Introduction
Renal carcinoma is a commonly diagnosed tumor in the
urinary system. In 2022, estimated new cases of kidney cancer
ranked 8th among all solid tumors in the United States, with 46,345
and 15,259 expected deaths in China and the United States,
respectively (1,2). Clear cell renal cell carcinoma
(ccRCC) is the most common pathological subtype and is highly
likely to metastasize (3).
Patients with distant metastasis often demonstrate unfavorable
prognoses even after radical nephrectomy (4).
F-box protein (FBP) 30 (FBXO30) is a member of the
FBP family. FBPs participate in the assembly of SKP1-CULLIN1-F-box
E3 ubiquitin ligase and act as key substrate-recognition subunits.
Based on the different types of substrate-binding domains, FBPs are
classified into three groups: FBXs, FBWs and FBLs (5,6).
FBXW7 mediates the ubiquitination degradation of c-myc, SOX9, mTOR
and cell cycle proteins, thereby enabling tumors to obtain
resistance to radiotherapy and chemotherapy (7,8).
FBL12 promotes the maturation of undifferentiated thymocytes by
targeting aldehyde dehydrogenase 3 for ubiquitination degradation
(9). In the FBX protein family,
FBXO3 interacts with Smurf1 and degrades it via the ubiquitination
pathway, and EP300-interacting inhibitor of differentiation 1 is
defined as a direct ubiquitination substrate of FBXO21 (10,11).
Despite the fact that the function of FBXO30 was previously
unknown, it has recently been indicated that FBXO30 is involved in
the intracellular ubiquitination regulation of multiple substrates
(12). FBXO30 targets Eg5 for
ubiquitinylation, and impairs mitosis and proliferation in the
mouse mammary gland (13). FBXO30
has been shown to use stem loop-binding protein as a substrate to
regulate chromosome segregation in mouse oocytes (14). In addition, FBXO30 is dysregulated
by BMP-Smad signaling and participates in muscle loss (15). In tumors, FBXO30 has been reported
to be hypermethylated in prostate cancer and has been identified as
a potential clinical biomarker (16,17).
In addition, FBXO30 is located near the D6S1581 gene and can
contribute to genetic susceptibility to nasopharyngeal carcinoma
(18). However, the role of FBXO30
in other types of cancer and the specific mechanisms are largely
unknown.
Hypoxia-inducible factor-1α (HIF-1α) is universally
enriched in solid tumors. Under normoxia, the classical degradation
pathway of HIF-1α occurs through the ubiquitin-proteasome pathway
after its binding to the Von Hippel-Lindau protein (pVHL) E3
ubiquitin ligase (19). An
increasing number of novel E3 ubiquitin ligases using HIF-1α as a
specific substrate have been reported in tumors. In breast cancer,
Parkin and TNF receptor-associated factor 6 have been shown to
target the ubiquitination of HIF-1α. In addition, GSK-3β can
mediate the degradation of HIF-1α in hepatoma cells through FBXW7
(20-22). These studies suggested that FBPs
may have a role in HIF-1α regulation.
As a member of the ZIP family, the main biological
function of human ZRT, IRT-like protein 1 (hZIP1), is to promote
the intracellular transport of Zn2+ to maintain zinc
homeostasis in and out of cells. In prostate cancer tissues, hZIP1
expression is markedly reduced. The decline in intracellular
Zn2+ levels triggered by the downregulation of hZIP1
abundance may be associated with the tumorigenesis of prostate
cancer (23-25). Zn2+ is an important
microelement that is involved in multiple metabolic processes, such
as sugar and lipid metabolism (26). Our previous studies indicated that
hZIP1 was downregulated in ccRCC tissues and that it inhibited
tumor progression by downregulating HIF-1α (27,28).
However, the specific mechanism through which hZIP1 decreases
HIF-1α and the role of Zn2+ remain unknown.
The present study aimed to verify whether HIF-1α was
a novel substrate of FBXO30 in ccRCC cells. In addition, the study
aimed to determine whether FBXO30 interacted with HIF-1α and
mediated its ubiquitination degradation, and whether hZIP1
downregulated HIF-1α by increasing FBXO30 protein abundance.
Finally, to the study explored whether Zn2+ served as a
crucial intermediary in this process.
Materials and methods
Bioinformatics analyses
UALCAN (http://ualcan.path.uab.edu/) was employed to obtain
the expression of FBXO30 in different sample types [533 ccRCC
tissues and 72 normal kidney tissues voluntarily donated by
patients with ccRCC; The Cancer Genome Atlas (TCGA) dataset: kidney
renal cell carcinoma (KIRC)], and to assess the relationship
between FBXO30 expression and different pathological grades and
clinical stages in patients with ccRCC based on data from TCGA
database [dataset (cancer name): KIRC]. The survival curves of
patients with ccRCC and with high or low FBXO30 expression levels
(TCGA dataset: KIRC) were produced using Gene Expression Profiling
Interactive Analysis (http://gepia.cancer-pku.cn/). Patients were split into
two groups based on FBXO30 expression: 50% were in the high
expression group and 50% were in the low expression group.
Cell culture and transfection
OS-RC-2 and 786-O cells were obtained from The Cell
Bank of Type Culture Collection of The Chinese Academy of Sciences
and were cultured in RPMI-1640 medium (HyClone; Cytiva)
supplemented with 10% fetal bovine serum (FBS; Biological
Industries; Sartorius AG). Cells were incubated under normoxia (5%
CO2 and 95% humidity) at 37°C.
The pLVX-Puro-FBXO30 and pLVX-Puro-hZIP1
overexpressing lentiviruses were purchased from Shanghai GeneChem
Co., Ltd., and control cells were transfected with empty vectors.
Lentivirus packaging was performed using a second generation
self-inactivated lentivirus packaging system [GV492 (containing
gcGFP, puromycin and the target gene) vector plasmid, 20 µg;
pHelper 1.0 vector plasmid, 15 µg; pHelper 2.0 vector
plasmid, 10 µg]. Briefly, 293T cells (The Cell Bank of Type
Culture Collection of The Chinese Academy of Sciences) were
cultured to 50% confluence in a six-well plate with 2 ml culture
medium and were transfected with the lentiviruses at a
concentration of 10×106 transducing units using HiTransG
(Shanghai GeneChem Co., Ltd.) according to the manufacturer's
instruction at 37°C for 48 h. After centrifugation at 1,500 × g for
15 min at 4°C, the virus suspension was collected and was then used
for transduction. OS-RC-2 and 786-O cells were transduced with
virus suspension at 37°C for 48 h (virus volume=cell counts x
MOI/virus titer). The multiplicity of infection for OS-RC-2 and
786-O cells was 5. The pGPU6/mCherry/Puro-Negative Control and
pGPU6/mCherry/Puro-FBXO30 short hairpin (sh)RNA plasmids were
constructed by Shanghai GenePharma Co., Ltd. and cells were
transfected using Lipofectamine® 3000 Reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol at room temperature for 8 h. After
selection with puromycin (2 µg/ml) over 72 h to construct
stable cell lines, stable cells transduced with overexpressing
lentiviruses and transfected with shRNA plasmids were harvested for
subsequent assays; puromycin (1 µg/ml) was used for
maintenance of stable cells transduced with overexpressing
lentiviruses and transfected with shRNA plasmids. Negative control,
FBXO30 and hZIP1 small interfering RNAs (siRNAs) to were purchased
from JTSBIO Co. The shRNA and siRNA sequences and the vector
transcript numbers are listed in Tables SI and SII.
Patient samples
All ccRCC tissues and corresponding adjacent normal
tissues (ANTs; ≥2 cm from the edge of the tumor) were obtained from
64 patients in the Urology Department, The First Hospital of China
Medical University (Shenyang, China) between January 2018 and
December 2021. The average age of the patients was 54 years (age
range, 45-63 years), the sex ratio was 1:1, and all of the patients
were diagnosed as ccRCC. A total of 20 pairs of tumor tissues and
ANTs were used for western blotting, 24 pairs underwent reverse
transcription-quantitative PCR (RT-qPCR) and 20 pairs were used for
immunohistochemistry. The groups were classified based on the
Fuhrman grade (29); T1 and T2
were defined as low grade, and T3 and T4 were defined as high
grade. The present study was approved by the Research Ethics
Committee of China Medical University, and the approval number was
[2019]2019-65-3; written informed consent was obtained from all
patients.
Western blotting
Antibody information is available in Table SIII. All antibodies were used
according to the manufacturers' protocols. First, total proteins of
OS-RC-2 and 786-O cells were extracted using RIPA solution (RIPA:
PMSF, 100:1; cat. no. P0013B; Beyotime Institute of Biotechnology).
Protein concentration was determined using Enhanced BCA Protein
Assay Kit (cat. no. P0010; Beyotime Institute of Biotechnology).
Proteins (40 µg/lane) were then separated by SDS-PAGE on 10%
gels. Subsequently, the proteins were transferred onto PVDF
membranes, which were blocked with skim milk for 1 h at 37°C. The
membranes were then incubated with primary antibodies (1:1,000)
overnight at 4°C and with secondary antibodies (1:4,000) for 1 h at
37°C. Finally, the ECL reagents (Beijing Transgen Biotech Co.,
Ltd.) on a MicroChemi Chemiluminescent Imaging System (DNR
Bio-Imaging Systems Ltd.). was used to visualize the blots. Image J
1.6.0 software (National Institutes of Health) was used to
determine the ratios of target protein/β-actin.
RNA isolation and RT-qPCR
RNAiso Plus and PrimeScript™ RT Master Mix (both
from Takara Biotechnology, Co., Ltd.) were used to isolate total
RNA of OS-RC-2 and 786-O cells and synthesize cDNA, respectively,
according to the manufacturer's protocols. qPCR was then performed
using SYBR® Premix Ex Taq™ (Tli RNaseH Plus) on a
LightCycler™ 480II system (Roche Diagnostics). The thermocycling
conditions were as follows: initial denaturation at 94°C for 3 min,
followed by 45 cycles of denaturation at 95°C for 20 sec, annealing
at 55°C for 20 sec and extension at 72°C for 20 sec, and a final
extension step at 72°C for 10 min. The 2−ΔΔCq method
(30) was performed to calculate
the relative expression levels. All of the aforementioned reagents
were obtained from Takara Biotechnology, Co., Ltd., unless
otherwise specified. Primer sequences are provided in Table SI.
Cell viability assay
OS-RC-2 or 786-O cells (3×103) were
seeded in each well of 96-well plates. After inoculated cells grew
to ~1×105, Cell Counting Kit-8 (CCK-8) working solution
(Bimake; CCK-8 total volume, 10:100) was then added to each well.
After incubation at 37°C for 1 h, the absorbance of the culture
solution was measured at 450 nm using a microplate reader (Model
680; Bio-Rad Laboratories, Inc.) at 24, 48 and 72 h.
5-Ethynyl-2′-deoxyuridine (EdU)
assay
An EdU Cell Proliferation Kit (cat. no. C0071S;
Beyotime Institute of Biotechnology) was used to assess
proliferation. After the cells grew to ~1×105, EdU
reagent (1:1,000) was added to each well and incubated at 37°C for
2 h. Subsequently, cells were successively treated with 4%
paraformaldehyde (5 min/time for three times) and 3% Triton X-100
(5 min/time for three times) at room temperature. The cells were
then treated with click reaction buffer at 37°C for 30 min, after
which Hoechst solution was added for 15 min at room temperature.
All reaction steps were performed in the dark. Finally, an
immunofluorescence microscope (Olympus Corporation) was used to
observe and capture images of the cells.
Wound-healing assay
After cells grew to 100% confluence in 24-well
plates, 1-ml pipette tips were used to make a scratch in the center
of the wells. The cells were then cultured with serum-free medium
at 37°C for 24 h. Cell images were acquired with an inverted
microscope (EVOS XL system; cat. no. AMEX12000; Thermo Fisher
Scientific, Inc.). ImageJ 1.6.0 software was used to calculate the
wound area, as follows: Average scratch width=scratch area/length;
cell migration rate=(0 h scratch width-24 h scratch width)/0 h
scratch width ×100.
Cell migration and invasion assays
Cells were suspended in medium supplemented with 2%
FBS (1×105 cells/well for migration assay and
2×105/well for invasion assay), and 200 µl cell
suspension was added to the upper chambers of Transwell plates
(pore size, 8.0 µm; Corning, Inc.). In the invasion assay,
chambers were covered with Matrigel (Matrigel:serum-free medium,
1:5; 20 µl/chamber; BD Biosciences) for 30 min at 37°C.
Subsequently, the chambers were placed into 24-well plates with 600
µl medium (10% FBS) in the lower chamber. After incubation
at 37°C for 48 h, the chambers were gently rinsed with PBS and the
cells were removed with cotton swabs. Crystal violet solution (cat.
no. C0121; Beyotime Institute of Biotechnology) was used for
staining (10 min at room temperature). Images were obtained under
an inverted light microscope at ×100 magnification.
MG132 inhibition and protein half-life
assays
For the MG132 (cat. no. HY-13259; MedChemExpress)
inhibition assay, cells were treated with MG132 (10 µM/well)
after growing to 70-80% confluence in six-well plates for 8 h at
37°C. Subsequently, total protein was extracted and subjected to
western blotting as aforementioned to detect the protein expression
levels of HIF-1α. For the protein half-life assay, cells were
cultured to 80-90% confluence in 100-mm culture dishes and were
treated with 10 µM cycloheximide (CHX, cat. no. HY-12320;
MedChemExpress) at 37°C. Cells were collected at 0, 20, 40 and 60
min. Subsequently, total protein was extracted and subjected to
western blotting as aforementioned to detect the protein expression
levels of HIF-1α. The half-life of HIF-1α protein was calculated by
scanning the density value of the blots using ImageJ 1.6.0
software.
Immunofluorescence staining assay
Cells were cultured to 80% density in 24-well plates
before fixation with 4% paraformaldehyde for 30 min at 37°C. Cells
were then incubated with 0.2% Triton X-100 for 30 min at 37°C,
followed by blocking with 5% FBS for 30 min at 37°C. Subsequently,
the cells were incubated with FBXO30 and HIF-1α antibodies (1:100)
at 4°C overnight. Alexa Fluor® 488 and 594 secondary
antibodies (cat. nos. ab150077 and ab150080; 1:200; Abcam) were
then added and incubated for 60 min in the dark at 37°C. Finally,
the cells were treated with DAPI solution [1:3; cat. no. abs9235,
Absin (Shanghai) Biotechnology Co., Ltd.] with agitation for 20 min
in the dark, and images were captured under a fluorescence
microscope.
Co-immunoprecipitation (Co-IP) assay
Cells were lysed in lysis buffer
(RIPA:PMSF:proteasome inhibitor, 100:1:1), then the lysate was
centrifuged at 13,201 × g for 20 min at 4°C. Appropriate amounts of
supernatant were separated for use as input. Subsequently, 2
µg FBXO30, HIF-1α or ubiquitin antibodies and IgG were added
to each 500 µl of lysate, with vertical mixing at 4°C
overnight. Afterwards, A/G beads (cat. no. B23202; Bimake) were
washed with TBS-0.1% Tween (cat. no. T1081; Beijing Solarbio
Science & Technology Co., Ltd.) for 1 h with vertical mixing at
4°C. Subsequently, 20 µl A/G beads was added to each 500
µl of lysate and vertically mixed at 4°C for 1 h. Each 20
µl of magnetic beads was isolated using a magnetic separator
(cat. no. B23803; Bimake). After being suspended in 10 µl 2X
SDS loading buffer (cat. no. P1040; Beijing Solarbio Science &
Technology Co., Ltd.), the magnetic beads were incubated at 100°C
for 20 min. Finally, the supernatant was aspirated and subjected to
western blotting.
Zinc supplementation assay
Cells were cultured to 70-80% confluence in six-well
plates. Subsequently, different concentration of ZnCl2
solution (cat no. 366374; MilliporeSigma) was added to the culture
medium for 2 h at 37°C. Total protein was extracted and subjected
to western blotting to detect the protein expression levels of
HIF-1α as aforementioned.
Animal experiments
The animal experiments performed in the present
study were approved by the Ethics Committee of China Medical
University (approval no. KT2022527). A total of 16 SPF/VAF female
BALB/c-nude mice (age, ~4 weeks) were obtained from Beijing Vital
River Laboratory Animal Technology Co., Ltd. for use in the in
vivo tumorigenesis experiments and were housed in the
Department of Laboratory Animal Science, China Medical University.
The nude mice were housed in individual ventilated cages under
suitable conditions (temperature, 25°C; humidity, 55%; ventilation
velocity, 0.20 m/sec; light/dark cycle, 12/12 h) and were given
free access to sterilized fodder and purified water via feeders on
the cages. The nude mice were euthanized through cervical
dislocation when they lost 20% of their original body weight. In
the tumor transplantation experiment, five mice were included in
each group. OS-RC-2 cells (2×106) transfected with empty
or FBXO30 vectors were resuspended in 150 µl serum-free
medium with 40% Matrigel and were then injected subcutaneously into
the flank of each nude mouse. The tumor mass was recorded after 30
days and tumor volume was recorded every 4 days. In the lung
metastasis experiment, each group contained three mice. OS-RC-2
cells (1×106) transfected with empty or FBXO30 vectors
were digested and resuspended in 150 µl sterile PBS, and
were then slowly injected into the tail vein of each nude mouse.
After ~45 days, the lung tissues were separated and the number of
metastatic tumors was counted.
Immunohistochemistry (IHC) assay and
hematoxylin and eosin (H&E) staining assay
Moderate-sized (1×1×0.2 cm) fresh ccRCC tissues from
human patients were immersed in 4% formalin for 48 h at room
temperature, and then dehydrated and embedded in paraffin. The
paraffin-embedded tissues were then cut into 4-µm sections.
At room temperature, the sections were deparaffinized in xylene I
and II for 10 min/time, and were then rehydrated in ethanol
gradient solution (from high to low: 100, 95, 80 and 70%, 2
min/time) and washed three times with PBS (3 min/wash). The
sections were soaked in sodium citrate antigen retrieval solution
(cat. no. C1032; Beijing Solarbio Science & Technology Co.,
Ltd.) and were put into a microwave for 10 min at 100°C. An
immunohistochemistry kit (cat. no. KIT-9720; Fuzhou Maxim
Biotechnology Development Co., Ltd.) was used for
immunohistochemistry. The sections were first incubated with
endogenous peroxidase blocker and non-specific dye blocking agent
for 10 min at room temperature. The sections were then incubated
with FBXO30 antibodies (1:100) at 4°C overnight, followed by
incubation with biotinylated anti-IgG antibody and Streptomyces
antibiotic protein-peroxidase (cat. no. KIT-9720; Fuzhou Maxim
Biotechnology Development Co., Ltd.) (10 min at room temperature).
Sections were stained with a DAB kit for 1 min at room temperature
(cat. no. DAB-0031; Fuzhou Maxim Biotechnology Development Co.,
Ltd.). Finally, sections were observed and images were captured
under an inverted light microscope.
For H&E staining, the whole lung tissues from
mice were sectioned after paraffin embedding. Subsequently, the
sections were dewaxed, rehydrated and stained with 1.0% hematoxylin
(cat. no. G4070; Beijing Solarbio Science & Technology Co.,
Ltd.) for 10 min at room temperature. The samples were then stained
with 0.5% eosin (cat. no. G1100; Beijing Solarbio Science &
Technology Co., Ltd.) for 2 min at room temperature. Finally,
sections were observed, and images were captured under an inverted
light microscope.
Statistical analysis
To ensure adequate power, all experiments were
performed independently at least three times. All data were
statistically analyzed using GraphPad Prism (version 9.0; GraphPad;
Dotmatics). The mean values of the groups were compared, and the
results were presented as the mean ± standard deviation (SD).
Differences between two paired groups were analyzed using the
nonparametric paired Student's t-test. Unpaired Student's t-test
(two-tailed) was used for differential analysis between groups.
One-way ANOVA followed by Dunnett's test was used to analyze the
differences between multiple experimental groups and a single
control group. Survival analysis was performed using the
Kaplan-Meier method and the log-rank test. P<0.05 was considered
to indicate a statistically significant difference.
Results
FBXO30 is expressed at low levels in
ccRCC patients and is associated with poor outcomes
To obtain a preliminary understanding of the
relationship between FBXO30 and ccRCC, TCGA database was used to
determine the differential expression of FBXO30 in ccRCC tissues
and its relevance to the prognosis of patients with ccRCC. The mRNA
expression levels of FBXO30 in 533 ccRCC tissues were significantly
lower than those in 72 ANTs from the 533 patients with ccRCC
(Fig. 1A). The mRNA expression
levels of FBXO30 were negatively associated with tumor grade and
with the individual cancer stages of ccRCC; the expression levels
of FBXO30 decreased with tumor progression and staging (Fig. 1B and C). Furthermore, a prognostic
analysis on FBXO30 and RCC was performed using Gene Expression
Profiling Interactive Analysis to plot Kaplan-Meier survival curves
based on data from TCGA database. The results showed that patients
with ccRCC and higher FBXO30 expression were inclined to have
better overall survival (Fig. 1D).
These data were then validated in clinical specimens acquired from
patients with ccRCC. Correspondingly, the results of western
blotting demonstrated a significant decrease in the protein
expression levels of FBXO30 in ccRCC tissues (Fig. 1E and F). The mRNA expression levels
of FBXO30 were also downregulated in ccRCC tissues, as determined
by RT-qPCR (Fig. 1G). In addition,
IHC results further revealed that FBXO30 exhibited deeper staining
in low-grade ccRCC tissues, indicating that FBXO30 protein
expression was negatively associated with the progression of ccRCC
(Fig. 1H).
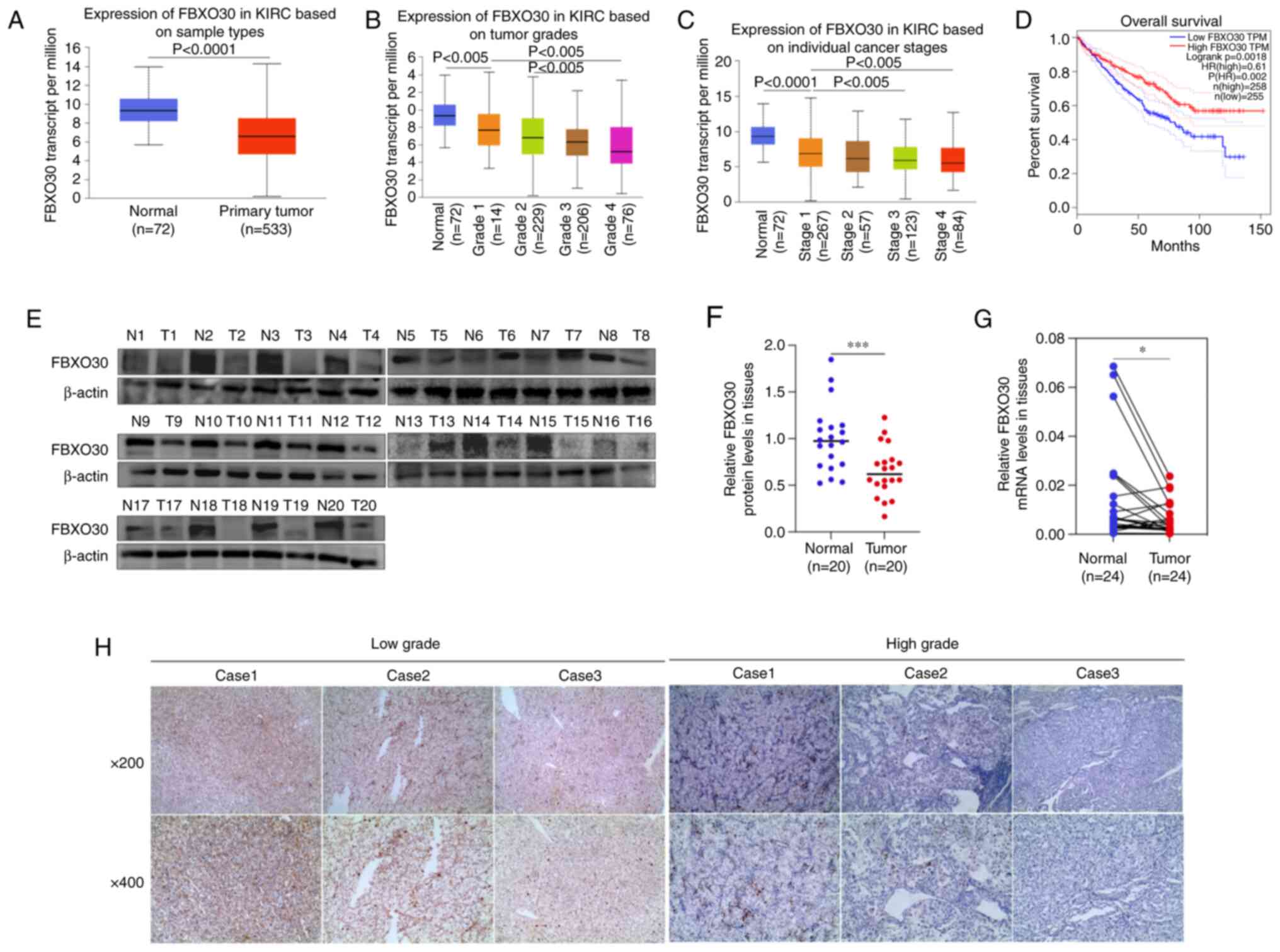 | Figure 1FBXO30 is expressed at low levels in
patients with ccRCC and with poor outcomes. (A) FBXO30 expression
levels in renal cancer tissues (n=253) and normal tissues (n=72)
based on TCGA database. FBXO30 expression levels according to (B)
different tumor grades and (C) individual tumor stages in TCGA
database. Data were statistically analyzed using one-way ANOVA and
Tukey's post hoc test. (D) Kaplan-Meier survival curves depicting
overall survival in patients with ccRCC stratified by FBXO30
protein expression based on TCGA database. Cut-off value, 50%. (E
and F) Western blotting was employed to detect the protein
expression levels of FBXO30 in 20 pairs of clinically derived ccRCC
tissues and corresponding ANTs. (G) mRNA expression levels of
FBXO30 in 24 pairs of ccRCC tissues and corresponding ANTs, as
determined by reverse transcription-quantitative PCR assay. (H)
Immunohistochemistry staining of FBXO30 in high-grade and low-grade
ccRCC tissues. The high and low groups were classified based on the
Fuhrman grade in patients with ccRCC. T1 and T2 were defined as low
grade, and T3 and T4 are defined as high grade. Images were
acquired with an inverted microscope at ×200 and ×400
magnification. *P<0.05, ***P<0.001, as
determined by paired Student's t-test. ANTs, adjacent normal
tissues; ccRCC, clear cell renal cell carcinoma; FBXO30, F-box
protein 30; KIRC, kidney renal clear cell carcinoma; N, normal; T,
tumor; TCGA, The Cancer Genome Atlas. |
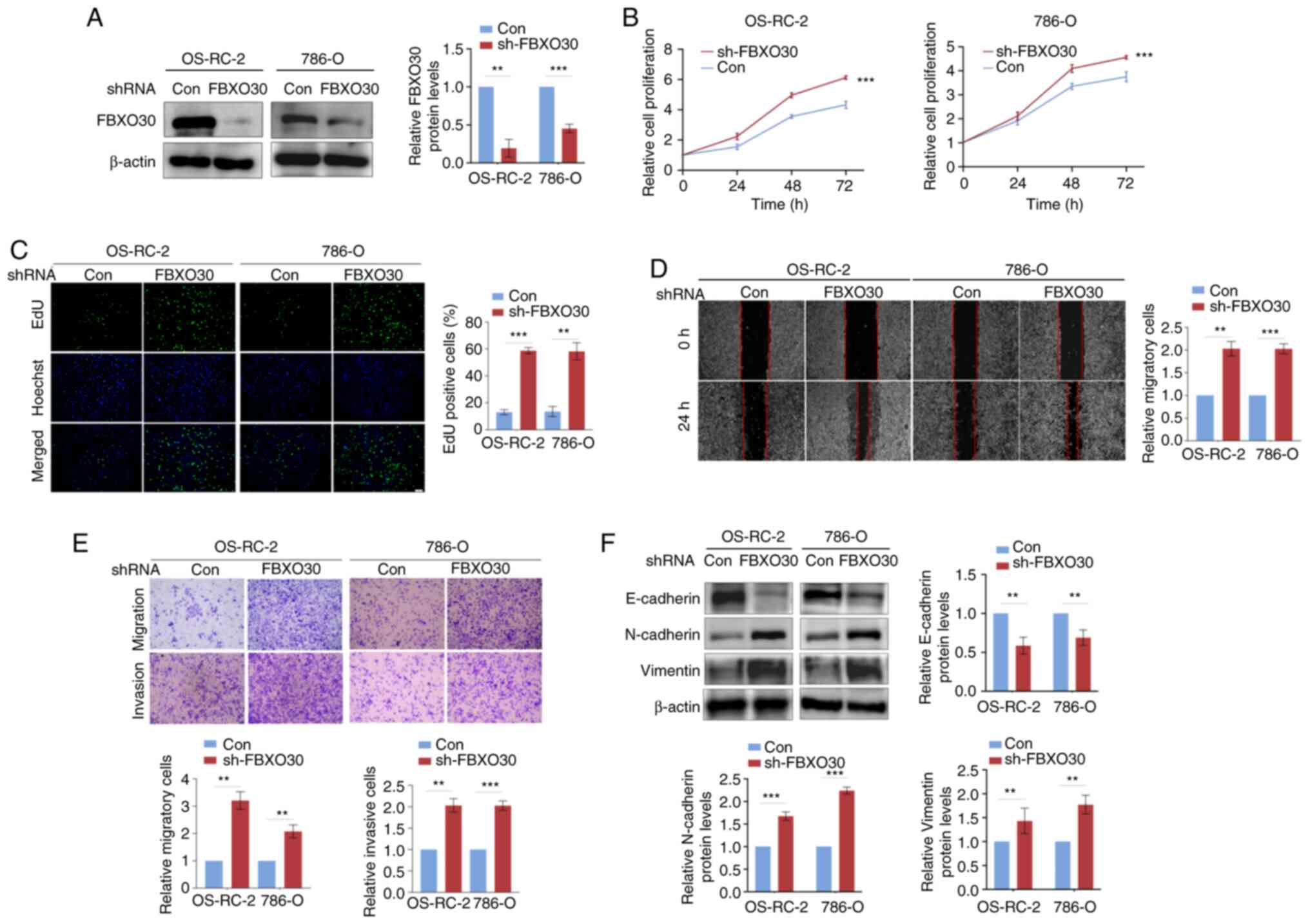
FBXO30 disruption fosters malignant
behaviors in ccRCC cells
To explore the biological function of FBXO30 in
ccRCC, FBXO30 was stably knocked down in OS-RC-2 and 786-O cells
and the knockdown efficiency was confirmed by western blotting
(Fig. 2A). CCK-8 and EdU assays
were then performed to measure the proliferation of cells. As
determined using the CCK-8 assay, knockdown of FBXO30 significantly
enhanced the proliferation of OS-RC-2 and 786-O cells (Fig. 2B). As determined using the EdU
assay, enhanced EdU fluorescent signals were observed in
FBXO30-knockdown ccRCC cells, which suggested that the ccRCC cells
had a stronger proliferative capacity after FBXO30 knockdown
(Fig. 2C). In the wound-healing
assay, FBXO30 knockdown increased the wound-healing area in the
center of the scratch, indicating that FBXO30 knockdown promoted
the migration of ccRCC cells (Fig.
2D). Consistently, in the Transwell assay, ccRCC cells with
FBXO30 knockdown more easily passed through the membrane in the
Transwell chamber, as more stained cells were observed (Fig. 2E). Furthermore, to detect the
change in the invasive ability of cells, Matrigel was used to coat
the Transwell chamber to simulate the extracellular matrix in
vivo. It was revealed that more cells passed through the
Matrigel and membrane in the FBXO30-knockdown group, and silencing
FBXO30 significantly augmented the invasiveness of OS-RC-2 and
786-O cells (Fig. 2E). Finally,
western blotting was performed to evaluate epithelial-mesenchymal
transition (EMT)-related proteins and it was revealed that
knockdown of FBXO30 upregulated N-cadherin and vimentin expression,
but downregulated E-cadherin expression, which indicated that
FBXO30 knockdown contributed to the EMT process in ccRCC cells
(Fig. 2F). These results suggested
that inhibition of FBXO30 could promote the proliferation,
migration, invasion and EMT progression of ccRCC cells.
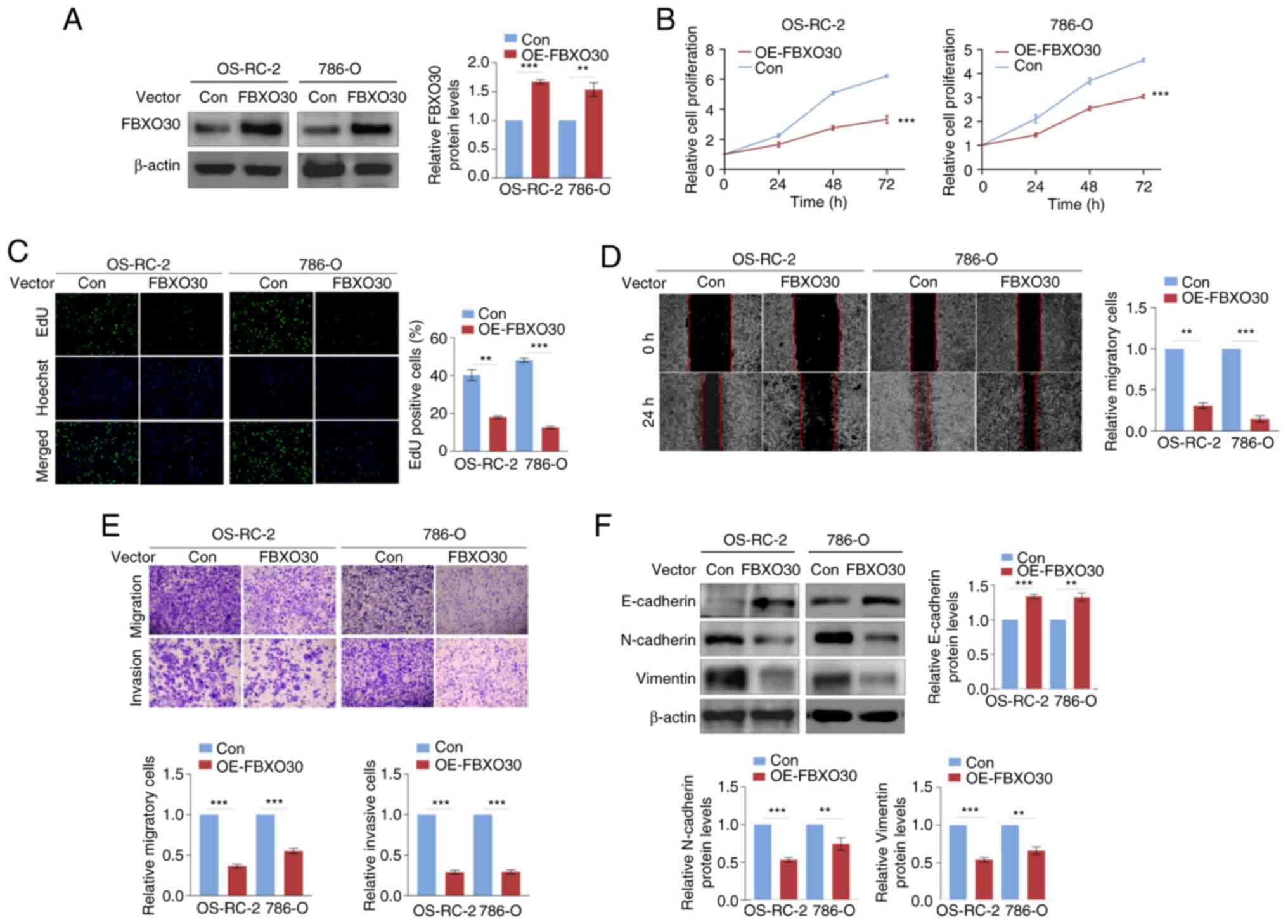
FBXO30 suppresses malignant behaviors in
ccRCC cells
The present study also constructed
FBXO30-overexpressing OS-RC-2 and 786-O cells, and the
overexpression efficiency was confirmed by western blotting
(Fig. 3A). Notably, FBXO30
overexpression limited the viability and proliferative capacity of
two ccRCC cell lines, as determined by the CCK-8 and EdU assays
(Fig. 3B and C). Furthermore,
FBXO30 overexpression significantly reduced the migratory capacity
of ccRCC cells in both the wound-healing and Transwell migration
assays (Fig. 3D and E). FBXO30
overexpression also suppressed the invasiveness of OS-RC-2 and
786-O cells in the Transwell invasion assay (Fig. 3E). Furthermore, FBXO30 inhibited
the EMT process, upregulating E-cadherin expression, but
downregulating N-cadherin and vimentin expression in ccRCC cells
(Fig. 3E and F). These data
further verified that FBXO30 might function as a tumor suppressor
gene in ccRCC.
FBXO30 mediates the ubiquitination and
degradation of HIF-1α in ccRCC cells
To study the relationship between FBXO30 and HIF-1α,
FBXO30 was first knocked down in the OS-RC-2 and 786-O cell lines
using siRNA and the transfection efficiency was tested by western
blotting. It was revealed that the protein expression levels of
HIF-1α increased (Fig. 4A).
Conversely, overexpression of FBXO30 in these cell lines decreased
the expression levels of the HIF-1α protein (Fig. 4A and B). However, there were no
significant alterations in the mRNA expression levels of HIF-1α in
RCC cells with both knockdown and overexpression of FBXO30
(Fig. 4C and D). These results
indicated that FBXO30 tended to post-translationally regulate
HIF-1α.
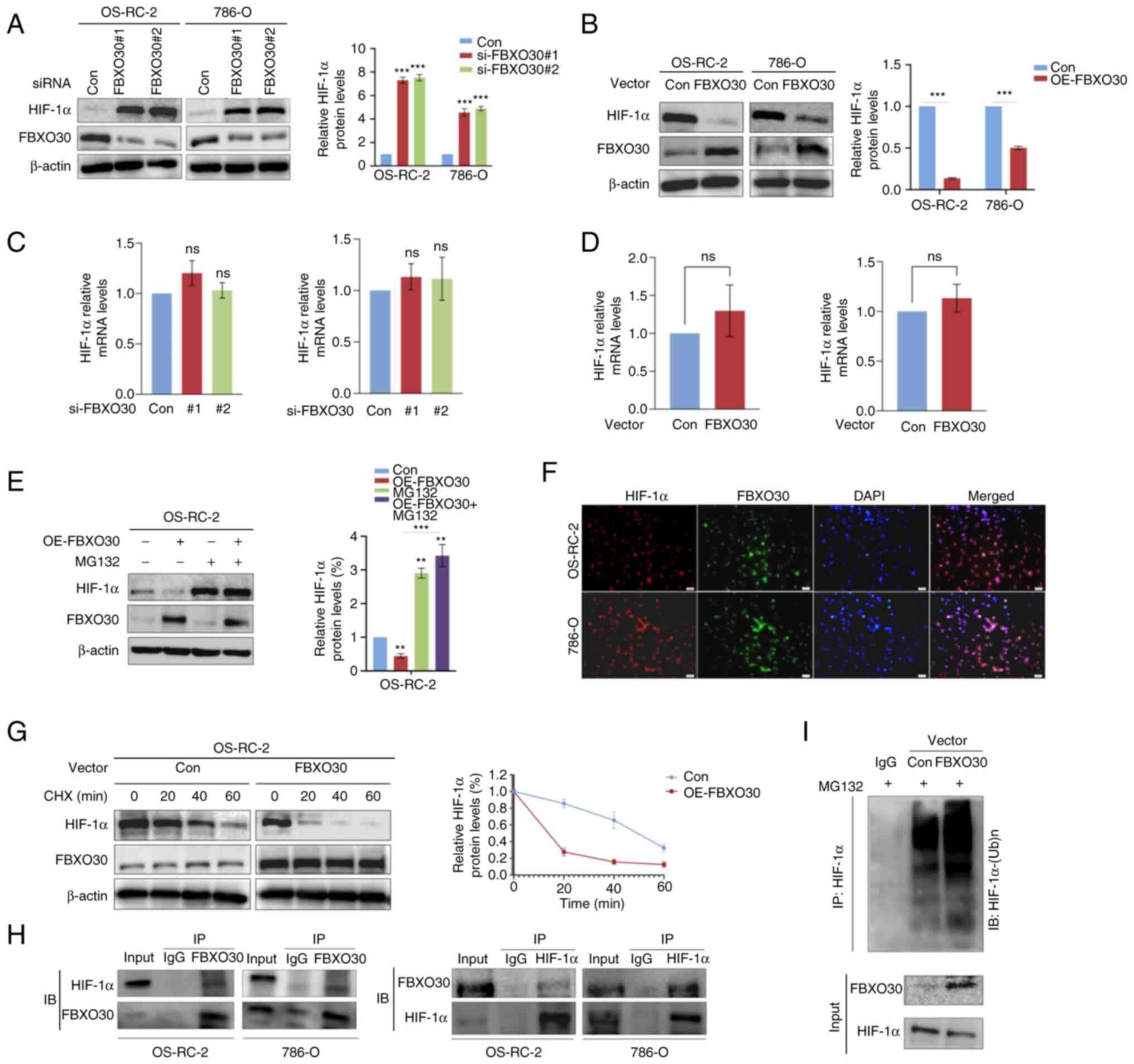 | Figure 4FBXO30 mediates the ubiquitination
and degradation of HIF-1α in ccRCC cells. After (A) knockdown and
(B) OE of FBXO30 in OS-RC-2 and 786-O cells, HIF-1α protein
expression was detected, and the efficiency of knockdown and
overexpression was examined by western blotting. Reverse
transcription-quantitative PCR was performed to detect the mRNA
expression levels of HIF-1α in (C) FBXO30-knockdown and (D)
FBXO30-OE OS-RC-2 and 786-O cells. (E) Western blotting was
performed to determine the effect of the proteasome inhibitor MG132
(10 µM, 8 h) on HIF-1α protein expression in OS-RC-2 cells,
and MG132 rescued the downregulation of HIF-1α in
FBXO30-overexpressing OS-RC-2 cells. (F) FBXO30 OE reduced the
half-life of HIF-1α protein. Under normoxia, OS-RC-2 cells were
treated with CHX (10 µM) at preset time points.
Subsequently, the cells were harvested and subjected to western
blotting. (G) Immunofluorescence colocalization assay demonstrated
that in OS-RC-2 and 786-O cells, HIF-1α (red) possessed the same
subcellular localization as FBXO30 (green), almost coinciding with
nuclear staining (blue). Images were captured under an
immunofluorescence microscope (×200 magnification). (H) Co-IP assay
indicated endogenous binding between FBXO30 and HIF-1α in both
OS-RC-2 and 786-O cells. (I) FBXO30 promoted the conjugation of
ubiquitin to HIF-1α in OS-RC-2 cells and targeted HIF-1α for
degradation. **P<0.01 and ***P<0.001 as
indicated or vs. control; ns, not significant. Data were analyzed
by (A, C and E) one-way ANOVA and Tukey's post hoc test, or (B and
D) unpaired Student's t-test. CHX, cycloheximide; FBXO30, F-box
protein 30; HIF-1α, hypoxia-inducible factor-1α; IB, immunoblot;
IP, immunoprecipitation; OE, overexpression; siRNA, small
interfering RNA. |
FBXO30-overexpressing OS-RC-2 cells were
subsequently treated with the proteasome inhibitor MG132. There was
a significant difference in HIF-1α protein expression levels before
and after treating FBXO30-overexpressing OS-RC-2 cells with MG132.
It was revealed that MG132 could reverse the negative regulation of
HIF-1α by FBXO30 (Fig. 4E),
indicating that the downregulation of HIF-1α mediated by FBXO30
was, at any rate, partially dependent on the proteasomal
degradation pathway. Therefore, the protein synthesis inhibitor CHX
was used to test the effect of FBXO30 on the degradation of HIF-1α.
Notably, FBXO30 prominently shortened the half-life of HIF-1α
protein (Fig. 4F).
Considering that FBXO30 may act as an E3 ubiquitin
ligase because it belongs to the FBP family, it was hypothesized
that FBXO30 was able to regulate the protein levels of HIF-1α
through the ubiquitin-proteasome degradation pathway. The results
of the immunofluorescence colocalization assay preliminarily showed
that the localization of FBXO30 and HIF-1α in OS-RC-2 cells almost
coincided, with these two proteins more likely to be located in the
nucleus (Fig. 4G). Furthermore,
co-IP experiments showed that FBXO30 directly bound to HIF-1α in
OS-RC-2 and 786-O cells (Fig. 4H).
Notably, overexpression of FBXO30 facilitated the conjugation of
ubiquitin to HIF-1α in coIP assay (anti-ubiquitin) (Fig. 4I). Taken together, these data
suggested that in ccRCC cells, FBXO30 was highly likely to
downregulate the protein expression levels, but not the mRNA
expression levels of HIF-1α through the ubiquitin proteasome
pathway.
hZIP1 regulates HIF-1α and FBXO30 in a
Zn2+-dependent manner in ccRCC cells
Given that our previous study reported that hZIP1
downregulated HIF-1α expression (25), the present study aimed to determine
whether FBXO30 had a role in this process. Thus, the present study
examined the expression levels of FBXO30 in hZIP1-overexpressing
OS-RC-2 and 786-O cells. The hZIP-1 overexpression efficiency was
tested by western blotting. Notably, overexpression of hZIP1
increased the protein expression levels of FBXO30 in OS-RC-2 and
786-O renal cancer cells (Fig.
5A). Furthermore, knockdown of FBXO30 rescued the
downregulation of HIF-1α protein expression levels induced by hZIP1
to some extent (Fig. 5A and B),
which verified the hypothesis that hZIP1 decreased HIF-1α protein
expression levels at least partially by downregulating FBXO30
expression.
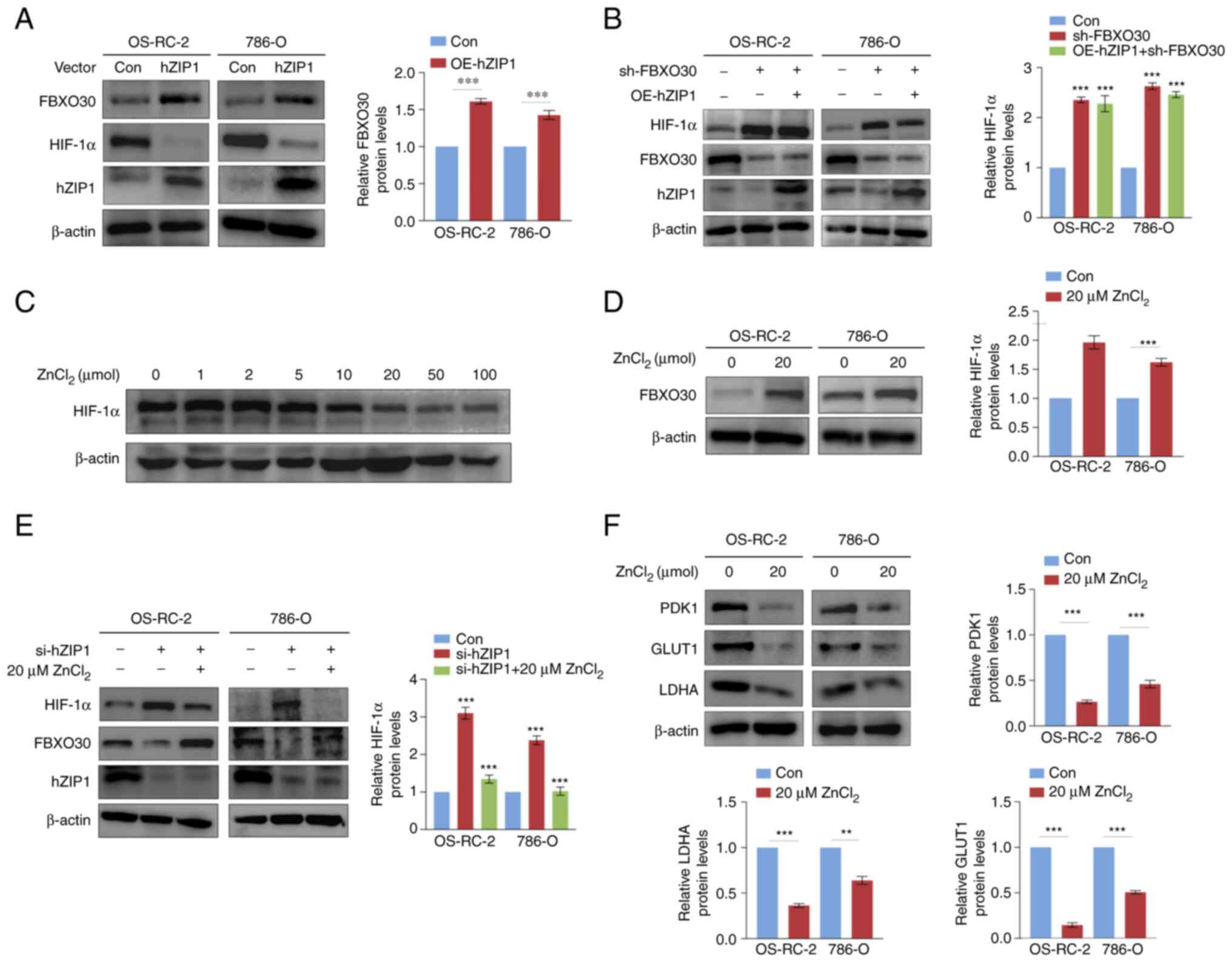 | Figure 5hZIP1 regulates HIF-1α and FBXO30 in
a Zn2+-dependent manner in ccRCC cells. (A) OE of hZIP1
in OS-RC-2 and 786-O cells inhibits the expression of HIF-1α and
upregulates the protein expression levels of FBXO30. (B) Western
blotting demonstrated that FBXO30 knockdown rescued HIF-1α
depletion in hZIP1-OE OS-RC-2 and 786-O cells. (C) Zinc
supplementation assay confirmed that zinc ions inhibited the
expression of HIF-1α in OS-RC-2 cells, and 20 µM was defined
as the optimal inhibitory concentration. After the addition of
increasing concentrations of ZnCl2 solution to the
culture medium for 2 h at 37°C in normoxia, the cells were
collected and lysed for western blotting. (D) Upregulation of
FBXO30 was induced by optimal concentrations of zinc ions in
OS-RC-2 and 786-O cells. (E) FBXO30 downregulation and HIF-1α
accumulation were observed after knockdown of hZIP1, whereas
supplementation with zinc ions counteracted this alteration in the
expression levels of FBXO30 and HIF-1α in hZIP1-knockdown OS-RC-2
and 786-O cells. ***P<0.001 in (B and E) was obtained
by comparing each experimental group with control group. (F) Zinc
ions downregulated the expression levels of glucose
metabolism-related enzymes (PDK1, GLUT1 and LDHA).
**P<0.01 and ***P<0.001 as indicated or
vs. control. Data were analyzed by (B and E) one-way ANOVA and
Dunnett's post hoc test, or (A, D and F) unpaired Student's t-test.
FBXO30, F-box protein 30; GLUT1, glucose transporter 1; HIF-1α,
hypoxia-inducible factor-1α; hZIP1, human ZRT, IRT-like protein 1;
LDHA, lactate dehydrogenase A; OE, overexpression; PDK1, pyruvate
dehydrogenase kinase 1; sh, short hairpin. |
It has previously been reported that hZIP1 serves as
a transport protein to transfer Zn2+ from the
extracellular zone to the intracellular area (22). To explore the potential function of
Zn2+, RCC cells were treated with progressively
increasing concentrations of ZnCl2. It was revealed that
the protein expression levels of HIF-1α were negatively associated
with the concentration of Zn2+, and the optimal
inhibiting concentration was likely to be ~20 µM (Fig. 5C). Correspondingly, treatment with
20 µM Zn2+ resulted in upregulation of FBXO30
protein expression levels in OS-RC-2 and 786-O cells (Fig. 5D). To ultimately identify the role
of Zn2+, hZIP1-knockdown RCC cells were treated with 20
µM Zn2+. Notably, Zn2+ rescued the
expression of FBXO30 and reduced the protein expression levels of
HIF-1α after treating hZIP1-knockdown OS-RC-2 and 786-O cells with
ZnCl2, which suggested that hZIP1 may regulate the
protein expression levels of FBXO30 and HIF-1α by altering the
concentration of Zn2+ in ccRCC cells (Fig. 5E).
To preliminarily understand the possible effects of
zinc ions on glycolysis in ccRCC cells, the expression levels of
some enzymes in glycolysis were detected, namely, pyruvate
dehydrogenase kinase 1 (PDK1), lactate dehydrogenase A (LDHA) and
glucose transporter 1 (GLUT1), which were proven to be regulated by
HIF-1α (31). The present study
revealed that Zn2+ significantly downregulated the
expression levels of PDK1, LDHA and GLUT1 (Fig. 5F). Overall, hZIP1 downregulated
HIF-1α through FBXO30, in which Zn2+ exerted an
essential intermediary role.
FBXO30 inhibits tumor growth in vivo
To further verify the tumor biological function of
FBXO30 in vivo, OS-RC-2 cells transfected with the empty and
FBXO30 vectors were subcutaneously injected into the flank of nude
mice. After 30 days, the nude mice were euthanized, and the tumors
were excised and weighed. FBXO30 overexpression markedly inhibited
tumor formation, as evidenced by the lower tumor weight (Fig. 6A and B).
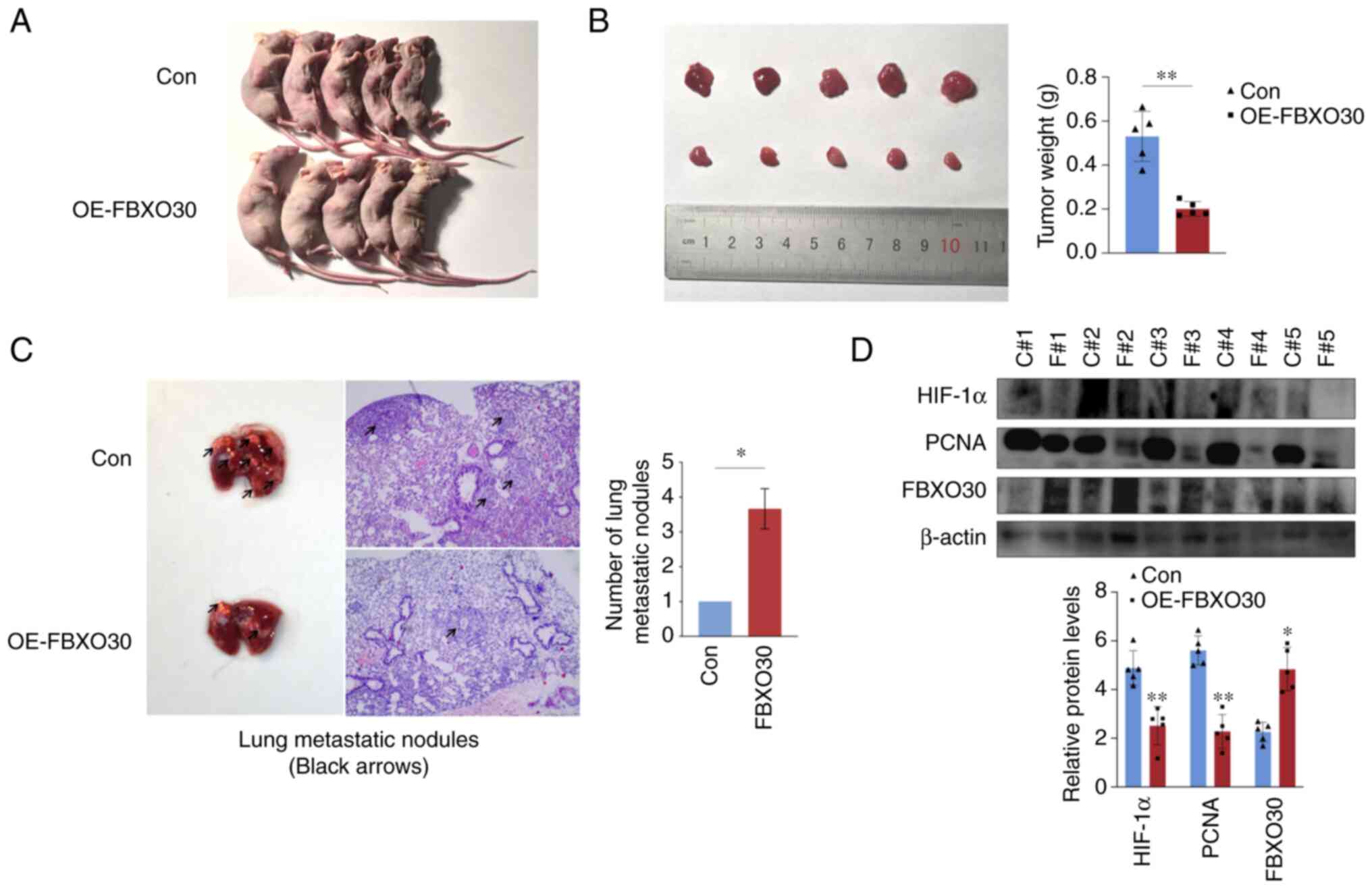 | Figure 6FBXO30 inhibits tumor growth in
vivo. (A) Representative images of BALB/c nude mice 30 days
after subcutaneous inoculation of OS-RC-2 cells transfected with
empty vector and FBXO30 vector. (B) Representative images of tumors
separated from the nude mice. (C) Representative images of the
lungs with tumor metastases excised from nude mice, which were then
subjected to hematoxylin and eosin staining. The numbers of
metastatic nodules were counted, with three lung metastasis
sections contained in each group. Images were captured under an
inverted microscope (×100 magnification). (D) Western blotting was
performed to detect the protein expression levels of HIF-1α, PCNA
and FBXO30. C#, Con; F#, OE-FBXO30. *P<0.05,
**P<0.01, as determined by unpaired Student's t-test
(two-tailed). FBXO30, F-box protein 30; HIF-1α, hypoxia-inducible
factor-1α; OE, overexpression; PCNA, proliferating cell nuclear
antigen. |
Subsequently, a lung metastasis model was
established in nude mice to evaluate the effect of FBXO30 on tumor
metastasis in vivo. Lung metastasis models using
immune-deficient mice can be used to effectively evaluate the level
of tumor invasion and metastasis, and are universally applied in
the construction of RCC cell tumor models with a high success rate
(32). ccRCC cells transfected
with the empty and FBXO30 vectors were injected via the tail vein
into nude mice, and the lung tissues were isolated after 45 days.
There were approximately six tumor metastases in the lung tissues
from the control group, but only two in the FBXO30 overexpression
group. This result indicated that FBXO30 markedly restricted the
formation of lung metastases. Subsequently, lung tissues from the
two groups were sectioned and subjected to H&E staining. The
number of tumor metastatic nodules per field of view was four in
the control group and one in the FBXO30 overexpression group. The
number of lung metastatic nodules following FBXO30 overexpression
was markedly reduced, suggesting that FBXO30 inhibited the
metastatic capacity of ccRCC cells in vivo (Fig. 6C). Finally, the subcutaneous tumors
of nude mice were lysed and proteins were extracted for western
blotting. It was revealed that in the subcutaneous tumor tissue
from the FBXO30 overexpression group, the expression levels of
HIF-1α and proliferating cell nuclear antigen were significantly
decreased (Fig. 6D). These results
indicated that FBXO30 significantly reduced the accumulation of
HIF-1α in tumors in vivo and decreased tumor proliferative
activity. These data suggested that FBXO30 could inhibit the
tumorigenic and metastatic capacity of ccRCC cells in
vivo.
Discussion
ccRCC is a solid tumor characterized by hypoxia,
which is possibly caused by long-term elevated oxygen consumption
and decreased oxygen diffusion due to the irregular distribution of
the vasculature system in tumors (33-35).
However, our previous studies confirmed that even without oxygen
content changes in the tumor extracellular microenvironment,
HIF-1α, which is supposed to be degraded via pVHL-mediated
ubiquitination, remained highly expressed in cultured ccRCC cell
lines under normoxia (8,28). Consistently, previous studies have
suggested that pVHL is frequently mutated and inactivated during
the development of ccRCC (19,33).
Therefore, it is still valuable to determine the potential
regulatory mechanism of HIF-1α in ccRCC cells with a sufficient
oxygen supply. The present study proposed that FBXO30, as a novel
E3 ubiquitin ligase, could promote HIF-1α degradation through the
ubiquitin-proteasome pathway in normoxia and directly inhibit ccRCC
tumor progression.
As a member of the FBP family, the role of FBXO30 in
ccRCC remains unclear. FBXO30 has been reported to target
intracellular ubiquitination to regulate cell mitosis and to
control muscle growth through the BMP signaling pathway (12,14,15).
In cancer, FBXO30 has been reported to be associated with the
occurrence of prostate cancer and nasopharyngeal cancer (16,17).
In the present study, FBXO30 expression was lower in ccRCC tissues
than in ANTs, at both the mRNA and protein levels. IHC assays also
suggested that FBXO30 was negatively associated with the malignancy
of ccRCC. These results were in accordance with data from TCGA
database. Subsequently, it was revealed that FBXO30 suppressed EMT
progression in ccRCC in vitro. HIF-1α is thought to enhance
the invasiveness of ccRCC cells by promoting glycolysis and
preventing tumor cells from undergoing apoptosis (36-38).
Based on our previous results, HIF-1α has also been reported to act
as an oncogene in ccRCC (28). In
the present study, FBXO30 reduced the upregulation of HIF-1α
protein expression in ccRCC cells under normoxia without affecting
its mRNA levels, which could be due to post-translational
regulation. FBXO30 possesses the potential to be classed as a novel
ubiquitin ligase (5,39). It is reasonable to hypothesize that
FBXO30 may act as a possible E3 ubiquitin ligase of HIF-1α. HIF-1α
has been reported to enhance EMT progression in peritoneal
epithelial cells by increasing the expression levels of VEGF,
Snail-1 and MMP-2 proteins (40).
In lung cancer, HIF-1α has been shown to increase the abundance of
angiopoietin-like 4 to promote tumor metastasis (41). In addition, HIF-1α can upregulate
lysyl oxidase expression in hepatoma cells, inducing cancer
metastasis and development (42).
Collectively, FBXO30 was able to reduce HIF-1α upregulation in
ccRCC cells through the ubiquitin-proteasome degradation pathway,
thus inhibiting tumor progression and metastasis.
Consistently with our previous study, FBXO30 was
knocked down in hZIP1-overexpressing ccRCC cells, and the depletion
of FBXO30 rescued HIF-1α downregulation triggered by hZIP1. Given
that hZIP1 is a known Zn2+ transport protein, the
function of Zn2+ was investigated. Zinc is involved in
immune system defense, and is associated with the development of
inflammation, metabolism and cancer (43). Zinc has been proven to restrict the
malignancy of prostate cancer cells (44). In addition, colorectal cancer and
adrenal cancer have been shown to be accompanied by zinc
deficiency, and Zn2+ also demonstrates cytotoxicity to
cancer cells (45). In ccRCC,
low-concentration Zn2+ treatment was capable of
effectively inhibiting cancer progression (46). In the present study, the protein
expression levels of HIF-1α were decreased in ccRCC cells with
increasing Zn2+ concentrations. Coincidentally, the
findings of Nardinocchi et al (47) showed that zinc also suppressed the
expression of HIF-1α and VEGF in prostate cancer and glioblastoma.
Notably, zinc supplementation rescued the decrease in FBXO30 and
HIF-1α upregulation in hZIP1-knockdown cells. Furthermore, a
previous study reported that Zn2+ could affect the
activity of key enzymes in the tricarboxylic acid cycle and
glycolytic pathway, thus improving the cytotoxicity of antitumor
drugs (48). HIF-1α often induces
the expression of glycolytic enzymes in tumors with abnormally
enhanced glycolysis (49). The
present study detected the protein expression levels of PDK1, GLUT1
and LDHA after zinc addition, and these three proteins were
revealed to be decreased by zinc. The present results suggested
that hZIP1 partially suppressed HIF-1α by upregulating FBXO30, and
that hZIP1 might directly impair the expression of HIF-1α by
enriching intracellular Zn2+. In addition, since
Zn2+ also upregulated FBXO30 protein expression levels,
it is possible that Zn2+ may be involved in the
ubiquitination process of HIF-1α mediated by FBXO30, which needs
further verification.
In conclusion, FBXO30 may be a novel E3 ubiquitin
ligase for HIF-1α that is involved in ccRCC progression. We also
verified the findings of our previous study that hZIP1 inhibits
HIF-1α protein expression (28).
In addition, hZIP1 may be upstream of FBXO30, promoting the
downregulation of HIF-1α via FBXO30. Furthermore, hZIP1 was
revealed to potentially recruit Zn2+ to regulate the
expression of FBXO30 and HIF-1α, thereby inhibiting glycolysis in
ccRCC. However, there were some limitations in the present study.
The binding domain in FBXO30 and HIF-1α, and the ubiquitination
site remain unknown. In-depth verification is required to determine
the effects of hZIP1, Zn2+ and FBXO30 on glycolysis at
the metabolite level. In addition, the experimental group design
needs improvements. There should have been a hZIP1 overexpression
group alone tested simultaneously with the hZIP1 overexpression +
FBXO30 knockdown group and the FBXO30 knockdown group, to support
the conclusion that knockdown of FBXO30 rescued the downregulation
of HIF-1α protein expression levels induced by hZIP1. ccRCC is
defined as a metabolic disease with highly abnormal glucose
metabolism, as well as crosstalk in lipid and amino acid metabolism
(50,51). Recently, targeted immunotherapies
to alleviate hypoxia and inhibit abnormal metabolic genes have
gained attention for application in the complex immune
microenvironment in ccRCC (52,53).
To a certain extent, the present study verified the probable
existence of the hZIP1/Zn2+/FBXO30/HIF-1α axis, which
may suppress EMT and glycolysis progression in ccRCC. The present
study not only provided novel insights into the occurrence and
development of ccRCC, but also identified potential biomarkers or
therapeutic targets for the clinical treatment of RCC.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YY and XD conceived the study and confirmed the
authenticity of all the raw data. YY participated in investigation,
data curation, visualization and wrote the original draft. ZiL, BL,
and ZG were involved in investigation and formal analysis. CP, ZhL
and ZZ contributed to the conception and design of the study, and
were accountable for all aspects of the work to ensure that
questions related to the accuracy or integrity of any part of the
work are appropriately investigated and resolved. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The animal experiments performed in the present
study were approved by the Ethics Committee of China Medical
University (approval no. KT2022527). The experiments using human
tissues in the present study were approved by the Research Ethics
Committee of China Medical University, and the approval number was
[2019]2019-65-3; written informed consent was obtained from all
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
This work was supported by a grant from the National Natural
Science Foundation of China (grant no. 81902591) and the Project of
Applied Basic Research Program of Liaoning province (grant no.
2022JH2/101300056).
References
|
1
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2022. CA Cancer J Clin. 72:7–33. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xia C, Dong X, Li H, Cao M, Sun D, He S,
Yang F, Yan X, Zhang S, Li N and Chen W: Cancer statistics in China
and United States, 2022: Profiles, trends, and determinants. Chin
Med J (Engl). 135:584–590. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hsieh JJ, Purdue MP, Signoretti S, Swanton
C, Albiges L, Schmidinger M, Heng DY, Larkin J and Ficarra V: Renal
cell carcinoma. Nat Rev Dis Primers. 3:170092017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cao H, Sun Z, Wu J, Hao C and Wang W:
Metastatic clear cell renal cell carcinoma to pancreas and distant
organs 24 years after radical nephrectomy: A case report and
literature review. Front Surg. 9:8942722022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bai C, Sen P, Hofmann K, Ma L, Goebl M,
Harper JW and Elledge SJ: SKP1 connects cell cycle regulators to
the ubiquitin proteolysis machinery through a novel motif, the
F-box. Cell. 86:263–274. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nguyen KM and Busino L: The biology of
F-box proteins: The SCF family of E3 ubiquitin ligases. Adv Exp Med
Biol. 1217:111–122. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shen W, Zhou Q, Peng C, Li J, Yuan Q, Zhu
H, Zhao M, Jiang X, Liu W and Ren C: FBXW7 and the hallmarks of
cancer: Underlying mechanisms and prospective strategies. Front
Oncol. 12:8800772022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhu H, Wang X, Zhou X, Lu S, Gu G and Liu
C: E3 ubiquitin ligase FBXW7 enhances radiosensitivity of non-small
cell lung cancer cells by inhibiting SOX9 regulation of CDKN1A
through ubiquitination. Lab Invest. 102:1203–1213. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nita A, Nishiyama M, Muto Y and Nakayama
KI: FBXL12 regulates T-cell differentiation in a cell-autonomous
manner. Genes Cells. 21:517–524. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Watanabe K, Yumimoto K and Nakayama KI:
FBXO21 mediates the ubiquitylation and proteasomal degradation of
EID1. Genes Cells. 20:667–674. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li D, Xie P, Zhao F, Shu J, Li L, Zhan Y
and Zhang L: F-box protein Fbxo3 targets Smurf1 ubiquitin ligase
for ubiquitination and degradation. Biochem Biophys Res Commun.
458:941–945. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tekcham DS, Chen D, Liu Y, Ling T, Zhang
Y, Chen H, Wang W, Otkur W, Qi H, Xia T, et al: F-box proteins and
cancer: An update from functional and regulatory mechanism to
therapeutic clinical prospects. Theranostics. 10:4150–4167. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sartori R, Schirwis E, Blaauw B,
Bortolanza S, Zhao J, Enzo E, Stantzou A, Mouisel E, Toniolo L,
Ferry A, et al: BMP signaling controls muscle mass. Nat Genet.
45:1309–1318. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jin Y, Yang M, Gao C, Yue W, Liang X, Xie
B, Zhu X, Fan S, Li R and Li M: Fbxo30 regulates chromosome
segregation of oocyte meiosis. Cell Mol Life Sci. 76:2217–2229.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cheng X, Pei P, Yu J, Zhang Q, Li D, Xie
X, Wu J, Wang S and Zhang T: F-box protein FBXO30 mediates retinoic
acid receptor γ ubiquitination and regulates BMP signaling in
neural tube defects. Cell Death Dis. 10:5512019. View Article : Google Scholar
|
|
16
|
Bjerre MT, Strand SH, Nørgaard M,
Kristensen H, Rasmussen AK, Mortensen MM, Fredsøe J, Mouritzen P,
Ulhøi B, Ørntoft T, et al: Aberrant DOCK2, GRASP, HIF3A and PKFP
hypermethylation has potential as a prognostic biomarker for
prostate cancer. Int J Mol Sci. 20:11732019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bjerre MT, Nørgaard M, Larsen OH, Jensen
SØ, Strand SH, Østergren P, Fode M, Fredsøe J, Ulhøi BP, Mortensen
MM, et al: Epigenetic analysis of circulating tumor DNA in
localized and metastatic prostate cancer: Evaluation of clinical
biomarker potential. Cells. 9:13622020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xiong W, Zeng ZY, Shen SR, Li XL, Li WF,
Wang R, Xiong F, Peng C, Zhang QH, Zhou M, et al: Studies on the
relationship between D6S1581, a high frequency allele imbalance
locus, and genetic susceptibility to nasopharyngeal carcinoma.
Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 20:311–314. 2003.In
Chinese.
|
|
19
|
Pezzuto A and Carico E: Role of HIF-1 in
cancer progression: Novel insights. A review. Curr Mol Med.
18:343–351. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li JN, Chen PS, Chiu CF, Lyu YJ, Lo C,
Tsai LW and Wang MY: TARBP2 suppresses ubiquitin-proteasomal
degradation of HIF-1α in breast cancer. Int J Mol Sci. 23:2082021.
View Article : Google Scholar
|
|
21
|
Liu J, Zhang C, Zhao Y, Yue X, Wu H, Huang
S, Chen J, Tomsky K, Xie H, Khella CA, et al: Parkin targets HIF-1α
for ubiquitination and degradation to inhibit breast tumor
progression. Nat Commun. 8:18232017. View Article : Google Scholar
|
|
22
|
Flügel D, Görlach A and Kietzmann T:
GSK-3β regulates cell growth, migration, and angiogenesis via Fbw7
and USP28-dependent degradation of HIF-1α. Blood. 119:1292–1301.
2012. View Article : Google Scholar
|
|
23
|
Bowers K and Srai SKS: The trafficking of
metal ion transporters of the Zrt- and Irt-like protein family.
Traffic. 19:813–822. 2018. View Article : Google Scholar
|
|
24
|
Costello LC and Franklin RB: A
comprehensive review of the role of zinc in normal prostate
function and metabolism; and its implications in prostate cancer.
Arch Biochem Biophys. 611:100–112. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jeong J and Eide DJ: The SLC39 family of
zinc transporters. Mol Aspects Med. 34:612–619. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dineley KE, Votyakova TV and Reynolds IJ:
Zinc inhibition of cellular energy production: Implications for
mitochondria and neurodegeneration. J Neurochem. 85:563–570. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dong X, Kong C, Zhang Z, Liu X, Zhan B,
Chen Z and Shi D: hZIP1 that is down-regulated in clear cell renal
cell carcinoma is negatively associated with the malignant
potential of the tumor. Urol Oncol. 32:885–892. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhan B, Dong X, Yuan Y, Gong Z and Li B:
hZIP1 inhibits progression of clear cell renal cell carcinoma by
suppressing NF-kB/HIF-1α pathway. Front Oncol. 11:7598182021.
View Article : Google Scholar
|
|
29
|
Delahunt B, Eble JN, Egevad L and
Samaratunga H: Grading of renal cell carcinoma. Histopathology.
74:4–17. 2019. View Article : Google Scholar
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
31
|
Ma X, Li C, Sun L, Huang D, Li T, He X, Wu
G, Yang Z, Zhong X, Song L, et al: Lin28/let-7 axis regulates
aerobic glycolysis and cancer progression via PDK1. Nat Commun.
5:52122014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Park JS, Lee ME, Kim SH, Jang WS and Ham
WS: Development of a highly pulmonary metastatic orthotopic renal
cell carcinoma murine model. Biol Open. 10:bio0585662021.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Schödel J, Grampp S, Maher ER, Moch H,
Ratcliffe PJ, Russo P and Mole DR: Hypoxia, hypoxia-inducible
transcription factors, and renal cancer. Eur Urol. 69:646–657.
2016. View Article : Google Scholar :
Wilson WR and Hay MP: Targeting hypoxia in
cancer therapy. Nat Rev Cancer. 11:393–410. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wigerup C, Påhlman S and Bexell D:
Therapeutic targeting of hypoxia and hypoxia-inducible factors in
cancer. Pharmacol Ther. 164:152–169. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Choi WSW, Boland J and Lin J:
Hypoxia-inducible factor-2α as a novel target in renal cell
carcinoma. J Kidney Cancer VHL. 8:1–7. 2021. View Article : Google Scholar :
|
|
36
|
Rashid M, Zadeh LR, Baradaran B, Molavi O,
Ghesmati Z, Sabzichi M and Ramezani F: Up-down regulation of HIF-1α
in cancer progression. Gene. 798:1457962021. View Article : Google Scholar
|
|
37
|
Doonachar A, Gallo MD, Doukas D, Pasricha
R, Lantsberg I and Schoenfeld AR: Differential effects of HIF-α
isoforms on apoptosis in renal carcinoma cell lines. Cancer Cell
Int. 15:232015. View Article : Google Scholar
|
|
38
|
Hoefflin R, Harlander S, Schäfer S,
Metzger P, Kuo F, Schönenberger D, Adlesic M, Peighambari A, Seidel
P, Chen CY, et al: HIF-1α and HIF-2α differently regulate tumour
development and inflammation of clear cell renal cell carcinoma in
mice. Nat Commun. 11:41112020. View Article : Google Scholar
|
|
39
|
Chen N, Kong X, Zhao S and Xiaofeng W:
Post-translational modification of baculovirus-encoded proteins.
Virus Res. 279:1978652020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Morishita Y, Ookawara S, Hirahara I, Muto
S and Nagata D: HIF-1α mediates hypoxia-induced
epithelial-mesenchymal transition in peritoneal mesothelial cells.
Ren Fail. 38:282–289. 2016. View Article : Google Scholar
|
|
41
|
Das B, Tsuchida R, Malkin D, Koren G,
Baruchel S and Yeger H: Hypoxia enhances tumor stemness by
increasing the invasive and tumorigenic side population fraction.
Stem Cells. 26:1818–1830. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ide T, Kitajima Y, Miyoshi A, Ohtsuka T,
Mitsuno M, Ohtaka K, Koga Y and Miyazaki K: Tumor-stromal cell
interaction under hypoxia increases the invasiveness of pancreatic
cancer cells through the hepatocyte growth factor/c-Met pathway.
Int J Cancer. 119:2750–2759. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Skrajnowska D and Bobrowska-Korczak B:
Role of zinc in immune system and anti-cancer defense mechanisms.
Nutrients. 11:22732019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
To PK, Do MH, Cho JH and Jung C: Growth
modulatory role of zinc in prostate cancer and application to
cancer therapeutics. Int J Mol Sci. 21:29912020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gelbard A: Zinc in cancer therapy
revisited. Isr Med Assoc J. 24:258–262. 2022.PubMed/NCBI
|
|
46
|
Liu L, Hou Y, Hu J, Zhou L, Chen K, Yang X
and Song Z: SLC39A8/zinc suppresses the progression of clear cell
renal cell carcinoma. Front Oncol. 11:6519212021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Nardinocchi L, Pantisano V, Puca R, Porru
M, Aiello A, Grasselli A, Leonetti C, Safran M, Rechavi G, Givol D,
et al: Zinc downregulates HIF-1α and inhibits its activity in tumor
cells in vitro and in vivo. PLoS One. 5:e150482010. View Article : Google Scholar
|
|
48
|
Olechnowicz J, Tinkov A, Skalny A and
Suliburska J: Zinc status is associated with inflammation,
oxidative stress, lipid, and glucose metabolism. J Physiol Sci.
68:19–31. 2018. View Article : Google Scholar :
|
|
49
|
Kierans SJ and Taylor CT: Regulation of
glycolysis by the hypoxia-inducible factor (HIF): Implications for
cellular physiology. J Physiol. 599:23–37. 2021. View Article : Google Scholar
|
|
50
|
Chakraborty S, Balan M, Sabarwal A,
Choueiri TK and Pal S: Metabolic reprogramming in renal cancer:
Events of a metabolic disease. Biochim Biophys Acta Rev Cancer.
1876:1885592021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wise DR and Thompson CB: Glutamine
addiction: A new therapeutic target in cancer. Trends Biochem Sci.
35:427–433. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lai Y, Tang F, Huang Y, He C, Chen C, Zhao
J, Wu W and He Z: The tumour microenvironment and metabolism in
renal cell carcinoma targeted or immune therapy. J Cell Physiol.
236:1616–1627. 2021. View Article : Google Scholar
|
|
53
|
Li F, Aljahdali IAM, Zhang R, Nastiuk KL,
Krolewski JJ and Ling X: Kidney cancer biomarkers and targets for
therapeutics: Survivin (BIRC5), XIAP, MCL-1, HIF1α, HIF2α, NRF2,
MDM2, MDM4, p53, KRAS and AKT in renal cell carcinoma. J Exp Clin
Cancer Res. 40:2542021. View Article : Google Scholar
|