1. Introduction
Pancreatic cancer (PC) is the most common type of
malignancy affecting the digestive system, which has the
characteristics of 'three low and three high'; that is, a low early
diagnosis rate, a low resection rate, a low 5-year survival rate,
and a high incidence, a high metastasis rate and a high mortality
rate. Owing to the deficiency of specific symptoms during the early
stages of the malignancy, the early diagnosis rate of PC is <5%.
At the first visit, only 15-20% of patients can undergo radical
resection, 30-40% of patients present with locally advanced cancer,
whereas 40-50% of patients have distant metastasis. The 5-year
survival rate has been found to be 7.2-9.0% (1,2). The
early detection, early diagnosis and early treatment of PC has
always been a pivotal and difficult challenge for PC. Hence, there
is an urgent need to elucidate the underlying mechanisms of its
pathogenesis, and to develop novel techniques with a greater
diagnostic efficacy.
A multitude of previously published articles in the
literature have demonstrated that N6-methyladenosine
(m6A) not only participates in physiological processes,
including neurogenesis, hematopoiesis, spermatogenesis, sex
determination, response to DNA damage or heat shock, circadian
rhythm, and the innate and adaptive immune response (3-7), but
that it is also tightly associated with the initiation, progression
(8,9), diagnosis and treatment of neoplasms
(10). Accordingly, the initial
part of the present review article will introduce the biological
functions of m6A; subsequently, the present review will
interpret the attributes of m6A in the carcinogenesis
and development of PC, and finally, the prospective clinical
applications of m6A in PC will be discussed.
2. Biology of m6A
Thus far, diverse chemical modifications have been
detected, and m6A remains the most abundant
post-transcriptional modification to be found in messenger RNAs
(mRNAs) (3). The modification of
m6A comprises methylation at the sixth N atom of adenine
in RNA, and approximately one third of mammalian mRNAs, with an
average of 3-5 m6A sites per mRNA, have been shown to
feature this modification (11,12).
m6A was first discovered in 1974 (13-15).
The concept of reversible m6A modification was proposed
in a stepwise manner owing to the discovery of the first
methyltransferase, namely methyltransferase-like protein 3 (METTL3)
in 1997 (16), and the first
demethylase, fat mass and obesity-associated protein (FTO), in 2011
(17).
In order to more clearly elucidate the functions of
m6A, scientists have been seeking improvements in the
available m6A-mapping methods. Meyer et al
(18) combined
m6A-specific methylated RNA immunoprecipitation with
next-generation sequencing (MeRIP-Seq) to depict the landscape of
m6A at the whole transcriptome level in 2012, which
heralded the creation of the 'epitranscriptome' (18). Since then, m6A has
attracted extensive attention in the research community. MeRIP-Seq
is able to discern the distribution of m6A at the single
transcript level, and identify which groups of transcripts are
modified by m6A. However, it has not been successful at
counting the precise stoichiometry of m6A sites; neither
has it been able to distinguish m6A from
6,2,-O-dimethyladenosine (m6Am). The molecular structure
of m6Am is similar to that of m6A: Compared
with m6A, the 2′-hydroxy group of adenylate is
methylated to produce this doubly methylated nucleotide.
M6A is conserved in eukaryotes (19-23),
and m6A sites are preferentially enriched near stop
codons, at 3′-untranslated regions (3′-UTRs) or in long exons, and
they are prone to recognize a typical consensus sequence, RRACH
(where 'R' is A/G, and 'H' is A/C/U) (4,16,18,21,24).
Notably, m6A is specifically located in genes associated
with development or cell fate; seldom are m6A sites to be found in
housekeeping genes (18,24-26).
Although the majority of research efforts thus far have focused on
mRNAs, researchers have also identified m6A sites on
ribosomal RNAs (rRNAs) (27,28)
and transfer RNAs (tRNAs) (29).
In addition, m6A sites have been identified in
non-coding RNAs (ncRNAs), which cannot be translated into proteins,
but have essential functions in gene expression and regulation, as
well as long ncRNAs (lncRNAs) (30,31),
microRNAs (miRNAs/miRs) (32,33),
circular RNAs (circRNAs) (34),
small nuclear RNAs (snRNAs) (35,36)
and small nucleolar RNAs (snoRNAs) (37).
M6A is added by methyltransferases
(termed 'writers'), removed by demethylases (termed 'erasers') and
identified by m6A-RNA-binding proteins (termed
'readers'), which are able to affect the fate of
m6A-RNAs, and then mediate post-transcriptional gene
expression (Fig. 1 and Table I).
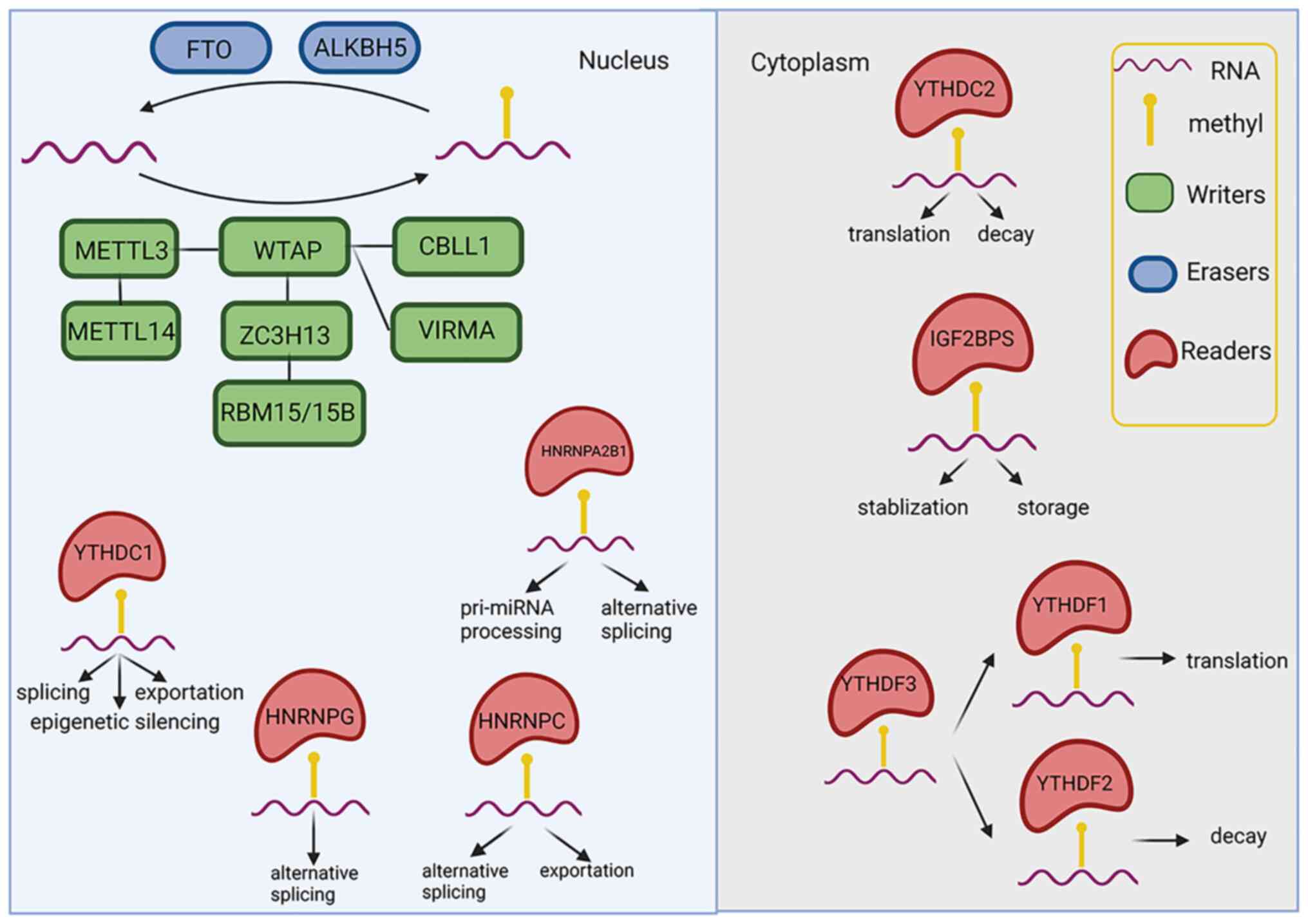 | Figure 1The biological roles of
N6-methyladenosine. FTO, fat mass and obesity-associated protein;
ALKBH5, alkB homolog 5, RNA demethylase; METTL,
methyltransferase-like protein; WTAP, Wilms' tumor 1-associating
protein; CBLL1, Casitas B lineage lymphoma transforming sequence
like protein 1; ZC3H13, zinc finger CCCH-type containing 13; VIRMA,
Vir-like m6A methyltransferase-associated; RBM15,
RNA-binding motif protein 15; YTHDC, YT521-B homology
domain-containing protein; HNRN, heterogeneous nuclear
ribonucleoprotein; IGF2BPS, insulin-like growth factor-2
mRNA-binding proteins. The figure was created using https://biorender.com. |
 | Table IOverview of m6A writers,
frasers andreaders. |
Table I
Overview of m6A writers,
frasers andreaders.
| Type of
m6 A and protein name | Location | function | (Refs.) |
|---|
| Writers | METTL3 | | Nuclear
speckle | Catalytic subunit,
transfer methyl group from SAM to target RNA | (16,26,39,40,43,44) |
| METTL14 | | Nuclear
speckle | Allosteric
activator, catalytically inactive recognize substrate RNA and
enhance METTL3 activity | (26,41-44) |
| WTAP | | Nucleus | METTL3 adaptor
anchor METTL3-METTL14 heterodimer complex to nuclear speckles and
target mRNA | (43,47,48) |
| VIRMA/KIAA1429 | | Nucleus | WATP interactor,
modification preference of m6A in 3,UTR and near stop
codon shorten 3,UTR | (49) |
| RBM15/15B | | Nucleus | WATP interactor,
regulate genetic transcription silencing | (50) |
| ZC3H13 | | Nucleus | WATP interactor,
bridge RBM15/15B toWTAPanchor ZC3H13-WTAP-VIRMA-HAKAI complex in
the nucleus to facilitate m6A modification | (51,52) |
| CBLL1/HAKAI | | Nucleus | WATP interactor,
modulate cell adhesion and EMT in cancer | (53,54) |
| Erasers | FTO | | Nucleus | Demethylate
m6A/m6Am | (17,55) |
| ALKBH5 | | Nucleus | Demethylate
m6Ain the testes and play a role in spermatogenesis | (56-58) |
| Readers | YTHs | YTHDF1 | Cytoplasm | Expedite
translation initiation DNA damage repair LLPS | (59-61) |
| | YTHDF2 | Cytoplasm | Degrade mRNA,
LLPS | (62-67) |
| | YTHDF3 | Cytoplasm | Assist YTHDF1 and
YTHDF2 LLPS | (68) |
| | YTHDC1 | Nucleus | mRNA splicing
nuclear exportation epigenetic silencing autophagy | (50,69-74) |
| | YTHDC2 |
Nucleus/cytoplasm | Spermatogenesis
elevate the translation efficiency decrease mRNA abundance | (75) |
| HNRNPs | HNRNPC | Nucleus | Regulate
alternative splicing mRNA,U snRNA exportation,
m6A-switch | (3,35,77-80) |
| | HNRNPG | Nucleus | Regulate
alternative splicing | (81,82) |
|
m6A-switch | |
| | HNRNPA2B1 | Nucleus | Pri-miRNA
processing regulate alternative splicing vesicular trafficking | (83,84) |
| IGF2BPs | IGF2BP1/2/3 | Nucleus and
cytoplasm | Stabilization and
storage of mRNA in physiological condition and stress response
human senescence | (80,85) |
Writers
S-adenosyl-l-methionine (SAM) has emerged as the
predominant methyl donor in vivo. Writers are considered to
participate in a methyltransferase complex (MTC), which transfers
methyl groups of SAM to target RNA in a highly specific manner. The
MTC comprises METTL3, methyltransferase-like protein 14 (METTL14),
an adaptor [Wilms' tumor 1-associating protein (WTAP)], a WTAP
interactor [Vir-like m6A methyltransferase-associated
(VIRMA; also known as KIAA1429)], RNA-binding motif protein
(RBM)15/15B, zinc finger CCCH-type containing 13 (ZC3H13) and
Casitas B lineage lymphoma transforming sequence like protein 1
[CBLL1/E3 ubiquitin-protein ligase (HAKAI)] (38). The members of this multi-subunit
complex will be introduced in turn.
METTL3 and METTL14
METTL3 is the catalytic subunit of the MTC that has
a SAM-binding motif and transfers methyl groups to RNA (16,39,40).
METTL14 is an allosteric activator that has an interrupted
SAM-binding motif, demonstrating that METTL14 is catalytically
inactive (41,42). Together, METTL14 and METTL3 form a
heterodimer complex (43), which
participates in recognizing substrate RNA and enhancing
methyltransferase activity (44).
In the absence of METTL3 or METTL14, mouse embryonic stem cells
have been shown to exhibit a decrease of 99% in their
m6A content (26). In
addition to the METTL3-METTL14 heterodimer complex, there are
several other critical methylation enzymes. For example, ZCCHC4
adds a methyl group on to adenylate located in the 28S rRNA subunit
(27), and METTL5-TRMT112
heterodimer complex modifies the 18S rRNA subunit (28). METTL16 independently catalyzes the
formation of m6A in the pre-mRNAs, U6 snRNA and
lncRNA.
WTAP
WTAP is the key METTL3 adaptor (43,45),
which anchors METTL3-METTL14 heterodimer complex to nuclear
speckles (46). Upon deletion of
WTAP, the affinity of METTL3 to bind RNA is clearly decreased,
indicating that WTAP recruits the METTL3-METTL14 heterodimer
complex to target mRNAs (45).
VIRMA/KIAA1429
VIRMA/KIAA1429 contributes towards the modification
preference of m6A for 3′-UTRs and in the vicinity of the
stop codon. A model has been proposed wherein VIRMA may function as
a scaffold to hold WTAP/HAKAI/ZC3H13 together and creates a
suitable pocket mainly through WTAP to accommodate METTL3/METTL14
in order to guide m6A modification in the 3′UTR and
around the stop codon. The 3′UTR has been shown to be shortened by
VIRMA through its interactions with the polyadenylation cleavage
factors, CPSF5 and CPSF6 (47).
RBM15/15B
lncRNA X-inactive specific transcript (XIST)
regulates epigenetic silencing on the X chromosome. RBM15 and its
paralogue, RBM15B, bind to and recruit the m6A
methylation complex to specific sites in lncRNAs, thereby mediating
the formation of m6A in XIST (48).
ZC3H13
ZC3H13 is considered to bridge RBM15/RBM15B to WTAP
(49). The ZC3H13-WTAP-VIRMA-HAKAI
complex has a profound effect on the regulation of m6A
in RNAs. ZC3H13 plays the role of anchoring this regulatory complex
in the nucleus to facilitate m6A modification (50).
CBLL1/HAKAI
In concert with E-cadherin, CBLL1 (also known as
HAKAI) modulates cell adhesion and epithelial-mesenchymal
transition (EMT) (51). The
knockdown of HAKAI has been shown to reduce m6A levels
(52). EMT provides a critical
foundation for tumor metastasis, suggesting that m6A may
be closely associated with tumor progression.
Erasers
Erasers are demethylases that catalyze the removal
of the methyl group, thereby converting m6A into
adenosine (A). At present, however, the roles of erasers under
physiological conditions appear to be limited. Erasers including
FTO and alkB homolog 5 (ALKBH5). ALKBH5 have been identified in a
limited number of tissues such as testicles. They are also active
under disease- and stress-associated conditions.
FTO
Consistently with ALKBs demethylating DNA, FTO
demethylates m6A in mRNA in a 2-oxoglutarate
(2OG)-Fe2+ oxygenase-dependent manner (17). Nevertheless, compared with ferrous
iron, the interaction with m6A is stronger, and
therefore, the demethylation effect of FTO in m6Am is
more efficient, which indicates that lower substrate concentrations
are required and the reaction times are shorter. Furthermore, FTO
exhibits differential substrate preferences: It can demethylate
m6Am at the 5′-cap or m6As in the inner
region of, mRNAs; m6Am at the 5′-cap or in the inner
regions of snRNAs; m6A in the inner regions of U6 RNA;
and N1-methyladenosine in tRNAs (53). It will be noteworthy to analyze
this divergence in greater depth, and to identify which
substrate(s) FTO truly catalyzes.
ALKBH5
ALKBH5 is also a 2OG-Fe2+-dependent
oxidative demethyltransferase that demethylates m6A in
mRNAs (54). Its expression is
particularly eriched in the testes, where is fulfills a role in
spermatogenesis (55). Moreover,
the m6A level in the mRNA of male mice wherein ALKBH5
has been knocked down has been shown to be increased (54). These mice have been shown to be
infertile, which is considered to have resulted from the apoptosis
of meiotic metaphase-stage spermatocytes (54). FTO and ALKBH5 share the same
sequential oxidative demethylation reaction process, which is
considered to be as follows: m6A to
N6-hydroxymethyladenosine to
N6-formyladenosine to A (56).
Readers
A myriad of writers and erasers can reversibly add
or remove methyl groups in target mRNA, which exerts a major
influence in the fate of target mRNAs. When m6A-mRNA
binds readers in the nucleus, this may have an impact on mRNA
splicing. Once exported to the cytoplasm, m6A-mRNA may
be recognized by various cytosolic readers, thereby influencing the
stability, translation and localization of mRNA (3). Readers comprise YTH N6
methyladenosine RNA binding proteins (YTHDF1/2/3), YT521-B homology
domain-containing proteins (YTHDC1/2), heterogeneous nuclear
ribonucleoproteins (HNRNPC/G/A2B1) and the insulin-like growth
factor-2 mRNA-binding proteins (IGF2BP)-1, -2 and -3.
YTHDF1
YTHDF1 has been reported to participate in
expediting the processes of translation initiation and DNA damage
repair. YTHDF1 promotes the loading of ribosomes to bound mRNAs to
enable the interactions with initiation factors (such as eIF3) to
occur, thereby promoting m6A-mRNA translation initiation
(57,58). Double-strand breaks (DSBs) are
responsible for the most severe type of DNA damage; the
METTL3-m6A-YTHDC1 axis has been shown to enhance the
deposition of DNA-RNA hybrids at DSB sites, which enables the
recruitment of breast cancer type 1 susceptibility protein and DNA
repair protein RAD51 homolog 1 (RAD51) for the homologous
recombination (HR)-mediated reparation of DSBs (59).
YTHDF2 and YTHDF3
YTHDF2 is the most abundant protein of domain family
(DF) paralogues purified from cells (60), and is crucial for the degradation
of mRNA. At present, there are two prevailing hypotheses concerning
the underlying mechanism. A large number of studies have confirmed
that m6A erects a scaffold that juxtaposes DF1/2/3,
enhancing the liquid-liquid phase separation (60-63)
of RNA-protein droplets, which are subsequently partitioned into
membrane-less compartments, such as processing bodies (P-bodies:
mRNA decay sites) (64), or into
stress granules during stress. In this manner, mRNAs may be either
degraded or stored (60-63). Furthermore, the N-terminal region
of YTHDF2 recruits and interacts with the superfamily homology (SH)
domain of the CCR4-NOT deadenylase complex to initiate the
deadenylation and degradation of target mRNAs (65). YTHDF3 functions in synergy with
YTHDF1 to facilitate translation, thereby assisting YTHDF2 in the
degradation of m6A-RNA (66).
YTHDC1
YTHDC1 fulfills a variety of roles in mRNA splicing,
nuclear exportation, genetic transcription silencing regulated by
the lncRNA XIST and autophagy. The nuclear export adaptor proteins,
SRSF3 and SRSF10, identified as pre-mRNA splicing factors,
competitively bind to YTHDC1. SRSF3 facilitates exon inclusion,
whereas SRSF10 promotes exon skipping. YTHDC1 promotes SRSF3,
whereas SRSF10 is antagonized by YTHDC1 with respect to their
localization, RNA-binding capabilities and associated splicing
events (67,68).
In terms of the more detailed mechanism, YTHDC1
interacts with SRSF3, thereby stimulating m6A-mRNA
binding to SRSF3 and the mRNA export receptor, nuclear RNA export
factor 1 (NXF1), which directs mRNAs into the nuclear export
pathway (69). SRSF3 stands at the
point of intersection between splicing and nuclear exportation,
indicating that there may be some connection between them. Another
study revealed that the m6A methylation complex
stimulates the recruitment of transcription export (TREX) to target
mRNAs, and TREX then additionally recruits YTHDC1 and NXF1 to
trigger the effective exportation of mRNA (70). As aforementioned, RBM15 and RBM15B
interact with the lncRNA XIST A-repeat region, targeting the
methylation complex to lncRNA XIST, and mediating the formation of
m6A in lncRNA XIST (48). M6A sites that are then
identified by YTHDC1, contributing towards expediting the process
of X-chromosome silencing (71).
Additionally, YTHDC1 modulates autophagy by modulating the
stability of nuclear mRNA of sequestosome-1, resulting in the
postponement of wound healing (72).
YTHDC2
The function of YTHDC2 is currently poorly
understood, although it may be instrumental to the process of
spermatogenesis, elevating translation efficiency and decreasing
mRNA abundance. YTHDC2 is enriched in the testes, and
Ythdc2−/− mice have been shown to be infertile
(73). Since YTHDC2 has been
detected in the 40-80S ribosomal fraction, it can be hypothesized
that YTHDC2 may interact with translation initiation elements to
accelerate translation efficiency. YTHDC2 may recruit
nonsense-mediated degradation machineries (including the helicases
UPF1 and MOV10, and the exoribonuclease XRN1) to promote the
degradation of certain mRNAs co- or post-translation (73). Further, in-depth studies are
required, however, to reveal the function of YTHDC2 in
m6A.
HNRNPC
HNRNPC has been implicated in alternative splicing,
mRNA and uridine-rich snRNA (U snRNA) exportation, and the
so-called 'm6A switch' mechanism, in which m6A induces
RNA unfolding and increases the accessibility of HNRNPS to
single-stranded RNA. HNRNPC controls pre-mRNA processing via the
regulation of alternative splicing. The individual-nucleotide
resolution UV-cross-linking and immunoprecipitation method has
revealed that the position of HNRNPC determines its impact on the
inclusion of alternative exons. Specifically, when HNRNPC binds to
exons, alternative exons are silenced; however, when HNRNPC binds
to introns, the inclusion of exons is enhanced (74). The exportation of mRNAs and U
snRNAs has also been shown to be associated with HNRNPC. HNRNPC
functions as a 'molecular ruler' to measure the length of the RNA
polymerase II transcripts, and is involved in the sorting of
transcripts into mRNA or U snRNA, depending on whether or not they
are longer than the threshold length (200-300 nucleotides), leading
to the export of these two different RNA species from the nucleus
(75). Researchers have discovered
that m6A modifies partial structures in mRNAs or
lncRNAs, which exposes buried RBMs and increases the accessibility
of HNRNPC to m6A-RNA, the process that is termed as the
'm6A-switch' (76).
This structural alteration has been shown to contribute to the
metastasis of PC cells, adding a new dimension to the manner in
which m6A regulates gene expression.
HNRNPG
HNRNPG also regulates alternative splicing and the
'structural switch'. HNRNPG depends upon the Arg-Gly-Gly repeat
sequence within its low-complexity region to interact with nascent
m6A-mediated pre-mRNA and the phosphorylated C-terminal
domain of RNA polymerase II, which ultimately regulates alternative
splicing (77). M6A
modification promotes the binding of HNRNPG to target RNAs in
virtue of the 'structural switch' (78).
HNRNPA2B1
HNRNPA2B1 stimulates primary miRNA (or pri-miRNA)
processing, participates in alternative splicing and has an
essential function in vesicular trafficking. DiGeorge syndrome
critical region 8 (DGCR8) serves as the miRNA microprocessor
complex protein, and HNRNPA2B1 interacts with DGCR8 in order to
facilitate pri-miRNA processing. In addition, HNRNPA2B1 is involved
in alternative splicing (79).
HNRNPA2B1 sorts specific miRNAs and lncRNAs into exosomes or
microvesicles, thereby fulfilling a pivotal role in the selection
of cancer-associated miRNAs and lncRNAs, emphasizing the function
of HNRNPA2B1 in cancer-associated vesicular trafficking (80).
IGF2BPs
IGF2BPs participate in the stabilization and storage
of mRNAs under various physiological conditions and during the
stress response. The interaction between the KH3-4 di-domain of
IGF2BPs and the 'GGAC' motif in m6A promotes the
stability and storage of certain mRNAs during normal or stress
conditions (81). In addition,
IGF2BP2 (also known as IMP2) plays a crucial role in human
senescence by recognizing and stabilizing MIS12 mRNA (82).
New readers
Recently, Wu et al (82) reported that proline-rich
coiled-coil 2A was discovered as a new m6A reader that
regulates myelination and oligodendroglial specification.
Furthermore, Fragile X messenger ribonucleoprotein 1 (FMR1) may
function as a sequence-context-dependent reader, negatively
regulating mRNA translation by stalling the process of ribosome
translocation. YTHDF1 and FMR1 competitively bind to m6A
sites on mRNA. The stress granule protein G3BP is an
anti-m6A reader that is repelled by m6A,
enhancing the stabilization of m6A-modified mRNA
(83).
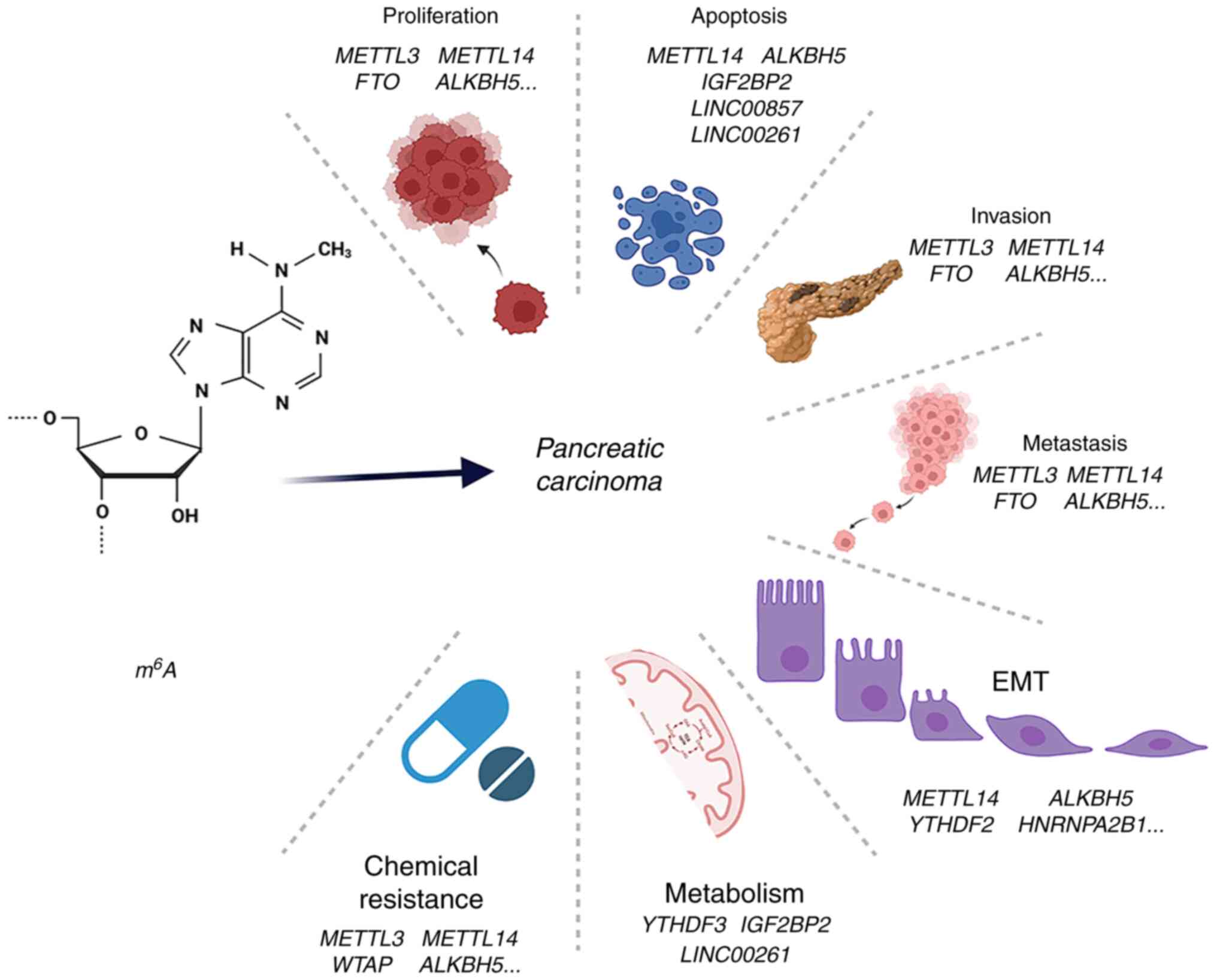
3. Roles of m6A in the
carcinogenesis and development of PC
As aforementioned, m6A is understood to
play a pivotal role in post-transcriptional modification,
modulating protein expression and ultimately affecting the
structure and function of modified proteins in key signaling
pathways. Numerous studies have confirmed that m6A is
implicated in the proliferation, apoptosis, invasion, metastasis,
EMT, metabolism and chemoresistance of PC. m6A
participates in the initialization and progression of PC through
altering the levels of m6A regulators and genetic
variants, and also serves an important role in aberrant alternative
splicing. Consequently, in the subsequent section of the review,
these studies are evaluated to disclose the function of
m6A in tumor onset and development (Fig. 2 and Table II).
 | Table IIRole of m6A in the
initiation and progression of pancreatic cancer. |
Table II
Role of m6A in the
initiation and progression of pancreatic cancer.
| Initiation
factor |
Oncogenicity/suppressor | Signaling or
target | Effect | (Refs.) |
|---|
| METTL3 | Oncogenicity↑ | | Proliferation↑
Metastasis↑
Invasion↑ | (103) |
| |
CSC-METTL3↑-miR-25-3p↑-PHLPP2↓-AKT-p70S6K↑ | Proliferation↑
Invasion↑
Metastasis↑ | (84) |
| | METTL3↑-NUCB1↓ | Proliferation↑
Chemical
Resistance↑ | (85) |
| | | Chemical
Resistance↑ | (112) |
| METTL14 | Oncogenicity↑ | METTL14↑-PERP↑ | Proliferation↑
Apoptosis↓
Invasion↑ Metastasis↑
EMT↑ | (86) |
| |
p65-METTL14↑-CDA↑ | Chemical
Resistance↑ | (113) |
| | METTL14↑-
apoptosis; METTL14↑-mTOR-autophagy↓ |
Apoptosis↓
Chemical Resistance↓ | (114) |
| METTLE 16 | Suppressor↓ | METTL 16↓-P21
pathway↓ | Proliferation↓ | (87) |
| METTL5 | Oncogenicity↑ | METTL5↑-c-myc
translation↑ | Proliferation↑
Invasion↑
Metastasis↑ | (88) |
| WTAP | Oncogenicity↑ | WTAP↑-mRNA↑ |
Metastasis↑
Chemical resistance↑ | (104) |
| FTO | Suppressor↓ |
FTO↓-YTHDF2-PJA2↓-Wnt signaling↑ | Proliferation↓
Invasion↓
Metastasis↓ | (89) |
| FTO | Oncogenicity↑ | | Proliferation↑ | (90) |
| ALKBH5 | Suppressor↓ |
ALKBH5↓-KCNK15-AS1↓ |
Apoptosis↑
Invasion↓Metastasis↓ | (92) |
| |
ALKBH5-↓lncRNA
KCNK15-AS1↓-MDM2-REST ubiquitination↓-PTEN↓-AKT pathway↑ |
Apoptosis↑
invasion↓
metastasis↓
EMT↓ | (93) |
| |
ALKBH5↓-PER1↓-ATM-CHK2-P53/CDC25C↓ | Proliferation↓
Invasion↓
Metastasis↓ | (91) |
| | ALKBH5↓-WIF-1↓-Wnt
signaling↑ |
Proliferation↓
Invasion↓ Metastasis↓
Chemical resistance↓ | (94) |
| |
ALKBH5↓-FBXL5↓-degradation↓ | Apoptosis↑ | (95) |
| YTHDF2 | Oncogenicity↑ | YTHDF2↑-YAP↓ | Proliferation↑
Invasion↓
Metastasis↓ EMT↓ | (105) |
| YTHDF3 | Oncogenicity↑ | YTHDF3↑-lncRNA
DICER1-AS1↓-DICER1↓-maturation of miR-5586-5p↓ |
Proliferation↑
Metastasis↑
Metabolism | (110) |
| HNRNPA2B1 | Oncogenicity↑ |
HNRNPA2B1-ERK/snail-E-cadherin↓N-cadherin
and vimentin↑ | Invasion↑ EMT↑ | (106) |
| IGF2BP2 | Oncogenicity↑ |
IGF2BP2↑-DANCR↑ | Proliferation↑ | (96) |
| | IGF2BP2↑-mRNA
GLUT1↑ | Proliferation↑
Metabolism | (111) |
| | IGF2BP2↑-
protein
IMP2↑ |
Apoptosis↑
Metastasis↑ EMT↑ | (107) |
| LINC00857 | Oncogenicity↑ |
LINC00857↑-miR-150-5p↓-E2F3↑ |
Proliferation↑
Apoptosis↓ | (97) |
| LINC00261 | Suppressor↓ |
LINC00261↓-IGF2BP1↑-c-Myc↑ |
Proliferation↑
Apoptosis↓ Metabolism | (98) |
| |
LINC00261↓-miR-222-3p/HIPK2/ |
Proliferation↑
Apoptosis↓ Metabolism | (98) |
| | ERK↑-c-myc↑ | | |
| rs7495 genetic
variant | | hnRNPC↑ | Proliferation↑ | (100) |
| | rs7495G
allele-has-miR- | Proliferation↑ | (99) |
| | 183-3p-hnRNPC↑ | | |
| rs142933486 genetic
variant | | PIK3CB↑ |
Proliferation↑
Metastasis↑ | (101) |
| CLK1↑ | | METTL14exon
10skipping↓ | Metastasis↑ | (108) |
| | Cyclin
L2exon6.3skipping↑ | Proliferation↑ | (108) |
| HNRNPC | Structural
switch | TAF8L↓ | Metastasis↑ | (109) |
| | TAF8S↑ | Metastasis↑ | (109) |
Proliferation and apoptosis
By maintaining proliferation and evading subsequent
apoptotic processes, m6A effectively stimulates the
progression of PC. Recently, there has been a growing understanding
that m6A modulates the proliferation and apoptosis of PC
cells via alterations in the levels of both m6A
regulators and lncRNAs, and genetic variants.
A large number of studies have focused on the
upregulation and oncogenic functions of METTL3 in PC. For example,
cigarette smoke condensate (CSC) has been shown to induce the
hypomethylation of the METTL3 promoter, which contributes to the
overexpression of METTL3. Enhanced m6A modifications
caused by elevated levels of METTL3 lead to excessive pri-miR-25
maturation. PH domain leucine-rich repeat protein phosphatase 2
(PHLPP2) is suppressed by miR-25-3p, which stimulates AKT-p70S6K
signaling and ultimately promotes the proliferation of PC cells
(84). In addition, the
overexpression of nucleobindin 1 (NUCB1; a calcium-binding protein)
has been shown to suppress cell proliferation, whereas METTL3
downregulates the expression of NUCB1 in a YTHDF2-dependent manner
(85). Taken together, these
studies reveal that METTL3 is implicated in promoting oncogenic
pri-miR-25 maturation and suppressing expression of the tumor
suppressor NUCB1 to exerts its oncogenic effects.
Reverse transcription-PCR and western blot analyses
have been used to reveal that, compared with adjacent normal tissue
samples, METTL14 expression is upregulated in PC samples. The
protein p53 effector related to PMP-22 (PERP) functions as a p53
effector, which regulates apoptosis induced by DNA damage. The
upregulation of METTL14 leads to an elevation in the m6A
level, which causes a decrease in the expression of PERP, promoting
cell growth and inhibiting apoptosis (86). Hence, METTL14 has been regarded as
an oncogenic regulator in PC. In addition to METTL3-METTL14, other
methylation enzymes are also involved in the modulation of PC
proliferation. METTL16 has been shown to suppress the proliferation
of PC cells through the P21 pathway (87). METTL5 specifically catalyzes the
methylation of A1832 of 18S rRNA, which is a crucial point in the
decoding mechanism, indicating its importance as a regulatory
protein in translation. METTL5 has been demonstrated to function as
an oncogene that promotes the proliferation of PC cells by
increasing c-Myc translation (88).
As regards the functions of demethylation enzymes,
the impact of FTO on the proliferation of PC cells remains
controversial. Zeng et al (89) identified that FTO was a tumor
suppressor protein. Reduced FTO expression led to the upregulation
of the m6A level of praja ringfinger ubiquitin ligase 2
(PJA2), and an enhanced rate of PJA2 degradation was shown to be
dependent on YTHDF2, which promoted Wnt signaling, ultimately
facilitating the proliferation of PC cells. By contrast, Tang et
al (90) demonstrated that,
when overexpressed, FTO functions as as oncogenic protein. The
knockdown of FTO resulted in compromised levels of cell
proliferation. The effects that are actually elicited by FTO in PC
remain an interesting topic that warrants further investigation.
Differently from FTO, another demethylation enzyme, ALKBH5, has
over time been shown to function as a tumor suppressor (91-95),
often involved in inhibiting the proliferation of PC. ALKBH5
demethylates period circadian regulator 1 (PER1), and it functions
in concert with YTHDF2 to activate PER1. The overexpression of PER1
activates the ATM-CHK2-P53/CDC25C signaling pathway, which leads to
the inhibition of tumor proliferation. p53 activates the
transcription of ALKBH5, which subsequently functions in a feedback
loop to regulate the m6A level (91). Furthermore, ALKBH5 has been shown
to exert an anticancer effect against PC by targeting regulators of
ferroptosis. Mechanistically, the ubiquitin ligase FBXL5 mediates
the degradation of PC cells via ferroptosis. A previous study
demonstrated that not only was ALKBH5 responsible for enhancing the
stability of FBXL5, but it also led to upregulation of the
expression of FBXL5 and the mitochondrial iron importer SLC25A28,
and alternative splicing of SLC25A37 was also increased (95).
Although numerous studies have focused on
alterations in the level of m6A regulators with respect
to their role as initiation factors in the proliferation and
apoptosis of PC, to date, relatively few studies have been
performed to assess the mechanisms through which m6A
functions in lncRNAs. Nevertheless, there is sufficient evidence to
suggest that lncRNAs also function as initiation factors in the
proliferation and apoptosis of PC. First, the lncRNA DANCR was
shown to promote the tumorigenesis and proliferation of PC; IGF2BP2
recognizes m6A modified at A664 of DANCR, which plays
the role of stabilizing DANCR (96). Secondly, LINC00857 has been shown
to be overexpressed, thereby promoting the tumorigenesis of PC in
an m6A-mediated manner (97). The deletion of LINC00857 clearly
hinders cell proliferation and facilitates cell apoptosis.
M6A is pre-eminently enriched within LINC00857, which
enhances its RNA stability, thereby suggesting a role for LINC00857
in modulating the miR-150-5p/E2F3 signaling axis. As a competitive
endogenous RNA, LINC00857 exerts its effects through sponging
miR-150-5p and upregulating the expression of E2F3 (97). Finally, LINC00261 has been reported
to function as a tumor suppressor that inhibits proliferation, and
accelerates the rate of apoptosis of PC tissues and cell lines. The
EZH2-mediated histone H3K27 trimethylation and DNA hypermethylation
both serve to downregulate LINC00261. Mechanistically, on the one
hand, the downregulation of LINC00261 leads to the upregulation of
the experession of c-Myc by binding to miR-222-3p, thereby
activating the HIPK2/ERK/c-Myc signaling pathway. On the other
hand, LINC00261 negatively regulates the expression of IGF2BP1,
where the overexpression of IGF2BP1 has been shown to enhance c-Myc
stability (98). From this
example, it may be noted that there are links among m6A,
DNA modification and histone modification.
Genetic variants, as another type of initiation
factor, may provide novel insight into the association between
m6A regulators and PC. For example, the genetic variant
rs7495 in the 3′-UTR region of hnRNPC has been shown to disrupt a
presumptive binding site for has-miR-183-3p that promotes the
expression of hnRNPC, and which contributes to the susceptibility
of PC (99). In another study,
cell viability assay indicated that the depletion of hnRNPC
hindered the proliferation of PC (100). Similarly, the rs142933486 variant
in the phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic
subunit beta (PIK3CB) has been observed to have decreased
m6A levels, and its expression is increased in a
YTHDF2-dependent manner. In addition, the variant of PIK3Cb[T] has
been shown to activate AKT signaling, accelerating the
proliferation of phosphatase and tensin homolog (PTEN)-deficient PC
cells (101).
Generally, it can be seen that alterations in the
levels of m6A regulators and lncRNAs, together with
genetic variants, are intimately involved in the proliferation and
apoptosis of PC.
Invasion, metastasis and EMT
As is currently understood, PC is characterized by
a high metastatic rate: 40-50% of patients present with distant
metastasis in their first hospital visit. Metastasis is a complex
process that comprises EMT, the migration of cancerous cells, their
infiltration into adjacent tissues, vascular transportation and
settlement in distant organs. EMT is defined as the transformation
from polar epithelial cells into interstitial cells under specific
physiological and pathological conditions. The process is mainly
characterized by the loss of epithelial phenotypic molecules (such
as E-cadherin) and the acquisition of interstitial cell
characteristics (such as vimentin), which enables tumor cells to
lose their intercellular adhesive properties and become more loose,
thereby gaining the abilities of invasion and metastasis (102). M6A regulates the
invasion, metastasis and EMT of PC through alterations in the
levels of m6A regulators, genetic variants and aberrant
alternative splicing.
In addition to sustaining proliferation, METTL3
promotes the invasion and migration of PC cells (103). The
CSC/METTL3/miR-25-3p/PHLPP2/AKT/p70S6K signaling axis has been
shown to accelerate invasion and metastasis (84). Furthermore, PERP serves as a
tetraspan plasma membrane protein, having multifaceted roles in EMT
and cell-cell adhesion. Of note, the tumor suppressor function of
PERP may involve inhibition of the invasion, metastasis and EMT of
PC cells. METTL14 downregulates the expression of PERP through the
METTL14/YTHDF2/PERP pathway (86).
Furthermore, METTL5 promotes the invasion and migration of PC
cells. METTL5 functions as an oncogenic regulator by increasing the
translation of c-Myc, whereas the knockdown of c-Myc has been shown
to abolish the oncogenic effects induced by the upregulation of
METTL5 (88). In addition, both
nuclear and cytoplasmic WTAP have been shown to be upregulated,
promoting metastasis by modulating the stability of downstream mRNA
(104).
Studies on the function of FTO in PC metastasis are
relatively limited, although one study documented that FTO
facilitates the invasion and metastasis of PC cells via the
FTO/YTHDF2/PJA2/Wnt signaling axis (89). Moreover, lncRNA KCNK15-AS1 inhibits
the invasion and migration of PC. ALKBH5 is responsible for the
demethylation and upregulation of KCNK15-AS1. In PC, ALKBH5 is
downregulated, and the expression of KCNK15-AS1, lying downstream
in the pathway, is subsequently also downregulated (92). He et al (92) also discovered that the
overexpression of KCNK15-AS1 suppresssed EMT and cell migration in
PC. Mechanistically, KCNK15-AS1 was shown to recruit the MDM2
proto-oncogene to promote the ubiquitination of RE1 silencing
transcription factor (REST), thereby inactivating the AKT pathway
via the transcriptional upregulation of PTEN (93). Furthermore, the upregulation of
ALKBH5 has been shown to inhibit tumoral invasive and migrative
activities in vitro, whereas the depletion of ALKBH5
promotes tumor progression. ALKBH5 demethylates and regulates PER1
in an m6A-dependent manner. PER1 itself has the role of
reactivating the ATM/CHK2/P53/CDC25C signaling axis, which inhibits
the processes of cell invasion and migration (91).
Additionally, the upregulation of YTHDF2 in PC
tissues facilitates cell proliferation, and suppresses invasion and
migration, a phenomenon that has been defined as the
'migration-proliferation dichotomy'. Moreover, deficiencies in
YTHDF2 have been shown to promote EMT, probably via the
upregulation of Yes-associated protein 1, and not via TGF-β/Smad
signaling in PC cells (105).
Furthermore, HNRNPA2B1 has been shown to influence EMT progression
in PC via the ERK/Snail signaling pathway. Both in vitro and
in vivo, HNRNPA2B1 downregulates E-cadherin, and upregulates
N-cadherin and vimentin to enhance EMT, thereby promoting the
invasion of PC cell lines (106).
In addition, the IGF2BP2 binding protein, IMP2, has been shown to
be upregulated in PC, particularly upon the treatment of Panc-1 PC
cells with TGF-β, where the induction of EMT indicates its
oncogenic effectivness (107).
Apart from alterations in m6A
regulators, the PIK3Cb[T] genetic variant has been shown to promote
metastasis of PTEN-deficient PC cells (101). Alternative splicing has also been
identified as a novel means through which m6A may
modulate gene expression. For example, Cdc2-like kinase 1 (CLK1) is
an alternative splicing-associated gene that is significantly
upregulated in PC tissues. An increased expression level of CLK1
has been shown to enhance phosphorylation on SR-like splicing
factor 5250-Ser (SRSF5250-Ser), which subsequently suppresses the
exon skipping of METTL14exon10, but facilitates the exon
skipping of cyclin L2exoN6.3. The alterative
skipping of METTL14exon 10 increases the adundance of
m6A and promotes metastasis. Alterative cyclin
L2exoN6.3 skipping promotes proliferation
(108). Moreover, HNRNPC
regulates alternative splicing via the 'm6A switch' to
accelerate the invasion and liver metastasis of PC. HNRNPC inhibits
the anti-metastatic isoform of TAF8 (TAF8L), but promotes the
pro-metastatic isoform of TAF8 (TAF8S). Mechanistically,
m6A increases the likelihood of pre-mRNA of TAF8 to form
a linear structure, which increases the accessibility of HNRNPC to
TAF8, and upregulates the expression of the pro-metastatic isoform,
TAF8S (109).
Above all, various studies, as aformentioned, have
described the crucial function of m6A in the invasion,
metastasis and EMT of PC, and these findings may provide novel
prospects for m6A therapy in the future.
Metabolism
The initiation and progression of carcinoma
requires the reprogramming of metabolism. Tumor cells automatically
transform their flux via diverse metabolic pathways to accommodate
an elevated bioenergetic and biosynthetic requirement. The
influence of m6A in altering metabolism has proven to be
a rich avenue for scientific exploration. Scientists have presented
several mechanisms to explain the mechanisms through which
m6A may promote metabolism transformation and tumor
progression.
For example, bioinformatics analysis has revealed
that DICER1 antisense RNA 1 (DICER1-AS1) is overexpressed in PC.
DICER1-AS1 recruits the transcription factor YY1 to the promoter of
DICER1 in order to expedite the transcription of DICER1. The
maturation of miR-5586-5p is promoted by DICER1 to inhibit the
expression of glycolytic genes, including LDHA, HK2, PGK1 and
SLC2A1. YTHDF3 forms a negative feedback loop with DICER1-AS1 to
facilitate glycolysis. Moreover, the upregulation of DICER1-AS1 has
been shown to inhibit the proliferation and metastasis of PC
(110).
In addition, in order to meet the enhanced
metabolic requirements, tumor cells are often observed to
upregulate the expression of glucose transporters (GLUTs), such as
GLUT1, to augment glucose uptake. IGF2BP2 overexpressed in PC has
been shown to increase the rate of aerobic glycolysis and PC cell
proliferation by stabilizing GLUT1 mRNA (111). Similarly, the epigenetic
silencing of LINC00261 has been found to modulate the
miR-222-3p/HIPK2/ERK signaling axis, and IGF2BP1 has been shown to
promote c-Myc-mediated aerobic glycolysis (98).
Taken together, the studies published to date in
this area have demonstrated that m6A plays a role of
paramount importance as a regulator of metabolic alterations, and
this function appears to play a crucial role in PC.
Chemoresistance
PC is characterized by a highly aggressive
progression: Only 15-20% of patients with PC are identified at an
early stage, whereas the majority of them (75-80%) exhibit either
locally advanced or distant metastasis, and should receive
chemotherapy as a first-line treatment. However, in spite of this,
numerous patients fail to benefit from the all the benefits
potentially afforded by the therapy due to rapid drug resistance
induced by the chemotherapeutic agents. Chemoresistance is a major
impediment in terms of the efficacy of therapeutic drugs.
A previous study revealed that the overexpression
of NUCB1 activated the antitumor effects of gemcitabine (GEM) by
inhibiting the GEM-induced unfolded protein response (UPR) and
autophagy; in PC, METTL3-YTHDF2 downregulated NUCB1 (85). The knockdown of METTL3 has been
shown to sensitize PC cells to irradiation and antitumor drugs
(namely, 5-fluorouracil, GEM and cisplatin). METTL3 targets various
pivotal pathways, including the ubiquitin-dependent pathway and the
MAPK signaling cascades. Alterations in these pathways trigger
aberrant biological behaviors, which may lead to chemotherapeutic
and radiation resistance (112).
Moreover, overexpression of METTL14 is associated
with resistance to GEM in PC. Cytidine deaminase (CDA) functions as
an enzyme which induces resistance to GEM by inactivating GEM.
METTL14 was overexpressed in PC via its promoter region binding to
the transcriptional factor p65, thereby increasing the expression
of CDA (113). By contrast, the
upregulation of METTL14 enhances apoptosis to sensitize PC cells to
cisplatin, and also promotes autophagy via an mTOR-dependent
signaling pathway (114). WTAP
has been identified as an oncogenic regulator, which modulate the
stability of downstream mRNA, thereby promoting chemoresistance to
GEM (104).
Furthermore, ALKBH5 has been shown to be
downregulated in a patient-derived xenograft model treated with
GEM. ALKBH5 upregulates the expression of Wnt inhibitory factor-1,
which subsequently suppresses the Wnt pathway. Wnt signaling boosts
the resistance of PC cells to GEM. The increased expression of
ALKBH5 has also been shown to sensitize PC cells to chemotherapy
(94). Taken together, the
upregulation of METTL3, METTL14 and WTAP, and the downregulation of
ALKBH5 have been shown to lead to resistance to GEM. On the other
hand, another study indicated that increased levels of METTL14
sensitized PC cells to cisplatin, and this apparent contradiction
is worthy of further exploration in future studies.
Although the various capabilities and properties of
m6A have been discussed separately in this review, it
should be noted that the functional roles of m6A in
terms of understanding the occurrence and progression of PC are all
intrinsically associated, and the process cannot be comprehended by
considering the individual capabilities of m6A entirely
in isolation.
4. Possible clinical applications of
m6A in PC
m6A is closely associated with every
stage in the development of PC, and is therefore predicted to be of
utmost importance in planning future diagnostic and therapeutic
directions for PC. However, to date, a large number of studies have
concentrated on the fundamental biological and physiological
aspects of m6A at the laboratory level, and few studies
have focused on translating these findings into clinical practice.
Thus, the final section of the present review summarizes the
possible clinical applications of m6A in PC, in an
attempt to broaden the current understanding of this
post-transcriptional modification (Table III).
 | Table IIIRole of m6A in the
pathological stage and poor prognosis of pancreatic cancer. |
Table III
Role of m6A in the
pathological stage and poor prognosis of pancreatic cancer.
| Initiation
factor |
Oncogenicity/suppressor | Signaling or
target | Effect | (Refs.) |
|---|
| METTL3 | Oncogenicity↑ | | Pathological
stage
Poor prognosis↑ | (103) |
| |
CSC-METTL3↑-miR-25-3p↑-PHLPP2↓-AKT-p70S6K↑ | Poor
prognosis↑ | (84) |
| WTAP | Oncogenicity↑ | WTAP↑- mRNA↑ | Poor
prognosis↑ | (104) |
| FTO | Suppressor↓ |
FTO↓-YTHDF2-PJA2↓-Wnt signaling↑ | Pathological
stage
Poor prognosis↓ | (89) |
| ALKBH5 | Suppressor↓ |
ALKBH5↓-PER1↓-ATM-CHK2-P53/CDC25C↓ | Poor
prognosis↓ | (91) |
| | ALKBH5↓-WIF-1↓-Wnt
signaling↑ | Poor
prognosis↓ | (94) |
| |
ALKBH5↓-FBXL5↓-degradation↓ | Poor
prognosis↓ | (95) |
| YTHDF2 | Oncogenicity↑ | YTHDF2↑-YAP↓ | Pathological
stage
Poor prognosis↑ | (105) |
| IGF2BP2 | Oncogenicity↑ |
IGF2BP2↑-DANCR↑ | Poor
prognosis↑ | (96) |
| | IGF2BP2↑-mRNA
GLUT1↑ | Poor
prognosis↑ | (111) |
| | IGF2BP2↑-protein
IMP2↑ | Poor
prognosis↑ | (107) |
| LINC00857 | Oncogenicity↑ |
LINC00857↑-miR-150-5p↓-E2F3↑ | Poor
prognosis↑ | (97) |
| CLK1↑□ |
Phosphorylationon
SRSF5250-Ser↑ | METTL14exon
10skipping↓ | Pathological
stage
Poor prognosis↑ | (108) |
| | Cyclin
L2exon6.3skipping↑ | | (108) |
| Structural | HNRNPC | TAF8L↓ | Pathological
stage | (109) |
| switch | | TAF8S↑ | Poor
prognosis↑ | (109) |
Pathological stage and the poor
prognosis
Different levels of m6A regulators
represent different tumor, node, metastasis (TNM) stages, and
therefore different prognoses of PC. Higher levels of expression of
the methylase METTL3 are associated with a higher pathological
stage (P=0.02) and a higher N stage (P=0.02) (103). METTL3 promotes pri-miR-25
maturation. Excessive levels of miR-25-3p have been detected in PC
tissues and smokers, and are associated with a worse prognosis
(84). Taken together, it has been
demonstrated that the survival rates are shorter in patients with a
high level of METTL3 expression (84,103). Furthermore, univariate analysis
has demonstrated that the overexpression of nuclear WTAP is
associated with a later N stage and poor overall survival
(P<0.001) (104).
The decreased expression of the demethylase FTO is
indicative of advanced carcinoma stages and the nodal metastasis
status. The median survival rate has bene found to be significantly
shorter within the low-level FTO expression group (16.90 vs. 48.67
months; P=0.0474), with a high level of LINC00857. In brief,
patients with decreased levels of FTO have poor survival outcomes
(89). In addition, survival
analysis has revealed that the lower the level of ALKBH5
expression, the shorter the overall survival rate. Reduced ALKBH5
levels have been shown to indicate the occurrence of PC, and poor
clinicopathological manifestations (91,94,95).
Emerging evidence suggests that readers, such as
YTHDF2 and IGF2BP2, are of vital importance for the prognosis of
PC. The overexpression of YTHDF2 in PC tissues is associated with
worse tumor stages (105).
Moreover, IGF2BP2 is characterized as an unfavorable prognostic
marker for PC. The overexpression of IGF2BP2 has been shown to be
closely associated with a poor overall survival and disease-free
survival rate (96,107,111).
As demonstrated in a previous study, the
upregulation of LINC00857 activates the miR-150-5p/E2F3 signaling
axis to facilitate the tumorigenesis of PC. In that previous study,
the overall survival rate [hazard ratio (HR), 1.6; P=0.034] and
disease-free survival rate (HR, 1.9; P=0.0046) in the
high-expression LINC00857 group were found to be significantly
lower (97).
The CLK1/SRSF5 signaling pathway promotes the
abnormal exon skipping of METTL14 and cyclin L2 to facilitate the
growth and metastasis of PC. In a previous study, univariate
analysis suggested that the overexpression of CLK1 was
representative of a severe TNM stage, lymphatic metastasis and
tumor size (108). An increased
CLK1 expression was also shown to be associated with a lower
overall survival rate (108).
HNRNPC impedes anti-metastatic alternative splicing events in an
m6A-dependent manner. The level of HNRNPC has been found
to be markedly higher in hepatic metastatic tissues compared with
non-metastatic tissues (P<0.01) (109). Kaplan-Meier curves were
previously employed to demonstrated that the overall survival rate
was significantly lower in the HNRNPC overexpression group (both
log-rank P<0.05). As aforementioned, the hepatic metastasis rate
was confirmed to be significantly higher in the HNRNPC
overexpression group (log-rank P=0.008) (109).
However, although there is a significant difference
in the abundance of m6A regulators between PC and normal
tissues, m6A regulators has not been widely utilized as
a diagnostic marker. However, it is considered that, with the
anticipated continued improvement of mapping methods,
m6A regulators will be able to precisely predict the
occurrence, TNM stage and prognosis of PC in the not-too-distant
future.
M6A inhibitors as potential
therapeutics in PC
A wealth of studies has confirmed the complex
functions and molecular mechanisms of m6A in PC.
Targeting m6A regulators as a means of therapeutic
invention therefore provides a promising prospect for the treatment
of cancer (Table IV).
 | Table IVN6 methyladenosine
inhibitors. |
Table IV
N6 methyladenosine
inhibitors.
| Target | Drug | Cancer type | (Refs.) |
|---|
| FTO | Rhein | Breast cancer | (115,116) |
| MA | Glioblastoma
stem | (117,118) |
| FB23/FB23-2 | Acute myeloid
leukemia | (119) |
| R-2HG | Leukemia cell | (120) |
| METTL3 | Adenosine (1-8) | | (121) |
| UZH1a, UZH2 and
UZH1a analogue | | (121) |
| STM2457 | Acute myeloid
leukemia | (121-123) |
| METTL14 | SPI1 | Acute myeloid
leukemia | (124) |
| WTAP | CA4 | Colon cancer | (125) |
| DNA double-strand
breaks | Fisetin | Pancreatic
cancer | (126) |
FTO inhibitors can be divided into selective or
non-selective types. Rhein is a natural product that was
demonstrated to be the first FTO inhibitor possessing cell activity
(115). Rhein has been shown to
restrict the occurrence of breast cancer cells (116). Meclofenamic acid is an example of
a selective FTO inhibitor (117),
which restrains the growth and self-renewal of glioblastoma stem
cells (118). FB23 and its
derivative (FB23-2) inhibit the proliferation, and increase the
rate of apoptosis and differentiation of acute myeloid leukemia
(AML) cells (119). Finally,
R-2-hydroxyglutarate has been shown to exert an anti-leukemic
effect by suppressing cell proliferation/viability, and enhancing
the apoptosis and cell-cycle arrest of leukemia cells (120).
METTL3 is a crucial catalytic subunit, which has
attracted extensive attention among all regulators. METTL3
inhibitors can be separated into nucleosides and non-nucleosides.
Inhibitors in both of these categories have been shown to function
as competitive substrate inhibitors of SAM. High-throughput
analysis identified adenosine (1-8) as
small-molecule METTL3 inhibitors which occupy the binding site of
SAM. The disadvantages of these adenosine (1-8)
inhibitors, however, are poor selectivity and poor cellular
permeability properties. The discovery of the METTL3 non-nucleoside
inhibitor UZH1a, UZH2 and UZH1a analogues, JMC-1, JMC-5, JMC-8 and
JMC-10, promoted the development of METTL3 inhibitors as
therapeutic targets (121). The
non-nucleoside METTL3 inhibitor, STM2457, has also been shown to be
effective against AML without exerting any significant effect on
normal hematopoiesis (121-123).
SPI1 has been shown to inhibit METTL14 expression
directly, and serves as a possible AML therapeutic target (124). Additionally, carbonic anhydrase
IV has been found to suppress the tumorigenesis of colon cancer by
inhibiting the WTAP-WT1-TBL1 pathway (125).
Currently, research regarding m6A
inhibitors in PC is still at its infancy. DNA damage repair
represents a prominent obstacle for evaluating the chemotherapy
efficacy of PC. The knockdown of PHF10 has been shown to result in
the elevated recruitment of γ-H2AX, RAD51 and 53BP1 to DSB sites,
and decreased HR repair efficiency. Fisetin induces DSBs, and
suppresses HR repair of DNA through impeding the ZC3H13-mediated
m6A regulation of PHF10. Fisetin treatment has also
provided insight into novel therapeutic strategies for PC (126).
Taken together, m6A inhibitors present a
frontier of research that may provide a novel direction in
m6A-targeted drug therapeutics. However, numerous
challenges lie ahead. The clinical application of m6A
inhibitors in PC remains insufficient at present, and the efficacy
and adverse reactions of the prospective inhibitors requires
further verification.
5. Conclusions and future perspectives
In the present review, the biological functions of
m6A were introduced, with a focus on its influence in
PC. There are, however, certain divergences and discrepancies that
have not been fully explored herein. Based on these unresolved
discrepancies, the authors' prediction is that the future
exploration of m6A will give attention to the following
topics. First, it is possible that certain m6A writers,
erasers and readers have not yet to be identified; therefore,
reforming m6A-mapping methods will be necessary to
promote the discovery of novel m6A regulators. Secondly,
FTO remains controversial with respect to whether it actually
targets m6A as a substrate, and what its precise role is
in the proliferation of PC. Thirdly, the question remains as to
whether erasers always function antagonistically with writers. For
example, if METTL3-METTL4 serves as an oncogene, FTO may function
as suppressor, although this would not be in agreement with the
experimental results described above. The detailed mechanisms that
account for how the actions of different regulators are coordinated
in the same tumor, and also how the same regulator may function
heterogeneously in distinct type of carcinomas, need to be further
elucidated. The current knowledge of m6A regulatory
networks only represent the tip of an iceberg, and the real
situation is far more complex than what has already been
discovered. Fourthly, the mechanisms through which m6A
functions in concert with DNA and histone modification, and whether
there are underlying connections between them, remain unclear.
Finally, the exploration of m6A inhibitors in PC is only
at the rudimentary experimental stages at present. An extensive
literature detailing their application in clinical trials does not
yet exist. Efficacious and safe m6A inhibitors, however,
do need to be developed and put forth into clinical practice.
Availability of data and materials
Not applicable.
Authors' contributions
GZ conceived the study. TY wrote the manuscript and
performed the literature search with assistance from JW, HZ and PL,
and GZ supervised the whole process. All authors have read and
approved the final manuscript. Data authentication is not
applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant no. 82070575).
References
|
1
|
Mizrahi JD, Surana R, Valle JW and Shroff
RT: Pancreatic cancer. Lancet. 395:2008–2020. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Martin-Perez E, Domínguez-Muñoz JE,
Botella-Romero F, Cerezo L, Matute Teresa F, Serrano T and Vera R:
Multidisciplinary consensus statement on the clinical management of
patients with pancreatic cancer. Clin Transl Oncol. 22:1963–1975.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zaccara S, Ries RJ and Jaffrey SR:
Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell
Biol. 20:608–624. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fu Y, Dominissini D, Rechavi G and He C:
Gene expression regulation mediated through reversible
m6A RNA methylation. Nat Rev Genet. 15:293–306. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhou Z, Lv J, Yu H, Han J, Yang X, Feng D,
Wu Q, Yuan B, Lu Q and Yang H: Mechanism of RNA modification
N6-methyladenosine in human cancer. Mol Cancer. 19:1042020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
He L, Li H, Wu A, Peng Y, Shu G and Yin G:
Functions of N6-methyladenosine and its role in cancer. Mol Cancer.
18:1762019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Frye M, Harada BT, Behm M and He C: RNA
modifications modulate gene expression during development. Science.
361:1346–1349. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sun T, Wu R and Ming L: The role of m6A
RNA methylation in cancer. Biomed Pharmacother. 112:1086132019.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen XY, Zhang J and Zhu JS: The role of
m6A RNA methylation in human cancer. Mol Cancer.
18:1032019. View Article : Google Scholar
|
|
10
|
Lan Q, Liu PY, Haase J, Bell JL,
Huttelmaier S and Liu T: The critical role of RNA m6A
methylation in cancer. Cancer Res. 79:1285–1292. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wei CM, Gershowitz A and Moss B:
Methylated nucleotides block 5′ terminus of HeLa cell messenger
RNA. Cell. 4:379–386. 1975. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rottman F, Shatkin AJ and Perry RP:
Sequences containing methylated nucleotides at the 5′ termini of
messenger RNAs: Possible implications for processing. Cell.
3:197–199. 1974. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Desrosiers R, Friderici K and Rottman F:
Identification of methylated nucleosides in messenger RNA from
Novikoff hepatoma cells. Proc Natl Acad Sci USA. 71:3971–3975.
1974. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Adams JM and Cory S: Modified nucleosides
and bizarre 5′-termini in mouse myeloma mRNA. Nature. 255:28–33.
1975. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schäfer KP: RNA synthesis and processing
reactions in a subcellular system from mouse L cells. Hoppe Seylers
Z Physiol Chem. 363:33–43. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bokar JA, Shambaugh ME, Polayes D, Matera
AG and Rottman FM: Purification and cDNA cloning of the
AdoMet-binding subunit of the human mRNA
(N6-adenosine)-methyltransferase. RNA. 3:1233–1247. 1997.PubMed/NCBI
|
|
17
|
Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang
Y, Yi C, Lindahl T, Pan T, Yang YG and He C: N6-methyladenosine in
nuclear RNA is a major substrate of the obesity-associated FTO. Nat
Chem Biol. 7:885–887. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Meyer KD, Saletore Y, Zumbo P, Elemento O,
Mason CE and Jaffrey SR: Comprehensive analysis of mRNA methylation
reveals enrichment in 3′ UTRs and near stop codons. Cell.
149:1635–1646. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schibler U, Kelley DE and Perry RP:
Comparison of methylated sequences in messenger RNA and
heterogeneous nuclear RNA from mouse L cells. J Mol Biol.
115:695–714. 1977. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rottman FM, Desrosiers RC and Friderici K:
Nucleotide methylation patterns in eukaryotic mRNA. Prog Nucleic
Acid Res Mol Biol. 19:21–38. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wei CM and Moss B: Nucleotide sequences at
the N6-methyladenosine sites of HeLa cell messenger ribonucleic
acid. Biochemistry. 16:1672–1676. 1977. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Krug RM: Influenza viral mRNA contains
internal N6-methyladenosine and 5′-terminal 7-methyl-guanosine in
cap structures. J Virol. 20:45–53. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Beemon K and Keith J: Localization of
N6-methyladenosine in the rous sarcoma viru genome. J Mol Bid.
113:165–179. 1977. View Article : Google Scholar
|
|
24
|
Dominissini D, Moshitch-Moshkovitz S,
Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K,
Jacob-Hirsch J, Amariglio N, Kupiec M, et al: Topology of the human
and mouse m6A RNA methylomes revealed by m6A-seq. Nature.
485:201–206. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schwartz S, Mumbach MR, Jovanovic M, Wang
T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N,
Cacchiarelli D, et al: Perturbation of m6A writers reveals two
distinct classes of mRNA methylation at internal and 5′ sites. Cell
Rep. 8:284–296. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Geula S, Moshitch-Moshkovitz S,
Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V,
Peer E, Mor N, Manor YS, et al: Stem cells. m6A mRNA methylation
facilitates resolution of naïve pluripotency toward
differentiation. Science. 347:1002–1006. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ma H, Wang X, Cai J, Dai Q, Natchiar SK,
Lv R, Chen K, Lu Z, Chen H, Shi YG, et al:
N6-methyladenosine methyltransferase ZCCHC4 mediates
ribosomal RNA methylation. Nat Chem Biol. 15:88–94. 2019.
View Article : Google Scholar
|
|
28
|
van Tran N, Ernst FGM, Hawley BR, Zorbas
C, Ulryck N, Hackert P, Bohnsack KE, Bohnsack MT, Jaffrey SR,
Graille M and Lafontaine DLJ: The human 18S rRNA m6A
methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids
Res. 47:7719–7733. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ueda Y, Ooshio I, Fusamae Y, Kitae K,
Kawaguchi M, Jingushi K, Hase H, Harada K, Hirata K and Tsujikawa
K: AlkB homolog 3-mediated tRNA demethylation promotes protein
synthesis in cancer cells. Sci Rep. 7:422712017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Luo H, Zhu G, Xu J, Lai Q, Yan B, Guo Y,
Fung TK, Zeisig BB, Cui Y, Zha J, et al: HOTTIP lncRNA promotes
hematopoietic stem cell self-renewal leading to AML-like disease in
mice. Cancer Cell. 36:645–659.e8. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zheng ZQ, Li ZX, Zhou GQ, Lin L, Zhang LL,
Lv JW, Huang XD, Liu RQ, Chen F, He XJ, et al: Long noncoding RNA
FAM225A promotes nasopharyngeal carcinoma tumorigenesis and
metastasis by acting as ceRNA to sponge miR-590-3p/miR-1275 and
upregulate ITGB3. Cancer Res. 79:4612–4626. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cesarini V, Silvestris DA, Tassinari V,
Tomaselli S, Alon S, Eisenberg E, Locatelli F and Gallo A:
ADAR2/miR-589-3p axis controls glioblastoma cell
migration/invasion. Nucleic Acids Res. 46:2045–2059. 2018.
View Article : Google Scholar :
|
|
33
|
Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu
HC, Yuan WB, Lu JC, Zhou ZJ, Lu Q, et al: METTL3 promote tumor
proliferation of bladder cancer by accelerating pri-miR221/222
maturation in m6A-dependent manner. Mol Cancer. 18:1102019.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li J, Huang C, Zou Y, Ye J, Yu J and Gui
Y: CircTLK1 promotes the proliferation and metastasis of renal cell
carcinoma by sponging miR-136-5p. Mol Cancer. 19:1032020.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pendleton KE, Chen B, Liu K, Hunter OV,
Xie Y, Tu BP and Conrad NK: The U6 snRNA m6A
methyltransferase METTL16 regulates SAM synthetase intron
retention. Cell. 169:824–835.e14. 2017. View Article : Google Scholar
|
|
36
|
Roundtree IA, Evans ME, Pan T and He C:
Dynamic RNA modifications in gene expression regulation. Cell.
169:1187–1200. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Linder B, Grozhik AV, Olarerin-George AO,
Meydan C, Mason CE and Jaffrey SR: Single-nucleotide-resolution
mapping of m6A and m6Am throughout the transcriptome. Nat Methods.
12:767–772. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Horiuchi K, Kawamura T, Iwanari H, Ohashi
R, Naito M, Kodama T and Hamakubo T: Identification of Wilms' tumor
1-associating protein complex and its role in alternative splicing
and the cell cycle. J Biol Chem. 288:33292–33302. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Narayan P and Rottman FM: An in vitro
system for accurate methylation of internal adenosine residues in
messenger RNA. Science. 242:1159–1162. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Schumann U, Shafik A and Preiss T: METTL3
gains R/W access to the epitranscriptome. Mol Cell. 62:323–324.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Śledź P and Jinek M: Structural insights
into the molecular mechanism of the m(6)A writer complex. Elife.
5:e184342016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang P, Doxtader KA and Nam Y: Structural
basis for cooperative function of Mettl3 and Mettl14
methyltransferases. Mol Cell. 63:306–317. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang
L, Jia G, Yu M, Lu Z, Deng X, et al: A METTL3-METTL14 complex
mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem
Biol. 10:93–95. 2014. View Article : Google Scholar :
|
|
44
|
Wang X, Feng J, Xue Y, Guan Z, Zhang D,
Liu Z, Gong Z, Wang Q, Huang J, Tang C, et al: Corrigendum:
Structural basis of N6-adenosine methylation by the METTL3-METTL14
complex. Nature. 542:2602017. View Article : Google Scholar
|
|
45
|
Ping XL, Sun BF, Wang L, Xiao W, Yang X,
Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al: Mammalian WTAP is
a regulatory subunit of the RNA N6-methyladenosine
methyltransferase. Cell Res. 24:177–189. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Schöller E, Weichmann F, Treiber T, Ringle
S, Treiber N, Flatley A, Feederle R, Bruckmann A and Meister G:
Interactions, localization, and phosphorylation of the m6A
generating METTL3-METTL14-WTAP complex. RNA. 24:499–512. 2018.
View Article : Google Scholar
|
|
47
|
Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang
Z, Cheng T, Gao M, Shu X, Ma H, et al: VIRMA mediates preferential
m6A mRNA methylation in 3′UTR and near stop codon and
associates with alternative polyadenylation. Cell Discov. 4:102018.
View Article : Google Scholar
|
|
48
|
Patil DP, Chen CK, Pickering BF, Chow A,
Jackson C, Guttman M and Jaffrey SR: m(6)A RNA methylation promotes
XIST-mediated transcriptional repression. Nature. 537:369–373.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Knuckles P, Lence T, Haussmann IU, Jacob
D, Kreim N, Carl SH, Masiello I, Hares T, Villaseñor R, Hess D, et
al: Zc3h13/Flacc is required for adenosine methylation by bridging
the mRNA-binding factor Rbm15/Spenito to the m6A machinery
component Wtap/Fl(2)d. Genes Dev. 32:415–429. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wen J, Lv R, Ma H, Shen H, He C, Wang J,
Jiao F, Liu H, Yang P, Tan L, et al: Zc3h13 regulates nuclear RNA
m6A methylation and mouse embryonic stem cell
self-renewal. Mol Cell. 69:1028–1038.e6. 2018. View Article : Google Scholar
|
|
51
|
Fujita Y, Krause G, Scheffner M, Zechner
D, Leddy HE, Behrens J, Sommer T and Birchmeier W: Hakai, a
c-Cbl-like protein, ubiquitinates and induces endocytosis of the
E-cadherin complex. Nat Cell Biol. 4:222–231. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bawankar P, Lence T, Paolantoni C,
Haussmann IU, Kazlauskiene M, Jacob D, Heidelberger JB, Richter FM,
Nallasivan MP, Morin V, et al: Hakai is required for stabilization
of core components of the m6A mRNA methylation
machinery. Nat Commun. 12:37782021. View Article : Google Scholar
|
|
53
|
Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC,
Shi H, Cui X, Su R, Klungland A, et al: Differential
m6A, m6Am, and m1A demethylation
mediated by FTO in the cell nucleus and cytoplasm. Mol Cell.
71:973–985.e5. 2018. View Article : Google Scholar
|
|
54
|
Zheng G, Dahl JA, Niu Y, Fedorcsak P,
Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, et al: ALKBH5
is a mammalian RNA demethylase that impacts RNA metabolism and
mouse fertility. Mol Cell. 49:18–29. 2013. View Article : Google Scholar :
|
|
55
|
Aik W, Scotti JS, Choi H, Gong L,
Demetriades M, Schofield CJ and McDonough MA: Structure of human
RNA N6-methyladenine demethylase ALKBH5 provides insights into its
mechanisms of nucleic acid recognition and demethylation. Nucleic
Acids Res. 42:4741–4754. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wang T, Kong S, Tao M and Ju S: The
potential role of RNA N6-methyladenosine in cancer progression. Mol
Cancer. 19:882020. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wang X, Zhao BS, Roundtree IA, Lu Z, Han
D, Ma H, Weng X, Chen K, Shi H and He C: N(6)-methyladenosine
modulates messenger RNA translation efficiency. Cell.
161:1388–1399. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y,
Cheng C, Li L, Pi J, Si Y, et al: The m6A reader YTHDF1 promotes
ovarian cancer progression via augmenting EIF3C translation.
Nucleic Acids Res. 48:3816–3831. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhang C, Chen L, Peng D, Jiang A, He Y,
Zeng Y, Xie C, Zhou H, Luo X, Liu H, et al: METTL3 and
N6-methyladenosine promote homologous recombination-mediated repair
of DSBs by modulating DNA-RNA hybrid accumulation. Mol Cell.
79:425–442.e7. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Liu SY, Feng Y, Wu JJ, Zou ML, Sun ZL, Li
X and Yuan FL: m6A facilitates YTHDF-independent phase
separation. J Cell Mol Med. 24:2070–2072. 2020. View Article : Google Scholar
|
|
61
|
Ries RJ, Zaccara S, Klein P,
Olarerin-George A, Namkoong S, Pickering BF, Patil DP, Kwak H, Lee
JH and Jaffrey SR: m6A enhances the phase separation
potential of mRNA. Nature. 571:424–428. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Gao Y, Pei G, Li D, Li R, Shao Y, Zhang QC
and Li P: Multivalent m6A motifs promote phase
separation of YTHDF proteins. Cell Res. 29:767–769. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Wang J, Wang L, Diao J, Shi YG, Shi Y, Ma
H and Shen H: Binding to m6A RNA promotes
YTHDF2-mediated phase separation. Protein Cell. 11:304–307. 2020.
View Article : Google Scholar
|
|
64
|
Sheth U and Parker R: Decapping and decay
of messenger RNA occur in cytoplasmic processing bodies. Science.
300:805–808. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M,
Ma J and Wu L: YTHDF2 destabilizes m(6)A-containing RNA through
direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun.
7:126262016. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu
PJ, Liu C and He C: YTHDF3 facilitates translation and decay of
N6-methyladenosine-modified RNA. Cell Res. 27:315–328.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Xiao W, Adhikari S, Dahal U, Chen YS, Hao
YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al: Nuclear m(6)A
reader YTHDC1 regulates mRNA splicing. Mol Cell. 61:507–519. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu
Y, Gregory BD, Schultz RM and Wang PJ: Nuclear m6A reader YTHDC1
regulates alternative polyadenylation and splicing during mouse
oocyte development. PLoS Genet. 14. pp. e10074122018, View Article : Google Scholar
|
|
69
|
Roundtree IA, Luo GZ, Zhang Z, Wang X,
Zhou T, Cui Y, Sha J, Huang X, Guerrero L, Xie P, et al: YTHDC1
mediates nuclear export of N6-methyladenosine methylated
mRNAs. Elife. 6:e313112017. View Article : Google Scholar
|
|
70
|
Lesbirel S, Viphakone N, Parker M, Parker
J, Heath C, Sudbery I and Wilson SA: The m6A-methylase
complex recruits TREX and regulates mRNA export. Sci Rep.
8:138272018. View Article : Google Scholar
|
|
71
|
Brockdorff N, Bowness JS and Wei G:
Progress toward understanding chromosome silencing by Xist RNA.
Genes Dev. 34:733–744. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Liang D, Lin WJ, Ren M, Qiu J, Yang C,
Wang X, Li N, Zeng T, Sun K, You L, et al: m6A reader
YTHDC1 modulates autophagy by targeting SQSTM1 in diabetic skin.
Autophagy. 18:1318–1337. 2022. View Article : Google Scholar
|
|
73
|
Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y,
Qi M, Lu Z, Shi H, Wang J, et al: Ythdc2 is an
N6-methyladenosine binding protein that regulates
mammalian spermatogenesis. Cell Res. 27:1115–1127. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
König J, Zarnack K, Rot G, Curk T, Kayikci
M, Zupan B, Turner DJ, Luscombe NM and Ule J: iCLIP reveals the
function of hnRNP particles in splicing at individual nucleotide
resolution. Nat Struct Mol Biol. 17:909–915. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
McCloskey A, Taniguchi I, Shinmyozu K and
Ohno M: hnRNP C tetramer measures RNA length to classify RNA
polymerase II transcripts for export. Science. 335:1643–1646. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Liu N, Dai Q, Zheng G, He C, Parisien M
and Pan T: N(6)-methyladenosine-dependent RNA structural switches
regulate RNA-protein interactions. Nature. 518:560–564. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Zhou KI, Shi H, Lyu R, Wylder AC, Matuszek
Ż, Pan JN, He C, Parisien M and Pan T: Regulation of
co-transcriptional pre-mRNA splicing by m6A through the
low-complexity protein hnRNPG. Mol Cell. 76:70–81.e9. 2019.
View Article : Google Scholar
|
|
78
|
Liu N, Zhou KI, Parisien M, Dai Q,
Diatchenko L and Pan T: N6-methyladenosine alters RNA structure to
regulate binding of a low-complexity protein. Nucleic Acids Res.
45:6051–6063. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Alarcón CR, Goodarzi H, Lee H, Liu X,
Tavazoie S and Tavazoie SF: HNRNPA2B1 is a mediator of
m(6)A-dependent nuclear RNA processing events. Cell. 162:1299–1308.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Fabbiano F, Corsi J, Gurrieri E, Trevisan
C, Notarangelo M and D'Agostino VG: RNA packaging into
extracellular vesicles: An orchestra of RNA-binding proteins? J
Extracell Vesicles. 10:e120432020. View Article : Google Scholar
|
|
81
|
Huang H, Weng H, Sun W, Qin X, Shi H, Wu
H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al: Recognition of RNA
N6-methyladenosine by IGF2BP proteins enhances mRNA
stability and translation. Nat Cell Biol. 20:285–295. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wu Z, Shi Y, Lu M, Song M, Yu Z, Wang J,
Wang S, Ren J, Yang YG, Liu GH, et al: METTL3 counteracts premature
aging via m6A-dependent stabilization of MIS12 mRNA. Nucleic Acids
Res. 48:11083–11096. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Edupuganti RR, Geiger S, Lindeboom RGH,
Shi H, Hsu PJ, Lu Z, Wang SY, Baltissen MPA, Jansen PWTC, Rossa M,
et al: N6-methyladenosine (m6A) recruits and
repels proteins to regulate mRNA homeostasis. Nat Struct Mol Biol.
24:870–878. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Zhang J, Bai R, Li M, Ye H, Wu C, Wang C,
Li S, Tan L, Mai D, Li G, et al: Excessive miR-25-3p maturation via
N6-methyladenosine stimulated by cigarette smoke promotes
pancreatic cancer progression. Nat Commun. 10:18582019. View Article : Google Scholar :
|
|
85
|
Hua YQ, Zhang K, Sheng J, Ning ZY, Li Y,
Shi WD and Liu LM: NUCB1 suppresses growth and shows additive
effects with gemcitabine in pancreatic ductal adenocarcinoma via
the unfolded protein response. Front Cell Dev Biol. 9:6418362021.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Wang M, Liu J, Zhao Y, He R, Xu X, Guo X,
Li X, Xu S, Miao J, Guo J, et al: Upregulation of METTL14 mediates
the elevation of PERP mRNA N6 adenosine methylation
promoting the growth and metastasis of pancreatic cancer. Mol
Cancer. 19:1302020. View Article : Google Scholar
|
|
87
|
Xie F, Liu S and Wang H: M6A
methyltransferase METTL16 suppresses pancreatic cancer
proliferation through p21 pathways. Pancreas. 49:14372020.
|
|
88
|
Huang H, Li H, Pan R, Wang S, Khan AA,
Zhao Y, Zhu H and Liu X: Ribosome 18S m6A
methyltransferase METTL5 promotes pancreatic cancer progression by
modulating c-Myc translation. Int J Oncol. 60:92022. View Article : Google Scholar
|
|
89
|
Zeng J, Zhang H, Tan Y, Wang Z, Li Y and
Yang X: m6A demethylase FTO suppresses pancreatic cancer
tumorigenesis by demethylating PJA2 and inhibiting Wnt signaling.
Mol Ther Nucleic Acids. 25:277–292. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Tang X, Liu S, Chen D, Zhao Z and Zhou J:
The role of the fat mass and obesity-associated protein in the
proliferation of pancreatic cancer cells. Oncol Lett. 17:2473–2478.
2019.PubMed/NCBI
|
|
91
|
Guo X, Li K, Jiang W, Hu Y, Xiao W, Huang
Y, Feng Y, Pan Q and Wan R: RNA demethylase ALKBH5 prevents
pancreatic cancer progression by posttranscriptional activation of
PER1 in an m6A-YTHDF2-dependent manner. Mol Cancer. 19:912020.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
He Y, Hu H, Wang Y, Yuan H, Lu Z, Wu P,
Liu D, Tian L, Yin J, Jiang K and Miao Y: ALKBH5 inhibits
pancreatic cancer motility by decreasing long non-coding RNA
KCNK15-AS1 methylation. Cell Physiol Biochem. 48:838–846. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
He Y, Yue H, Cheng Y, Ding Z, Xu Z, Lv C,
Wang Z, Wang J, Yin C, Hao H and Chen C: ALKBH5-mediated
m6A demethylation of KCNK15-AS1 inhibits pancreatic
cancer progression via regulating KCNK15 and PTEN/AKT signaling.
Cell Death Dis. 12:11212021. View Article : Google Scholar
|
|
94
|
Tang B, Yang Y, Kang M, Wang Y, Wang Y, Bi
Y, He S and Shimamoto F: m6A demethylase ALKBH5 inhibits
pancreatic cancer tumorigenesis by decreasing WIF-1 RNA methylation
and mediating Wnt signaling. Mol Cancer. 19:32020. View Article : Google Scholar
|
|
95
|
Huang R, Yang L, Zhang Z, Liu X, Fei Y,
Tong WM, Niu Y and Liang Z: RNA m6A demethylase ALKBH5
protects against pancreatic ductal adenocarcinoma via targeting
regulators of iron metabolism. Front Cell Dev Biol. 9:7242822021.
View Article : Google Scholar
|
|
96
|
Hu X, Peng WX, Zhou H, Jiang J, Zhou X,
Huang D, Mo YY and Yang L: IGF2BP2 regulates DANCR by serving as an
N6-methyladenosine reader. Cell Death Differ. 27:1782–1794. 2020.
View Article : Google Scholar :
|
|
97
|
Meng X, Deng Y, He S, Niu L and Zhu H:
m6A-mediated upregulation of LINC00857 promotes
pancreatic cancer tumorigenesis by regulating the miR-150-5p/E2F3
axis. Front Oncol. 11:6299472021. View Article : Google Scholar
|
|
98
|
Zhai S, Xu Z, Xie J, Zhang J, Wang X, Peng
C, Li H, Chen H, Shen B and Deng X: Epigenetic silencing of LncRNA
LINC00261 promotes c-myc-mediated aerobic glycolysis by regulating
miR-222-3p/HIPK2/ERK axis and sequestering IGF2BP1. Oncogene.
40:277–291. 2021. View Article : Google Scholar :
|
|
99
|
Geng Y, Guan R, Hong W, Huang B, Liu P,
Guo X, Hu S, Yu M and Hou B: Identification of m6A-related genes
and m6A RNA methylation regulators in pancreatic cancer and their
association with survival. Ann Transl Med. 8:3872020. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Ying P, Li Y, Yang N, Wang X, Wang H, He
H, Li B, Peng X, Zou D, Zhu Y, et al: Identification of genetic
variants in m6A modification genes associated with
pancreatic cancer risk in the Chinese population. Arch Toxicol.
95:1117–1128. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Tian J, Zhu Y, Rao M, Cai Y, Lu Z, Zou D,
Peng X, Ying P, Zhang M, Niu S, et al:
N6-methyladenosine mRNA methylation of PIK3CB regulates
AKT signalling to promote PTEN-deficient pancreatic cancer
progression. Gut. 69:2180–2192. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Brabletz T, Kalluri R, Nieto MA and
Weinberg RA: EMT in cancer. Nat Rev Cancer. 18:128–134. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Xia T, Wu X, Cao M, Zhang P, Shi G, Zhang
J, Lu Z, Wu P, Cai B, Miao Y and Jiang K: The RNA m6A
methyltransferase METTL3 promotes pancreatic cancer cell
proliferation and invasion. Pathol Res Pract. 215:1526662019.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Li BQ, Huang S, Shao QQ, Sun J, Zhou L,
You L, Zhang TP, Liao Q, Guo JC and Zhao YP: WT1-associated protein
is a novel prognostic factor in pancreatic ductal adenocarcinoma.
Oncol Lett. 13:2531–2538. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Chen J, Sun Y, Xu X, Wang D, He J, Zhou H,
Lu Y, Zeng J, Du F, Gong A and Xu M: YTH domain family 2
orchestrates epithelial-mesenchymal transition/proliferation
dichotomy in pancreatic cancer cells. Cell Cycle. 16:2259–2271.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Dai S, Zhang J, Huang S, Lou B, Fang B, Ye
T, Huang X, Chen B and Zhou M: HNRNPA2B1 regulates the
epithelial-mesenchymal transition in pancreatic cancer cells
through the ERK/snail signalling pathway. Cancer Cell Int.
17:122017. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Dahlem C, Barghash A, Puchas P, Haybaeck J
and Kessler SM: The insulin-like growth factor 2 mRNA binding
protein IMP2/IGF2BP2 is overexpressed and correlates with poor
survival in pancreatic cancer. Int J Mol Sci. 20:32042019.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Chen S, Yang C, Wang ZW, Hu JF, Pan JJ,
Liao CY, Zhang JQ, Chen JZ, Huang Y, Huang L, et al: CLK1/SRSF5
pathway induces aberrant exon skipping of METTL14 and Cyclin L2 and
promotes growth and metastasis of pancreatic cancer. J Hematol
Oncol. 14:602021. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Huang XT, Li JH, Zhu XX, Huang CS, Gao ZX,
Xu QC, Zhao W and Yin XY: HNRNPC impedes m6A-dependent
anti-metastatic alternative splicing events in pancreatic ductal
adenocarcinoma. Cancer Lett. 518:196–206. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Hu Y, Tang J, Xu F, Chen J, Zeng Z, Han S,
Wang F, Wang D, Huang M, Zhao Y, et al: A reciprocal feedback
between N6-methyladenosine reader YTHDF3 and lncRNA DICER1-AS1
promotes glycolysis of pancreatic cancer through inhibiting
maturation of miR-5586-5p. J Exp Clin Cancer Res. 41:692022.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Huang S, Wu Z, Cheng Y, Wei W and Hao L:
Insulin-like growth factor 2 mRNA binding protein 2 promotes
aerobic glycolysis and cell proliferation in pancreatic ductal
adenocarcinoma via stabilizing GLUT1 mRNA. Acta Biochim Biophys Sin
(Shanghai). 51:743–752. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Taketo K, Konno M, Asai A, Koseki J,
Toratani M, Satoh T, Doki Y, Mori M, Ishii H and Ogawa K: The
epitranscriptome m6A writer METTL3 promotes chemo- and
radioresistance in pancreatic cancer cells. Int J Oncol.
52:621–629. 2018.PubMed/NCBI
|
|
113
|
Zhang C, Ou S, Zhou Y, Liu P, Zhang P, Li
Z, Xu R and Li Y: m6A methyltransferase METTL14-mediated
upregulation of cytidine deaminase promoting gemcitabine resistance
in pancreatic cancer. Front Oncol. 11:6963712021. View Article : Google Scholar
|
|
114
|
Kong F, Liu X, Zhou Y, Hou X, He J, Li Q,
Miao X and Yang L: Downregulation of METTL14 increases apoptosis
and autophagy induced by cisplatin in pancreatic cancer cells. Int
J Biochem Cell Biol. 122:1057312020. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Chen B, Ye F, Yu L, Jia G, Huang X, Zhang
X, Peng S, Chen K, Wang M, Gong S, et al: Development of
cell-active N6-methyladenosine RNA demethylase FTO inhibitor. J Am
Chem Soc. 134:17963–17971. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Niu Y, Lin Z, Wan A, Chen H, Liang H, Sun
L, Wang Y, Li X, Xiong XF, Wei B, et al: RNA N6-methyladenosine
demethylase FTO promotes breast tumor progression through
inhibiting BNIP3. Mol Cancer. 18:462019. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Huang Y, Yan J, Li Q, Li J, Gong S, Zhou
H, Gan J, Jiang H, Jia GF, Luo C and Yang CG: Meclofenamic acid
selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic
Acids Res. 43:373–384. 2015. View Article : Google Scholar :
|
|
118
|
Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, Sun
G, Lu Z, Huang Y, Yang CG, et al: m6A RNA methylation
regulates the self-renewal and tumorigenesis of glioblastoma stem
cells. Cell Rep. 18:2622–2634. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu
H, Ni T, Zhang ZS, Zhang T, Li C, et al: Small-molecule targeting
of oncogenic FTO demethylase in acute myeloid leukemia. Cancer
Cell. 35:677–691.e10. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Su R, Dong L, Li C, Nachtergaele S,
Wunderlich M, Qing Y, Deng X, Wang Y, Weng X, Hu C, et al: R-2HG
exhibits anti-tumor activity by targeting
FTO/m6A/MYC/CEBPA signaling. Cell. 172:90–105.e23. 2018.
View Article : Google Scholar
|
|
121
|
Xu P and Ge R: Roles and drug development
of METTL3 (methyltransferase-like 3) in anti-tumor therapy. Eur J
Med Chem. 230:1141182022. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Forino NM, Hentschel J and Stone MD:
Cryo-EM structures tell a tale of two telomerases. Nat Struct Mol
Biol. 28:457–459. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Yankova E, Blackaby W, Albertella M, Rak
J, De Braekeleer E, Tsagkogeorga G, Pilka ES, Aspris D, Leggate D,
Hendrick AG, et al: Small-molecule inhibition of METTL3 as a
strategy against myeloid leukaemia. Nature. 593:597–601. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Weng H, Huang H, Wu H, Qin X, Zhao BS,
Dong L, Shi H, Skibbe J, Shen C, Hu C, et al: METTL14 inhibits
hematopoietic stem/progenitor differentiation and promotes
leukemogenesis via mRNA m6A modification. Cell Stem
Cell. 22:191–205.e9. 2018. View Article : Google Scholar
|
|
125
|
Zhang J, Tsoi H, Li X, Wang H, Gao J, Wang
K, Go MY, Ng SC, Chan FK, Sung JJ and Yu J: Carbonic anhydrase IV
inhibits colon cancer development by inhibiting the Wnt signalling
pathway through targeting the WTAP-WT1-TBL1 axis. Gut.
65:1482–1493. 2016. View Article : Google Scholar
|
|
126
|
Huang C, Zhou S, Zhang C, Jin Y, Xu G,
Zhou L, Ding G, Pang T, Jia S and Cao L: ZC3H13-mediated
N6-methyladenosine modification of PHF10 is impaired by fisetin
which inhibits the DNA damage response in pancreatic cancer. Cancer
Lett. 530:16–28. 2022. View Article : Google Scholar : PubMed/NCBI
|