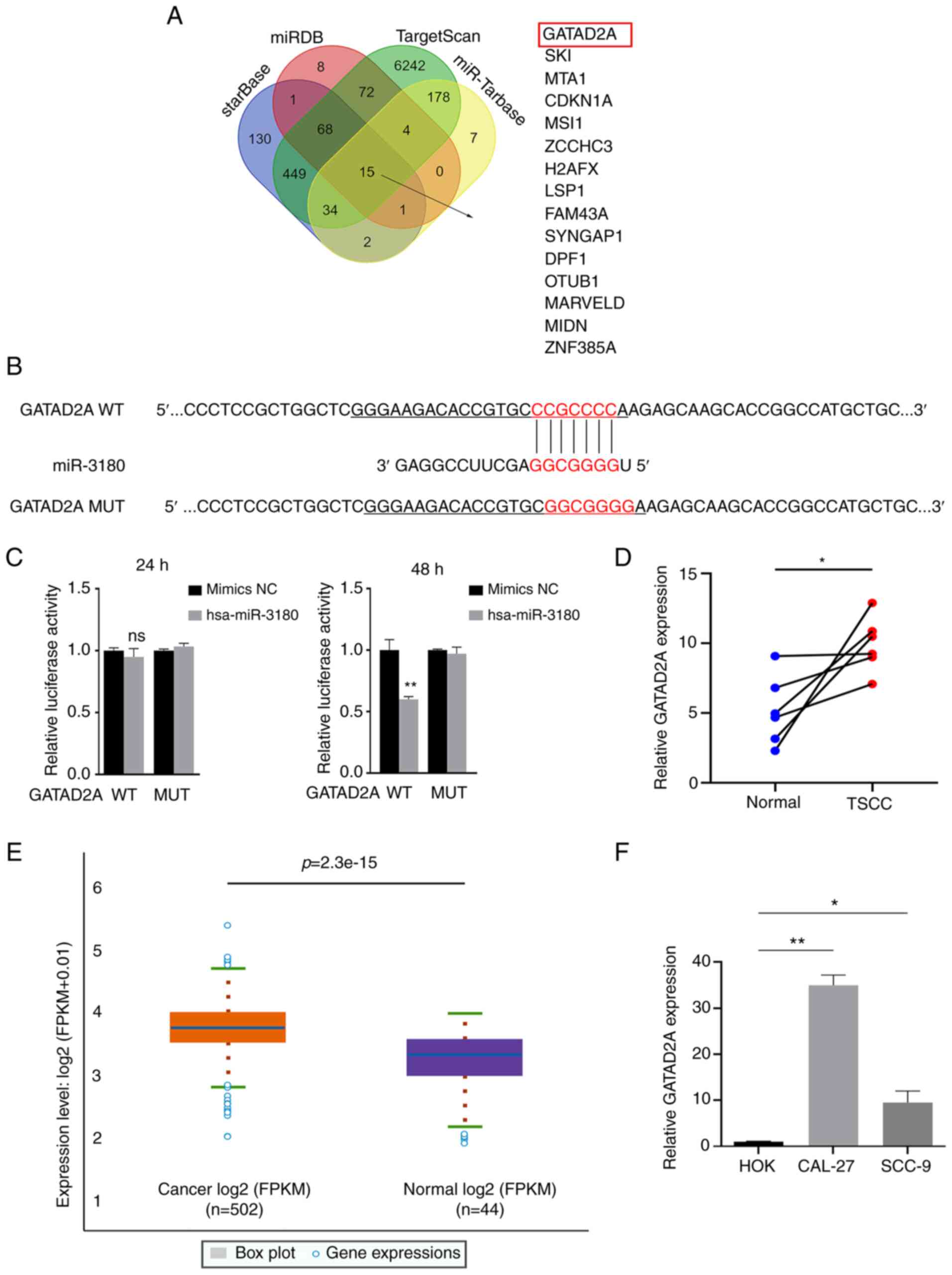 | Figure 12GATAD2A is a direct target of
hsa-miR-3180. (A) Schematic showing the overlap between miR-3180
target mRNAs predicted by the ENCORI, miRDB, TargetScan and
miRTarBase databases. (B) The binding sites between GATAD2A and
miR-3180 were predicted by the miRDB database. (C) The
dual-luciferase reporter assay detected the luciferase activities
of 293(T) cells cotransfected with a luciferase reporter construct
containing WT or MUT GATAD2A and the miR-3180 mimic. (D)
High-throughput sequencing of GATAD2A in six pairs of TSCC and
adjacent samples. (E) Expression of GATAD2A in HNSC in the ENCORI
database. (F) Reverse transcription-quantitative PCR analysis was
used to compare GATAD2A expression in tongue cancer and
noncancerous cell lines. All data are presented as the mean ±
standard error of the mean of three independent experiments.
*P<0.05, **P<0.01. GATAD2A, GATA zinc
finger domain containing 2A; miR, microRNA; sh, short hairpin; lnc,
long noncoding RNA; WT, wild type; MUT, mutant; TSCC, tongue
squamous cell carcinoma; HNSC, head and neck squamous cell
carcinoma; NC, negative control. |
Inhibiting GATAD2A reverses the
tumor-promoting effects of hsa-miR-3180 silencing in TSCC
cells
Furthermore, rescue experiments were performed to
verify whether miR-3180 serves a role in TSCC by interacting with
GATAD2A. CAL-27 and SCC-9 cells were stably cotransfected with the
miR-3180 inhibitor and GATAD2A siRNAs. The transfection efficiency
of miR-3180 inhibitor, miR-3180 mimics and GATAD2A siRNAs were
verified in CAL-27 and SCC-9 cells (Fig. S2). One (si-2) of the two siRNAs
against GATAD2A was used in the subsequent experiments after
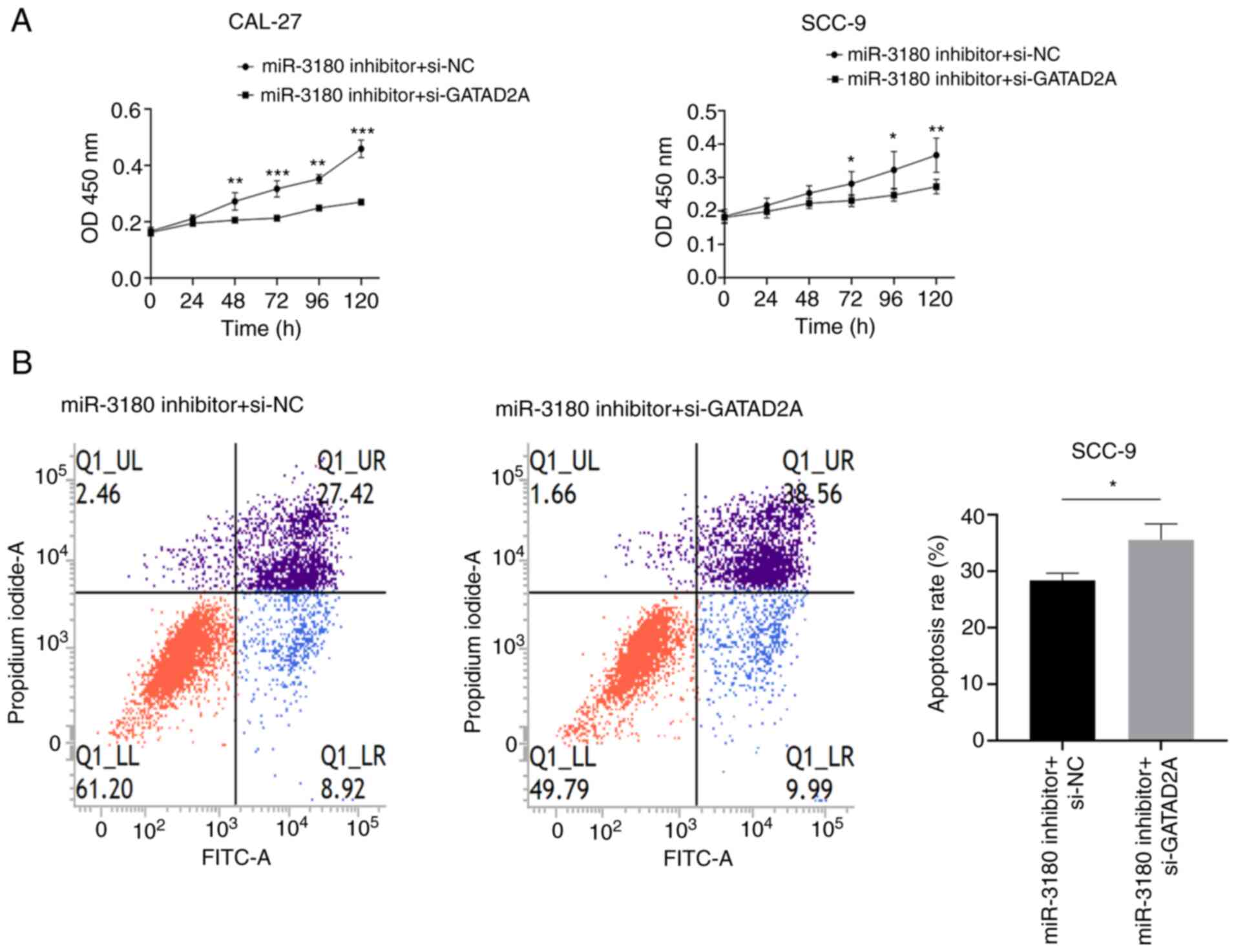
verifying the transfection efficiency. The inhibitory effect on
GATAD2A may reverse the increase in cell viability induced by the
miR-3180 inhibitor, as demonstrated by CCK-8 assays (Fig. 13A). Flow cytometric analysis
showed that the proportion of apoptotic cells was significantly
increased following si-GATAD2A/miR-3180 inhibitor cotreatment
(Fig. 13B). In addition,
Transwell invasion and migration assays and colony formation assays
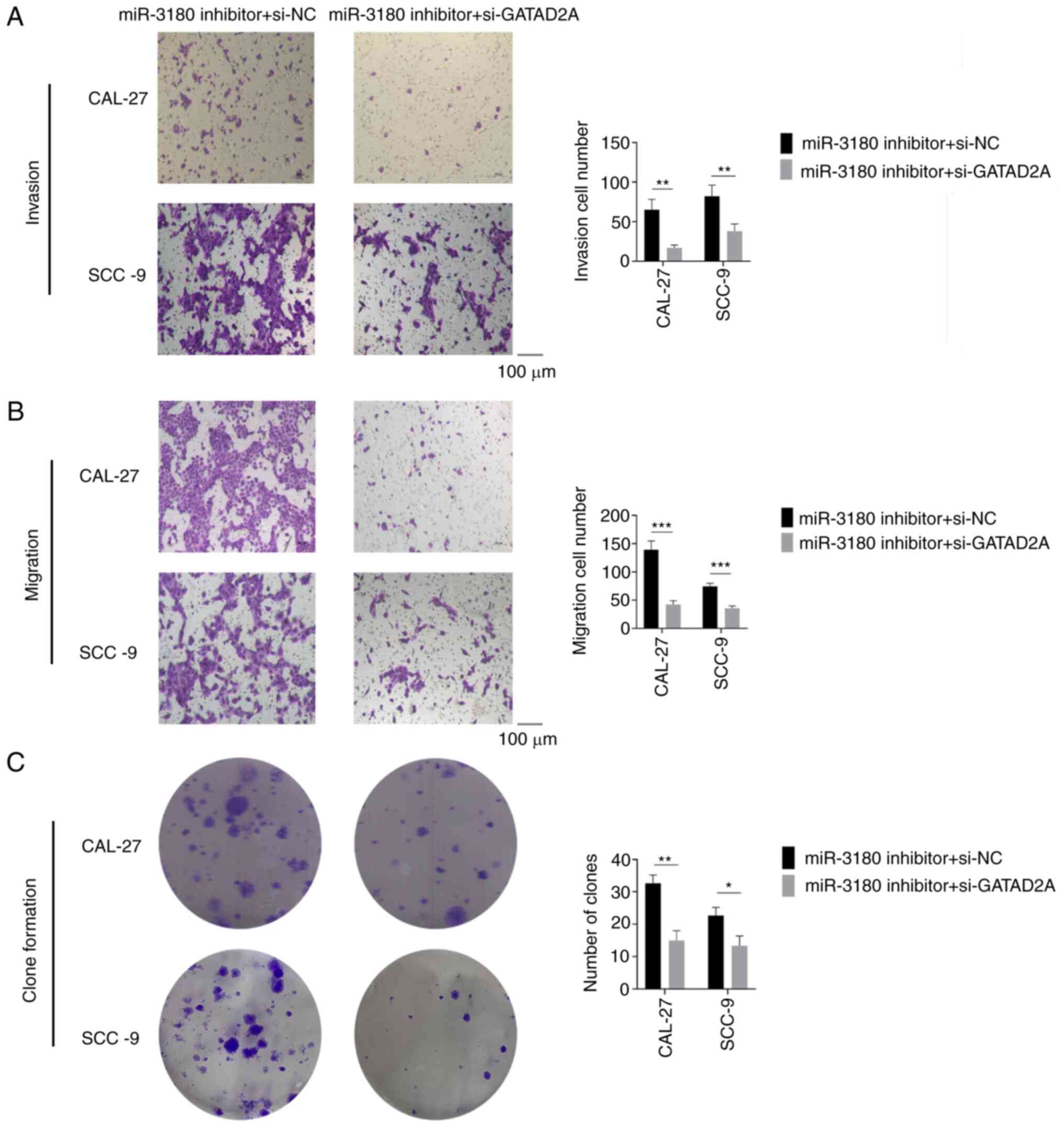
demonstrated that GATAD2A partially attenuated the increases in
promotion of migration and invasion caused by miR-3180 depletion
(Fig. 14). These findings
indicated that miR-3180 inhibits TSCC progression by interacting
with GATAD2A.
lncKRT16P6 acts as a decoy of
hsa-miR-3180 to upregulate GATAD2A
Further experiments were performed to confirm the
regulatory relationship among lncKRT16P6, GATAD2A and miR-3180 and
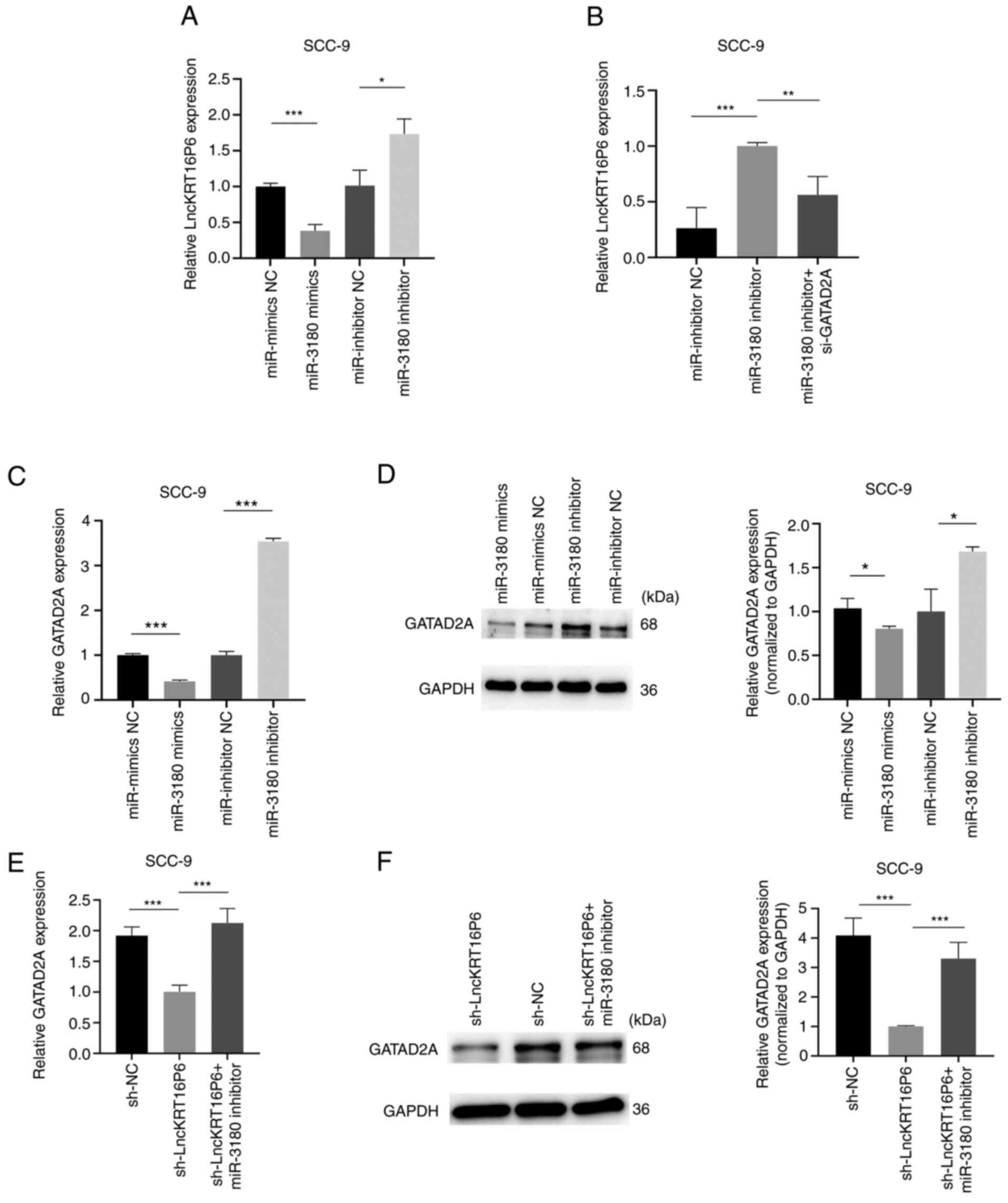
found that the miR-3180 mimic downregulated lncKRT16P6 expression,
but knockdown of miR-3180 rescued lncKRT16P6 expression (Fig. 15A). In parallel, lncKRT16P6
expression was decreased after GATAD2A knockdown and was increased
when miR-3180 was inhibited (Fig.
15B). Moreover, it was verified that the miR-3180 mimic
downregulated GATAD2A expression, but the knockdown of miR-3180
rescued GATAD2A expression (Fig. 15C
and D). GATAD2A expression was decreased after lncKRT16P6
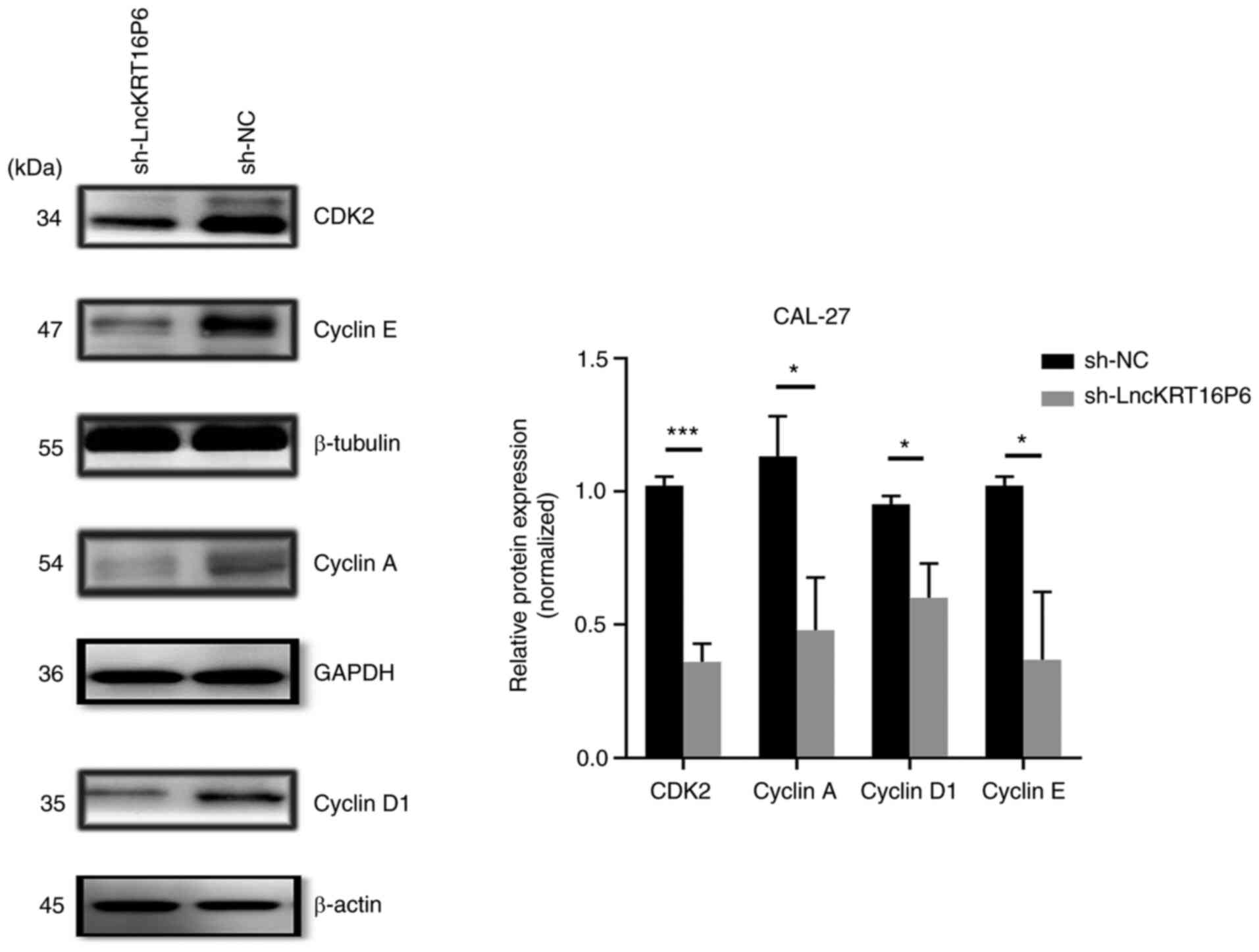
knockdown and was increased when miR-3180 was inhibited (Fig. 15E and F). Moreover, in experiments
to confirm the effect of lncKRT16P6 on the cell cycle in TSCC, it
was found that the expression of CDK2, Cyclin A, Cyclin D1 and
Cyclin E decreased following lncKRT16P6 knockdown (Fig. 16). Taken together, these data
revealed that lncKRT16P6 serves as a ceRNA and sponges miR-3180,
leading to enhanced GATAD2A expression to regulate tongue cancer
progression.
Discussion
Recently, noncoding RNAs have been gradually used to
elucidate the mechanisms of tumorigenesis and tumor progression.
lncRNAs are a type of noncoding RNA that mediate gene expression
and they are closely associated with a variety of tumors, including
lung, gastric, colorectal and bladder tumors (27-31).
The differential expression of lncRNAs is significantly associated
with tumor proliferation, differentiation and metastasis, TNM stage
and other clinical characteristics (32). Therefore, lncRNAs may provide new
insights for exploring tumor pathogenesis and serve as potential
biomarkers for multiple types of cancers. While lncRNAs have been
widely studied in the development of TSCC, the pathogenic mechanism
is not completely clear.
The present study screened dysregulated lncRNAs
associated with TSCC and identified a novel lncRNA, lncKRT16P6. The
evidence suggested that lncKRT16P6 is an oncogenic lncRNA and is
closely associated with tumor stage and differentiation grade. The
present study used two cell lines commonly employed in most
research and other tongue cancer cell lines will be used in
follow-up research to further verify the results of the present
study and ultimately improve the experimental conclusions. The
oncogenic role of lncKRT16P6 revealed in vitro and in
vivo in the present study highlights the potential implication
of this lncRNA as a predictive biomarker and therapeutic target for
TSCC.
CeRNAs mediate gene expression and are assumed to
function through MREs to form a transcriptional regulatory network
in which mRNAs, pseudogenes and lncRNAs may 'interact' with each
other, which may affect the progression of certain diseases
(33,34). Studies have shown that ceRNAs serve
pivotal roles in the invasion, proliferation and metastasis of
cancer cells (35,36) and have confirmed that lncRNAs are
also involved in regulating tumor ceRNA networks (37-40).
The present study found that lncKRT16P6 was located mainly in the
cytoplasm of TSCC cells and acted as a sponge for miR-3180. In
addition, the findings revealed the significance of the interaction
between lncKRT16P6 and miR-3180 in tumorigenesis. The present study
found that miR-3180 partially reversed the oncogenic role of
lncKRT16P6.
In general, as ceRNAs, the functions of lncRNAs
depend on the miRNA target (41-43).
Using an online database, GATAD2A was predicted a potential target
of miR-3180, which was confirmed by a luciferase reporter assay.
Furthermore, overexpression of miR-3180 inhibited GATAD2A mRNA and
protein expression. GATAD2A is located on chromosome 19 in humans
and is a key member of the MBD2-NuRD complex (44). GATAD2A interacts with MBD2 to
recruit the deacetylase core of the NuRD complex to repress target
genes (45,46). Several studies have verified that
GATAD2A promotes the progression of thyroid, breast and papillary
thyroid cancers (47-49) and confirmed that the NuRD complex
is closely associated with the G1/S cell cycle
checkpoint (50). The present
study demonstrated that lncKRT16P6 upregulated GATAD2A expression
by competitively sponging miR-3180, thus affecting the
proliferation and metastasis of TSCC cells. Furthermore, it
indicated that lncKRT16P6 can activate the expression of CDK2,
Cyclin A, Cyclin D1 and Cyclin E, thus promoting the activity of
the G1/S checkpoint signaling pathways, which promote
malignant behavior.
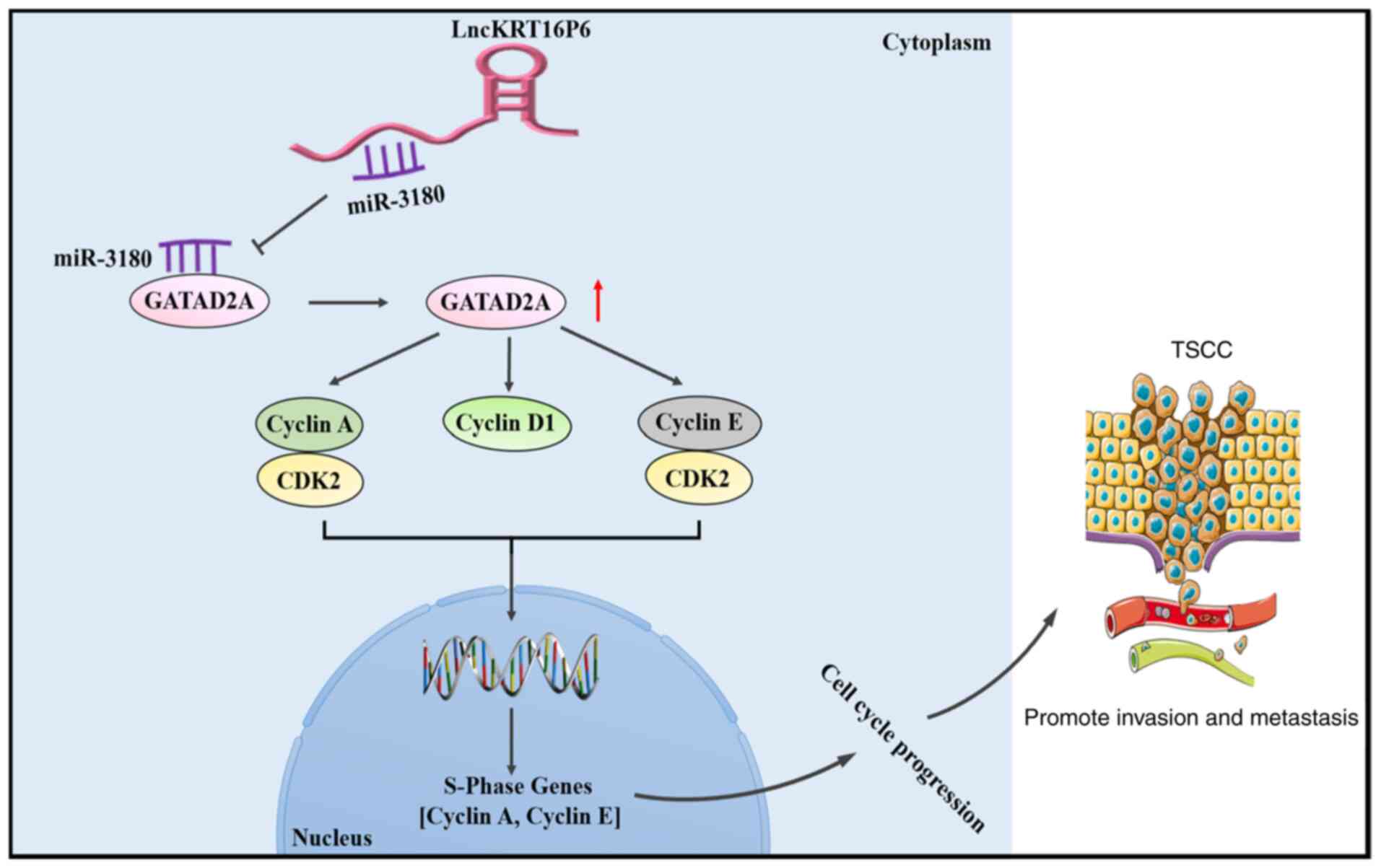
In conclusion, the present study identified
lncKRT16P6 as an oncogenic lncRNA involved in TSCC development and
progression. Functional and mechanistic analyses revealed that
lncKRT16P6 promoted the malignant behavior of TSCC cells by acting
as a ceRNA that sponges miR-3180, leading to enhanced GATAD2A
expression. Moreover, depletion of lncKRT16P6 inactivated
G1/S checkpoint signaling (Fig. 17). The present study demonstrated
that lncKRT16P6 serves an essential role in TSCC tumorigenesis and
progression and highlighted its importance as a prognostic
indicator. The identification of the lncKRT16P6/miR-3180/GATAD2A
axis provides new strategies for targeted therapies for TSCC. The
mechanisms by which lncKRT16P6 simultaneously affects the
expression of cyclins to regulate tumor progression warrant further
investigation. Furthermore, RNAscope experiments could allow highly
specific quantitative analysis of nucleic acid in situ
hybridization at the single-cell and single-molecule levels; this
experiment will be conducted in follow-up research to continuously
improve on the present results.
The present study also had certain limitations.
First, the reason for the increase in lncKRT16P6 has not been
clarified in the current study. Furthermore, the molecular
mechanism by which GATAD2A and cyclin interact has not been
clarified. These unanswered questions will be the focus of future
research and will be addressed by complementary experiments in the
future.
Supplementary Data
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
MZ and LW confirm the authenticity of all the raw
data. Research design and implementation: MZ and LW designed and
implemented the study and performed data analysis and
interpretation. MZ, LW and XW performed statistical analysis. MZ
wrote the manuscript, JC revised the manuscript and supervised the
study and helped to solve various problems in the research. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Ethics
Committee of the First Affiliated Hospital of Fujian Medical
University [approval number: 159 (2020) of the First Affiliated
Hospital of Fujian Medical University]. Written informed consent
was obtained from all participants in accordance with the
Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors thank the Public Technology Service
Center of Fujian Medical University for their support.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant no. 81771126) and the Science and
Technology Planning Project of Fujian Province (grant no.
2019Y9031).
References
|
1
|
Mannelli G, Arcuri F, Agostini T,
Innocenti M, Raffaini M and Spinelli G: Classification of tongue
cancer resection and treatment algorithm. J Surg Oncol.
117:1092–1099. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sagheb K, Kumar V, Rahimi-Nedjat R,
Dollhausen M, Ziebart T, Al-Nawas B and Walter C: Cervical
metastases behavior of T1-2 squamous cell carcinoma of the tongue.
J Maxillofac Oral Surg. 16:300–305. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen X, Xie R, Gu P, Huang M, Han J, Dong
W, Xie W, Wang B, He W, Zhong G, et al: Long noncoding RNA LBCS
inhibits self-renewal and chemoresistance of bladder cancer stem
cells through epigenetic silencing of SOX2. Clin Cancer Res.
25:1389–1403. 2019. View Article : Google Scholar
|
|
4
|
Yue B, Cai D, Liu C, Fang C and Yan D:
Linc00152 functions as a competing endogenous RNA to confer
oxaliplatin resistance and holds prognostic values in colon cancer.
Mol Ther. 24:2064–2077. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Djebali S, Davis CA, Merkel A, Dobin A,
Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F,
et al: Landscape of transcription in human cells. Nature.
489:101–108. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chu C, Spitale RC and Chang HY:
Technologies to probe functions and mechanisms of long noncoding
RNAs. Nat Struct Mol Biol. 22:29–35. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Batista PJ and Chang HY: Long noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang G, Li S, Lu J, Ge Y, Wang Q, Ma G,
Zhao Q, Wu D, Gong W, Du M, et al: LncRNA MT1JP functions as a
ceRNA in regulating FBXW7 through competitively binding to
miR-92a-3p in gastric cancer. Mol Cancer. 17:872018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yi W, Li J, Zhu X, Wang X, Fan L, Sun W,
Liao L, Zhang J, Li X, Ye J, et al: CRISPR-assisted detection of
RNA-protein interactions in living cells. Nat Methods. 17:685–688.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Long Y, Wang X, Youmans DT and Cech TR:
How do lncRNAs regulate transcription? Sci Adv. 3:eaao21102017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang J, Zhang X, Chen W, Hu X, Li J and
Liu C: Regulatory roles of long noncoding RNAs implicated in cancer
hallmarks. Int J Cancer. 146:906–916. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang S, Ma H, Zhang D, Xie S, Wang W, Li
Q, Lin Z and Wang Y: LncRNA KCNQ1OT1 regulates proliferation and
cisplatin resistance in tongue cancer via miR-211-5p mediated
Ezrin/Fak/Src signaling. Cell Death Dis. 9:7422018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Su SC, Yeh CM, Lin CW, Hsieh YH, Chuang
CY, Tang CH, Lee YC and Yang SF: A novel melatonin-regulated lncRNA
suppresses TPA-induced oral cancer cell motility through
replenishing PRUNE2 expression. J Pineal Res. 71:e127602021.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang F, Wang H, Yu J, Yao X, Yang S, Li
W, Xu L and Zhao L: LncRNA CRNDE attenuates chemoresistance in
gastric cancer via SRSF6-regulated alternative splicing of PICALM.
Mol Cancer. 20:62021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kristensen LS, Andersen MS, Stagsted LV,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu J, Mayekar MK, Wu W, Yan M, Guan H,
Wang J, Zaman A, Cui Y, Bivona TG, Choudhry H, et al: Long
non-coding RNA ESCCAL-1 promotes esophageal squamous cell carcinoma
by down regulating the negative regulator of APOBEC3G. Cancer Lett.
493:217–227. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zheng ZQ, Li ZX, Zhou GQ, Lin L, Zhang LL,
Lv JW, Huang XD, Liu RQ, Chen F, He XJ, et al: Long noncoding RNA
FAM225A promotes nasopharyngeal carcinoma tumorigenesis and
metastasis by acting as ceRNA to Sponge miR-590-3p/miR-1275 and
upregulate ITGB3. Cancer Res. 79:4612–4626. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sheng X, Dai H, Du Y, Peng J, Sha R, Yang
F, Zhou L, Lin Y, Xu S, Wu Y, et al: LncRNA CARMN overexpression
promotes prognosis and chemosensitivity of triple negative breast
cancer via acting as miR143-3-p host gene and inhibiting DNA
replication. J Exp Clin Cancer Res. 40:2052021. View Article : Google Scholar
|
|
21
|
Jia B, Xie T, Qiu X, Sun X, Chen J, Huang
Z, Zheng X, Wang Z and Zhao J: Long noncoding RNA FALEC inhibits
proliferation and metastasis of tongue squamous cell carcinoma by
epigenetically silencing ECM1 through EZH2. Aging (Albany NY).
11:4990–5007. 2019. View Article : Google Scholar
|
|
22
|
Zhang H, Zhao L, Wang YX, Xi M, Liu SL and
Luo LL: Long non-coding RNA HOTTIP is correlated with progression
and prognosis in tongue squamous cell carcinoma. Tumour Biol.
36:8805–8809. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fang Z, Zhang S, Wang Y, Shen S, Wang F,
Hao Y, Li Y, Zhang B, Zhou Y and Yang H: Long non-coding RNA
MALAT-1 modulates metastatic potential of tongue squamous cell
carcinomas partially through the regulation of small proline rich
proteins. BMC Cancer. 16:7062016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang M, Chen Z, Zhang S, Wu L, Jie Y,
Liao Y, Huang Y, Chen J and Shi B: Analysis of differentially
expressed long non-coding RNAs and the associated TF-mRNA network
in tongue squamous cell carcinoma. Front Oncol. 10:14212020.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu Y, Hu Q, Pan Y, Wang Y, Jiang L, Lin
H, Lin D and Cheng H: The apoptotic and autophagic effects of cast
Au-Pt, and differently manufactured Co-Cr and cp-Ti on
three-dimensional oral mucosal model. Mater Sci Eng C Mater Biol
Appl. 120:1116722021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
27
|
Chen Z, Chen X, Lu B, Gu Y, Chen Q, Lei T,
Nie F, Gu J, Huang J, Wei C, et al: Up-regulated LINC01234 promotes
non-small-cell lung cancer cell metastasis by activating VAV3 and
repressing BTG2 expression. J Hematol Oncol. 13:72020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shuai Y, Ma Z, Liu W, Yu T, Yan C, Jiang
H, Tian S, Xu T and Shu Y: TEAD4 modulated LncRNA MNX1-AS1
contributes to gastric cancer progression partly through
suppressing BTG2 and activating BCL2. Mol Cancer. 19:62020.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu TP, Ma P, Wang WY, Shuai Y, Wang YF, Yu
T, Xia R and Shu YQ: KLF5 and MYC modulated LINC00346 contributes
to gastric cancer progression through acting as a competing
endogeous RNA and indicates poor outcome. Cell Death Differ.
26:2179–2193. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen L, He M, Zhang M, Sun Q, Zeng S, Zhao
H, Yang H, Liu M, Ren S, Meng X and Xu H: The role of non-coding
RNAs in colorectal cancer, with a focus on its autophagy. Pharmacol
Ther. 226:1078682021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen C, Zheng H, Luo Y, Kong Y, An M, Li
Y, He W, Gao B, Zhao Y, Huang H, et al: SUMOylation promotes
extracellular vesicle-mediated transmission of lncRNA ELNAT1 and
lymph node metastasis in bladder cancer. J Clin Invest.
131:e1464312021. View Article : Google Scholar
|
|
32
|
Wu P, Mo Y, Peng M, Tang T, Zhong Y, Deng
X, Xiong F, Guo C, Wu X, Li Y, et al: Emerging role of
tumor-related functional peptides encoded by lncRNA and circRNA.
Mol Cancer. 19:222020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Abdollahzadeh R, Daraei A, Mansoori Y,
Sepahvand M, Amoli MM and Tavakkoly-Bazzaz J: Competing endogenous
RNA (ceRNA) cross talk and language in ceRNA regulatory networks: A
new look at hallmarks of breast cancer. J Cell Physiol.
234:10080–10100. 2019. View Article : Google Scholar
|
|
36
|
Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y,
Chen W, Liu F, Sun W, Li XF, et al: Exosome-transmitted lncARSR
promotes Sunitinib resistance in renal cancer by acting as a
competing endogenous RNA. Cancer Cell. 29:653–668. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li A, Mallik S, Luo H, Jia P, Lee DF and
Zhao Z: H19, a long non-coding RNA, mediates transcription factors
and target genes through interference of micrornas in pan-cancer.
Mol Ther Nucleic Acids. 21:180–191. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xu J, Xiao Y, Liu B, Pan S, Liu Q, Shan Y,
Li S, Qi Y, Huang Y and Jia L: Exosomal MALAT1 sponges miR-26a/26b
to promote the invasion and metastasis of colorectal cancer via
FUT4 enhanced fucosylation and PI3K/Akt pathway. J Exp Clin Cancer
Res. 39:542020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ji W, Diao YL, Qiu YR, Ge J, Cao XC and Yu
Y: LINC00665 promotes breast cancer progression through regulation
of the miR-379-5p/LIN28B axis. Cell Death Dis. 11:162020.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhu X, Bu F, Tan T, Luo Q, Zhu J, Lin K,
Huang J, Luo C and Zhu Z: Long noncoding RNA RP11-757G1.5 sponges
miR-139-5p and upregulates YAP1 thereby promoting the proliferation
and liver, spleen metastasis of colorectal cancer. J Exp Clin
Cancer Res. 39:2072020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jia Y, Tian C, Wang H, Yu F, Lv W, Duan Y,
Cheng Z, Wang X, Wang Y, Liu T, et al: Long non-coding RNA
NORAD/miR-224-3p/MTDH axis contributes to CDDP resistance of
esophageal squamous cell carcinoma by promoting nuclear
accumulation of beta-catenin. Mol Cancer. 20:1622021. View Article : Google Scholar
|
|
42
|
He Y, Jiang X, Duan L, Xiong Q, Yuan Y,
Liu P, Jiang L, Shen Q, Zhao S, Yang C and Chen Y: LncRNA PKMYT1AR
promotes cancer stem cell maintenance in non-small cell lung cancer
via activating Wnt signaling pathway. Mol Cancer. 20:1562021.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li ZY, Xie Y, Deng M, Zhu L, Wu X, Li G,
Shi NX, Wen C, Huang W, Duan Y, et al: c-Myc-activated intronic
miR-210 and lncRNA MIR210HG synergistically promote the metastasis
of gastric cancer. Cancer Lett. 526:322–334. 2022. View Article : Google Scholar
|
|
44
|
Sher F, Hossain M, Seruggia D,
Schoonenberg VA, Yao Q, Cifani P, Dassama LM, Cole MA, Ren C,
Vinjamur DS, et al: Rational targeting of a NuRD subcomplex guided
by comprehensive in situ mutagenesis. Nat Genet. 51:1149–1159.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gnanapragasam MN, Scarsdale JN, Amaya ML,
Webb HD, Desai MA, Walavalkar NM, Wang SZ, Zu Zhu S, Ginder GD and
Williams DC Jr: p66Alpha-MBD2 coiled-coil interaction and
recruitment of Mi-2 are critical for globin gene silencing by the
MBD2-NuRD complex. Proc Natl Acad Sci USA. 108:7487–7492. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Desai MA, Webb HD, Sinanan LM, Scarsdale
JN, Walavalkar NM, Ginder GD and Williams DC Jr: An intrinsically
disordered region of methyl-CpG binding domain protein 2 (MBD2)
recruits the histone deacetylase core of the NuRD complex. Nucleic
Acids Res. 43:3100–3113. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang Z, Kang J, Deng X, Guo B, Wu B and
Fan Y: Knockdown of GATAD2A suppresses cell proliferation in
thyroid cancer in vitro. Oncol Rep. 37:2147–2152. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lu D, Song J, Lu Y, Fall K, Chen X, Fang
F, Landén M, Hultman CM, Czene K, Sullivan P, et al: A shared
genetic contribution to breast cancer and schizophrenia. Nat
Commun. 11:46372020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yao Y, Chen X, Yang H, Chen W, Qian Y, Yan
Z, Liao T, Yao W, Wu W, Yu T, et al: Hsa_circ_0058124 promotes
papillary thyroid cancer tumorigenesis and invasiveness through the
NOTCH3/GATAD2A axis. J Exp Clin Cancer Res. 38:3182019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
van den Heuvel S and Dyson NJ: Conserved
functions of the pRB and E2F families. Nat Rev Mol Cell Biol.
9:713–724. 2008. View Article : Google Scholar : PubMed/NCBI
|