Introduction
Prostate cancer is a prevalent malignancy and has
the highest incidence rate of all cancer types in men over the age
of 65 years both in China and in the USA (1,2).
During the initial stages of prostate cancer, androgen deprivation
therapy (ADT) is the first line of treatment (3). Combined with radical prostatectomy or
radiation therapy, ADT can effectively reduce the level of serum
prostate specific antigen and suppress tumor growth (3). The majority of patients with
hormone-sensitive prostate cancer (HSPC) who are initially
sensitive to ADT gradually develop resistance to anti-androgen
drugs such as enzalutamide or bicalutamide after 18-24 months of
treatment (4). Consequently, HSPC
progresses to castration-resistant prostate cancer (CRPC), which
exhibits notably increased proliferation and metastasis (4). A previous study indicated that 90% of
patients exhibit bone metastases when the CRPC further progresses
to the metastatic CRPC stage, and those with distant metastasis
exhibit poor 5-year survival rates of <3% (5). Therefore, there is an urgent need to
determine the detailed mechanisms underlying the development of ADT
resistance and to identify novel therapeutic targets.
Numerous studies have confirmed that non-coding RNAs
serve important roles in the carcinogenesis and progression of
several types of cancer (6,7).
Long non-coding RNAs (lncRNAs) are a novel class of endogenous
non-coding RNAs, which are pervasively transcribed from the human
genome and are >200 nucleotides in length (8). Previously, lncRNAs were considered to
be the byproducts of RNA splicing errors with no or limited
physiological and pathological functions (9). With the rapid development of
high-throughput sequencing technologies, an increasing number of
lncRNAs have been found and identified as key regulators of
biological and/or pathological behaviors during the occurrence and
progression of numerous diseases, including prostate cancer,
glioma, gastric cancer and cholangiocarcinoma (7,9-11).
For example, it has been reported that lncRNA 01614 promoted
pancreatic cancer progression by suppressing GSK-3β (12). Zhang et al (13) reported that LncKRT16P6 promoted
tongue squamous cell carcinoma progression by functioning as a
competing endogenous RNA. In prostate cancer, the majority of
previous studies have paid attention to the effects of lncRNAs on
the proliferation and metastasis of cancer cells (10,14,15),
while the expression patterns and functions of lncRNAs in the
progression from HSPC to CRPC remain overlooked.
A previous study demonstrated that there were 134
differentially expressed lncRNAs between LNCaP (an established
androgen-dependent prostate cancer cell line typically used as an
in vitro model for HSPC) and C4-2 cells (an established
androgen-independent prostate cancer cell line typically used as an
in vitro model for CRPC) using high-throughput lncRNA
sequencing (16). This suggested
that these lncRNAs may be involved in the process of ADT
resistance. Furthermore, the expression profiles of the four most
upregulated lncRNAs (plncRNA-1, Linc00963, SNHG17 and VIM-AS1) were
confirmed in LNCaP and C4-2 cells to verify the lncRNA sequencing
results. The follow-up studies successively identified that these
four lncRNAs accelerated progression by sponging microRNAs
(miRNAs/miRs) or mediating epithelial-mesenchymal transition (EMT)
(17-19). However, the specific mechanisms by
which VIM-AS1 increased proliferation and promoted the acquisition
of enzalutamide resistance in prostate cancer requires further
elucidation.
In the present study, it was demonstrated that
VIM-AS1 was highly expressed in CRPC and this was indicative of
poor disease-free survival. It was also revealed that VIM-AS1
promoted proliferation and induced enzalutamide resistance in
vitro and in vivo. Notably, VIM-AS1 was demonstrated to
interact with insulin like growth factor 2 mRNA binding protein 2
(IGF2BP2) protein to enhance the mRNA stability of
3-hydroxy-3-methylglutaryl-CoA synthase 1 (HMGCS1), which further
resulted in malignant proliferation and enzalutamide
resistance.
Materials and methods
Cell culture
LNCaP (cat. no. TCHu73), VCaP (cat. no. TCHu220) and
PC-3 (cat. no. TCHu158) human prostate cancer cell lines were
obtained from The Cell Bank of Type Culture Collection of The
Chinese Academy of Sciences. DU145 (cat. no. HTB-81) human prostate
cancer cells were obtained from American Type Culture Collection.
C4-2 (cat. no. CL-0046) human prostate cancer cells were obtained
from Procell Life Science & Technology Co., Ltd. LNCaP, VCaP,
C4-2 and DU145 cells were maintained in DMEM (HyClone; Cytiva)
supplemented with 10% FBS (Cellmax) and 1% penicillin-streptomycin
(Gibco; Thermo Fisher Scientific, Inc.). PC-3 cells were cultured
with DMEM/F12 (HyClone; Cytiva) with 10% FBS (Cellmax) and 1%
penicillin-streptomycin (Gibco; Thermo Fisher Scientific, Inc.).
All cells were kept in a humidified incubator supplied with 5%
CO2 at 37°C.
Transfection
VIM-AS1 overexpression vector (VIM-AS1 OE; cat. no.
PG1-21050014; plasmid backbone, pcDNA3.1), IGF2BP2 overexpression
vector (IGF2BP2 OE; cat. PB1-20100010; plasmid backbone, pcDNA3.1)
and HMGCS1 overexpression vector (HMGCS1 OE; cat. no. PB1-20100011;
plasmid backbone, pcDNA3.1), and pcDNA3.1 empty vector (Vector;
cat. no. CL-0046) were purchased from Genecreate. VIM-AS1 short
hairpin RNA (shRNA/sh) (sh-VIM-AS1; target sequence, 5′-GCT CCC TTT
GGA TGA CAT AGA-3′; plasmid backbone, GV344) and normal scramble
short hairpin RNA (sh-NC; control sequence, 5′-TTC TCC GAA CGT GTC
ACG T-3′; plasmid backbone, GV344) were purchased from Shanghai
GeneChem Co., Ltd. IGF2BP2 small interfering RNA (si-IGF2BP2),
HMGCS1 small interfering RNA (si-HMGCS1) and non-targeting control
small interfering RNA (siRNA/si) (si-NC; sequence) were purchased
from Guangzhou RiboBio Co., Ltd and the sequences of siRNAs are
listed in Table SI. The vector
and siRNA transfections were performed as described previously
(20). Briefly, ~1×106
prostate cancer cells, which were seeded in six-well plates, were
transfected with 0.5 µg vector or siRNAs using 2.5 µl
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) at
37°C for 48 h according to the manufacturer's protocol. Cells were
collected for reverse transcription-quantitative PCR (RT-qPCR),
western blotting, Cell Counting Kit-8 (CCK-8) assays,
5-ethynyl-2′-deoxyuridine (EdU) assays, colony formation assays,
chemosensitivity assays, TUNEL assays and flow cytometry analysis
48 h after siRNA transfection. shRNA infections were performed as
described previously (21).
sh-VIM-AS1 (lentiviral plasmid, GV344; GeneChem, Inc.) and sh-NC
were packaged in 293T cells (cat. no. CRL-3216; American Type
Culture Collection) using a 2nd generation system with the ratio of
lentiviral construct, packaging plasmid (Helper 1.0; GeneChem,
Inc.) and envelope plasmid (Helper 2.0; GeneChem, Inc.) (20
µg:15 µg:10 µg). The culture medium was
centrifugated at 1,000 × g for 5 min at 4°C after 293T cells were
cultured for 48 h at 37°C. The supernatant containing viral
particles was collected. After C4-2 cells were infected with
sh-VIM-AS1 and sh-NC (multiplicity of infection, 50) at 37°C for 48
h, the efficiency was initially validated by assessing VIM-AS1
expression using RT-qPCR. The cells were then selected using
puromycin at a concentration of 2 µg/ml for 2 weeks to
obtain C4-2 cells with stable knockdown of VIM-AS1. Subsequently,
the cells were maintained in complete medium with puromycin at a
concentration of 0.5 µg/ml and collected for RT-qPCR,
western blotting, CCK-8 assays, EdU assays, colony formation
assays, chemosensitivity assays, TUNEL assays and flow cytometry
analysis 48 h after shRNA infection.
RNA extraction and RT-qPCR
A Cytoplasmic & Nuclear RNA Purification Kit
(Norgen Biotek Corp.) was used to isolate cytoplasmic and nuclear
RNA of C4-2 cells according to the manufacturer's protocol. Total
RNA from prostate cancer cells and resected tumor tissues was
isolated using TRIzol® solution (Invitrogen; Thermo
Fisher Scientific, Inc.). For VIM-AS1, HMGCS1 mRNA and IGF2BP2 mRNA
expression analysis, the aforementioned RNA extracts were reverse
transcribed using a RevertAid First Strand cDNA Synthesis Kit
(Beijing Solarbio Science & Technology Co., Ltd.) according to
the manufacturer's protocol, followed by amplification and
quantification using a 2× SYBR Green PCR MasterMix Kit (Beijing
Solarbio Science & Technology Co., Ltd.) according to the
manufacturer's protocol. CFX96 real time PCR detection system
(Bio-Rad Laboratories, Inc.) was used for quantitative detection
with the following thermocycling conditions: Initial denaturation
at 95°C for 2 min, followed by 25 cycles of denaturation at 95°C
for 15 sec, annealing at 60°C for 30 sec and extension at 72°C for
1 min. The relative expression levels were determined using the
2−ΔΔCq method (22).
GAPDH was used as the internal control. The primers used in the
present study were purchased from Sangon Biotech Co., Ltd. and are
shown in Table SI.
Protein extraction and western
blotting
Total protein from prostate cancer cells was
extracted using RIPA lysis buffer (cat. no. G2002; Wuhan Servicebio
Technology Co., Ltd.) supplemented with protease inhibitors
(Beijing Solarbio Science & Technology Co., Ltd.). After
determining the concertation of each proteins using a BCA protein
assay kit (cat. no. G2026; Wuhan Servicebio Technology Co., Ltd.),
a total of 30 µg protein per lane was loaded on a 10%
SDS-gel, resolved using SDS-PAGE and transferred to a PVDF
membrane. The membranes were blocked with 5% nonfat dry milk at
room temperature for 2 h, and then incubated with anti-HMGCS1 (cat.
no. ab155787; 1:500; Abcam), anti-IGF2BP2 (cat. no. ab117809;
1:500; Abcam) or anti-GAPDH (cat. no. ab9485; 1:500; Abcam) primary
antibodies overnight at 4°C, and subsequently incubated with an
HRP-labeled goat-anti-rabbit (cat. no. 7074; 1:5,000; Cell
Signaling Technology, Inc.) secondary antibody for 2 h at room
temperature. The bands were visualized using Immobilon™ Western
Chemilum HRP Substrate (cat. no. WBKLS0100, MilliporeSigma) and
analyzed using Quantity One software version 4.6.6 (Bio-Rad
Laboratories, Inc.).
Chemosensitivity assay
A total of 3×103 cells/well were plated
into a 96-well plate. After adherence, cells were treated with
different concentrations (0, 1, 1.5, 2, 2.5, 3, 3.5, 4, 5, 10, 20,
30, 40, 50 µM) of enzalutamide at 37°C for 72 h.
Subsequently, 10 µl/well CCK-8 solution (cat. no. K1018;
Dojindo Molecular Technologies, Inc.) was added to the culture
medium. The cells were incubated with CCK-8 reagent at 37°C for 1 h
and the optical density values at 450 nm were measured using a
microplate reader. The IC50 value was calculated as
previously described (23).
Colony formation assay
A total of 500 C4-2 cells infected with sh-NC and
sh-VIM-AS1 and LNCaP cells transfected with Vector and VIM-AS1 OE
per well were plated into a 6-well plate. After 10 days of
incubation at 37°C, the colonies were washed with PBS, fixed with
90% methanol at room temperature for 10 min and stained with
crystal violet for 15 min at room temperature. Subsequently, the
colonies (>50 cells) were imaged using a digital camera (version
D3200; Nikon Corporation) at ×4 magnification and quantified using
Image J software (version 1.5; National Institutes of Health).
TUNEL assay
TUNEL assays were performed using an In Situ
Cell Death Detection Kit (cat. no. 11684817910; Roche Diagnostics
GmbH) according to the manufacturer's instructions. Briefly,
1×105 cells/well were seeded in 96-well plates and
cultured with complete medium for 48 h at 37°C. Subsequently, cells
were stained with 50 µl terminal deoxynucleotidyl
transferase for 1 h in the dark at 37°C and 450 µl
fluorescein-labeled deoxyuridine triphosphate solution for 1 h in
the dark at 37°C after cell fixation by 4% paraformaldehyde for 30
min at room temperature and permeabilization by 0.5% TRITON X-100
for 90 sec at 4°C. Next, cells were stained with TUNEL reaction
mixture and DAPI-containing mounting media (0.5 mg/ml; cat. no.
S2110; Beijing Solarbio Science & Technology Co., Ltd.) for 1 h
in the dark at 37°C. Three visual fields were randomly selected for
observation using a laser scanning confocal microscope (FV1000;
Olympus Corporation) and corresponding software (FV10-ASW Viewer;
version 4.2; Olympus Corporation) at ×200 magnification.
EdU staining
EdU assays were conducted using a Cell-Light™ EdU
DNA Cell Proliferation Kit (cat. no. C10310-1; Guangzhou RiboBio
Co., Ltd.) according to the manufacturer's instructions. Briefly,
1×105 cells/well were seeded in 96-well plates and
cultured with complete medium for 48 h at 37°C. Cells were stained
with 100 µl 50 µM EdU solution for 2 h in the dark at
room temperature. The cells were stained with Apollo®567
(Guangzhou RiboBio Co., Ltd.) for 30 min at 4°C and DAPI for 10 min
at 4°C after cell fixation by 4% paraformaldehyde for 30 min at
room temperature and permeabilization by 0.5% TRITON X-100 for 90
sec at 4°C. Three visual fields were randomly selected for
observation using a laser scanning confocal microscope (FV1000;
Olympus Corporation) and corresponding software (FV10-ASW Viewer;
version 4.2; Olympus Corporation) at ×200 magnification.
Flow cytometry assay
Cell apoptosis and cell cycle analyses were
performed using an Annexin V Alexa fluor 488/PI Cell Apoptosis Kit
and DNA Content Quantitation Kit (cat. no. CA1040; Beijing Solarbio
Science & Technology Co., Ltd.) according to the manufacturer's
instructions. Briefly, 1×106 cells/well were seeded in
6-well plates and cultured for 48 h at 37°C. For cell apoptosis,
1×106 cells were harvested and stained with annexin-V
and PI for 30 min at 4°C. For cell cycle analysis, 1×106
cells were harvested and stained with PI for 30 min at 4°C after
fixing with 70% ethanol for 30 min at 4°C. Early cell apoptosis
(lower right quadrant-prophase apoptosis; Beijing Solarbio Science
& Technology Co., Ltd.) and cell cycle (Beijing Solarbio
Science & Technology Co., Ltd.) distribution analyses were
performed using a flow cytometer (CytoFLEX LX; Beckman Coulter,
Inc.). All data were analyzed with Flowjo version 10.0.6 (Becton,
Dickinson and Company).
High-throughput sequencing analysis
Total RNA was isolated from transfected LNCaP cells.
Subsequently, 1.3 µg total RNA was sent for RNA sequencing
(RNA-seq; Genecreate Co., Ltd.). RNA libraries for RNA-seq were
prepared using a NEBNext® Ultra™ RNA Library Prep Kit
for Illumina® (cat. no. NEB+e7770; New England BioLabs,
Inc.) according to the manufacturer's protocols (New England
BioLabs, Inc.). The loading concentration of the constructed
library was detected using the Agilent High Sensitivity DNA kit
(cat. no. 5067-4626; Agilent Technologies, Inc.) and an Agilent
2100 bioanalyzer (Agilent Technologies, Inc), and was 6 pM for
RNA-seq. Libraries which consisted of cDNA fragments of 200 bp in
length were sequenced on the Illumina NovaSeq 6000 (150 base pairs;
paired ends; Illumina, Inc.) using a MiniSeq High Output Reagent
Kit (cat. no. FC-420-1002; Illumina, Inc.). Sequence reads were
trimmed for adaptor sequence/low-quality sequence using fastp
(version 0.12.0; https://github.com/OpenGene/fastp) (24). Trimmed sequence reads were mapped
to hg38 using Hisat2 software (version 2.2.1; http://daehwankimlab.github.io/hisat2/)
(25). Reads per kilobase of exon
per megabase of library size were calculated using featureCounts
(version 1.5.3; http://bioinf.wehi.edu.au/featureCounts/) (26). Differentially expressed genes were
identified according to |log2(fold change)|>1 and P<0.05
using the R program (version 4.03; http://www.r-project.org). Next, Gene Ontology (GO;
http://geneontology.org/) (27,28)
analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG; https://www.kegg.jp/) (29) analysis, and Gene Set Enrichment
Analysis (GSEA; http://www.gsea-msigdb.org/gsea/index.jsp) were used
to further analyze the differentially expressed genes. The
significance cult-off level was P<0.05.
RNA pull-down
Cells were lysed in NP40 lysis buffer (cat. no.
N8030; Beijing Solarbio Science & Technology Co., Ltd.), and 1
mg cell extracts were incubated with a biotin-labeled VIM-AS1
(Genecreate Co., Ltd.; sequence, 5′-UCC CUG AGA UGA UGA AGA GGA CCA
GUG CCC AUU CCA GGA-3′) or a normal scrambled control (NC;
Genecreate Co., Ltd.; sequence, 5′-UAU CAC GUA GCC GUU GCA UUU GCC
GUA GCC CUG UGG GCC-3′) probe at 4°C for 6 h. Subsequently, cell
extracts, biotin-labeled VIM-AS1 or NC, and 40 µl
streptavidin agarose beads were mixed and incubated on a rotator
overnight at 4°C. After washing with PBS, precipitates were pulled
down by centrifugation at 3,000 × g for 5 min at 4°C. The potential
binding proteins in the retrieved precipitates were identified by
high performance liquid chromatography-mass spectrometry (HPLC-MS)
detection (Genecreate Co., Ltd.). HPLC-MS analysis was performed on
a Orbitrap Exploris 480-mass spectrometer (Thermo Fisher
Scientific, Inc.) that was coupled to a Nanospray Flex™ (Thermo
Fisher Scientific, Inc.) for 60 min. The mass spectrometer was
operated in positive ion mode to monitor the m/z transitions for
all peptides. Target peptides (two for each protein) were measured
in multiple reaction monitoring (MRM). Peptide ions between 350 and
1,200 m/z were scanned in the Orbitrap detector (Thermo Fisher
Scientific, Inc.) every 3 sec with a resolution of
1.2×105 (maximum fill time 50 msec; automatic gain
control target 4×105). A scheduled MRM acquisition
method was constructed using manually optimized decluttering
potentials, collision energies, collision cell entrance and exit
potentials.
mRNA stability assay
A total of 1×106 cells were seeded in
6-well plates and cultured overnight. The following day, 5
µg/ml actinomycin D (cat. no. HY-17559; MedChemExpress) was
added to cells to inhibit gene transcription for 2, 4, 6 or 8 h at
4°C. At the indicated times, the mRNA levels of HMGCS1 were
determined using RT-qPCR as aforementioned.
RNA immunoprecipitation (RIP)
RIP assays were performed using a Magna RIP™
RNA-Binding Protein Immunoprecipitation Kit (cat. no. 17-704;
MilliporeSigma) according to the manufacturer's instructions.
Briefly, a total of 1×107 cells were collected,
centrifuged at 4°C for 5 min at 1,000 × g, washed in pre-cooled
PBS, and then lysed using complete RIP Lysis buffer (cat. no.
R0010; Beijing Solarbio Science & Technology Co., Ltd.). A
total of 5 µg anti-IGF2BP2 (cat. no. ab117809; 1:500; Abcam)
or lgG (cat. no. ab172730; 1:500; Abcam) was added to 50 µl
protein A/G magnetic bead suspension for 30 min at room
temperature. Subsequently, 100 µl of the aforementioned
lysates were incubated with the beads-antibody complex overnight at
4°C. After washing with RIP wash buffer (part of the Magna RIP™
RNA-Binding Protein Immunoprecipitation Kit) three times, Protease
K buffer (cat. no. P1120; Beijing Solarbio Science & Technology
Co., Ltd.) was added to the immunoprecipitated product, followed by
incubation at 55°C for 30 min. Following centrifugation at 4°C for
5 min at 1,000 × g, the immunoprecipitated RNA was isolated using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and analyzed using RT-qPCR as aforementioned.
Fluorescence in situ hybridization
(FISH)
FISH assays were performed using a RiboTM lncRNA
FISH probe Mix Kit (cat. no. c10910; Guangzhou RiboBio Co., Ltd.)
according to the manufacturer's instructions. Oligonucleotide
modified Cy-3-labeled probes for VIM-AS1 (5′-TAG GAC TTC CTA GTA
CTT CTG A-3), GAPDH and U6 were designed and synthesized by
Genecreate. C4-2 cells were seeded on 20-mm confocal dishes
(Corning, Inc). After overnight incubation, C4-2 cells were fixed
with 4% paraformaldehyde for 20 min at 4°C and permeabilized using
Triton X-100 for 90 sec at 4°C. Next, 250 µl
prehybridization solution with 1% blocking solution (Guangzhou
RiboBio Co., Ltd.) was added to C4-2 cells and cells were incubated
at 42°C for 1 h. Subsequently, C4-2 cells were incubated with 100
µl hybridization buffer (Guangzhou RiboBio Co., Ltd.)
supplemented with 1% blocking solution and 2.5 µl 20
µM 22-nucleotide CY-3-labeled-VIM-AS1, CY-3-labeled-GAPDH or
CY-3-labeled-U6 FISH probe at 37°C overnight in a dark moist
chamber. The following day, cells were washed three times in 2X
sodium citrate buffer for 5 min at 42°C and stained with DAPI at
4°C for 10 min. Images were acquired using a laser scanning
confocal microscope (FV1000; Olympus Corporation) and corresponding
software (FV10-ASW Viewer; version 4.2; Olympus Corporation) at a
magnification of ×400.
In vivo experiments
All animal experiments were performed in accordance
with the relevant national ethical regulations and were approved by
the Animal Care and Use Committee of the Air Force Medical
University (approval no: 20220967; Xi'an, China). All mice were
purchased from Beijing Vital River Laboratory Animal Technology
Co., Ltd., and housed in an animal room at a controlled temperature
of 22°C and 40% humidity, with a 12 h light/dark cycle, and ad
libitum access to food and water. Female BALB/c nude mice (6
weeks old; n=12; weight, ~20 g) were used to establish the C4-2
xenograft models. 100 µl C4-2 cells (5×106/100
µl PBS) infected with sh-VIM-AS1 or sh-NC were injected
subcutaneously into the left upper limb of the nude mice. For the
following 5 weeks, the health and behavior of the mice were
monitored twice a week, and tumor growth was monitored using a
vernier caliper twice a week and an in vivo imaging system
once a week. When the humane endpoints were reached or the tumor
volume of mice in the sh-VIM-AS1 or sh-NC groups was closed 1,000
mm3, the mice would be euthanized. According to the
requirements of the Animal Center of Air Force Medical University
(Xi'an, China), humane endpoints were reached when the xenograft
tumor diameter was >20 mm, or signs of unrelieved pain or
distress without recovery were observed. At the end of 35 days of
observation, all 12 mice had not reached human endpoints but the
largest tumor volume was 892.4 mm3. Therefore, all 12
mice were sacrificed by acute exsanguination following isoflurane
inhalation (5% for induction and 2% for maintenance of 5 min),
cardiac arrest was then used to verify death based on a lack of a
heartbeat. The tumors were excised and assessed using
immunohistochemistry and RT-qPCR, and the tumor volumes and weights
were measured.
Immunohistochemistry
Resected tumor tissues were fixed with 4%
paraformaldehyde at 4°C for 24 h. The tissues were embedded in
paraffin and sectioned (4 µm). After conventional dewaxing
and rehydration with descending alcohol series at room temperature,
the sections were incubated with 3% H2O2 for
15 min. The sections were incubated with 5% normal goat serum
(SL038; Beijing Solarbio Science & Technology Co., Ltd.) at
room temperature for 1 h. Subsequently, Ki-67 antibody (cat. no.
GB121141; 1:1,000; Wuhan Servicebio Technology Co., Ltd.) or HMGCS1
antibody (cat. no. ab155787; 1:500; Abcam) were added for
incubation at 4°C overnight after antigen retrieval with citric
acid buffer (pH 6.0) for 1 min 40 sec at 100°C in a pressure
cooker. Subsequently, the sections were incubated with
HRP-conjugated secondary antibody (cat. no. G1214; 1:200; Wuhan
Servicebio Technology Co., Ltd.) at room temperature for 1 h. The
sections were rinsed three times using TBS with 0.1% Tween-20 for 5
min and visualized using DAB solution (cat. no. G1212; 1:1,000;
Wuhan Servicebio Technology Co., Ltd.) for 10 min at room
temperature. Images were acquired using a light microscope at a
magnification of ×200.
Bioinformatics analysis
RNA-seq expression profiles and corresponding
clinical information for VIM-AS1 in prostate cancer were downloaded
from The Cancer Genome Atlas (TCGA; https://portal.gdc.cancer.gov/projects/TCGA-PRAD;
project ID, TCGA-PRAD, mRNA sequencing data) and the Gene
Expression Omnibus (GEO) database (dataset no. 32269, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE32269)
(30) and analyzed using the R
program (version 4.03; http://www.r-project.org). The m6A modification status
was predicted using Whistle (http://180.208.58.19/whistle/index.html). The gene
(HMGCS1) was entered in the search box. m6A predicted modification
sites were listed. P<0.05 was considered to indicate a
statistically significant difference.
Statistical analysis
GraphPad Prism version 8.02 (GraphPad Software;
Dotmatics) and SPSS version 22.0 (IBM Corp.) were used to perform
statistical analysis. Data are presented as the mean ± SD or as
scatter dot plots. All experiments were repeated three times.
Unpaired Student's t-test was used to compare the difference
between two groups, and one-way ANOVA followed by post hoc tests
(least significant difference test for three groups and Bonferroni
test for more than three groups) was used for comparisons among
multiple groups. A Kruskal-Wallis test followed by a Bonferroni
test was used to compare the difference of VIM-AS1 expression in
patients with prostate cancer with different T stage and normal
controls. Kaplan-Meier survival curves and the log-rank test were
used to compare the differences in disease-free survival of
patients with prostate cancer with different VIM-AS1 expression in
TCGA dataset (project ID, TCGA-PRAD; mRNA data; cutoff-high, 75%;
cutoff-low, 25%). The following parameters were selected:
Disease-free survival, split patients by lower quartile, follow-up
threshold: 14 years. Log rank P-values <0.05 for the
Kaplan-Meier plots of patients with prostate cancer with different
VIM-AS1 expression were considered to indicate a statistically
significant difference. P-values were determined using a two-sided
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
VIM-AS1 expression is upregulated in
patients with advanced prostate cancer, and C4-2, PC-3 and DU145
cells
A previous study indicated that VIM-AS1 was
upregulated in C4-2 cells compared with LNCaP cells (14). To further assess the differential
expression profiles of VIM-AS1 during the progression of prostate
cancer, VIM-AS1 expression was analyzed using TCGA and GEO
(GSE32269). VIM-AS1 expression was consistently increased in
advanced prostate cancer based on the data from TCGA (P<0.001;
Fig. 1A). In addition, VIM-AS1 was
upregulated in the tissues of patients with CRPC compared with
those of patients in the HSPC group based on the GSE32269 dataset
from GEO (P<0.001; Fig. 1B).
Furthermore, Kaplan-Meier survival data indicated that patients
with prostate cancer with high VIM-AS1 expression exhibited worse
disease-free survival compared with patients with low VIM-AS1
expression (P=0.003; Fig. 1C).
Increased VIM-AS1 expression was also observed in
androgen-independent prostate cancer cell lines (DU145, PC-3 and
C4-2) compared with LNCaP and VCaP cell lines, which are
androgen-dependent prostate cancer cell lines (Fig. 1D). These data indicated that
VIM-AS1 expression was upregulated in advanced prostate cancer.
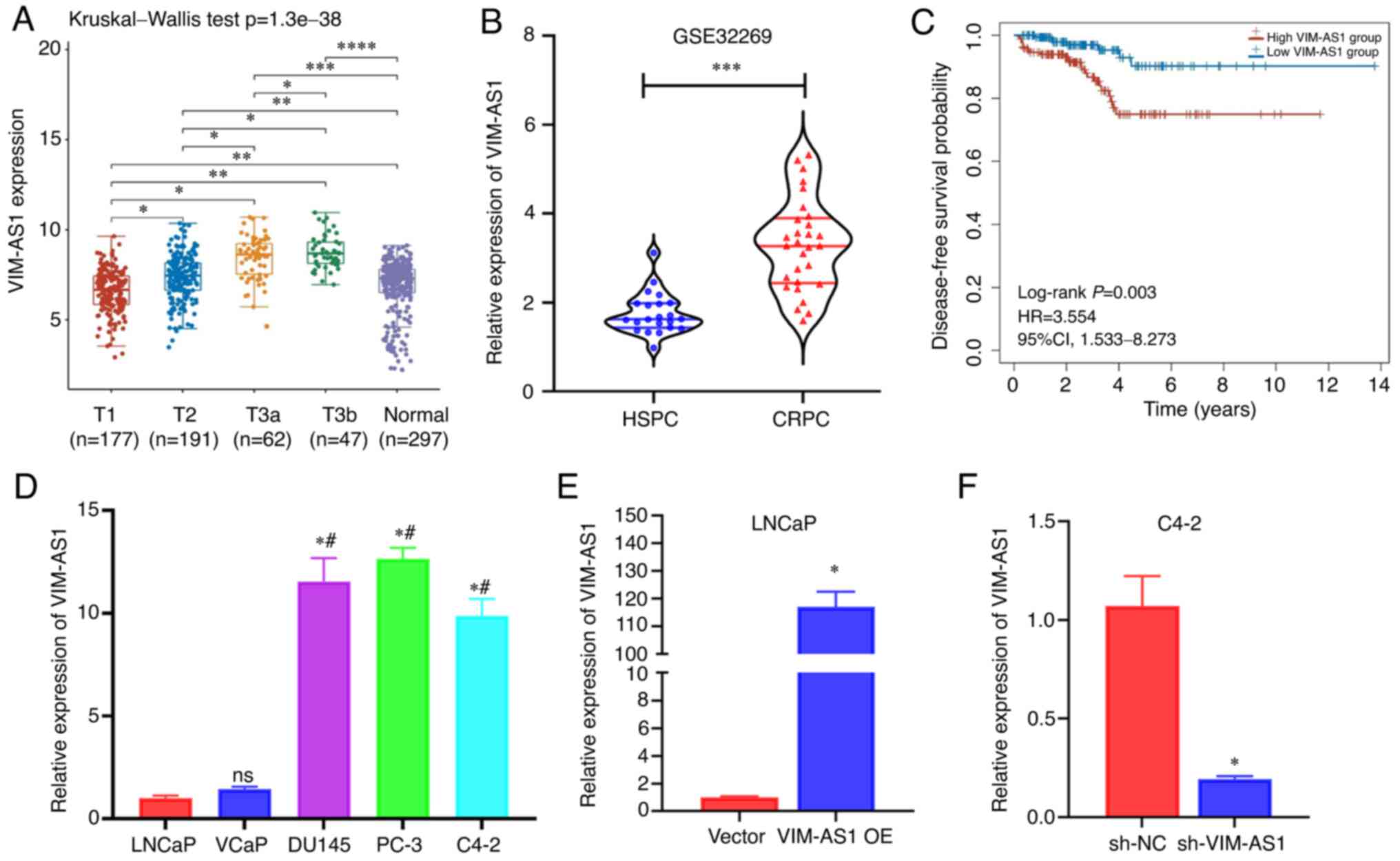 | Figure 1VIM-AS1 expression is associated with
tumor stage and androgen deprivation therapy resistance. (A) TCGA
data of VIM-AS1 expression in patients with prostate cancer with
different T stages. (B) Gene Expression Omnibus data of VIM-AS1
expression in patients with CRPC and HSPC. (C) Kaplan-Meier
survival curves of disease-free survival probability in patients
with prostate cancer based on data obtained from TCGA. Patients
were stratified into low and high VIM-AS1 expression group. (D)
VIM-AS1 expression in two HSPC cell lines (LNCaP and VCaP) and
three CRPC cell lines (DU145, PC-3 and C4-2). (E) VIM-AS1 levels in
LNCaP cells transfected with VIM-AS1 OE or vector. (F) Detection of
VIM-AS1 levels in C4-2 cells transfected with sh-VIM-AS1 or sh-NC.
*P<0.05 vs. LNCaP or as indicated,
#P<0.05 vs. VCaP, nsP>0.05 vs. LNCaP,
**P<0.01,
***P<0.001,****P<0.0001. CRPC,
castration-resistant prostate cancer; HR, hazard ratio; HSPC,
hormone-sensitive prostate cancer; ns, not significant; sh-NC,
normal scramble short hairpin RNA; TCGA, The Cancer Genome
Atlas. |
VIM-AS1 enhances proliferation and
induces enzalutamide resistance in prostate cancer cells in
vitro
To examine the effects of VIM-AS1, VIM-AS1 was
stably overexpressed in LNCaP cells and VIM-AS1 expression was
knocked down in C4-2 cells. RT-qPCR demonstrated that transfection
with the VIM-AS1 overexpression vector increased VIM-AS1 expression
in LNCaP cells (P<0.05; Fig.
1E), whereas sh-VIM-AS1 transfection stably knocked down
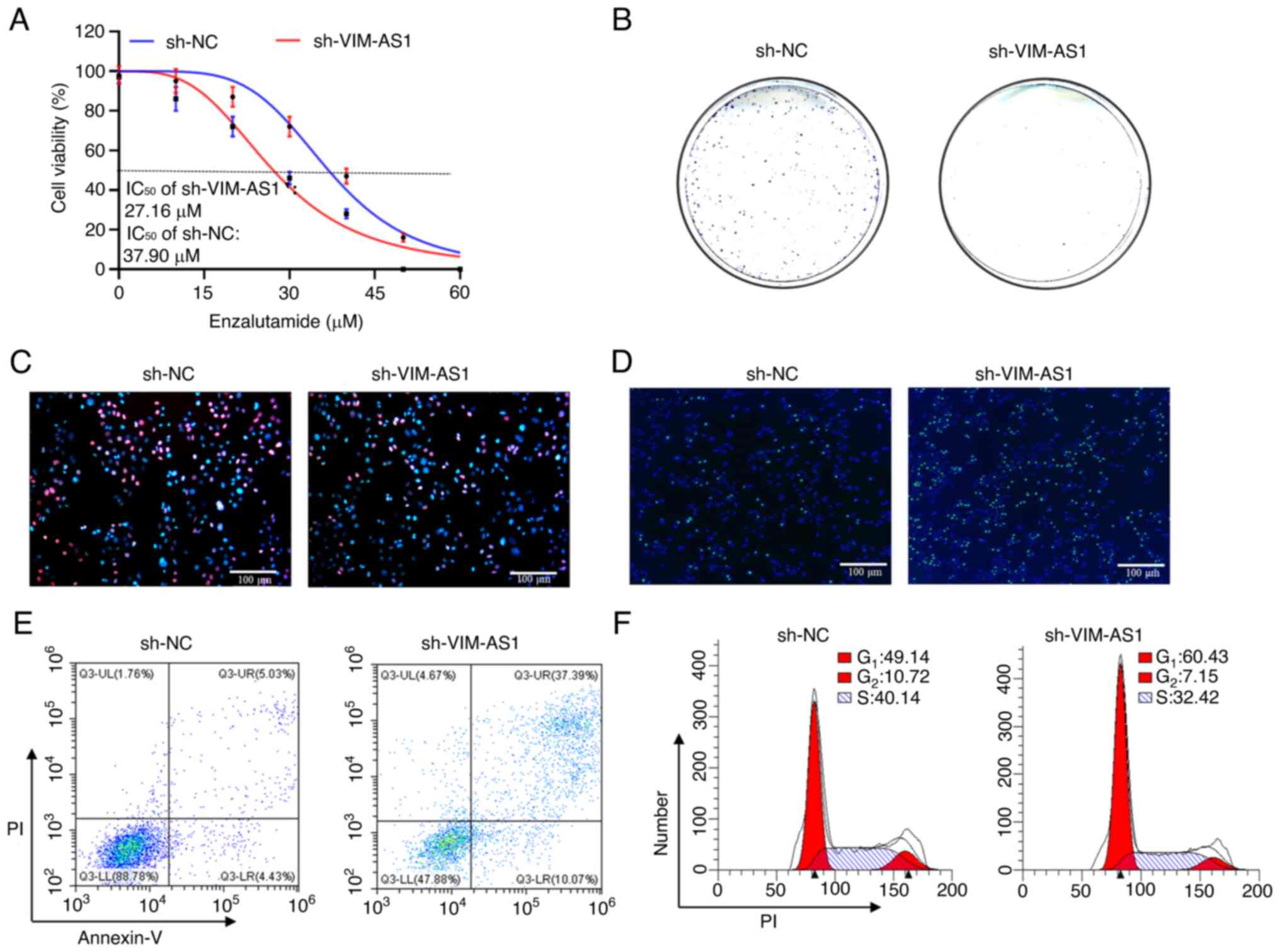
VIM-AS1 expression in C4-2 cells (P<0.05; Fig. 1F). Furthermore, it was demonstrated
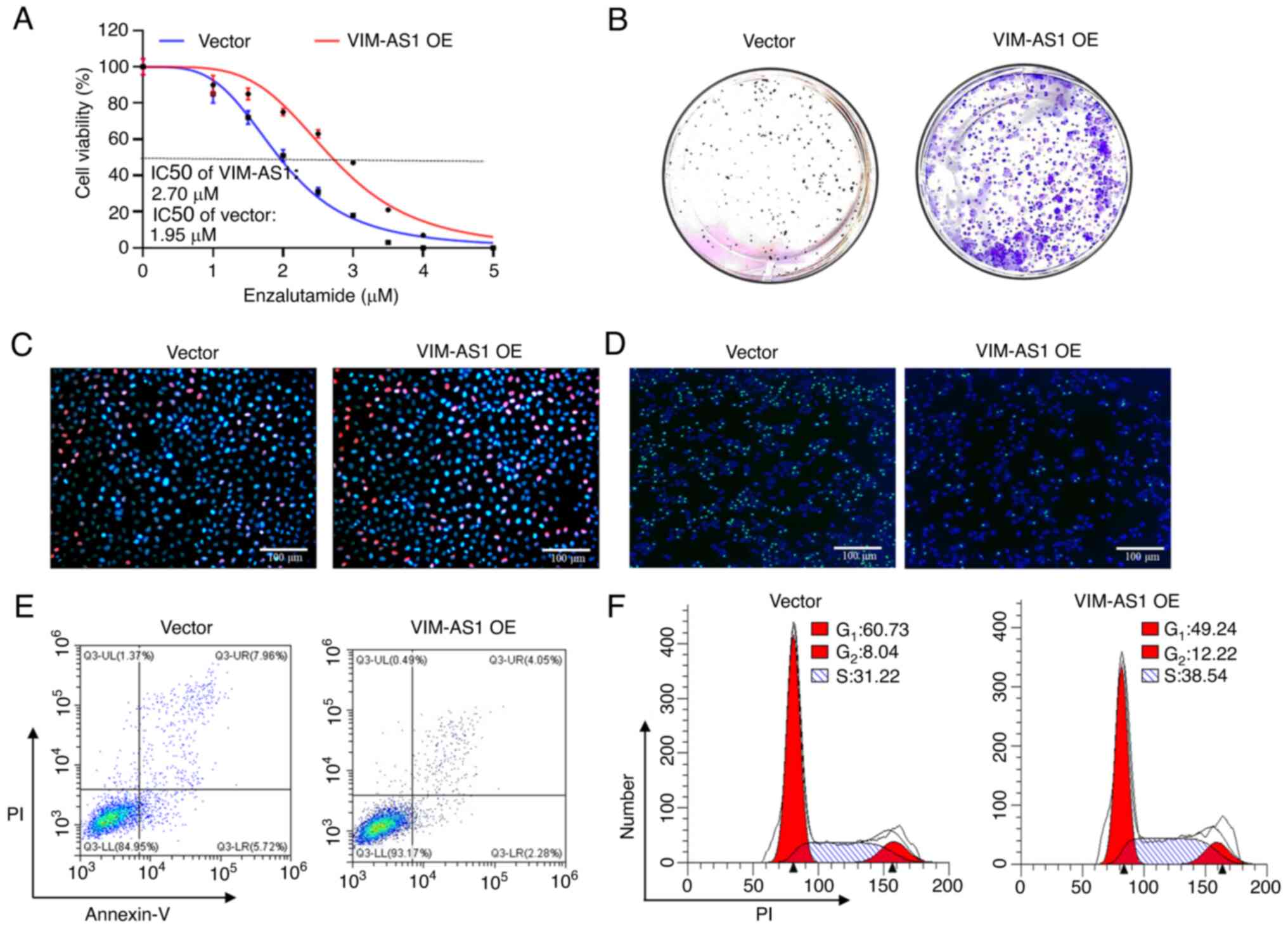
that the IC50 of enzalutamide was increased after
overexpression of VIM-AS1 in LNCaP cells (P<0.05; Figs. 2A and S1A). However, the IC50 of
enzalutamide was decreased after the knockdown of VIM-AS1 in C4-2
cells (P<0.05; Figs. 3A and
S1A). Colony formation and EdU
assays indicated that the proliferation of VIM-AS1-overexpressing
LNCaP cells was significantly increased compared with that of the
control group (P<0.05; Figs. 2B and
C, and S1B and C), and
sh-VIM-AS1-infected C4-2 cells exhibited significantly reduced
proliferation compared with the sh-NC-infected cells (P<0.05;
Figs. 3B and C, and S1B and C). In addition, TUNEL and flow
cytometry analyses demonstrated that overexpression of VIM-AS1
significantly reduced apoptosis, increased the percentage of cells
in the S stage, and decreased the percentage of cells in the G1
stage in LNCaP cells (P<0.05; Figs.
2D-F and S1D-F), whereas cell
apoptosis and the percentage of cells in the G1 stage were markedly
enhanced and the percentage of cells in the S stage was decreased
following VIM-AS1 knockdown in C4-2 cells (P<0.05; Figs. 3D-F and S1D-F). Collectively, these data
suggested that aberrant upregulation of VIM-AS1 served an important
role in promoting proliferation and driving enzalutamide resistance
in prostate cancer in vitro.
Effects of VIM-AS1 on tumor growth in a
prostate cancer xenograft model
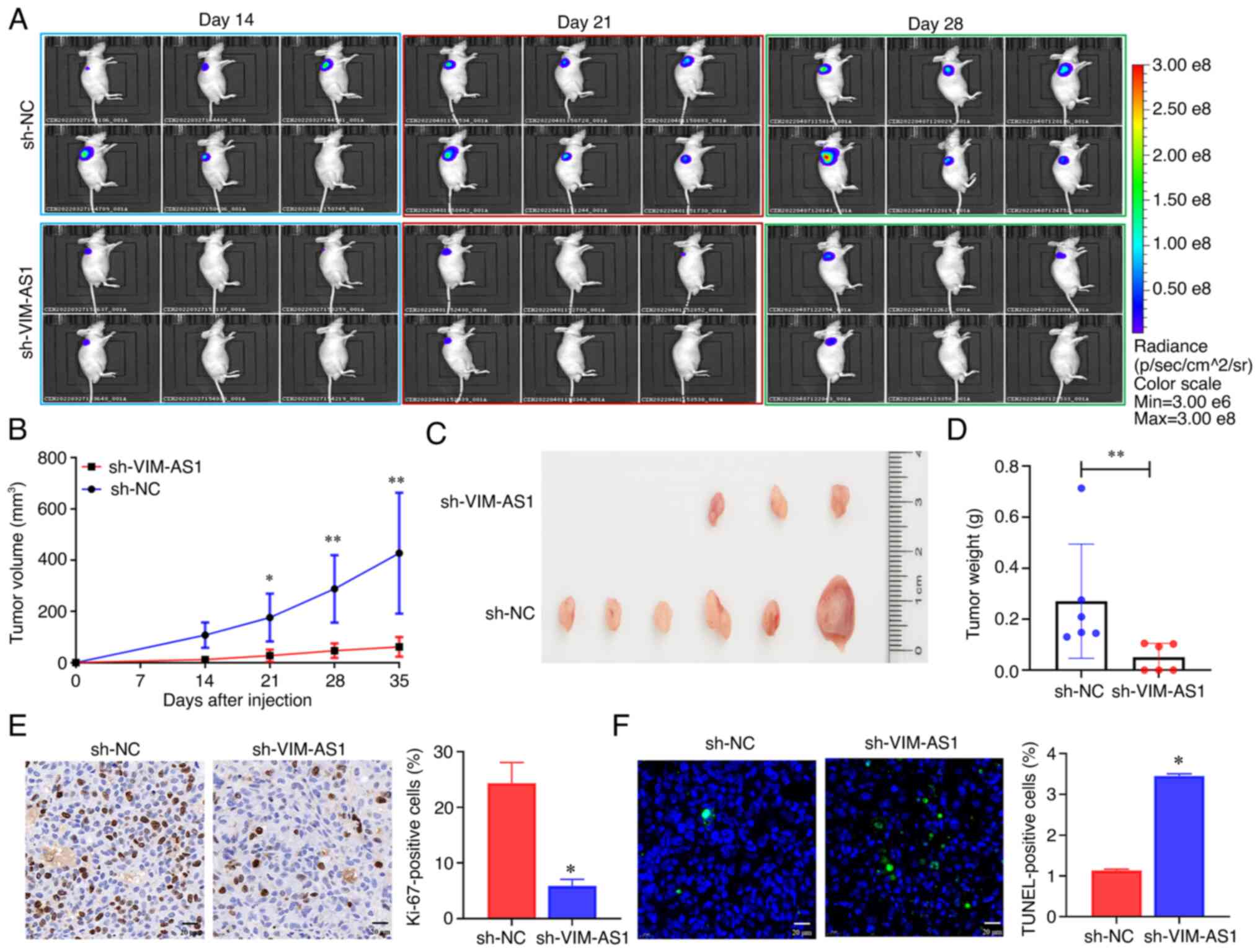
A nude mouse model was used to validate the effects
of knockdown of VIM-AS1 on the proliferation of prostate cancer
cells in vivo. C4-2 cells with stable knockdown of VIM-AS1
were injected into nude mice via subcutaneous injection. Small
animal in vivo imaging and growth curves indicated that the
mice injected with the VIM-AS1 knockdown C4-2 cells exhibited
slower tumor growth than the mice in the sh-NC group (Fig. 4A and B). Similarly, the sizes and
weights of the dissected tumors from nude mice indicated that
knockdown of VIM-AS1 suppressed tumor growth of prostate cancer
cells in vivo (Fig. 4C and
D). VIM-AS1 expression was lower in the resected tissues from
the sh-VIM-AS1 group compared with the sh-NC group (Fig. S2A). Ki-67 and TUNEL assays
indicated that the subcutaneous tumors in the VIM-AS1 knockdown
group had fewer Ki-67-positive cells (P<0.05; Fig. 4E) and more TUNEL-positive cells
(P<0.05; Fig. 4F) compared with
those in the sh-NC group. Overall, these results indicated that
knockdown of VIM-AS1 expression in C4-2 cells markedly suppressed
tumor growth of prostate cancer in vivo.
HMGCS1 is involved in the functions of
VIM-AS1 in prostate cancer cells
Next, it was attempted to determine the mechanism
through which VIM-AS1 regulated the proliferation and enzalutamide
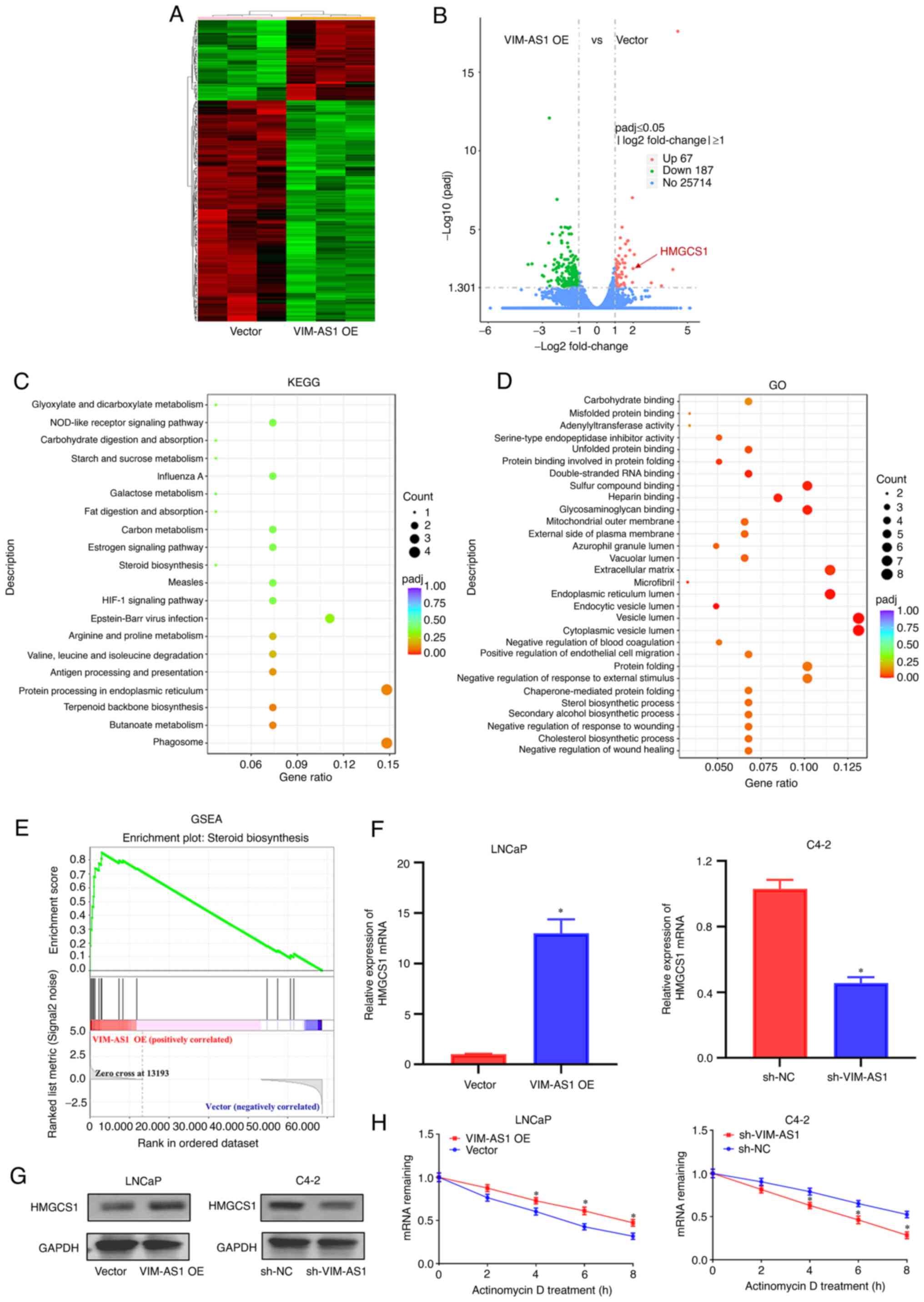
sensitivity of prostate cancer cells. First, RNA-seq was used to
identify the target genes in VIM-AS1-overexpressing LNCaP cells and
vector-transfected cells. A total of 67 genes were found to be
statistically upregulated by |log2(fold change)|>1 and 187 genes
were found to be statistically downregulated by |log2(fold
change)|>1 in VIM-AS1-overexpressing LNCaP cells (Fig. 5A). Notably, HMGCS1 was one of the
most significantly upregulated genes in VIM-AS1-overexpressing
cells (Fig. 5B). KEGG analysis
showed that Steroid biosynthesis, Fat digestion and absorption and
HIF-1 signaling pathway etc. were among the top 20 most enriched
pathways (Fig. 5C). GO analysis
demonstrated the 'steroid biosynthetic process', 'unfolded protein
binding' and 'double-stranded RNA binding' were among the top 20
most enriched functions (Fig. 5D).
Furthermore, GSEA demonstrated that the gene signatures of 'Steroid
biosynthesis' were enriched in VIM-AS1-overexpressing cells
(Fig. 5E). Notably, a previous
study has reported that HMGCS1 was actively involved in steroid
biosynthesis (31). As indicated
by IHC analyses (Fig. S2B),
RT-qPCR (Figs. 5F and S2C), and western blotting (Fig. 5G), the mRNA and protein levels of
HMGCS1 were upregulated in LNCaP cells transfected with the VIM-AS1
overexpression vector in vitro (Fig. 5F and G), but downregulated in C4-2
cells transfected with sh-VIM-AS1 compared with the
vector-transfected cells in vitro (Fig. 5F and G) and in vivo
(Fig. S2B and C) (all P<0.05).
Next, it was assessed whether VIM-AS1 regulated HMGCS1 expression
by stabilizing HMGCS1 mRNA. To test this hypothesis, prostate
cancer cells were treated with actinomycin D to measure the
degradation of HMGCS1 mRNA. VIM-AS1 overexpression enhanced HMGCS1
mRNA stability, and VIM-AS1 knockdown significantly reduced HMGCS1
mRNA stability (at 4, 6 and 8 h; P<0.05; Fig. 5H). These results indicated that
HMGCS1 may be involved in the functions of VIM-AS1 in prostate
cancer cells.
VIM-AS1 specifically interacts with
IGF2BP2 in prostate cancer cells
To investigate the regulatory mechanisms of VIM-AS1
in prostate cancer, the subcellular localization of VIM-AS1 was
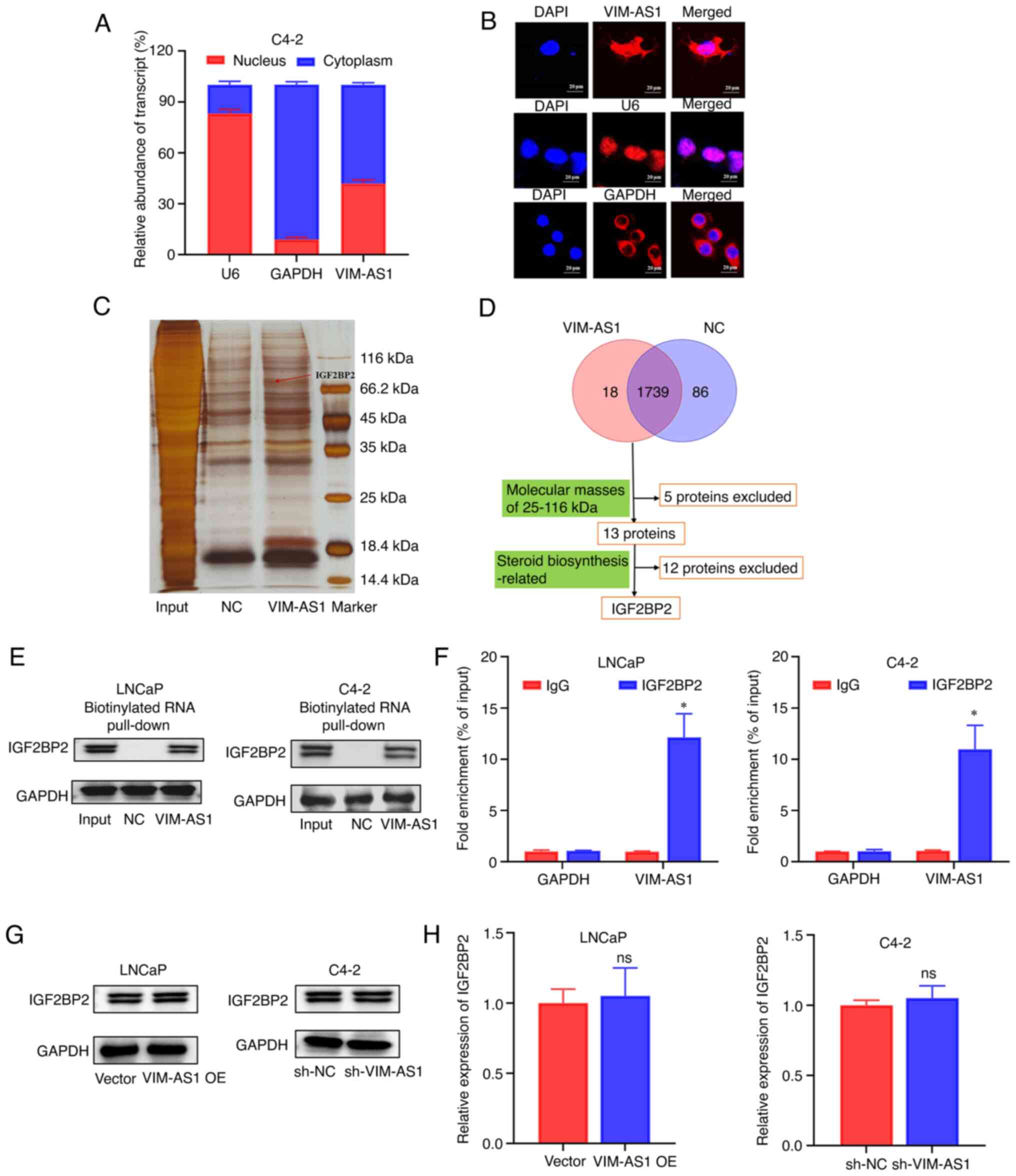
further explored. RT-qPCR analysis of the nuclear and cytoplasmic
fractions (Fig. 6A) and FISH
analysis (Fig. 6B) showed that
VIM-AS1 was present in both the cytoplasm and the nucleus of C4-2
cells. RNA pull-down assays were performed to identify
VIM-AS1-interacting proteins. Several bands were identified as
potential binding proteins that could be pulled down by
biotinylated VIM-AS1 transcripts (Fig.
6C). By searching the functions of 13 potential VIM-AS1 binding
proteins in published studies, IGF2BP2, a major differential band
precipitated in LNCaP lysates of RNA pulldown (Fig. 6D) and mass spectrometry (Fig. S3), was confirmed as the only
steroid biosynthesis-related protein with a molecular mass of
25-116 kDa (32). RNA pull-down
assays and western blotting further confirmed that VIM-AS1
interacted with IGF2BP2 in LNCaP and C4-2 cells (Fig. 6E). The enrichment of VIM-AS1 in the
precipitates of the IGF2BP2 antibody but not IgG antibody were
further confirmed by RIP assays (Fig.
6F). VIM-AS1 knockdown or overexpression exhibited no influence
on IGF2BP2 mRNA and protein levels (Fig. 6G and H). In summary, these results
indicated that VIM-AS1 directly bound to IGF2BP2 protein but did
not regulate its expression in prostate cancer cells.
VIM-AS1/IGF2BP2 complex mediates the
regulation of HMGCS1 mRNA stability
As IGF2BP2 is a known N6-methyladenosine
(m6A) reader and serves a pivotal role in downstream mRNA
stabilization (33), it was next
assessed whether IGF2BP2 was involved in the regulation of HMGCS1
expression. First, the HMGCS1 mRNA m6A modification status was
predicted using Whistle (34). The
results demonstrated that the 3′-untranslated region (3′-UTR) of
HMGCS1 mRNA contained three m6A modification sites (Fig. S4). RT-qPCR and western blotting
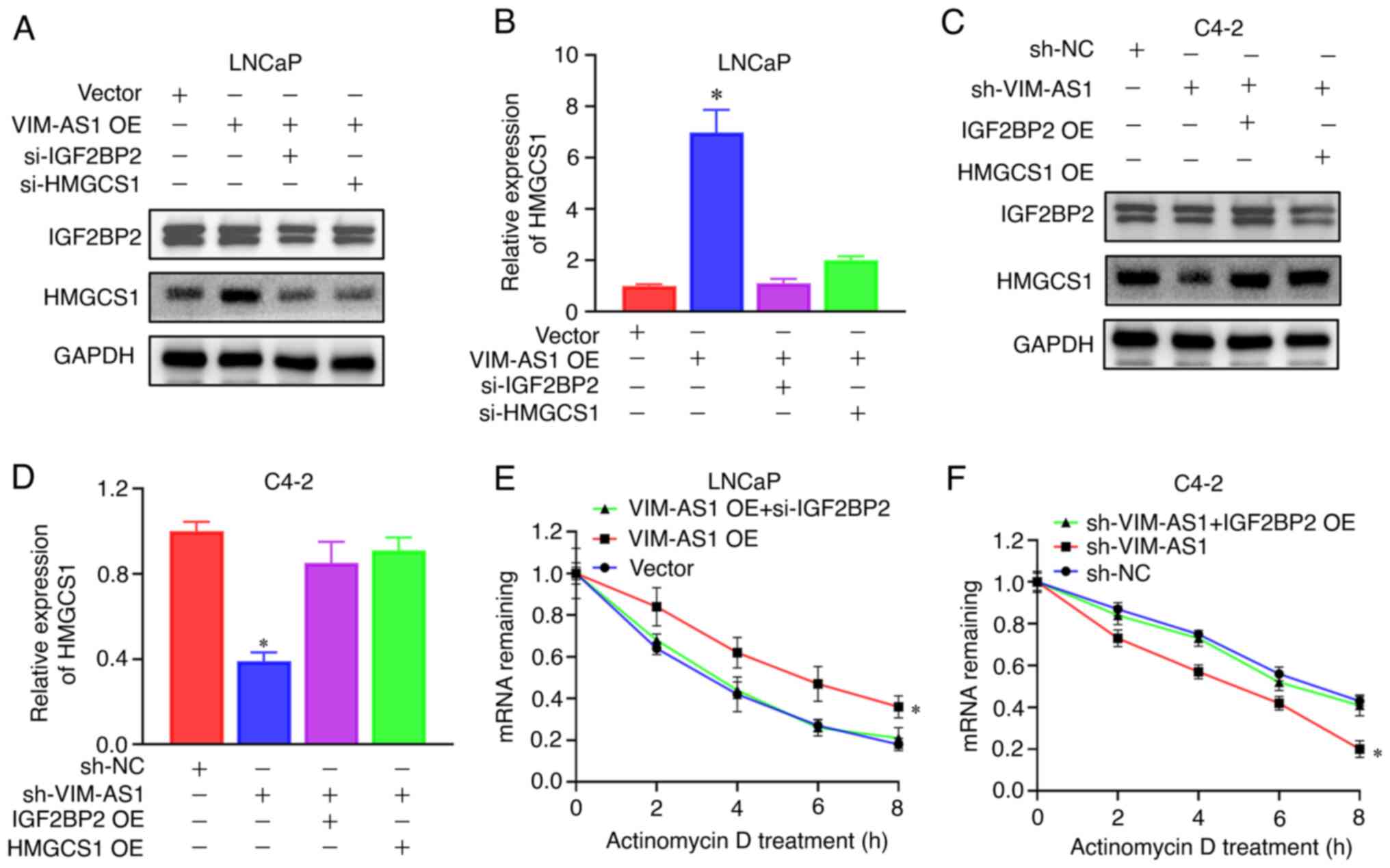
revealed that knockdown of IGF2BP2 or transfection with si-HMGCS1
inhibited VIM-AS1 overexpression vector transfection-induced HMGCS1
upregulation (Figs. 7A and B, and
S5A and B). Overexpression of
IGF2BP2 or HMGCS1 reversed sh-VIM-AS1-induced HMGCS1 downregulation
(Figs. 7C and D, and S5C and D). mRNA stability assays
demonstrated that overexpression of VIM-AS1 enhanced HMGCS1 mRNA
stability, and enhancement of HMGCS1 mRNA stability was decreased
following IGF2BP2 knockdown (Fig.
7E). Furthermore, knockdown of VIM-AS1 decreased HMGCS1 mRNA
stability, and this was reversed by overexpression of IGF2BP2
(Fig. 7F). Collectively, these
data suggested that the VIM-AS1/IGF2BP2 axis may exert roles in
HMGCS1 post-transcriptional mRNA stabilization.
HMGCS1 is a functional mediator of
VIM-AS1 in the regulation of proliferation and enzalutamide
sensitivity in prostate cancer
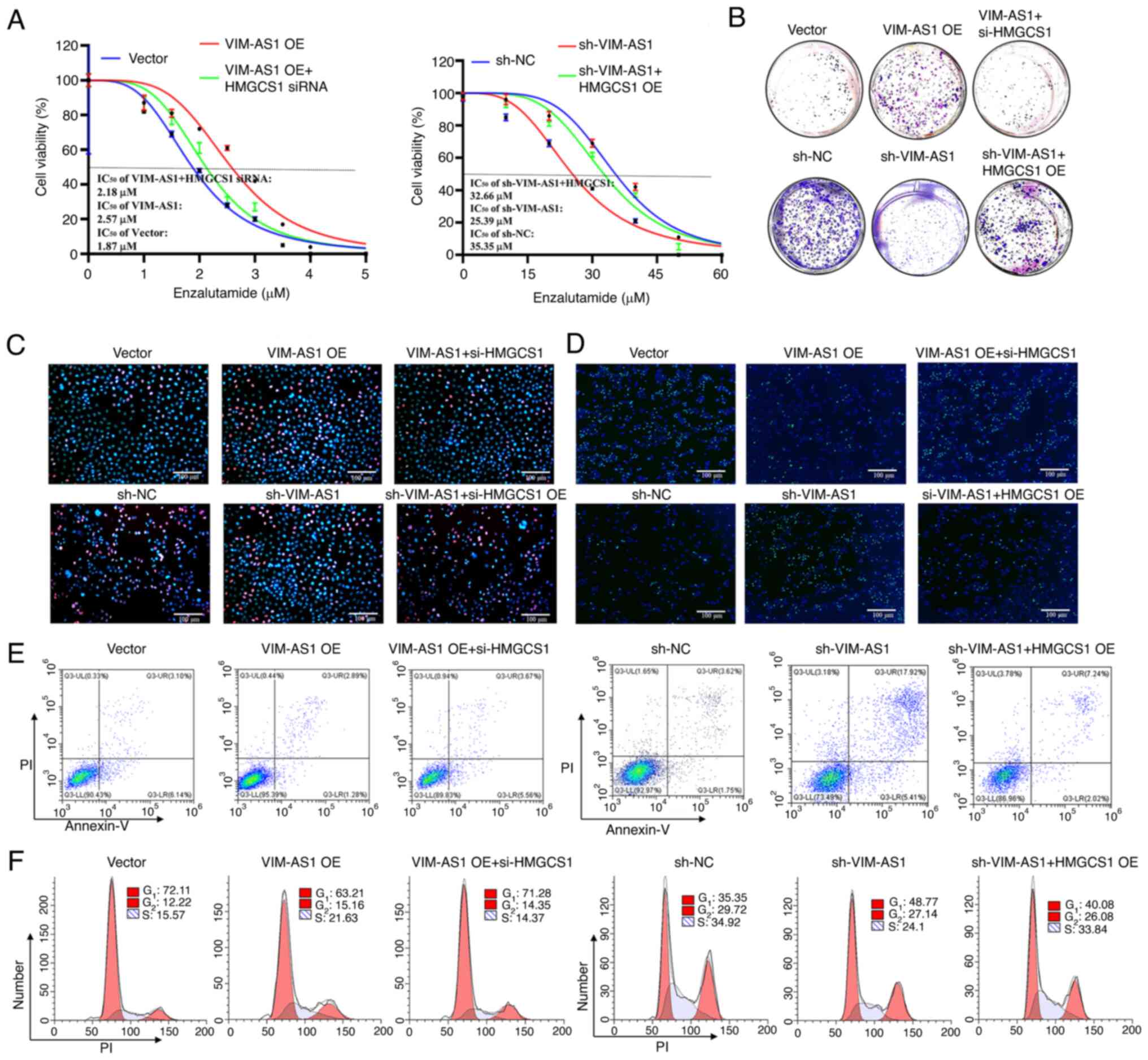
To further elucidate the functional interplay
between HMGCS1 and VIM-AS1 in the regulation of proliferation and
enzalutamide sensitivity of prostate cancer, HMGCS1 expression was
knocked down in VIM-AS1-overexpressing LNCaP cells and HMGCS1 was
overexpressed in the VIM-AS1 knockdown C4-2 cells. Functional
experiments revealed that knockdown of HMGCS1 expression abrogated
the increase in proliferation (Figs.
8B and C, and S5F and G) and
cells at S stage (Figs. 8F and
S5J), and decrease in
enzalutamide sensitivity (Figs. 8A
and S5E), apoptosis levels
(Figs. 8E and S5I) and cells at G1 stage (Figs. 8F and S5J) induced by VIM-AS1 overexpression in
LNCaP cells. Overexpression of HMGCS1 ameliorated the increase in
enzalutamide sensitivity (Figs. 8A
and S5E), apoptosis level
(Figs. 8E and S5I) and cells at G1 stage (Figs. 8F and S5J), and reduced the increase in
proliferation (Figs. 8B and C, and
S5F and G) and cells at S stage
(Figs. 8F and S5J) induced by VIM-AS1 knockdown in C4-2
cells. These results indicated that HMGCS1 mediated the
VIM-AS1-induced increase in proliferation and enzalutamide
resistance in prostate cancer cells.
Discussion
The development of high throughput sequencing
technologies, together with the advances in computational
pipelines, has led to an explosion in the discovery of
disease-related lncRNAs. An increasing number of lncRNAs have been
identified as crucial regulators of multiple diseases, including
prostate cancer (9-11,35-37).
For example, Zhang et al (35) found that lncRNA DSCAM-AS1 was
upregulated in prostate cancer, and promoted cancer progression by
forming a positive feedback loop with forkhead box A1. Wen et
al (36) also reported that
lncRNA NEAT1 expression was higher in patients with prostate cancer
with bone metastases, and it induced cancer cell metastasis to the
lungs and bones via m6A. A recent study demonstrated that lncRNA
NXTAR was downregulated in prostate cancer, and restoration of
NXTAR expression could suppress prostate cancer cell proliferation
and abrogate enzalutamide-resistant prostate xenograft tumor growth
(37). However, the expression
patterns and roles of lncRNAs in the progression from HSPC to CRPC
should be further elucidated.
In previous study, the first expression profile of
dysregulated lncRNAs between cell models of HSPC and CRPC was
mapped, and it was found that there were 134 dysregulated lncRNAs
in C4-2 cells compared with LNCaP cells (14). In addition, the expression of the
four most upregulated lncRNAs, including plncRNA-1, Linc00963,
SNHG17 and VIM-AS1, was validated in C4-2 cells (16). However, the study did not further
explore the functions and mechanisms of the four lncRNAs in the
progression of prostate cancer (16). Consistent with the findings of this
study, a series of follow-up studies also revealed that these four
lncRNAs were dysregulated and served pivotal roles in the
development of prostate cancer (17,19,38-42).
Linc00963 has been reported to promote prostate cancer progression
by modulating miR-655/tripartite motif containing 24 and the
miR-542-3p/NOP2 nucleolar protein axes (17,38).
SNHG17 has been demonstrated to enhance the acquisition of
malignant phenotypes in prostate cancer cells by activating the
β-catenin signaling pathway (39)
or by targeting miR-144/CD51 (40). plncRNA-1 has been demonstrated to
accelerate the progression of prostate cancer by inducing EMT
(41) or by modulating the
PTEN/AKT signaling pathway (42).
Zhang et al (19) reported
that VIM-AS1 expression was upregulated in prostate cancer tissues
and promoted the proliferation and invasion of CRPC PC-3 cells by
regulating EMT. Consistent with previous literature (17,19,38-42),
our previous study also revealed that the knockdown of VIM-AS1
inhibited proliferation and restored the sensitivity to
bicalutamide in CRPC C4-2 cells (43). However, the underlying mechanism by
which VIM-AS1 modulates the development of prostate cancer has not
yet been elucidated.
The current study further demonstrated that VIM-AS1
expression was upregulated in tumor tissues from patients with CRPC
compared with patients with HSPC, which further indicated that
VIM-AS1 may be involved in the progression from HSPC to CRPC.
Subsequently, the effects of VIM-AS1 on proliferation and
enzalutamide sensitivity were assessed using gain-and-loss
functional experiments. VIM-AS1 markedly promoted cell
proliferation and induced enzalutamide resistance in vitro.
These findings were consistent with the expression patterns of
VIM-AS1 in HSPC and CRPC tissues, and further indicated that
VIM-AS1 acted as a pivotal regulator of tumor growth and
sensitivity to anti-androgen therapy during the progression from
HPSC to CRPC, and may thus be considered a potential therapeutic
target for management of CRPC.
To elucidate the underlying mechanism by which
VIM-AS1 regulated proliferation and enzalutamide sensitivity in
prostate cancer cells, RNA-seq analysis was performed. HMGCS1 was
found to be a potential downstream target and functional mediator
of VIM-AS1 as it was one of the most upregulated genes in
VIM-AS1-overexpressing LNCaP cells. Additionally, rescue assays
indicated that upregulation of HMGCS1 reversed the effects of
VIM-AS1 knockdown on the proliferation and enzalutamide sensitivity
of prostate cancer cells. Previous studies have demonstrated that
the mevalonate pathway was frequently dysregulated and involved in
the carcinogenesis and progression of several types of cancer by
modulating inflammation, cell proliferation and steroidogenesis
(44,45). Consistently, HMGCS1, an established
regulatory node in the mevalonate pathway, has also been found to
be highly expressed in several types of cancer (30) and serves an important role in
regulating cancer progression by mediating cholesterol
biosynthesis, including in gastric cancer (46), colorectal cancer (47) and breast cancer (48). Notably, a previous study indicated
HMGCS1 was upregulated in CRPC PC-3 and 22Rv1 cells compared with
HSPC LNCaP cells, and knockdown of HMGCS1 in 22Rv1 cells resulted
in a reduction in cell viability and colony formation (49). In addition, Cheng et al
(50) reported that HMGCS1 was
associated with poor overall survival, and genetic variants of
HMGCS1 were shown to act as protective factors for prostate cancer.
Although several studies have highlighted HMGCS1 as a potential
mediator of the progression of prostate cancer (49,50),
to the best of our knowledge, the regulatory mechanism by which
HMGCS1 exerts its effects on prostate cancer is largely unknown.
The results of the present study not only further identified the
important roles of HMGCS1 in the regulation of proliferation and
enzalutamide sensitivity in prostate cancer cells but also found
that VIM-AS1 was the potential upstream regulator of HMGCS1 in
prostate cancer cells.
Mechanistically, the functional pattern of lncRNAs
depends on their subcellular location. If lncRNAs are distributed
primarily in the cytoplasm, they are likely to enhance target gene
expression at the post-transcriptional level by binding with miRNAs
or directly enhancing mRNA stability (51,52).
For example, LINC00680, which is primarily located in the
cytoplasm, promotes esophageal squamous cell carcinoma progression
by functioning as a competing endogenous RNA to bind with
miR-423-5p (51). Furthermore,
Lang et al (52) found that
lncRNA PCAT6, which is evenly distributed between the cytoplasm and
nucleus, enhanced IGF1R mRNA stabilization by biding with the
IGF2BP2 protein. However, if lncRNAs are primarily localized to the
nucleus, they typically bind with proteins to regulate target gene
expression at the transcriptional levels (53). For example, Chen et al
(53) reported that lncRNA LBCS,
which is primarily located in the nucleus suppressed SOX2
transcription by binding with heterogeneous nuclear
ribonucleoprotein K and enhancer of zeste 2 polycomb repressive
complex 2 subunit to mediate H3K27 tri-methylation. In the present
study, it was revealed that VIM-AS1 was located in both the
cytoplasm and the nucleus of prostate cancer cells and could
directly interact with the IGF2BP2 protein. Previous studies have
demonstrated that IGF2BP2 was both an important RNA-binding protein
and an m6A reader (54,55). Numerous studies have indicated that
IGF2BP2 could enhance mRNA stability by recognizing m6A
modification sites (56,57). In the present study, it was shown
that the 3′-UTR of HMGCS1 has three potential m6A modification
sites. Furthermore, the present study revealed that the knockdown
of IGF2BP2 abrogated the effects of VIM-AS1 on the mRNA stability
of HMGCS1. Therefore, the results of the present study combined
with those of previous studies (55-58)
further elucidated the pivotal roles of IGF2BP2 in the regulation
of HMGCS1 expression in prostate cancer.
In conclusion, it was demonstrated that VIM-AS1 was
upregulated in patients with CRPC compared with patients with HSPC,
and its upregulation induced enzalutamide resistance, promoted cell
proliferation, and inhibited cell apoptosis in prostate cancer
cells Furthermore, HMGCS1 was revealed to be the downstream target
and functional mediator of VIM-AS1 in prostate cancer cells.
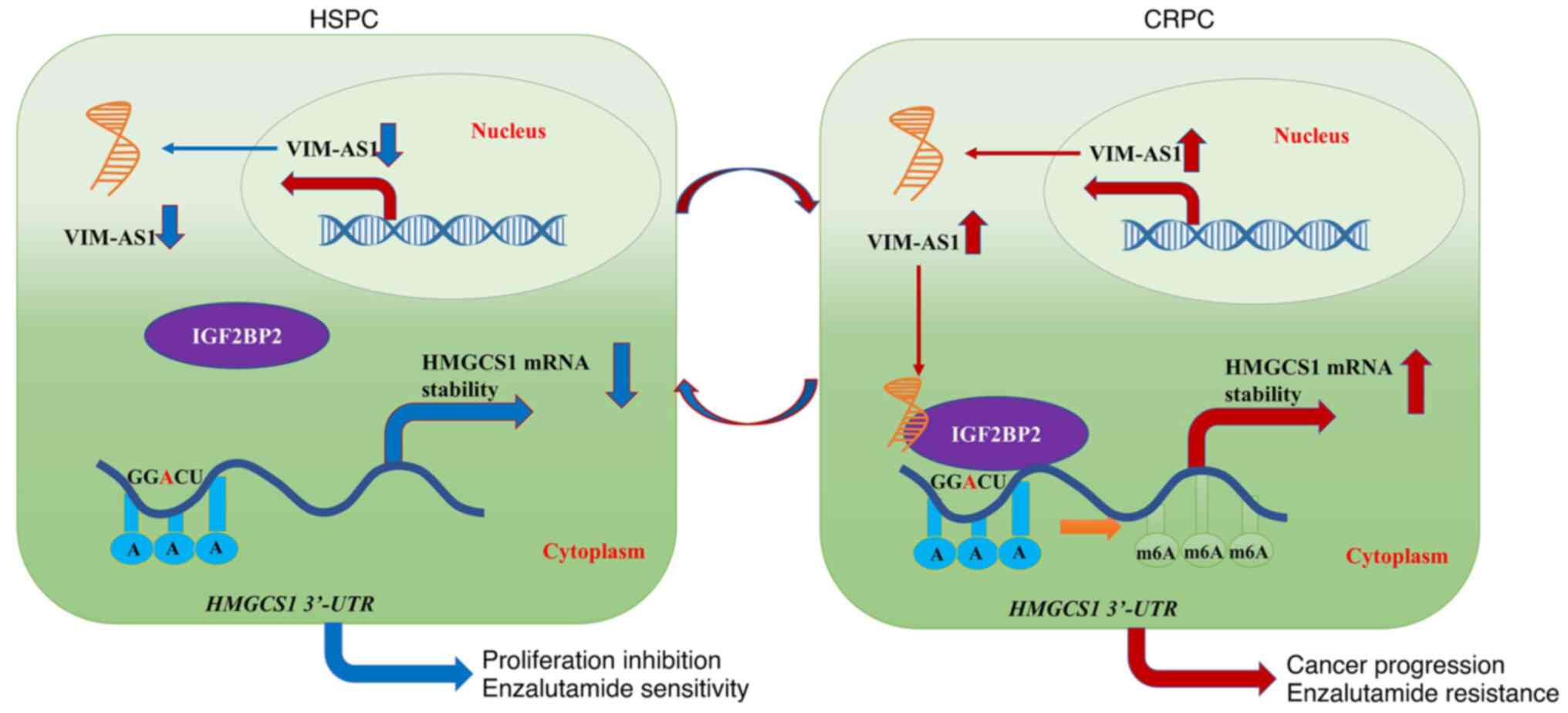
Finally, it was also demonstrated that VIM-AS1 enhanced HMGCS1 mRNA
stability and accelerated the progression of prostate cancer by
binding with the IGF2BP2 protein (Fig.
9). Therefore, the present study contributed to the
understanding of the regulatory mechanism by which VIM-AS1 and
HMGCS1 exert their effects on prostate cancer and may provide novel
biomarkers for the prediction of enzalutamide sensitivity.
Supplementary Data
Availability of data and materials
The high-throughput sequencing datasets generated
and/or analyzed during the current study are available in the GEO
database repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE220250).
The other datasets used and/or analyzed during the current study
are available from the corresponding author on reasonable
request.
Authors' contributions
SJS, DHH, JLZ and YL performed the functional and
animal experiments. DHH, JLZ and YL performed the RNA pulldown and
RIP experiments. SJS and RZ confirm the authenticity of all the raw
data. AGY and RZ designed the research study. SJS, DHH and JLZ
assembled the figures. SJS analyzed the data. SJS wrote the paper.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
All animal experiments complied with ethical
regulations and were approved by the Animal Care and Use Committee
of the Air Force Medical University (approval no. 20220967; Xi'an,
China)
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
This work was supported by the Nature Science Foundation of
Shaanxi Province (grant no. 2022JQ-774), China Postdoctoral Science
Foundation funded project (grant no. 2021MD703953) and Fundamental
Research Funds for the Central Universities (grant no.
xzy012021077).
Abbreviations:
|
lncRNA
|
long non-coding RNA
|
|
CRPC
|
castration-resistant prostate
cancer
|
|
HSPC
|
hormone-sensitive prostate cancer
|
|
ADT
|
androgen deprival therapy
|
|
EMT
|
epithelial-mesenchymal transition
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
GSEA
|
Gene Set Enrichment Analysis
|
|
RIP
|
RNA immunoprecipitation
|
References
|
1
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2022. CA Cancer J Clin. 72:7–33. 2022.
View Article : Google Scholar
|
|
2
|
Xia C, Dong X, Li H, Cao M, Sun D, He S,
Yang F, Yan X, Zhang S, Li N and Chen W: Cancer statistics in China
and United States, 2022: Profiles, trends, and determinants. Chin
Med J (Engl). 135:584–590. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Desai K, McManus JM and Sharifi N:
Hormonal therapy for prostate cancer. Endocr Rev. 42:354–373. 2021.
View Article : Google Scholar
|
|
4
|
Teo MY, Rathkopf DE and Kantoff P:
Treatment of advanced prostate cancer. Annu Rev Med. 70:479–499.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lin SR, Mokgautsi N and Liu YN: Ras and
Wnt interaction contribute in prostate cancer bone metastasis.
Molecules. 25:23802020. View Article : Google Scholar :
|
|
6
|
Tan YT, Lin JF, Li T, Li JJ, Xu RH and Ju
HQ: LncRNA-mediated posttranslational modifications and
reprogramming of energy metabolism in cancer. Cancer Commun (Lond).
41:109–120. 2021. View Article : Google Scholar
|
|
7
|
Li S, Xie X, Peng F, Du J and Peng C:
Regulation of temozolomide resistance via lncRNAs: Clinical and
biological properties of lncRNAs in gliomas (Review). Int J Oncol.
61:1012022. View Article : Google Scholar
|
|
8
|
Yang G, Wu Y, Wan R, Sang H, Liu H and
Huang W: The role of noncoding RNAs in the regulation, diagnosis,
prognosis and treatment of osteosarcoma (Review). Int J Oncol.
59:692021. View Article : Google Scholar
|
|
9
|
Lv Y, Wang Z, Zhao K, Zhang G, Huang S and
Zhao Y: Role of noncoding RNAs in cholangiocarcinoma (Review). Int
J Oncol. 57:7–20. 2020.PubMed/NCBI
|
|
10
|
Sun X, Xin S, Zhang Y, Jin L, Liu X, Zhang
J, Mei W, Zhang B, Ma W and Ye L: Long noncoding RNA CASC11
interacts with YBX1 to promote prostate cancer progression by
suppressing the p53 pathway. Int J Oncol. 61:1102022. View Article : Google Scholar
|
|
11
|
Chen C, Tang X, Liu Y, Zhu J and Liu J:
Induction/reversal of drug resistance in gastric cancer by
non-coding RNAs (Review). Int J Oncol. 54:1511–1524. 2019.
|
|
12
|
Chen LJ, Wu L, Wang W, Zhai LL, Xiang F,
Li WB and Tang ZG: Long non-coding 01614 hyperactivates
WNT/β-catenin signaling to promote pancreatic cancer progression by
suppressing GSK-3β. Int J Oncol. 61:1162022. View Article : Google Scholar
|
|
13
|
Zhang M, Wu L, Wang X and Chen J:
LncKRT16P6 promotes tongue squamous cell carcinoma progression by
sponging miR-3180 and regulating GATAD2A expression. Int J Oncol.
61:1112022. View Article : Google Scholar :
|
|
14
|
Xie H, Zhao J, Wan J, Zhao J, Wang Q, Yang
X, Yang W, Lin P and Yu X: Long noncoding RNA AC245100.4 promotes
prostate cancer tumorigenesis via the microRNA1455p/RBBP5 axis.
Oncol Rep. 45:619–629. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huo W, Qi F and Wang K: Long noncoding RNA
BCYRN1 promotes prostate cancer progression via elevation of
HDAC11. Oncol Rep. 44:1233–1245. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang L, Han S, Jin G, Zhou X, Li M, Ying
X, Wang L, Wu H and Zhu Q: Linc00963: A novel, long non-coding RNA
involved in the transition of prostate cancer from
androgen-dependence to androgen-independence. Int J Oncol.
44:2041–2049. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bai M, He C, Shi S, Wang M, Ma J, Yang P,
Dong Y, Mou X and Han S: Linc00963 promote cell proliferation and
tumor growth in castration-resistant prostate cancer by modulating
miR-655/TRIM24 axis. Front Oncol. 11:6369652021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hu CY, Wu KY, Lin TY and Chen CC: The
crosstalk of long non-coding RNA and MicroRNA in
castration-resistant and neuroendocrine prostate cancer: Their
interaction and clinical importance. Int J Mol Sci. 23:3922021.
View Article : Google Scholar
|
|
19
|
Zhang Y, Zhang J, Liang S, Lang G, Liu G,
Liu P and Deng X: Long non-coding RNA VIM-AS1 promotes prostate
cancer growth and invasion by regulating epithelial-mesenchymal
transition. J BUON. 24:2090–2098. 2019.PubMed/NCBI
|
|
20
|
Yin H, Zhang X, Yang P, Zhang X, Peng Y,
Li D, Yu Y, Wang Y, Zhang J, Ding X, et al: RNA m6A methylation
orchestrates cancer growth and metastasis via macrophage
reprogramming. Nat Commun. 12:13942021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sun J, Xiong Y, Jiang K, Xin B, Jiang T,
Wei R, Zou Y, Tan H, Jiang T, Yang A, et al: Hypoxia-sensitive long
noncoding RNA CASC15 promotes lung tumorigenesis by regulating the
SOX4/beta-catenin axis. J Exp Clin Cancer Res. 40:122021.
View Article : Google Scholar
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
23
|
Feng T, Wei D, Zhao J, Li Q, Guo P, Yang
X, Li M, Jiang Y and Luo Y: Construction of enzalutamide-resistant
cell model of prostate cancer and preliminary screening of
potential drug-resistant genes. Exp Bio Med (Maywood).
15:1776–1787. 2021. View Article : Google Scholar
|
|
24
|
Chen S, Zhou Y, Chen Y and Gu J: fastp: An
ultra-fast all-in-one FASTQ preprocessor. Bioinformatics.
34:i884–i890. 2018. View Article : Google Scholar :
|
|
25
|
Kim D, Langmead B and Salzberg SL: HISAT:
A fast spliced aligner with low memory requirements. Nat Methods.
12:357–360. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liao Y, Smyth GK and Shi W: featureCounts:
An efficient general purpose program for assigning sequence reads
to genomic features. Bioinformatics. 30:923–930. 2014. View Article : Google Scholar
|
|
27
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
The Gene Ontology Consortium: The gene
ontology resource: 20 years and still GOing strong. Nucleic Acids
Res. 47:D330–D338. 2019. View Article : Google Scholar :
|
|
29
|
Kanehisa M: Post-Genome Informatics.
Oxford University Press; New York, NY: 2000
|
|
30
|
Cai C, Wang H, He HH, Chen S, He L, Ma F,
Mucci L, Wang Q, Fiore C, Sowalsky AG, et al: ERG induces androgen
receptor-mediated regulation of SOX9 in prostate cancer. J Clin
Invest. 123:1109–1122. 2013. View Article : Google Scholar :
|
|
31
|
Zhou C, Wang Z, Cao Y and Zhao L:
Pan-cancer analysis reveals the oncogenic role of
3-hydroxy-3-methylglutaryl-CoA synthase 1. Cancer Rep (Hoboken).
5:e15622021.
|
|
32
|
Yang M, Gallo-Ebert C, Hayward M, Liu W,
McDonough V and Nickels JT Jr: Human insulin growth factor 2 mRNA
binding protein 2 increases MicroRNA 33a/b inhibition of liver
ABCA1 expression and alters low-density apolipoprotein levels in
mice. Mol Cell Biol. 40:e00058–e00020. 2020. View Article : Google Scholar :
|
|
33
|
Xu K, Dai X, Wu J and Wen K:
N(6)-methyladenosine (m(6)A) reader IGF2BP2 stabilizes HK2
stability to accelerate the Warburg effect of oral squamous cell
carcinoma progression. J Cancer Res Clin Oncol. 148:3375–3384.
2022. View Article : Google Scholar
|
|
34
|
Xu Q, Chen K and Meng J: WHISTLE: A
functionally annotated high-accuracy map of human m(6)a
epitranscriptome. Methods Mol Biol. 2284:519–529. 2021. View Article : Google Scholar
|
|
35
|
Zhang Y, Huang YX, Wang DL, Yang B, Yan
HY, Lin LH, Li Y, Chen J, Xie LM, Huang YS, et al: LncRNA DSCAM-AS1
interacts with YBX1 to promote cancer progression by forming a
positive feedback loop that activates FOXA1 transcription network.
Theranostics. 10:10823–10837. 2020. View Article : Google Scholar :
|
|
36
|
Wen S, Wei Y, Zen C, Xiong W, Niu Y and
Zhao Y: Long non-coding RNA NEAT1 promotes bone metastasis of
prostate cancer through N6-methyladenosine. Mol Cancer. 19:1712020.
View Article : Google Scholar :
|
|
37
|
Ghildiyal R, Sawant M, Renganathan A,
Mahajan K, Kim EH, Luo J, Dang HX, Maher CA, Feng FY and Mahajan
NP: Loss of long noncoding RNA NXTAR in prostate cancer augments
androgen receptor expression and enzalutamide resistance. Cancer
Res. 82:155–168. 2022. View Article : Google Scholar
|
|
38
|
Sun F, Wu K, Yao Z, Mu X, Zheng Z, Sun M,
Wang Y, Liu Z and Zhu Y: Long noncoding RNA LINC00963 induces NOP2
expression by sponging tumor suppressor miR-542-3p to promote
metastasis in prostate cancer. Aging (Albany NY). 12:11500–11516.
2020. View Article : Google Scholar
|
|
39
|
Zhao H, Dong H, Wang P and Zhu H: Long
non-coding RNA SNHG17 enhances the aggressiveness of C4-2 human
prostate cancer cells in association with β-catenin signaling.
Oncol Lett. 21:4722021. View Article : Google Scholar
|
|
40
|
Bai M, Lei Y, Wang M, Ma J, Yang P, Mou X,
Dong Y and Han S: Long Non-coding RNA SNHG17 promotes cell
proliferation and invasion in castration-resistant prostate cancer
by targeting the miR-144/CD51 Axis. Front Genet. 11:2742020.
View Article : Google Scholar
|
|
41
|
Jin Y, Cui Z, Li X, Jin X and Peng J:
Upregulation of long non-coding RNA PlncRNA-1 promotes
proliferation and induces epithelial-mesenchymal transition in
prostate cancer. Oncotarget. 8:26090–26099. 2017. View Article : Google Scholar
|
|
42
|
Cui Z, Gao H, Yan N, Dai Y, Wang H, Wang
M, Wang J, Zhang D, Sun P, Qi T, et al: LncRNA PlncRNA-1
accelerates the progression of prostate cancer by regulating
PTEN/Akt axis. Aging (Albany NY). 13:12113–12128. 2021. View Article : Google Scholar
|
|
43
|
Shi SJ, Zhang X, Han DH, Yang F, LI Y and
Wang LJ: Long non-coding RNA VIM-AS1 promote proliferation and
invasion of castrition-resistant prostate cancer C4-2 cells. Chin J
Cell Mol Imm. 36:1083–1088. 2020.
|
|
44
|
Gobel A, Riffel RM, Hofbauer LC and
Rachner TD: The mevalonate pathway in breast cancer biology. Cancer
Lett. 542:2157612022. View Article : Google Scholar
|
|
45
|
Laka K, Makgoo L and Mbita Z:
Cholesterol-lowering phytochemicals: Targeting the mevalonate
pathway for anticancer interventions. Front Genet. 13:8416392022.
View Article : Google Scholar
|
|
46
|
Wang IH, Huang TT, Chen JL, Chu LW, Ping
YH, Hsu KW, Huang KH, Fang WL, Lee HC, Chen CF, et al: Mevalonate
pathway enzyme HMGCS1 contributes to gastric cancer progression.
Cancers (Basel). 12:10882020. View Article : Google Scholar
|
|
47
|
Yao W, Jiao Y, Zhou Y and Luo X: KLF13
suppresses the proliferation and growth of colorectal cancer cells
through transcriptionally inhibiting HMGCS1-mediated cholesterol
biosynthesis. Cell Biosci. 10:762020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Walsh CA, Akrap N, Garre E, Magnusson Y,
Harrison H, Andersson D, Jonasson E, Rafnsdottir S, Choudhry H,
Buffa F, et al: The mevalonate precursor enzyme HMGCS1 is a novel
marker and key mediator of cancer stem cell enrichment in luminal
and basal models of breast cancer. PLoS One. 15:e02361872020.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ashida S, Kawada C and Inoue K: Stromal
regulation of prostate cancer cell growth by mevalonate pathway
enzymes HMGCS1 and HMGCR. Oncol Lett. 14:6533–6542. 2017.PubMed/NCBI
|
|
50
|
Cheng Y, Meng Y, Li S, Cao D, Ben S, Qin
C, Hua L and Cheng G: Genetic variants in the cholesterol
biosynthesis pathway genes and risk of prostate cancer. Gene.
774:1454322021. View Article : Google Scholar
|
|
51
|
Xue ST, Zheng B, Cao SQ, Ding JC, Hu GS,
Liu W and Chen C: Long non-coding RNA LINC00680 functions as a
ceRNA to promote esophageal squamous cell carcinoma progression
through the miR-423-5p/PAK6 axis. Mol Cancer. 21:692022. View Article : Google Scholar :
|
|
52
|
Lang C, Yin C, Lin K, Li Y, Yang Q, Wu Z,
Du H, Ren D, Dai Y and Peng X: m(6) A modification of lncRNA PCAT6
promotes bone metastasis in prostate cancer through
IGF2BP2-mediated IGF1R mRNA stabilization. Clin Transl Med. 11. pp.
e4262021, View Article : Google Scholar
|
|
53
|
Chen X, Xie R, Gu P, Huang M, Han J, Dong
W, Xie W, Wang B, He W, Zhong G, et al: Long noncoding RNA LBCS
inhibits self-renewal and chemoresistance of bladder cancer stem
cells through epigenetic silencing of SOX2. Clin Cancer Res.
25:1389–1403. 2019. View Article : Google Scholar
|
|
54
|
Li S, Wu Q, Liu J and Zhong Y:
Identification of Two m6A Readers YTHDF1 and IGF2BP2 as immune
biomarkers in head and neck squamous cell carcinoma. Front Genet.
13:9036342022. View Article : Google Scholar
|
|
55
|
Zhang Z, Xing Y, Gao W, Yang L, Shi J,
Song W and Li T: N(6)-methyladenosine (m(6)A) reader IGF2BP2
promotes gastric cancer progression via targeting SIRT1.
Bioengineered. 13:11541–11550. 2022. View Article : Google Scholar
|
|
56
|
Yao B, Zhang Q, Yang Z, An F, Nie H, Wang
H, Yang C, Sun J, Chen K, Zhou J, et al: CircEZH2/miR-133b/IGF2BP2
aggravates colorectal cancer progression via enhancing the
stability of m(6) A-modified CREB1 mRNA. Mol Cancer. 21:1402022.
View Article : Google Scholar
|
|
57
|
Liu Y, Shi M, He X, Cao Y, Liu P, Li F,
Zou S, Wen C, Zhan Q, Xu Z, et al: LncRNA-PACERR induces pro-tumour
macrophages via interacting with miR-671-3p and m6A-reader IGF2BP2
in pancreatic ductal adenocarcinoma. J Hematol Oncol. 15:522022.
View Article : Google Scholar
|
|
58
|
Han L, Lei G, Chen Z, Zhang Y, Huang C and
Chen W: IGF2BP2 regulates MALAT1 by serving as an
N6-methyladenosine reader to promote NSCLC proliferation. Front Mol
Biosci. 8:7800892021. View Article : Google Scholar
|