1. Introduction
Due to environmental pollution and tobacco use, lung
cancer has become the most common cause of cancer-related death
worldwide, accounting for 18.4% of total cancer mortalities
(1). Of note, female patients have
a better prognosis than males (2).
Nearly 85% of lung cancers are non-small cell lung cancers
(NSCLCs). The remainder are SCLCs, of which lung adenocarcinoma is
the most common subtype, accounting for 50% of NSCLC cases,
followed by squamous cell carcinoma (3). Lung cancer, one of the most common
cancer types, is usually detected only at a late stage, as it has
no obvious symptoms in its early stages. Early implementation of
screening programs is one of the main tools for patients to reduce
lung cancer-associated mortality (3). Early-stage lung cancer may be
surgically resected, depending on the size and location of the
tumor (4). Advanced non-resectable
NSCLCs may be treated with dual chemotherapy using cisplatin or
carboplatin along with chest radiotherapy (5). However, the overall survival rate of
patients with lung cancer is generally low and most lung cancers
are metastatic at the time of diagnosis, which in turn contributes
to patients' low overall survival rate. There remains an urgent
need to explore new biomolecules and pathways involved in lung
cancer formation, which may provide new prognostic predictors and
therapeutic targets. Doing so may provide a new direction for the
treatment of lung cancer.
Recent research has found that in the process of the
occurrence and development of lung cancer, neural precursor cell
expressed, developmentally down-regulated 8 (NEDD8) is abnormally
expressed. NEDD8 is a ubiquitin-like protein that is involved in
the post-translational modification of proteins, also known as
neddylation. Neddylation regulates not only ubiquitination
modification, but also a variety of life activities, thus playing
an important role in the formation and prognosis of lung
cancer.
2. NEDD8 in post-translational modifications
of proteins
The NEDD8 molecule
NEDD8 is a ubiquitin-like, highly conserved protein
cloned from mouse embryonic brain tissue, consisting of 81 amino
acids and containing one α-helix and three β-lamellar structures.
NEDD8 is most highly expressed in cardiac and skeletal tissues and
is mainly expressed in the nucleus with relatively low expression
in the cytoplasm. The NEDD8 molecule itself is a nonfunctional
precursor protein that becomes fully-fledged and functional only
when the precursor protein is hydrolyzed by specific proteases of
its C-terminal amino acids. The presence of NEDD8 precursors
prevents overexpression of neddylation modifications and serves as
a reserve pool of NEDD8, ensuring that NEDD8 levels are within
normal limits. Following the hydrolysis of amino acids, NEDD8
undergoes regulation in a manner similar to ubiquitination-through
a multistage enzymatic reaction and binding to target proteins, but
with a specific binding mode unlike that of ubiquitin (6,7).
The neddylation process
The NEDD8 modification process, neddylation, is
similar to ubiquitination and requires NEDD8-activating enzyme
(NAE) E1, NAE E2 and NAE E3. NAE is the only enzyme found to be
able to perform neddylation. NAE consists of β-amyloid precursor
protein binding protein 1 APPBP1 (NAE1) and ubiquitin-like modifier
activating enzyme 3 (UBA3), which form the N-terminal and
C-terminal ends of ubiquitin activating enzymes (8). The two types of E2 ligases that have
been identified are ubiquitin-binding enzyme UBE2M (also known as
UBC12) and UBE2F. UBE2M is a dual E2 enzyme that may act both as a
NEDD8 ligase E2 for neddylation and as a ubiquitination ligase E2
to ubiquitinate and degrade UBE2F (9). The RING class includes ring-box 1
(RBX1), RBX2 (10), murine double
minute 2 (MDM2) (11), F-box
protein 11 (FBXO11) (12),
c-casitas B-lineage lymphoma (c-CBL) (13), inhibitor of apoptosis proteins
(14), and defective in cullin
neddylation 1-like protein 1-5 (DCNL1-5), also known as DCUN1D1-5
or SCCRO1-5 (15), among others.
Defective in cullin neddylation 1 (DCN1) is yeast's E3 ligase. In
mammalian cells, however, at least five DCN1-like proteins act as
E3 ligases (16). HECT types
include SMAD ubiquitylation regulatory factor 1 (Smurf1) and Smurf2
(17). Most of the NEDD8 E3
ligases may also act as ubiquitin ligases (18).
Various specific protease 1 isozymes such as NEDD8
protease 1 (NEDP1), ubiquitin-specific peptidase 21 (USP21) and
ubiquitin C-terminal hydrolase-LC3 (UCH-LC3) expose the NEDD8
precursor's glycine residue at position 76 through hydrolysis.
Subsequently, NAE catalyzes the formation of a NAE-S-NEDD8
high-energy thioester bond at the C-terminal in combination with
the cysteine active site of UBA3 in the presence of ATP and
Mg2+. This process may be referred to as the activation
of NEDD8. Activated NEDD8 binds to ligase E2 UBE2M or UBE2F to form
another thioester bond, while NAE departs. Ligase E3 instantly
interacts with E2, which carries NEDD8, to form an isopeptide bond
between the glycine at the C-terminal of NEDD8's position 76 and
the lysine residue on the substrate. This process involves
transferring NEDD8 to the substrate and completing neddylation.
After NEDD8 binds to the substrate, ligase E3 releases binding
enzyme E2, which rebinds to the next activated NEDD8 for further
neddylation (6,8,19)
(Fig. 1).
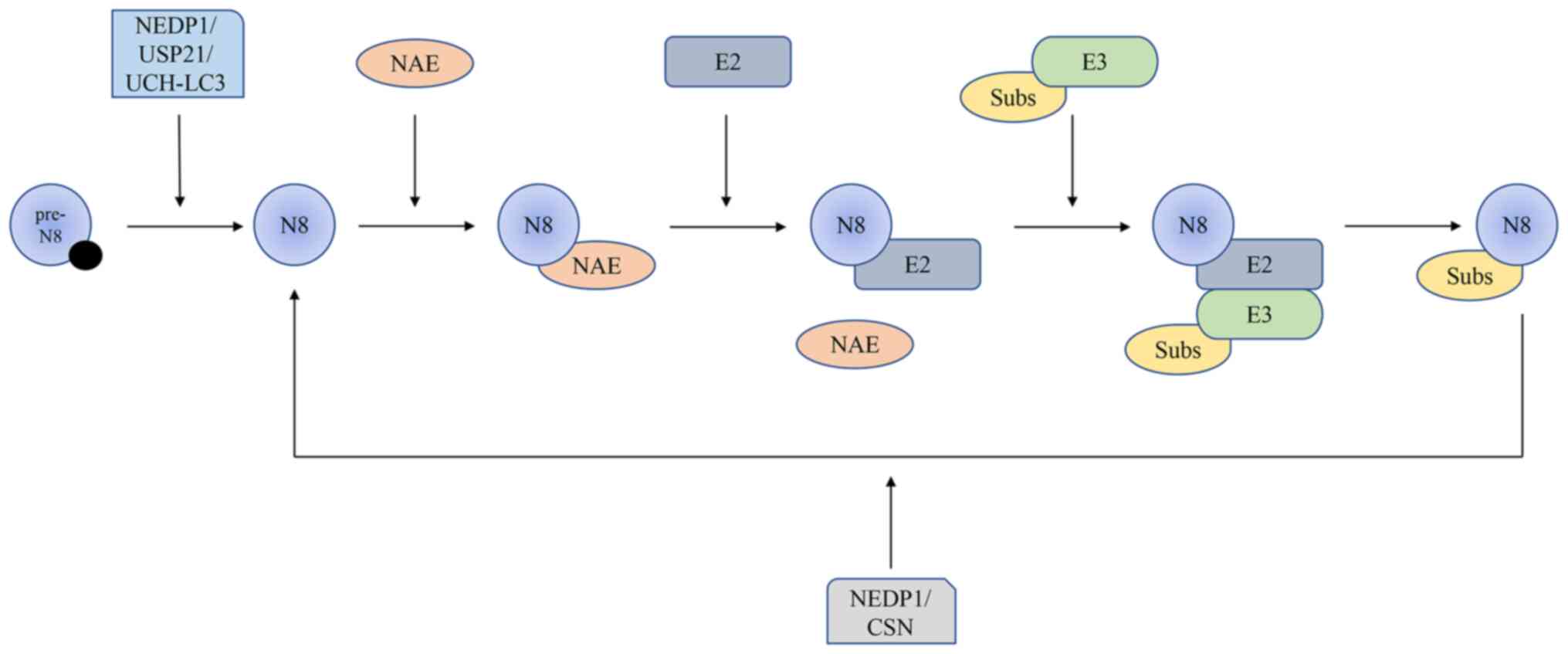 | Figure 1The neddylation modification process
and the deneddylation modification process. First, enzymes such as
NEDP1 are involved in the hydrolysis of NEDD8 precursors. NAE is
then involved in the activation of NEDD8, thus binding to enzyme
E2, at which time NAE leaves. Ligase E3 binds to E2, which carries
the substrate and transfers NEDD8 to the substrate. CSN or NEDP1
participates in the depolymerization of NEDD8 from the substrate,
known as deneddylation. NEDD8, neural precursor cell expressed
developmentally downregulated 8; NAE, NEDD8-activating enzyme;
NEDP1, NEDD8 protease 1; CSN, constitutive photomorphogenesis
signalosome; USP21, ubiquitin-specific peptidase 21; UCH-LC3,
ubiquitin C-terminal hydrolase-LC3. |
The deneddylation process
Neddylation is a reversible dynamic modification
process. Deneddylation occurs when NEDD8 is separated from the
substrate by NEDD8 depolymerase. The current study indicated that
the zinc metalloprotease COP signalosome [constitutive
photomorphogenesis signalosome (CSN)] and the cysteine protease
NEDP1, also known as SUMO peptidase family member NEDD8 specific,
catalyzes deneddylation (8). In
vivo, CSN is mainly responsible for cullin family
deneddylation, while NEDP1 is responsible for the deneddylation of
other proteins (19). Enzymes
involved in ubiquitination modifications, such as USP21, Ataxin-3,
UCH-L1 and UCH-L3, also display deneddylation activity.
The main depolymerase involved in the deneddylation
process is the NEDD8-specific enzyme CSN. CSN has 8 subunits, CSN1
through to CSN8; when the 8 subunits are complete, the main
catalytic subunit, CSN5, makes the CSN begin deneddylation to
remove NEDD8 from cullin and non-cullin proteins (8). NEDP1 is involved in both the
activation of NEDD8 and the separation of NEDD8 from the substrate
during deneddylation, meaning that NEDP1 has dual enzymatic
activity. However, if the catalytic unit of NEDP1 is mutated into
alanine, it is no longer able to remove NEDD8 from non-cullin
proteins (20) (Fig. 1).
3. Substrates and signaling pathways
associated with NEDD8
Unlike ubiquitination, neddylation mainly relies on
interactions with proteins and on signal transduction to exert
biological effects. Neddylation also has an important role in
signal transduction, participates in DNA repair, promotes cell
cycle regulation and protein degradation, and may be used as a key
entry point for targeted cancer therapy (Fig. 2).
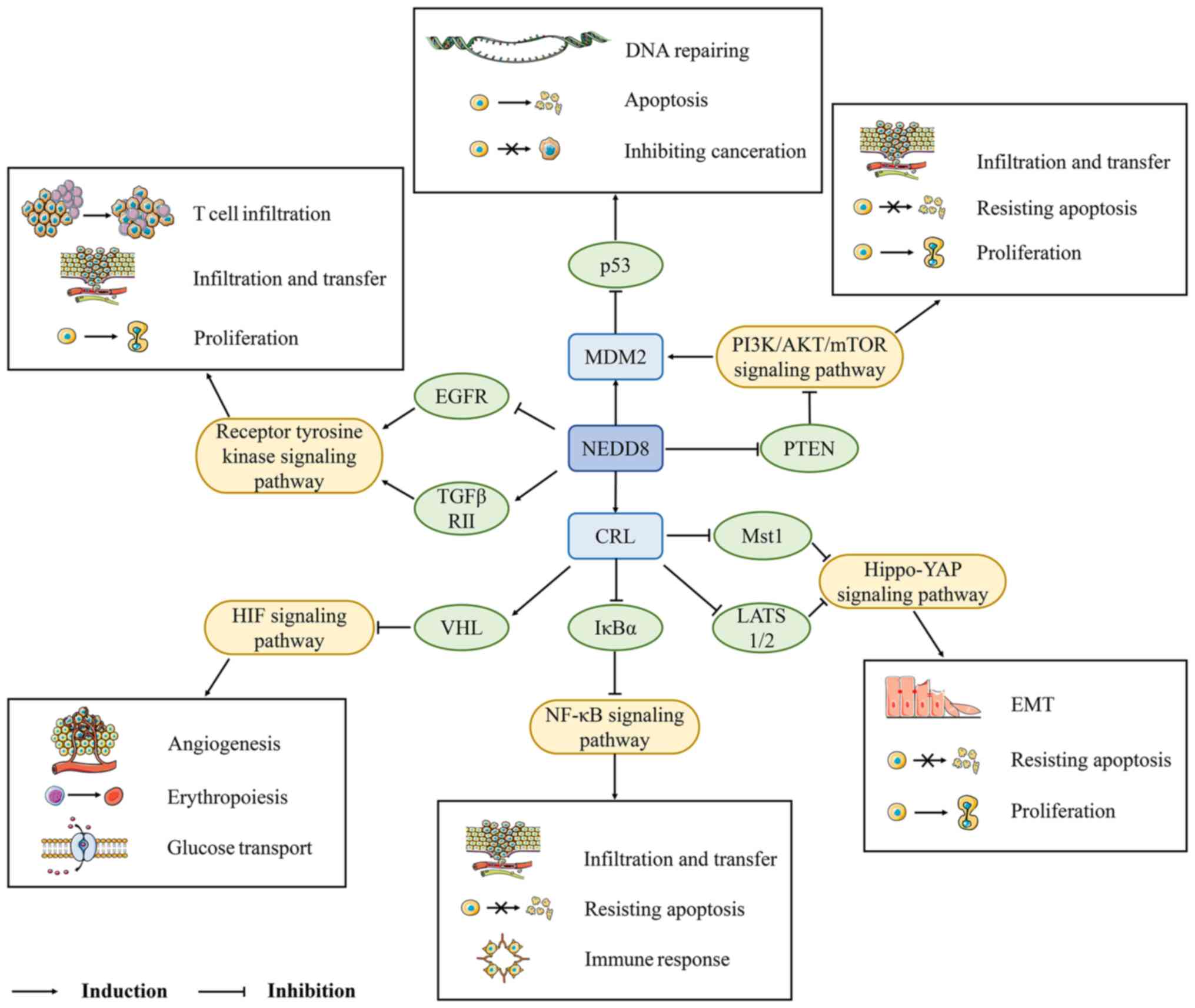 | Figure 2NEDD8 regulates the
ubiquitin-proteasome degradation pathways related to CRL and MDM2,
and it realizes the regulation of proteins by promoting the NF-κB,
PI3K/AKT/mTOR, Hippo-YAP and receptor tyrosine kinase signaling
pathways. NEDD8 also inhibits MDM2-p53, HIF and other signaling
pathways, which in turn affects DNA damage repair, the cell cycle
and tumor development. HIF, hypoxia-inducible factor; CRL,
cullin-ring ligase; EMT, epithelial to mesenchymal transition;
NEDD8, neural precursor cell expressed developmentally
downregulated 8; NAE, NEDD8-activating enzyme; NEDP1, NEDD8
protease 1; CSN, constitutive photomorphogenesis signalosome; PTEN,
phosphatase and tensin homolog; VHL, von Hippel-Lindau; Mst1/2,
mammalian sterile 20-like kinase 1/2; YAP, Yes-associated protein;
MDM2, murine double minute 2; PI3K, phosphatidylinositol-3-kinase;
AKT, protein kinase B; mTOR, mammalian target of rapamycin;
LATS1/2, large tumor suppressor homolog 1/2; EGFR, epidermal growth
factor receptor; TGFβRII, TGF-β type II receptor. |
Regulation of cullin family proteins by
neddylation
Cullin family proteins are well-validated
neddylation substrates and are important components of ubiquitin
cullin-Ring ligase (CRL), having an important role in neddylation.
Furthermore, inhibiting neddylation leads to the accumulation of a
variety of key CRL substrates, causing apoptosis and aging, in turn
inhibiting the development of tumors. Mammalian CRLs contain CUL1,
2, 3, 4A, 4B, 5, 7 and 9, and RBX1 (also known as ROC1) or RBX2
(15). In mammalian cells, the
binding enzyme UBE2M functions via RBX1 to mediate CUL1-4
neddylation, while the binding enzyme UBE2F pairs with RBX2 to
control CUL5 neddylation (9,21-23).
In this process, the CRL3 complex may be used as ligase E3 in the
ubiquitin proteasome degradation pathway; it also has an important
regulatory role in neddylation and deneddylation. Furthermore, this
indicates that NEDD8-mediated neddylation affects the
ubiquitination degradation of proteins (9). Cullin and the RING protein RBX
constitute ubiquitin E3 ligase and the binding of the CUL1
C-terminal to RBX1 may promote the accumulation of CUL1 in the
nucleus, thereby activating CUL1 ubiquitin ligase activity and
promoting CUL1 neddylation in the nucleus. Neddylation promotes
CUL1 ubiquitin ligase activity; however, disrupting this process
has no effect on RBX1 binding to CUL1, meaning that there is no
effect on the nuclear localization of CUL1 (24). CRL controls ~20% of
proteasome-regulated proteins to regulate protein degradation
(21).
In mammals, neddylation affects CRL activity through
at least two mechanisms. On the one hand, CUL1, 2, 3, 4A and 5 bind
to the assembly inhibitor cullin associated and neddylation
dissociated 1 (CAND1) when the CRL without neddylation is
inactivated, thereby inhibiting the assembly of functional CRL
(25,26). CRL activation requires to be guided
by NEDD8 ligase E3, which separates CAND1 from cullin by
substitution and results in conformational changes that regulate
CRL activity. The assembly of functional CRL is also promoted and
its ubiquitinated ligase is activated. The Skp1-CUL1-F-box protein
(SCF) complex is one such CRL (27,28).
CUL1 is a scaffold element of the SCF complex and the SCF complex
may only be formed through the interaction of CUL1 with the linker
protein Skp1 (29). This
interaction is involved in proteasomal degradation regulation of
various proteins in the cell cycle, such as Wee1 (30), p27 (31), nuclear factor erythroid 2-related
factor 2 (32) and SCF, for which
expression abnormalities also contribute to tumorigenesis. However,
decreased CAND1 levels only have a small effect on the binding of
CUL1 to NEDD8, suggesting that the two may be independent processes
(33). The dissociation of CAND1
from cullin provides a spatial site for Skp1 and RBX to bind to the
cullin so that the substrate recognition protein F-box is able to
recognize Skp1 cullin. Subsequently, the RBX protein binds to UBE2
and transfers the ubiquitin tag on UBE2 to the substrate, acting as
an E3 ligase for the CRL complex (34). In lung cancer cells, CAND1
regulates proliferation and migration, which may affect CRL
neddylation. However, CAND1 overexpression may also have adverse
effects, including excessive centrosomal replication and mitotic
defects that promote malignant tumor progression (35). On the other hand, after NEDD8
modifies cullin, it changes the β chain of cullin to connect with
RBX1, which facilitates RBX1 linkage to ubiquitin ligase E2
(18).
Deneddylation of CRLs is mediated by CSNs (8,19),
which specifically bind to NEDD8 on CRLs to control their activity.
After the dissociation between NEDD8 and the CRL complex,
interferon-induced protein NEDD8 ultimate buster 1 mediates CRL
complex proteasomal degradation following NEDD depolymerization and
the degradation of the substrate with the ubiquitin tag.
Studies have indicated that the NAE inhibitor
MLN4924 completely inhibits the neddylation of cullin, thus
inactivating CRL and causing a build-up of CRL substrates,
including cell cycle inhibitors p21, p27 and WEE1; NF-κB inhibitor
IκB-α; and DNA replication-licensed proteins chromatin and DNA
replication factor 1 (Cdt1) and replication initiation recognition
complex, subunit 1 (36).
In addition to the cullin family of proteins,
proteins associated with the ubiquitin-proteasome pathway that may
serve as neddylation substrates include MDM2 and SMAD
ubiquitination regulator factor 1 (Smurf1). MDM2 serves as both the
ubiquitination ligase E3 and the neddylation ligase E3. Neddylation
modification enhances MDM2 stability and promotes ubiquitination
degradation of p53 (37). Multiple
lysine sites of Smurf1 may enhance both the recruitment of
ubiquitin-binding enzyme E2 and its own ubiquitin ligase activity
after neddylation (38).
Regulation of the RPL11-MDM2-p53
signaling pathway by neddylation
Nucleoli are at the center of ribosome genesis.
Ribosomal proteins generally enter the nucleoli and bind to
ribosomal RNA and other proteins to form ribosome-sized subunits
(39). p53 is an important tumor
suppressor gene that has a role in inhibiting cell cycle
progression and inducing cell senescence or apoptosis, responding
to a variety of emergency signals, including DNA damage, hypoxia
and ribosomal stress. Normally, neddylation inhibits the release of
RPL11 from the nucleoli and protects it from degradation. MDM2 then
binds to p53 at the promoter site, inhibits the transcription of
p53 and promotes p53 degradation to reduce p53 expression. During
the nucleolar stress response, the neddylation of L11 in the
nucleoli is inhibited. In turn, L11 is released from the nucleolus.
L11 released into the nucleoplasm binds to MDM2 at the promoter to
inhibit the ubiquitination of MDM2 to p53, which promotes the
transcriptional activation of p53. p53 expression increases and
promotes p53-dependent cell cycle arrest. L11 is also able to
undergo neddylation in the cytoplasm before entering the nucleus,
and this process is mediated by MDM2. The cytoplasm contains NEDP1,
which mediates the deneddylation of L11 (39,40).
In addition, the neddylation of human p53 depends on HDM2, the
homologous protein of MDM2. Ribosomal protein S14 may also induce
p53 activation during nucleolar stress. The signaling between the
two is connected by HDM2, wherein the process is similar to that of
RPL11 (41). Mutations in p53
frequently occur in lung cancer and their expression is closely
related to lung cancer development and progression. In wild-type
p53-preserved tumors, MDM2 overexpression may block the
transcriptional activity of p53 and thus reduce the expression
levels of p53 (39,42).
MDM2 may reduce the expression of p53 in two
aspects. First, MDM2 degrades p53 with ubiquitin ligase E3 through
the ubiquitin-proteasome pathway; furthermore, MDM2 may be used as
NEDD8 ligase E3 to neddylate p53 in order to inhibit nuclear
translocation and transcriptional activity. These processes are
inhibited by cyclin-dependent kinase inhibitor p14 and histone
acetyltransferase TIP60, respectively (11). However, p53 neddylation by MDM2 may
be weakened by heat stress, thus reducing the inhibition of p53
activity. Another NEDD8 ligase, FBXO11, has also been found to
enable p53 neddylation, thereby inhibiting p53 function and its
transcriptional activity without affecting stability (12) (Fig.
3).
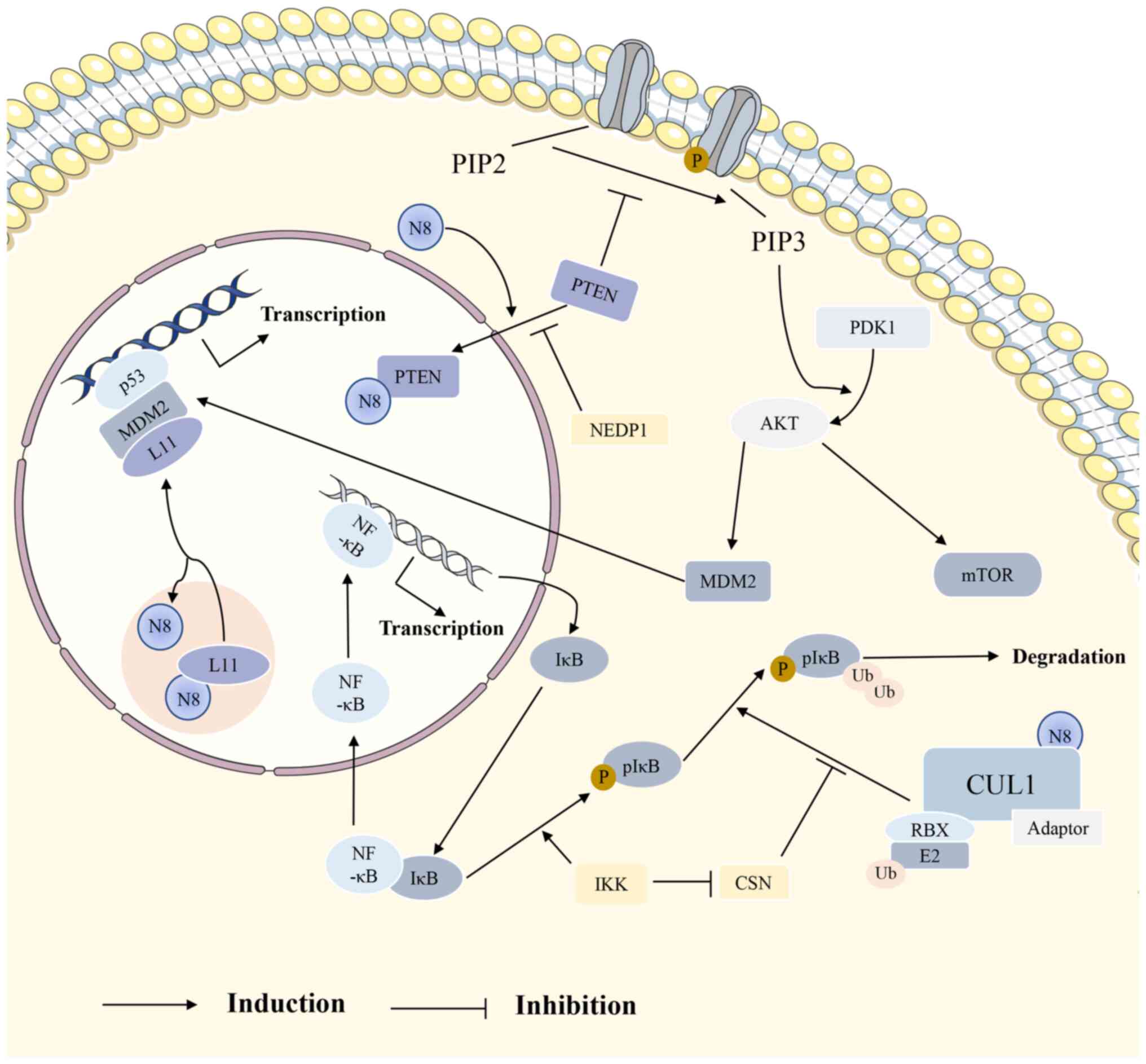 | Figure 3Several signaling pathways are
closely related to NEDD8. L11 is modified by NEDD8 and remains in
the nucleolus; in contrast, unmodified L11 enters the nucleoplasm
to bind to MDM2, thereby promoting the transcriptional activation
of p53. IκB, the inhibitor of NF-κB, is recognized by
NEDD8-modified SCF after phosphorylation, thereby degrading it and
activating the NF-κB signaling pathway. In the PI3K/AKT/mTOR
signaling pathway, PTEN is modified by NEDD8 and moves into the
nucleus, promoting the phosphorylation of PIP2 and activating
downstream pathways. NEDD8, neural precursor cell expressed
developmentally downregulated 8; PTEN, phosphatase and tensin
homolog; CSN, constitutive photomorphogenesis signalosome; MDM2,
murine double minute 2; SCF, Skp1-CUL1-F-box protein; PI3K,
phosphatidylinositol-3-kinase; AKT, protein kinase B; mTOR,
mammalian target of rapamycin; PDK1, phosphoinositide-dependent
kinase 1; RBX, ring-box; NEDP1, NEDD8 protease 1. |
Regulation of the NF-κB signaling pathway
by neddylation
NF-κB is a dimer composed of p50 and p65, as well as
a transcription factor that inhibits apoptosis. It is also related
to immune and inflammatory responses, may induce the expression of
a variety of pro-inflammatory mediators and is closely related to
tumor occurrence, growth and metastasis. It binds specifically to
the enhancer B sequence of the immunoglobulin κ light chain gene
GGGACTTTCC, thereby promoting κ light chain gene expression. NF-κB
is widely distributed in mammalian cells, and the Rel homology
domains at the N-terminus contained in NF-κB. The NF-κB family
contains homologous or heterologous dimers-regions specific to the
DNA sequence of NF-κB, including NF-κB1 (p50 and its precursor
p105), NF-κB2 (p52 and its precursor p100), c-Rel, RelA (p65) and
RelB (43).
p100, p105, IκBα, IκBβ, IκBγ, IκBε, Bcl-3 and IκB-R
are inhibitory units of the IκB family that both bind to NF-κB and
impede its transfer to the nucleus. Thus, NF-κB is usually present
in the cytoplasm in an inactive form (43). When IκB is phosphorylated to pIκB
by the activated IκB phosphokinase (IKK) complex, it may be
recognized by the ubiquitin-binding enzyme SCF, and it may be
degraded by ubiquitination and proteases. CUL1 may be activated by
enhancing SCF activity after neddylation (44). Activated NF-κB may be transferred
to the nucleus, specifically binding to the κB sequence to induce
the transcription of related genes, such as tumor necrosis
factor-α, interleukin-6 (IL-6) and other pro-inflammatory factors,
leading to inflammation and immune response (45). However, it also rapidly encodes
inhibitor IκB transcription and the newly synthesized IκB binds to
NF-κB to prevent it from moving to the nucleus and performing
feedback regulation. In lung cancer, neddylation inactivation
inhibits SCF activity and induces substrate IκBα accumulation to
block NF-κB transcriptional activity and chemotactic cytokine
ligand 2 (CCL2) transactivation, affecting the invasion of
tumor-associated macrophages. This in turn affects the development
of lung cancer (46). IL-6 also
enhances the invasion and metastasis of lung cancer cells via the
NF-κB pathway (47).
In addition to phosphorylating IκB, IKK may
phosphorylate CSN, thereby ubiquitinating and degrading it
(48). The main role of CSN is to
promote deneddylation. Thus, it is inferred that CSN degradation
maintains neddylation such that CRL is activated to promote the
NF-κB pathway (Fig. 3).
Regulation of the PI3K/AKT/mTOR signaling
pathway by neddylation
The PI3K/AKT/mTOR signaling pathway is able to
regulate cell proliferation, differentiation, apoptosis and
migration, and it has an important role in cancer development.
Phosphatidylinositol (PI)-3-kinase (PI3K) is a dimer composed of
the regulatory subunit p85 and the catalytic subunit p110;
furthermore, it is a downstream effector for G protein-coupled
receptors and receptor tyrosine kinase (RTK) (49). PI generates PIP and PIP2 by
phosphorylation, while RTK activates PI3K. PI3K may phosphorylate
PIP2 to generate a second messenger, PIP3, and stay in the plasma
membrane to be used as a docking site for intracellular proteins,
thereby recruiting protein kinase B (AKT) and
phosphoinositide-dependent kinase 1 to the plasma membrane to
further activate AKT. In mammals, the mammalian target of rapamycin
(mTOR) may be used as a downstream target of AKT, with two
complexes, mTORC1 and mTORC2. mTORC1 mainly regulates cell growth
and metabolism, and mTORC2 mainly regulates cell proliferation and
cytoskeletal remodeling. The downstream transcription factors of
mTOR include hypoxia-inducible factor 1 (HIF-1α) and c-Myc. MDM2 is
also one of the substrates of AKT and may activate itself by
mediating MDM2 phosphorylation, therefore causing MDM2 to enter the
nucleus and bind to p53. The tumor suppressor protein phosphatase
and tensin homolog (PTEN) may act as a phosphatase to
dephosphorylate PIP3 to form PIP2, thereby inhibiting this pathway
(49,50). Recent studies have indicated that
neddylation may promote the nuclear translocation of PTEN and
subsequently reduce cytoplasmic PI3K/AKT/mTOR signaling inhibition.
As such, neddylation may enhance the PI3K/AKT/mTOR signaling
pathway to promote tumor cell proliferation and infiltration. The
neddylation of PTEN relies on the activating enzyme E1 UBA3, the
binding enzyme E2 UBE2M and the ligase XIAP, whose deneddylation is
mediated by NEDP1. This process is also affected by the glucose
concentration; the modification of PTEN by neddylation is promoted
under conditions of high glucose concentrations (51).
Activation of the PI3K/AKT/mTOR signaling pathway
may promote the morphological transformation of SCLC cells.
Adherent SCLC cells with an activated PI3K/AKT/mTOR signaling
pathway are more susceptible to chemotherapy resistance (52). This suggests that the PI3K/AKT/mTOR
signaling pathway may be inhibited by inhibiting neddylation, thus
affecting lung cancer treatment (Fig.
3).
Regulation of the HIF signaling pathway
by neddylation
HIF is regulated by oxygen and its own expression;
HIF includes HIF-1 and HIF-2, of which HIF-1 is the most important
and is composed of two subunits, HIF-1α and HIF-1β. In a study of
NSCLC, it was found that the HIF pathway is involved in the early
stages of tumor progression. Cancer cells grow without restriction
when oxygen-supplying blood vessels are not generated, creating a
hypoxic environment that induces upregulation of HIF-1α and its
binding to HIF-1β. The bound heterodimer enters the nucleus and
activates the transcriptional expression of related genes. This
provides ideal cancerous conditions and the formation of blood
vessels for cancer cells growing under hypoxic conditions; it does
so by activating glycolytic pathways and increasing glucose
transport (53). Activation of
cancer-associated fibroblasts (CAFs) is critical for establishing a
tumor-promoting microenvironment, and HIF-1α is highly expressed in
lung cancer in hypoxic conditions, inducing the transformation of
normal fibroblasts into CAFs (54). Hyperthermia is one of the clinical
treatments for lung cancer; however, recurrence is common because
the thermal effect at the time of treatment may promote the growth
of residual tumors by upregulating the expression of HIF-1α
(55).
HIF-1α's main ubiquitin ligase E3 is a complex, the
most important component of which is the von Hippel-Lindau (VHL)
protein. CUL2 is an upstream protein of VHL that regulates the
degradation of HIF-1α by VHL. Both VHL and CUL2 may be neddylated.
When CUL2 is modified by neddylation, it may bind to VHL and
activate the E3 complex of HIF-1α; CUL2 then recognizes and binds
to HIF-1α and multi-ubiquitinates it. The labeled HIF-1α may be
degraded by the 26s proteasome; therefore, inhibition of
neddylation with MLN4924 enhances HIF transcription and stabilizes
HIF levels, while upregulating the expression of HIF-1α-mediated
UBE2M (9,56). VHL contains three NEDD8 receptor
sites at lysines 159, 171 and 196, and it binds to the NEDD8 ligase
MDM2 (57,58). When VHL is modified by neddylation,
it prevents binding to CUL2. The ubiquitination and degradation of
HIF-1α are inhibited and stabilized, thereby promoting
tumorigenesis (57). As such,
homeostasis of neddylation and deneddylation may regulate the
degradation of HIF-1α (Fig.
4).
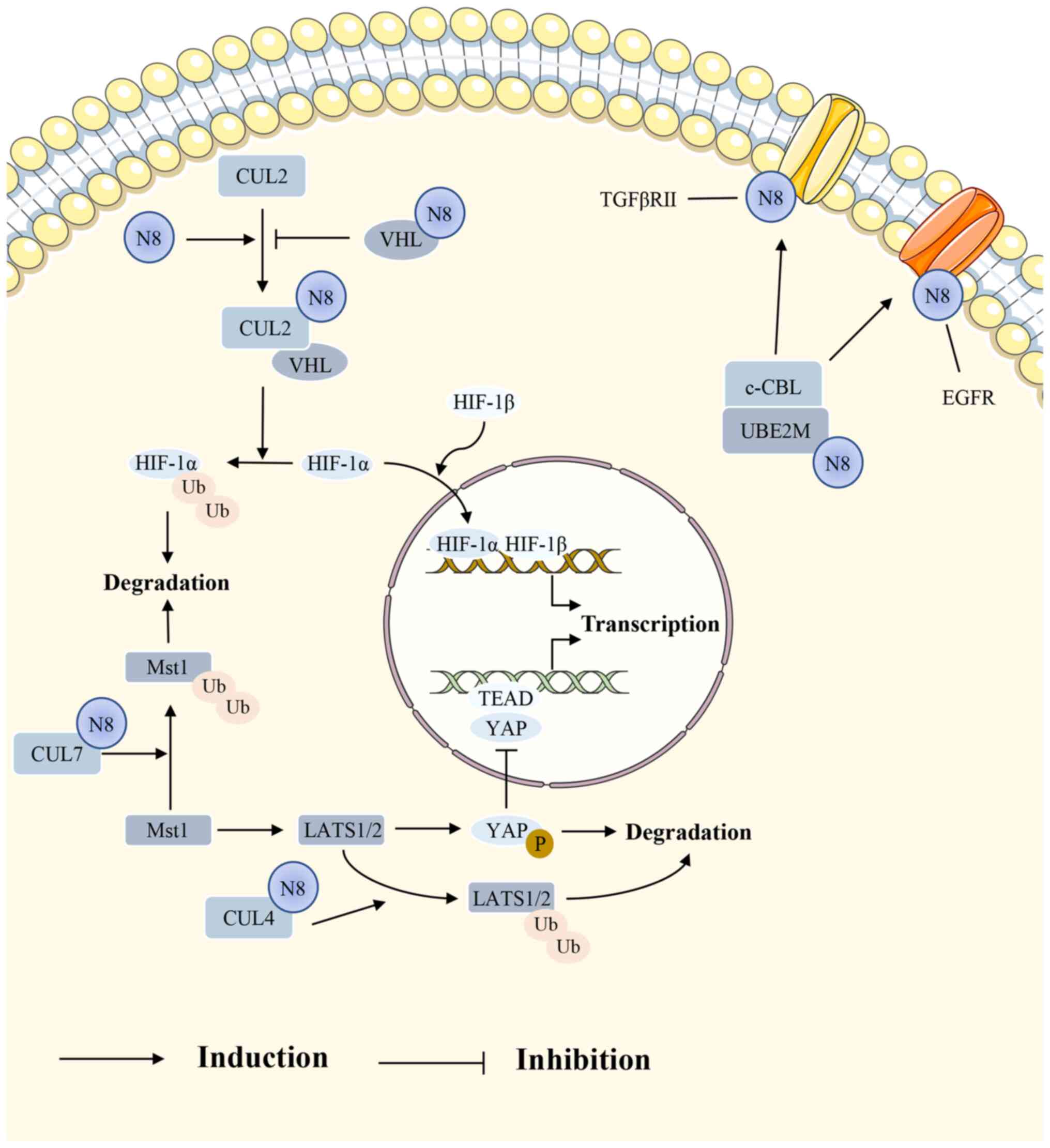 | Figure 4Several signaling pathways are
closely related to NEDD8. In the HIF signaling pathway, VHL binds
to NEDD8-modified CUL2, thereby ubiquitinating HIF-1α;
NEDD8-modified VHL inhibits this process. The Hippo-YAP pathway
requires NEDD8 for a series of phosphorylation reactions. For
instance, NEDD8-modified CUL7 promotes Mst1 degradation and
NEDD8-modified CUL4 promotes LATS1/2 degradation. In the RTK
signaling pathway, TGFβRII and EGFR are also modified by NEDD8 to
remain stable. HIF, hypoxia-inducible factor; NEDD8, neural
precursor cell expressed developmentally downregulated 8; Mst1/2,
mammalian sterile 20-like kinase 1/2; YAP, Yes-associated protein;
RTK, receptor tyrosine kinase; TGFβRII, TGF-β type II receptor;
EGFR, epidermal growth factor receptor; TEAD, transcriptional
enhanced associate domain; c-CBL, c-casitas B-lineage lymphoma;
VHL, von Hippel-Lindau; LATS1/2, large tumor suppressor homolog
1/2. |
Regulation of the Hippo-YAP signaling
pathway by neddylation
The Hippo signaling pathway is involved in cancer
cell genesis, invasion, migration and treatment; its core effector
Yes-associated protein (YAP) acts as a multifunctional
intracellular connexin and transcriptional coactivator that has a
role in both intracellular signal transduction and gene
transcriptional regulation (59).
Hippo kinases include neurofibromatosis 2, mammalian sterile
20-like kinase 1/2 (Mst1/2) and large tumor suppressor homolog 1/2
(LATS1/2). In the cytoplasm, once signaled by extracellular growth
inhibition, a series of kinase cascade phosphorylation reactions
are activated. This process activates the Hippo kinase and
eventually phosphorylates YAP, causing YAP to remain in the
cytoplasm and degrade. The YAP signaling pathway is subsequently
inhibited. Conversely, when kinase cascade phosphorylation
reactions are inhibited, YAP in the cytoplasm is transferred to the
nucleus and binds to transcriptional enhanced associate domain,
thereby activating downstream gene transcription and regulating
cell proliferation, epithelial-mesenchymal transformation (EMT),
metastasis, proliferation and differentiation of cancer stem cells,
all of which promote tumor development. NEDD8 substrate CUL7, as a
ubiquitin ligase, is able to promote Mst1 ubiquitination, and CUL4,
as a ubiquitin ligase, may promote LATS1/2 ubiquitination. Both
activate YAP signaling, indicating that neddylation is able to
regulate the Hippo-YAP pathway (59-61).
Of note, the function of YAP may differ among different lung cancer
types. YAP activation promotes the growth of NSCLCs, but inhibits
SCLCs; the specific reasons for this require to be further studied
(59,62) (Fig.
4).
Regulation of the receptor tyrosine
kinase (RTK) signaling pathway by neddylation
RTKs are the largest class of enzyme-linked
receptors and are receptors for both growth factors and enzymes to
catalyze phosphorylation of downstream target proteins. They have
an important role in mediating cell growth, movement,
differentiation and metabolism; in addition, they interact with
transforming growth factor-β (TGF-β), PI3K/AKT and other signaling
pathways. The most common RTKs include epidermal growth factor
receptor (EGFR) and fibroblast growth factor receptor (63,64).
TGF-β type II receptor (TGFβRII) and EGFR are known examples of RTK
signaling regulated by NEDD8 (65).
TGFβRII upregulates the expression of interferon-β
in tumor-associated macrophages after radiotherapy; it also
facilitates T-cell infiltration. For advanced lung cancer, TGFβRII
promotes tumor development; inhibition of TGFβRII may be used to
treat lung cancer (66). c-CBL is
a proto-oncogene encoding ubiquitin ligase E3 that may bind UBE2M
to neddylate TGFβRII, thereby inhibiting the ubiquitination
degradation of TGFβRII. In other words, TGFβRII may be stabilized
by neddylation to promote TGF-β signaling (prolonging TGFβRII
signaling in clathrin-mediated cellular endocytosis). Therefore,
c-CBL may be used as the NEDD8 ligase E3 of TGFβRII. At the same
time, MLN4924 may block the promotion of TGF-β by c-CBL, promote
the ubiquitination and degradation of TGFβRII and induce cell cycle
arrest (65,67).
The receptor tyrosine kinase EGFR binds to and is
activated by extracellular growth hormone. This process activates a
signaling cascade within the cell. c-CBL has dual enzymatic
activity that may both ubiquitinate EGFR as a ubiquitin E3 ligase,
as well as recruit NEDD8 binding enzyme UBE2M. Neddylation may
further enhance the ubiquitination of EGFR, which promotes the
degradation and endocytosis of EGFR (65,68).
In the treatment of lung cancer, most drugs target EGFR; however,
the drug-resistant mutation of EGFR is a significant barrier to
treatment (63,69). EGFR gene fusion also affects the
therapeutic effect of targeting EGFR in patients with lung cancer
(70). Therefore, studying the
process and function of EGFR neddylation modification may also help
in the exploration of treatments targeting EGFR (Fig. 4).
4. Effects of neddylation on lung
cancer
NEDD8-mediated neddylation is closely related to the
occurrence and development of tumors. Overexpression of
neddylation-related proteins has been seen in lung cancer (30), breast cancer (51), liver cancer (71) and colorectal cancer (72) and may significantly reduce the
overall survival rate of patients. To date, it has been found that
the expression pattern of NEDD8-related proteins has changed in
cancerous lung tissues, including large-cell neuroendocrine
carcinoma of the lung (LCNEC), lung adenocarcinoma (LUAD) and lung
squamous cell carcinoma (LUSC) (73).
Clinicopathological features
Real-time quantitative PCR and microarray testing of
mRNA have indicated significantly elevated NAE1 mRNA expression in
LUAD and LUSC compared with adjacent normal tissues (36). The mRNA levels of UBE2M were
significantly higher in LUAD, LUSC and LCNEC than in normal
tissues, and the expression of UBE2M mRNA was higher in
poorly-differentiated tumors within lung adenocarcinoma (30). The ligase E3 DCNL and its paragenes
are generally dysregulated in human cancers and are associated with
the neddylation of the cullin family, where the most common
dysregulated ligase E3 is DCNL5. DCNL5 is overexpressed in LUSC and
LUAD (74). This suggests that
NEDD8 may be used as a serum metabolic fingerprint (SMF) for the
diagnosis and treatment of lung cancer, and there are numerous
platforms and methods based on SMF for the early detection of lung
cancer (75,76).
Immunohistochemical staining and western blot
analysis revealed that primary tumor protein neddylation is
over-activated and that NAE1, UBA3, UBE2M and UBE2F are
overexpressed in LUAD and LUSC tissues compared with adjacent
normal tissues (36,77,78).
Particularly in SCLCs, western blot analysis indicated bands with a
higher migration form than CUL1 and detected neddylation-modified
fragments. This affects the activity of SCF complexes.
Immunohistochemical staining suggested a negative correlation with
CAND1 protein levels in SCLCs. This, however, is not observed in
normal tissues, NSCLCs or carcinoid tissues (29).
In addition, Kaplan-Meier single-gene survival
analysis indicated that patients with lung cancer with high UBE2M
expression had a poor prognosis and low overall survival. Analysis
also indicated that patients with elevated neddylation had lower
overall survival than patients with low expression (30,36,73,79);
furthermore, patients with high UBE2M and NEDD8 mRNA levels had
lower overall survival than patients with low expression. However,
there is no significant association between NAE1 - a component of
NEDD8-activating enzyme E1 - and UBA3 mRNA levels with the overall
survival rates of patients with lung cancer (30).
During the development and progression of lung
cancer, related microRNAs (miRNAs/miRs) also change. Efficient,
sensitive and specific detection methods and systems for miRNA may
improve the accuracy of diagnosis and prognosis of cancers
(80). For instance, miR-155
expression is gradually elevated in the development and progression
of lung cancer, suggesting a target for early screening and
tracking of lung cancer (81).
miR-155 is positively regulated by NF-κB, while NEDD8 promotes the
NF-κB signaling pathway. This results in high miR-155 expression
and association with certain aggressive diseases (82).
Experiments at the cellular level
Excessive activation of neddylation in human lung
cancer cells (H1299, A549 and H460) compared with normal lung
fibroblasts (WI38 and MRC-5) (36)
suggests that the development and progression of lung cancer may be
closely related to neddylation. After knockout of UBE2M or NEDD8,
A549 and H1299 cell proliferation, colony formation,
transportation, migration and invasion were all inhibited, thus
affecting the malignant phenotype of lung cancer cells (30,73).
Propidium iodide staining and fluorescence-assisted cell sorting
analysis of NEDD8-knockout cells revealed cell cycle arrest in
G2/M phase. G2/M phase conversion inhibitor
Wee1 accumulation and M-phase marker phospho-histone h3
downregulation have been discovered by western blot, further
indicating that NEDD8 leads to G2 phase arrest in LUAD
cells by inhibiting CRL activation. p21, p27 and Wee1 are
substrates for NEDD8-modified CRL; as such, the protein stability
and half-life of these cell cycle-associated proteins were analyzed
with cycloheximide chase. The protein stability and half-life of
these proteins increased significantly in NEDD8-knockdown A549
cells, indicating that they are prevented from degradation and
thus, G2 phase arrest was induced (73). Furthermore, these cells appear
enlarged and flattened in shape, suggesting that NEDD8 deletion
triggers senescence in A549 cells. This is further confirmed by the
β-galactosidase staining associated with senescence (73). It was also indicated that
inhibition of NEDD8 expression targeted with small inhibitory siRNA
promotes H1299, HCC-44, NCI-H1650 and NCI-H292 cell migration,
while having no effect on A549 migration (13). It is important to note that in A549
cells, the degree of neddylation may differ from other cell
lines.
5. Lung cancer treatment targeting
neddylation
Mouse tumor models established with A549 cells
indicated significant inhibition of growth and metastasis in the
NEDD8-knockout group (46), and
IHC staining suggested that there were fewer Ki67 (tumor
proliferation marker)-positive cells in the NEDD8 knockout group
(73). This suggests that NEDD8
may be used as a potential target for the treatment of lung cancer.
At the same time, this targeted therapy may also be used as an
adjunct to radiotherapy (77),
chemotherapy (83) and pulsed
electromagnetic therapy (84).
Basic information and the mechanism of
MLN4924
MLN4924, also known as Pevonedistat, is an AMP-like
adenosine sulfamate analog with a similar position in the NEDD8
structure to ATP (85). Therefore,
it may specifically bind to the activating enzyme NAE of NEDD8 and
inhibit its activity to prevent the formation of thioester bonds
between UBA3 and NEDD8 on NAE, which is required to activate NEDD8.
Inhibiting NEDD8 inhibits neddylation and CRL activity, resulting
in CRL substrate accumulation. Lung metastases in mouse models
using wild-type LLC cells have resulted in reduced intrapulmonary
metastases in the MLN4924-treated group (46). Clinical trials of MLN4924 in the
treatment of solid tumors have also been performed. In the Phase Ib
study of MLN4924, it was preliminarily proved that MLN4924 has no
additional toxicity for the treatment of solid tumors, which
ensures a certain safety. It also has a pharmacodynamic effect in
solid tumors such as melanoma, gastric, ovarian, head and neck,
adrenal and breast cancer (86,87).
In vivo and in vitro experiments have demonstrated
that MLN4924 has an inhibitory effect on lung cancer (36,46,88,89).
MLN4924 induces cell death
MLN4924 effectively inhibits cancer cell growth by
inducing three common types of cell death: Apoptosis, senescence
and autophagy (Fig. 5).
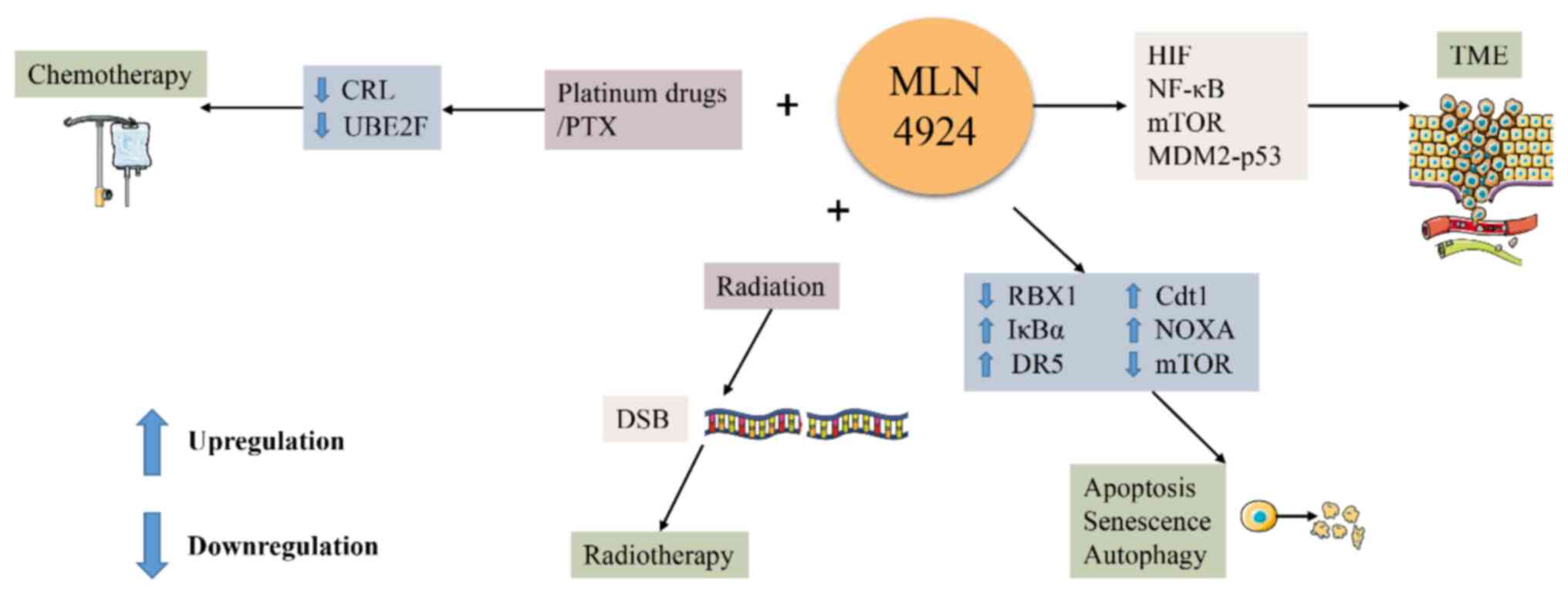 | Figure 5The NAE inhibitor MLN4924 has utility
in the treatment of lung cancer. It may affect the formation of the
TME and promote tumor cell death by regulating a variety of
proteins and various signaling pathways, such as the HIF pathway
and NF-κB pathway. MLN4924 may also be combined with certain drugs
to make chemotherapy and radiotherapy more effective. TME, tumor
microenvironment; HIF, hypoxia-inducible factor; DSB, double-strand
break; NAE, NEDD8-activating enzyme; mTOR, mammalian target of
rapamycin; MDM2, murine double minute 2; RBX1, ring-box 1; Cdt1,
chromatin and DNA replication factor 1; NOXA,
phorbol-12-myristate-13-acetate-induced protein 1; DR5, death
receptor 5; PTX, paclitaxel; CRL, cullin-ring ligase. |
Apoptosis
MLN4924 inactivates CRL1 and CRL4 by inhibiting
neddylation. This results in an increase in substrate Cdt1 levels,
inducing DNA re-replication, and triggering a DNA damage response.
As such, apoptosis and aging processes that inhibit tumor growth
are activated (90-92), and different sensitivities in
liver, ovarian, prostate, colon and lymphoma tissues were observed
(90).
The activity of the ubiquitin-binding enzyme SCF is
also inhibited, and its substrate-NF-κB inhibitory unit
IκBα-accumulates, thereby blocking NF-κB activation and inducing
apoptosis. MLN4924 also causes the accumulation of CRL substrates
such as the pro-apoptotic proteins
phorbol-12-myristate-13-acetate-induced protein 1 (NOXA) and
activating transcription factor 4, leading to apoptosis (34,36,93).
Senescence
RBX1 may mediate the neddylation of CUL1-4. Studies
have indicated that RBX1 is overexpressed in a variety of human
tumors, such as lung, liver, breast, colon and ovarian cancers.
RBX1 silencing inhibits neddylation, upregulates pro-apoptotic
proteins such as the substrate p53 up-regulated modulator of
apoptosis, downregulates anti-apoptotic proteins such as Bcl-2 and
triggers a DNA damage response. Accordingly, RBX1 silencing may
induce apoptosis and senescence. This aging process is not related
to p53 and p16 (94). The
percentage of senescence-associated-β -galactose staining may also
be increased in cells following brief treatment with MLN4924,
suggesting that the same therapeutic effect may be achieved even if
the MLN4924 treatment is short in duration (92,95).
Another study suggested that p21-mediated aging is associated with
growth inhibition induced by low-dose MLN4924. Furthermore, the
study suggested that following drug removal, p21 continues to
accumulate; DNA damage responses continue to activate, making
MLN4924-induced aging irreversible. Thus, it is possible to use
low-dose drugs to achieve the desired therapeutic effect (91).
Autophagy
mTOR inactivation and reactive oxygen species (ROS)
excess under MLN4924 are the main causes of autophagy.
mTOR is the main regulatory molecule of cell growth
and metabolism; it promotes anabolic processes and inhibits
catabolic processes such as autophagy. MLN4924 blocks the
degradation of the cullin family substrate mTOR inhibitory protein
Deptor, resulting in mTORC1 inactivation and autophagy (34,71,96).
Furthermore, HIF-1α is also significantly accumulated due to the
inhibition of CUL2 neddylation, which may negatively regulate
mTORC1 through the HIF1-regulated in development and DNA damage
responses-1-TSC1 axis and in turn trigger autophagy (96). MLN4924 also enhances ROS production
and induces oxidative stress in cancer cells; ROS inhibits the
activity of the downstream protein mTOR and induces autophagy
(71,97).
MLN4924 affects tumor microenvironments
and metastasis
Abnormal angiogenesis is an important feature of
malignancy (98). In pancreatic
cancer, MLN4924 is found to significantly inhibit capillary
formation and cell migration, reduce vascular branch points, and
inhibit tumor angiogenesis and growth. Mediating the inactivation
of RBX2 in CRL, which accumulates substrates the Ras homolog gene
family, member A (99),
neurofibromatosis type 1 (100)
and p27 (101), impairs cell
migration and angiogenic processes.
The HIF signaling pathway also affects the tumor
microenvironment. The early stages of tumor progression are
frequently accompanied by hypoxia, so the expression of HIF-1α and
HIF-2α is found to be upregulated in most lung cancer cases.
Furthermore, their high expression may be closely related to the
expression of vascular endothelial growth factor, thymidine
phosphorylase and basic fibroblast growth factor. Overexpression of
HIF-2α is associated with a poor prognosis for patients (53). MLN4924 has a negative role here,
but it may be degraded by ubiquitination of HIF-1α to promote tumor
development (56).
Immune evasion is considered one of the hallmarks of
cancer; inflammation is involved in almost all stages of
tumorigenesis and promotes tumor development. Tumor-infiltrating
leukocytes include dendritic cells (DCs), T-cells and macrophages
(102); these have an important
role in generating tumor-promoting immune microenvironments.
MLN4924 significantly attenuates the inflammatory response by
reducing the expression of pro-inflammatory cytokines and
chemokines, such as IL-1β, IL-6 and C-X-C motif chemokine ligand 1,
all of which are induced by IL-17A (103-105). MLN4924 accumulates Deptor and
inactivates the mTOR signaling pathway, while also inhibiting the
biological function of DCs (106). In addition, it inhibits the
production of pro-inflammatory factors in DCs by inhibiting the
NF-κB signaling pathway (107),
further curbing the production of a pro-tumor microenvironment.
Tumor-associated macrophages (TAMs) are the most abundant tumor
stromal cells and they provide a suitable microenvironment for
tumor development by inducing growth factors, angiogenesis
regulators and inflammatory mediators. In tumors, TAMs are
recruited from the bone marrow primarily by CCL2. In LUAD, NEDD8
expression levels are positively correlated with a high expression
of CCL2, leading to a decrease in overall survival. Partial
inactivation of neddylation may block transcriptional activation of
the NF-κB-regulated CCL2, thereby exerting anticancer effects to
inhibit monocyte chemotaxis and TAM invasion, and ultimately, tumor
metastasis (46). RBX2-CUL5 may
also modulate certain functions of TAMs to control lung
inflammation (22,108).
Metastasis is one of the leading causes of death in
cancer patients, and cancer metastasis involves a series of
processes. Neddylation may be used as a target for anti-metastatic
therapy. In non-small cell carcinomas, MLN4924 has been found to
reduce the number of intravascular cancer cells and inhibit their
extravasation. MLN4924 inhibits EMT by enhancing p53 activity,
inhibiting lung cancer metastasis (109,110) (Fig.
5).
Effect of MLN4924 on chemotherapy
NSCLC is the most common type of lung cancer, and
cytotoxic drugs such as cisplatin and carboplatin may be used in
the clinical treatment of NSCLC (83). However, resistance to platinum
drugs may lead to cancer recurrence and treatment failure, and the
prognosis is poor. Upregulation of the NEDD8-binding enzyme UBE2F
is an important pathway for lung cancer cells to evade
platinum-induced apoptosis. After platinum-based drug treatment,
UBE2F as a substrate has a weakened ability to bind to CUL3,
resulting in the accumulation of UBE2F. However, the accumulation
of UBE2F combined with RBX2 promotes the neddylation of CUL5, which
in turn promotes the degradation of substrate NOXA. The oxidative
stress capacity of cells and cell survival are subsequently
reduced. This also suggests that UBE2F may be used as a new
therapeutic target. MLN4924 may inhibit neddylation, thereby
indirectly inhibiting UBE2F and allowing NOXA to further promote
apoptosis (111).
Conventional chemotherapy also uses paclitaxel
(PTX). Resistance to chemotherapy drugs such as PTX is a major
cause of chemotherapy failure in patients with NSCLC. MLN4924
significantly inhibits the proliferation of lung cancer cells, as
well as tumor formation and metastasis, by increasing CRL substrate
levels. The combination of MLN4924 and PTX does not have any
synergistic effects in PTX-resistant NSCLC cells, indicating that
MLN4924 may be used as a drug for the treatment of PTX-resistant
NSCLC (36,112) (Fig.
5).
Effects of MLN4924 on radiotherapy
At present, radiotherapy is the main treatment
method for different stages of lung cancer and may significantly
prolong the survival time of patients. The use of ionizing
radiation and radiation therapy induces cancer cell death,
primarily by producing ROS through water and oxygen reactions;
however, high levels of ROS increase the oxidative stress response
and induce DNA damage as well as apoptosis. Compared with SCLC,
NSCLC exhibited greater tolerance to radiotherapy. Neddylation may
be the key to anti-cancer therapy.
After 48 h of irradiation, the expression of UBE2F
in A549 and H1299 cells gradually increased. N-acetylcysteine, a
typical reactive oxygen species scavenger, completely blocks
radiation-induced increases of UBE2F. In cells treated with
different concentrations of H2O2, UBE2F
increased significantly in a dose-dependent manner, further
suggesting that radiation-induced upregulation of UBE2F may be
associated with increased ROS levels. Staining with trypan blue
after UBE2F knockout indicated that the mortality rate of UBE2F
knockout cells in the experimental group increased under
irradiation and western blot analysis demonstrated an increase in
the concentration of CRL5 substrate pro-apoptotic protein NOXA in
cells of the experimental group (77,78).
This suggests that the resistance effect may be eliminated by
MLN4924.
DNA double strand break (DSB) is a severe form of
DNA damage that, if not repaired, may lead to cell carcinogenesis
or death. There are two main mechanisms of DSB repair in mammalian
cells, namely homologous recombination and non-homologous end
joining (NHEJ). FBXW7 is an important tumor suppressor gene and may
act as a target protein of the ubiquitin ligase SCF complex,
thereby regulating its downstream genes such as cyclin E. In
addition, it may enhance NHEJ by promoting the recruitment of DSB
repair factor, which is one of the reasons for cancer cell
survival. MLN4924 inhibits the activity of CUL1 and thus inhibits
the activity of the SCF complex. This prevents the binding of FBXW7
to the SCF complex, impairing NHEJ and subsequently inducing
radio-sensitization of cancer cells (113) (Fig.
5).
Negative effects of MLN4924
In related experiments, it has been found that
MLN4924 has different effects depending on serum and drug
concentrations, which may inhibit cancer cell survival and may also
stimulate tumorigenesis and cell migration (13,114).
In the case of MLN4924 blocking neddylation, c-Myc
gradually accumulates as a CRL substrate and c-Myc may induce
fibroblasts to transform into pluripotent stem cells (115). EGFR dimerization may be induced
to activate and prolong EGFR signaling that is frequently caused by
mutation or overexpression of EGFR (114). MLN4924 also activates the
PI3K/Akt/mTOR pathway, thereby inducing HIF-1α expression and
promoting cancer cell migration (56,116). Furthermore, neddylation
restriction increases cancer cell migration and Slug expression in
cells lacking p53, thereby promoting EMT (110). NEDD8 is expressed at low levels
in A549 cells, which may also be the reason why the migration of
A549 cells after MLN4924 exposure is not affected (13,117).
Other medications
HA-1141 (Ui5-8-11-41), a small molecule compound,
may induce autophagy by blocking the neddylation of cullin and
triggering endoplasmic reticulum (ER) stress, thus inhibiting
cancer cell growth. On the one hand, HA-1141 inactivates the
activation enzyme NAE. This blocks neddylation and allows cullin
family substrates to accumulate for a short duration. However, it
may trigger atypical ER stress and integration stress response. It
may also produce ROS, inactivate mTORC1 and inhibit protein
synthesis at the translation level, therefore prompting autophagy
(97).
The antihypertensive drug candesartan cilexetil, as
a neddylation inhibitor, also competitively inhibits NAE and
inhibits tumor growth (118).
Candesartan cilexetil is a derivative of benzimidazole and recent
studies have evaluated different derivatives of benzimidazole to
improve certain shortcomings of candesartan cilexetil by inhibiting
neddylation efficacy to fight cancer (119).
Another factor targeting NAE is activity-based
probe (ABP) A3. ABP A3 is a dual inhibitor of ubiquitin and
NEDD8-activating enzyme NAE and is more effective in inhibiting
substrate ubiquitination. It induces the accumulation of
intracellular misfolded proteins, stressing the ER, and stimulating
apoptosis to inhibit tumor development (120).
Further drugs, including MLN4924, are being used to
target NAE. However, during treatment, MLN4924's target UBA3
mutates to result in drug resistance (121-123), as MLN4924 becomes less impactful.
Thus, it is necessary to look for other molecules in neddylation as
therapeutic targets. Studies have indicated that DI-591 and its
derivatives, piperidinyl ureas, may also competitively bind to
ligase DCN1 with NEDD8-binding enzyme UBE2M. This blocks the
neddylation of CUL1 and CUL3 and achieves therapeutic effects
(124-127). However, the flaw of DI-591,
compared to MLN4924, is that it cannot halt the neddylation of the
entire cullin family.
In addition to drugs targeting NEDD8, there are
others that may block the neddylation process by knocking out its
substrates. Biomolecules such as DNA, proteins and lipids may be
assembled into nanoparticles that may be used to diagnose and treat
tumors (128). For instance,
polydopamine (PDA) may work to prepare PDA nanoparticles. PDA
nanodrugs may be used as carriers of siRNA, targeting tumors and
knocking out the neddylation substrate RBX1 according to the pH of
tumors and adjacent tissues, thereby promoting tumor cell
apoptosis. In liver cancer, it was found that in combination with
photothermal therapy, PDA nanodrugs modified by folic acid are able
to better target liver cells and improve the therapeutic effect
(129). However, there are
currently no studies on this in lung cancer. Of note, it may be
possible to block neddylation through PDA nanomedicine.
6. Conclusion
The main function of NEDD8 is to mediate the
participation of neddylation in a variety of signaling pathways.
Neddylation may affect CRL activity and the ubiquitination process.
It may also participate in tumorigenesis, growth and metastasis by
regulating multiple signaling pathways. There is a strong
association between NEDD8 overactivation and cancer progression in
lung cancer, breast cancer (51),
liver cancer (71) and colorectal
cancer (72). NEDD8-mediated
upregulation of neddylation has been found in lung cancer,
affecting the prognosis and treatment of patients. Numerous studies
have revealed a potential link between lung cancer and NEDD8, which
encourages the study of the molecular biological mechanisms of lung
cancer. In vivo and in vitro experiments also suggest
that NEDD8 as a therapeutic target is a feasible approach. MLN4924
as a selective NEDD8-activating enzyme inhibitor has entered phase
III clinical trials in myelodysplastic syndrome, chronic myeloid
leukemia and acute myeloid leukemia (130). This suggests that MLN4924
targeting NEDD8 may have therapeutic effects in lung cancer.
In summary, NEDD8 is expected to become an early
diagnostic or prognostic index and drug development target for
tumors as well as other related diseases.
Availability of data and materials
Not applicable.
Authors' contributions
ZT and JL contributed equally to this work; they
were involved in the conception and design of the study and drafted
the manuscript. RM and TL were involved in the analysis of the
literature. ZS and SH conceived and supervised the project and
directed the writing. All the authors have read and approved the
final version of this review. Data authentication is not
applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Figures of this review were created with Servier
Medical Art (https://smart.servier.com/).
Funding
This research was supported by the National Natural Science
Foundation of China (grant no. 81971088) and the Innovation Project
of Shandong Academy of Medical Sciences, Shandong Provincial
Natural Science Foundation, China (grant no. ZR2018MC008).
Abbreviations:
|
CAFs
|
cancer-associated fibroblasts
|
|
CAND1
|
cullin-associated and
neddylation-dissociated 1
|
|
c-CBL
|
c-casitas B-lineage lymphoma
|
|
CCL2
|
chemotactic cytokine ligand 2
|
|
Cdt1
|
chromatin and DNA replication factor
1
|
|
CRL
|
cullin-ring ligase
|
|
CSN
|
constitutive photomorphogenesis
signalosome
|
|
DCNL1-5
|
defective in cullin neddylation
1-like protein 1-5
|
|
EGFR
|
epidermal growth factor receptor
|
|
FBXO11
|
F-box protein 11
|
|
HIF
|
hypoxia-inducible factor
|
|
LATS1/2
|
large tumor suppressor homolog
1/2
|
|
LCNEC
|
large-cell neuroendocrine carcinoma
of the lung
|
|
LUAD
|
lung adenocarcinoma
|
|
LUSC
|
lung squamous cell carcinoma
|
|
MDM2
|
murine double minute 2
|
|
Mst1/2
|
mammalian sterile 20-like kinase
1/2
|
|
mTOR
|
mammalian target of rapamycin
|
|
NAE
|
NEDD8-activating enzyme
|
|
NEDD8
|
neural precursor cell expressed
developmentally downregulated 8
|
|
NEDP1
|
NEDD8 protease 1
|
|
NSCLC
|
non-small cell lung cancer
|
|
PI3K
|
phosphatidylinositol-3-kinase
|
|
RBX1
|
ring-box 1
|
|
ROS
|
reactive oxygen species
|
|
SCF
|
Skp1-CUL1-F-box protein
|
|
Smurf1
|
SMAD ubiquitylation regulatory factor
1
|
|
TGF
|
transforming growth factor
|
|
UBA3
|
ubiquitin-like modifier activating
enzyme 3
|
|
UBE2
|
ubiquitin-binding enzyme E2
|
|
YAP
|
Yes-associated protein
|
References
|
1
|
Oudkerk M, Liu S, Heuvelmans MA, Walter JE
and Field JK: Lung cancer LDCT screening and mortality
reduction-evidence, pitfalls and future perspectives. Nat Rev Clin
Oncol. 18:135–151. 2021. View Article : Google Scholar
|
|
2
|
Sachs E, Sartipy U and Jackson V: Sex and
Survival After Surgery for Lung Cancer: A Swedish Nationwide
Cohort. Chest. 159:2029–2039. 2021. View Article : Google Scholar :
|
|
3
|
Thai AA, Solomon BJ, Sequist LV, Gainor JF
and Heist RS: Lung cancer. Lancet. 398:535–554. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Howington JA, Blum MG, Chang AC, Balekian
AA and Murthy SC: Treatment of stage I and II non-small cell lung
cancer: Diagnosis and management of lung cancer, 3rd ed: American
College of Chest Physicians evidence-based clinical practice
guidelines. Chest. 143(5 Suppl): e278S–e313S. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hirsch FR, Scagliotti GV, Mulshine JL,
Kwon R, Curran WJ Jr, Wu YL and Paz-Ares L: Lung cancer: Current
therapies and new targeted treatments. Lancet. 389:299–311. 2017.
View Article : Google Scholar
|
|
6
|
Watson IR, Irwin MS and Ohh M: NEDD8
pathways in cancer, Sine quibus non. Cancer Cell. 19:168–176. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kamitani T, Kito K, Nguyen HP and Yeh ET:
Characterization of NEDD8, a developmentally down-regulated
ubiquitin-like protein. J Biol Chem. 272:28557–28562. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chung D and Dellaire G: The Role of the
COP9 Signalosome and Neddylation in DNA damage signaling and
repair. Biomolecules. 5:2388–2416. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou W, Xu J, Tan M, Li H, Li H, Wei W and
Sun Y: UBE2M Is a Stress-Inducible Dual E2 for neddylation and
ubiquitylation that promotes targeted degradation of UBE2F. Mol
Cell. 70:1008–1024.e6. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang DT, Ayrault O, Hunt HW, Taherbhoy
AM, Duda DM, Scott DC, Borg LA, Neale G, Murray PJ, Roussel MF and
Schulman BA: E2-RING expansion of the NEDD8 cascade confers
specificity to cullin modification. Mol Cell. 33:483–495. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xirodimas DP, Saville MK, Bourdon JC, Hay
RT and Lane DP: Mdm2-mediated NEDD8 conjugation of p53 inhibits its
transcriptional activity. Cell. 118:83–97. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Abida WM, Nikolaev A, Zhao W, Zhang W and
Gu W: FBXO11 promotes the Neddylation of p53 and inhibits its
transcriptional activity. J Biol Chem. 282:1797–1804. 2007.
View Article : Google Scholar
|
|
13
|
Lee GW, Park JB, Park SY, Seo J, Shin SH,
Park JW, Kim SJ, Watanabe M and Chun YS: The E3 ligase C-CBL
inhibits cancer cell migration by neddylating the proto-oncogene
c-Src. Oncogene. 37:5552–5568. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Broemer M, Tenev T, Rigbolt KT, Hempel S,
Blagoev B, Silke J, Ditzel M and Meier P: Systematic in vivo RNAi
analysis identifies IAPs as NEDD8-E3 ligases. Mol Cell. 40:810–822.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Keuss MJ, Thomas Y, Mcarthur R, Wood NT,
Knebel A and Kurz T: Characterization of the mammalian family of
DCN-type NEDD8 E3 ligases. J Cell Sci. 129:1441–1454.
2016.PubMed/NCBI
|
|
16
|
Meyer-Schaller N, Chou YC, Sumara I,
Martin DD, Kurz T, Katheder N, Hofmann K, Berthiaume LG, Sicheri F
and Peter M: The human Dcn1-like protein DCNL3 promotes Cul3
neddylation at membranes. Proc Natl Acad Sci USA. 106:12365–12370.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
He S, Cao Y, Xie P, Dong G and Zhang L:
The Nedd8 Non-covalent binding region in the smurf HECT domain is
critical to its ubiquitn ligase function. Sci Rep. 7:413642017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Deshaies RJ and Joazeiro CA: RING domain
E3 ubiquitin ligases. Annu Rev Biochem. 78:399–434. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Soucy TA, Dick LR, Smith PG, Milhollen MA
and Brownell JE: The NEDD8 conjugation pathway and its relevance in
cancer biology and therapy. Genes Cancer. 1:708–716. 2010.
View Article : Google Scholar
|
|
20
|
Mendoza HM, Shen LN, Botting C, Lewis A,
Chen J, Ink B and Hay RT: NEDP1, a highly conserved cysteine
protease that deNEDDylates Cullins. J Biol Chem. 278:25637–25643.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhou L, Zhu J, Chen W, Jiang Y, Hu T, Wang
Y, Ye X, Zhan M, Ji C, Xu Z, et al: Induction of NEDD8-conjugating
enzyme E2 UBE2F by platinum protects lung cancer cells from
apoptosis and confers to platinum-insensitivity. Cell Death Dis.
11:9752020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhou L, Jiang Y, Luo Q, Li L and Jia L:
Neddylation: A novel modulator of the tumor microenvironment. Mol
Cancer. 18:772019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Duda DM, Scott DC, Calabrese MF, Zimmerman
ES, Zheng N and Schulman BA: Structural regulation of cullin-RING
ubiquitin ligase complexes. Curr Opin Struct Biol. 21:257–264.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Furukawa M, Zhang Y, McCarville J, Ohta T
and Xiong Y: The CUL1 C-terminal sequence and ROC1 are required for
efficient nuclear accumulation, NEDD8 modification, and ubiquitin
ligase activity of CUL1. Mol Cell Biol. 20:8185–8197. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Duda DM, Borg LA, Scott DC, Hunt HW,
Hammel M and Schulman BA: Structural insights into NEDD8 activation
of cullin-RING ligases: Conformational control of conjugation.
Cell. 134:995–1006. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kim AY, Bommelje CC, Lee BE, Yonekawa Y,
Choi L, Morris LG, Huang G, Kaufman A, Ryan RJ, Hao B, et al: SCCRO
(DCUN1D1) is an essential component of the E3 complex for
neddylation. J Biol Chem. 283:33211–33220. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Goldenberg SJ, Cascio TC, Shumway SD,
Garbutt KC, Liu J, Xiong Y and Zheng N: Structure of the
Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the
assembly of the multisubunit cullin-dependent ubiquitin ligases.
Cell. 119:517–528. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Feng S, Shen Y, Sullivan JA, Rubio V,
Xiong Y, Sun TP and Deng XW: Arabidopsis CAND1, an unmodified
CUL1-interacting protein, is involved in multiple developmental
pathways controlled by ubiquitin/proteasome-mediated protein
Degradation. Plant Cell. 16:1870–1882. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Salon C, Brambilla E, Brambilla C,
Lantuejoul S, Gazzeri S and Eymin B: Altered pattern of Cul-1
protein expression and neddylation in human lung tumours:
Relationships with CAND1 and cyclin E protein levels. J Pathol.
213:303–310. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li L, Kang J, Zhang W, Cai L, Wang S,
Liang Y, Jiang Y, Liu X, Zhang Y, Ruan H, et al: Validation of
NEDD8-conjugating enzyme UBC12 as a new therapeutic target in lung
cancer. EBioMedicine. 45:81–91. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Podust VN, Brownell JE, Gladysheva TB, Luo
RS, Wang C, Coggins MB, Pierce JW, Lightcap ES and Chau V: A Nedd8
conjugation pathway is essential for proteolytic targeting of
p27Kip1 by ubiquitination. Proc Natl Acad Sci USA. 97:4579–4584.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Iso T, Suzuki T, Baird L and Yamamoto M:
Absolute Amounts and Status of the Nrf2-Keap1-Cul3 Complex within
Cells. Mol Cell Biol. 36:3100–3112. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bennett EJ, Rush J, Gygi SP and Harper JW:
Dynamics of cullin-RING ubiquitin ligase network revealed by
systematic quantitative proteomics. Cell. 143:951–965. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhao Y, Morgan MA and Sun Y: Targeting
Neddylation pathways to inactivate cullin-RING ligases for
anticancer therapy. Antioxid Redox Signal. 21:2383–2400. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kang M, Li Y, Zhao Y, He S and Shi J:
MiR-33a inhibits cell proliferation and invasion by targeting CAND1
in lung cancer. Clin Transl Oncol. 20:457–466. 2018. View Article : Google Scholar
|
|
36
|
Li L, Wang M, Yu G, Chen P, Li H, Wei D,
Zhu J, Xie L, Jia H, Shi J, et al: Overactivated neddylation
pathway as a therapeutic target in lung cancer. J Natl Cancer Inst.
106:dju0832014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Watson IR, Li BK, Roche O, Blanch A, Ohh M
and Irwin MS: Chemotherapy induces NEDP1-mediated destabilization
of MDM2. Oncogene. 29:297–304. 2010. View Article : Google Scholar
|
|
38
|
Xie P, Zhang M, He S, Lu K, Chen Y, Xing
G, Lu Y, Liu P, Li Y, Wang S, et al: The covalent modifier Nedd8 is
critical for the activation of Smurf1 ubiquitin ligase in
tumorigenesis. Nat Commun. 5:37332014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mahata B, Sundqvist A and Xirodimas DP:
Recruitment of RPL11 at promoter sites of p53-regulated genes upon
nucleolar stress through NEDD8 and in an Mdm2-dependent manner.
Oncogene. 31:3060–3071. 2012. View Article : Google Scholar
|
|
40
|
Sundqvist A, Liu G, Mirsaliotis A and
Xirodimas DP: Regulation of nucleolar signalling to p53 through
NEDDylation of L11. EMBO Rep. 10:1132–1139. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang J, Bai D, Ma X, Guan J and Zheng X:
hCINAP is a novel regulator of ribosomal protein-HDM2-p53 pathway
by controlling NEDDylation of ribosomal protein S14. Oncogene.
33:246–254. 2014. View Article : Google Scholar
|
|
42
|
Deben C, Deschoolmeester V, Lardon F,
Rolfo C and Pauwels P: TP53 and MDM2 genetic alterations in
non-small cell lung cancer: Evaluating their prognostic and
predictive value. Crit Rev Oncol Hematol. 99:63–73. 2016.
View Article : Google Scholar
|
|
43
|
Karin M and Ben-Neriah Y: Phosphorylation
meets ubiquitination: The control of NF-[Kappa]B activity. Annu Rev
Immunol. 18:621–663. 2000. View Article : Google Scholar
|
|
44
|
Read MA, Brownell JE, Gladysheva TB,
Hottelet M, Parent LA, Coggins MB, Pierce JW, Podust VN, Luo RS,
Chau V and Palombella VJ: Nedd8 Modification of Cul-1 Activates
SCF(beta(TrCP)-Dependent Ubiquitination of IkappaBalpha. Mol Cell
Biol. 20:2326–2333. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yu H, Lin L, Zhang Z, Zhang H and Hu H:
Targeting NF-κB pathway for the therapy of diseases: Mechanism and
clinical study. Signal Transduct Target Ther. 5:2092020. View Article : Google Scholar
|
|
46
|
Zhou L, Jiang Y, Liu X, Li L, Yang X, Dong
C, Liu X, Lin Y, Li Y, Yu J, et al: Promotion of tumor-associated
macrophages infiltration by elevated neddylation pathway via
NF-κB-CCL2 signaling in lung cancer. Oncogene. 38:5792–5804. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu W, Wang H, Bai F, Ding L, Huang Y, Lu
C, Chen S, Li C, Yue X, Liang X, et al: IL-6 promotes metastasis of
non-small-cell lung cancer by up-regulating TIM-4 via NF-κB. Cell
Prolif. 53:e127762020. View Article : Google Scholar
|
|
48
|
Orel L, Neumeier H, Hochrainer K, Binder
BR and Schmid JA: Crosstalk between the NF-kappaB activating
IKK-complex and the CSN signalosome. J Cell Mol Med. 14:1555–1568.
2010. View Article : Google Scholar
|
|
49
|
Ediriweera MK, Tennekoon KH and Samarakoon
SR: Role of the PI3K/AKT/mTOR signaling pathway in ovarian cancer:
Biological and therapeutic significance. Semin Cancer Biol.
59:147–160. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chibaya L, Karim B, Zhang H and Jones SN:
Mdm2 phosphorylation by Akt regulates the p53 response to oxidative
stress to promote cell proliferation and tumorigenesis. Proc Natl
Acad Sci USA. 118:e20031931182021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Xie P, Peng Z, Chen Y, Li H, Du M, Tan Y,
Zhang X, Lu Z, Cui CP, Liu CH, et al: Neddylation of PTEN regulates
its nuclear import and promotes tumor development. Cell Res.
31:291–311. 2021. View Article : Google Scholar :
|
|
52
|
Li X, Li C, Guo C, Zhao Q, Cao J, Huang
HY, Yue M, Xue Y, Jin Y, Hu L and Ji H: PI3K/Akt/mTOR signaling
orchestrates the phenotypic transition and chemo-resistance of
small cell lung cancer. J Genet Genomics. 48:640–651. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Giatromanolaki A, Koukourakis MI, Sivridis
E, Turley H, Talks K, Pezzella F, Gatter KC and Harris AL: Relation
of hypoxia inducible factor 1 alpha and 2 alpha in operable
non-small cell lung cancer to angiogenic molecular profile of
tumours and survival. Br J Cancer. 85:881–890. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhang Y, Bian Y, Wang Y, Wang Y, Duan X,
Han Y, Zhang L, Wang F, Gu Z and Qin Z: HIF-1α is necessary for
activation and tumour-promotion effect of cancer-associated
fibroblasts in lung cancer. J Cell Mol Med. 25:5457–5469. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wan J and Wu W: Hyperthermia induced
HIF-1a expression of lung cancer through AKT and ERK signaling
pathways. J Exp Clin Cancer Res. 35:1192016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Curtis VF, Ehrentraut SF, Campbell EL,
Glover LE, Bayless A, Kelly CJ, Kominsky DJ and Colgan SP:
Stabilization of HIF through inhibition of Cullin-2 neddylation is
protective in mucosal inflammatory responses. FASEB J. 29:208–215.
2015. View Article : Google Scholar :
|
|
57
|
Russell RC and Ohh M: NEDD8 acts as a
'molecular switch' defining the functional selectivity of VHL. EMBO
Rep. 9:486–491. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wolf ER, Mabry AR, Damania B and Mayo LD:
Mdm2-mediated neddylation of pVHL blocks the induction of
antiangiogenic factors. Oncogene. 39:5228–5239. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Hsu PC, Yang CT, Jablons DM and You L: The
Crosstalk between Src and Hippo/YAP signaling pathways in non-small
cell lung cancer (NSCLC). Cancers (Basel). 12:13612020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zou J, Ma W, Li J, Littlejohn R, Zhou H,
Kim IM, Fulton DJR, Chen W, Weintraub NL, Zhou J and Su H:
Neddylation mediates ventricular chamber maturation through
repression of Hippo signaling. Proc Natl Acad Sci USA.
115:E4101–E4110. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Cooper J, Xu Q, Zhou L, Pavlovic M, Ojeda
V, Moulick K, de Stanchina E, Poirier JT, Zauderer M, Rudin CM, et
al: Combined Inhibition of NEDD8-Activating Enzyme and mTOR
Suppresses NF2 Loss-Driven Tumorigenesis. Mol Cancer Ther.
16:1693–1704. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Maehama T, Nishio M, Otani J, Mak TW and
Suzuki A: The role of Hippo-YAP signaling in squamous cell
carcinomas. Cancer Sci. 112:51–60. 2021. View Article : Google Scholar
|
|
63
|
Du Z and Lovly CM: Mechanisms of receptor
tyrosine kinase activation in cancer. Mol Cancer. 17:582018.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Shi Q and Chen YG: Interplay between TGF-β
signaling and receptor tyrosine kinases in tumor development. Sci
China Life Sci. 60:1133–1141. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Enchev RI, Schulman BA and Peter M:
Protein neddylation: Beyond cullin-RING ligases. Nat Rev Mol Cell
Biol. 16:30–44. 2015. View Article : Google Scholar
|
|
66
|
Hamon P, Thoré MGD, Classe M, Signolle N,
Liu W, Bawa O, Meziani L, Clémenson C, Milliat F, Deutsch E and
Mondini M: TGFβ receptor inhibition unleashes interferon-β
production by tumor-associated macrophages and enhances
radiotherapy efficacy. J Immunother Cancer. 10:e0035192022.
View Article : Google Scholar
|
|
67
|
Zuo W, Huang F, Chiang YJ, Li M, Du J,
Ding Y, Zhang T, Lee HW, Jeong LS, Chen Y, et al: c-Cbl-mediated
neddylation antagonizes ubiquitination and degradation of the TGF-β
type II receptor. Mol Cell. 49:499–510. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Oved S, Mosesson Y, Zwang Y, Santonico E,
Shtiegman K, Marmor MD, Kochupurakkal BS, Katz M, Lavi S, Cesareni
G and Yarden Y: Conjugation to Nedd8 instigates ubiquitylation and
down-regulation of activated receptor tyrosine kinases. J Biol
Chem. 281:21640–21651. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Tumbrink HL, Heimsoeth A and Sos ML: The
next tier of EGFR resistance mutations in lung cancer. Oncogene.
40:1–11. 2021. View Article : Google Scholar
|
|
70
|
Konduri K, Gallant JN, Chae YK, Giles FJ,
Gitlitz BJ, Gowen K, Ichihara E, Owonikoko TK, Peddareddigari V,
Ramalingam SS, et al: EGFR fusions as novel therapeutic targets in
lung cancer. Cancer Discov. 6:601–611. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Luo Z, Yu G, Lee HW, Li L, Wang L, Yang D,
Pan Y, Ding C, Qian J, Wu L, et al: The Nedd8-activating enzyme
inhibitor MLN4924 induces autophagy and apoptosis to suppress liver
cancer cell growth. Cancer Res. 72:3360–3371. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Invrea F, Punzi S, Petti C, Minelli R,
Peoples MD, Bristow CA, Vurchio V, Corrado A, Bragoni A, Marchiò C,
et al: Synthetic lethality screening highlights colorectal cancer
vulnerability to concomitant blockade of NEDD8 and EGFR pathways.
Cancers (Basel). 13:38052021. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Jiang Y, Cheng W, Li L, Zhou L, Liang Y,
Zhang W, Chen W, Wang S, Zhao H, Chen G, et al: Effective targeting
of the ubiquitin-like modifier NEDD8 for lung adenocarcinoma
treatment. Cell Biol Toxicol. 36:349–364. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Bommelje CC, Weeda VB, Huang G, Shah K,
Bains S, Buss E, Shaha M, Gönen M, Ghossein R, Ramanathan SY and
Singh B: Oncogenic function of SCCRO5/DCUN1D5 requires its
Neddylation E3 activity and nuclear localization. Clin Cancer Res.
20:372–381. 2014. View Article : Google Scholar :
|
|
75
|
Wang L, Zhang M, Pan X, Zhao M, Huang L,
Hu X, Wang X, Qiao L, Guo Q, Xu W, et al: Integrative serum
metabolic fingerprints based multi-modal platforms for lung
adenocarcinoma early detection and pulmonary nodule classification.
Adv Sci (Weinh). 9:e22037862022. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Yang J, Yin X, Zhang L, Zhang X, Lin Y,
Zhuang L, Liu W, Zhang R, Yan X, Shi L, et al: Defective Fe
metal-organic frameworks enhance metabolic profiling for
high-accuracy diagnosis of human cancers. Adv Mater.
34:e22014222022. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Zhou L, Dong C, Xu Z, Wang X, Zhang L,
Chen S, Chen J and Zhu Y: NEDD8-conjugating enzyme E2 UBE2F confers
radiation resistance by protecting lung cancer cells from
apoptosis. J Zhejiang Univ Sci B. 22:959–965. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Zhou W, Xu J, Li H, Xu M, Chen ZJ, Wei W,
Pan Z and Sun Y: Neddylation E2 UBE2F promotes the survival of lung
cancer cells by activating CRL5 to Degrade NOXA via the K11
Linkage. Clin Cancer Res. 23:1104–1116. 2017. View Article : Google Scholar :
|
|
79
|
Guo ZP, Hu YC, Xie Y, Jin F, Song ZQ, Liu
XD, Ma T and Zhou PK: MLN4924 suppresses the BRCA1 complex and
synergizes with PARP inhibition in NSCLC cells. Biochem Biophys Res
Commun. 483:223–229. 2017. View Article : Google Scholar
|
|
80
|
Meng F, Yu W, Chen C, Guo S, Tian X, Miao
Y, Ma L, Zhang X, Yu Y, Huang L, et al: A versatile electrochemical
biosensor for the detection of circulating MicroRNA toward
non-small cell lung cancer diagnosis. Small. 18:e22007842022.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zhu HZ, Hou J, Guo Y, Liu X, Jiang FL,
Chen GP, Pang XF, Sun JG and Chen ZT: Identification and imaging of
miR-155 in the early screening of lung cancer by targeted delivery
of octreotide-conjugated chitosan-molecular beacon nanoparticles.
Drug Deliv. 25:1974–1983. 2018. View Article : Google Scholar
|
|
82
|
Khalife J, Radomska HS, Santhanam R, Huang
X, Neviani P, Saultz J, Wang H, Wu YZ, Alachkar H, Anghelina M, et
al: Pharmacological targeting of miR-155 via the NEDD8-activating
enzyme inhibitor MLN4924 (Pevonedistat) in FLT3-ITD acute myeloid
leukemia. Leukemia. 29:1981–1992. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Ardizzoni A, Boni L, Tiseo M, Fossella FV,
Schiller JH, Paesmans M, Radosavljevic D, Paccagnella A, Zatloukal
P, Mazzanti P, et al: Cisplatin-versus carboplatin-based
chemotherapy in first-line treatment of advanced non-small-cell
lung cancer: An individual patient data meta-analysis. J Natl
Cancer Inst. 99:847–857. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Xu W, Xie X, Wu H, et al: Pulsed
electromagnetic therapy in cancer treatment: Progress and outlook.
View. 3:202200292022. View Article : Google Scholar
|
|
85
|
Brownell J E, Sintcha k MD, Gavin JM, Liao
H, Bruzzese FJ, Bump NJ, Soucy TA, Milhollen MA, Yang X, Burkhardt
AL, et al: Substrate-assisted inhibition of ubiquitin-like
protein-activating enzymes: The NEDD8 E1 inhibitor MLN4924 forms a
NEDD8-AMP mimetic in situ. Mol Cell. 37:102–111. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Lockhart AC, Bauer TM, Aggarwal C, Lee CB,
Harvey RD, Cohen RB, Sedarati F, Nip TK, Faessel H, Dash AB, et al:
Phase Ib study of pevonedistat, a NEDD8-activating enzyme
inhibitor, in combination with docetaxel, carboplatin and
paclitaxel, or gemcitabine, in patients with advanced solid tumors.
Invest New Drugs. 37:87–97. 2019. View Article : Google Scholar :
|
|
87
|
Sarantopoulos J, Shapiro GI, Cohen RB,
Clark JW, Kauh JS, Weiss GJ, Cleary JM, Mahalingam D, Pickard MD,
Faessel HM, et al: Phase I study of the investigational
NEDD8-Activating enzyme inhibitor pevonedistat (TAK-924/MLN4924) in
patients with advanced solid tumors. Clin Cancer Res. 22:847–857.
2016. View Article : Google Scholar
|
|
88
|
Yin Y, Xie CM, Li H, Tan M, Chen G, Schiff
R, Xiong X and Sun Y: The FBXW2-MSX2-SOX2 axis regulates stem cell
property and drug resistance of cancer cells. Proc Natl Acad Sci
USA. 116:20528–20538. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Norton JP, Augert A, Eastwood E, Basom
Rudin CM and MacPherson D: Protein neddylation as a therapeutic
target in pulmonary and extrapulmonary small cell carcinomas. Genes
Dev. 35:870–887. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Soucy TA, Smith PG, Milhollen MA, Berger
AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP,
Critchley S, et al: An inhibitor of NEDD8-activating enzyme as a
new approach to treat cancer. Nature. 458:732–736. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Jia L, Li H and Sun Y: Induction of
p21-dependent senescence by an NAE inhibitor, MLN4924, as a
mechanism of growth suppression. Neoplasia. 13:561–569. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Lin JJ, Milhollen MA, Smith PG, Narayanan
U and Dutta A: NEDD8-targeting drug MLN4924 elicits DNA
rereplication by stabilizing Cdt1 in S phase, triggering checkpoint
activation, apoptosis, and senescence in cancer cells. Cancer Res.
70:10310–10320. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Chen P, Hu T, Liang Y, Li P, Chen X, Zhang
J, Ma Y, Hao Q, Wang J, Zhang P, et al: Neddylation inhibition
activates the extrinsic apoptosis pathway through ATF4-CHOP-DR5
axis in human esophageal cancer cells. Clin Cancer Res.
22:4145–4157. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Jia L, Soengas MS and Sun Y: ROC1/RBX1 E3
ubiquitin ligase silencing suppresses tumor cell growth via
sequential induction of G2-M arrest, apoptosis, and senescence.
Cancer Res. 69:4974–4982. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Rizzardi LF and Cook JG: Flipping the
switch from g1 to s phase with e3 ubiquitin ligases. Genes Cancer.
3:634–648. 2012. View Article : Google Scholar
|
|
96
|
Zhao Y, Xiong X, Jia L and Sun Y:
Targeting Cullin-RING ligases by MLN4924 induces autophagy via
modulating the HIF1-REDD1-TSC1-mTORC1-DEPTOR axis. Cell Death Dis.
3:e3862012. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Li Y, Wang C, Xu T, Pan P, Yu Q, Xu L,
Xiong X, Hou T, Cui S and Sun Y: Discovery of a small molecule
inhibitor of cullin neddylation that triggers ER stress to induce
autophagy. Acta Pharm Sin B. 11:3567–3584. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Weis SM and Cheresh DA: Tumor
angiogenesis: Molecular pathways and therapeutic targets. Nat Med.
17:1359–1370. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Yao WT, Wu JF, Yu GY, Wang R, Wang K, Li
LH, Chen P, Jiang YN, Cheng H, Lee HW, et al: Suppression of tumor
angiogenesis by targeting the protein neddylation pathway. Cell
Death Dis. 5:e10592014. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Tan M, Zhao Y, Kim SJ, Liu M, Jia L,
Saunders TL, Zhu Y and Sun Y: SAG/RBX2/ROC2 E3 ubiquitin ligase is
essential for vascular and neural development by targeting NF1 for
degradation. Dev Cell. 21:1062–1076. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Tan M, Li H and Sun Y: Endothelial
deletion of Sag/Rbx2/Roc2 E3 ubiquitin ligase causes embryonic
lethality and blocks tumor angiogenesis. Oncogene. 33:5211–5220.
2014. View Article : Google Scholar :
|
|
102
|
Conway EM, Pikor LA, Kung SH, Hamilton MJ,
Lam S, Lam WL and Bennewith KL: Macrophages, inflammation, and lung
cancer. Am J Respir Crit Care Med. 193:116–130. 2016. View Article : Google Scholar
|
|
103
|
Deng Q, Zhang J, Gao Y, She X, Wang Y,
Wang Y and Ge X: MLN4924 protects against bleomycin-induced
pulmonary fibrosis by inhibiting the early inflammatory process. Am
J Transl Res. 9:1810–1821. 2017.PubMed/NCBI
|
|
104
|
Zhou L, Zhang W, Sun Y and Jia L: Protein
neddylation and its alterations in human cancers for targeted
therapy. Cell Signal. 44:92–102. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Hao R, Song Y, Li R, Wu Y, Yang X, Li X,
Qian F, Ye RD and Sun L: MLN4924 protects against
interleukin-17A-induced pulmonary inflammation by disrupting
ACT1-mediated signaling. Am J Physiol Lung Cell Mol Physiol.
316:L1070–L1080. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Cheng M, Hu S, Wang Z, Pei Y, Fan R, Liu
X, Wang L, Zhou J, Zheng S, Zhang T, et al: Inhibition of
neddylation regulates dendritic cell functions via Deptor
accumulation driven mTOR inactivation. Oncotarget. 7:35643–35654.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Mathewson N, Toubai T, Kapeles S, Sun Y,
Oravecz-Wilson K, Tamaki H, Wang Y, Hou G, Sun Y and Reddy P:
Neddylation plays an important role in the regulation of murine and
human dendritic cell function. Blood. 122:2062–2073. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Zhu Z, Sun L, Hao R, Jiang H, Qian F and
Ye RD: Nedd8 modification of Cullin-5 regulates
lipopolysaccharide-induced acute lung injury. Am J Physiol Lung
Cell Mol Physiol. 313:L104–L114. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Jiang Y, Liang Y, Li L, Zhou L, Cheng W,
Yang X, Yang X, Qi H, Yu J, Jeong LS, et al: Targeting neddylation
inhibits intra-vascular survival and extravasation of cancer cells
to prevent lung-cancer metastasis. Cell Biol Toxicol. 35:233–245.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Kim Y, Park JB, Fukuda J, Watanabe M and
Chun YS: The effect of neddylation blockade on slug-dependent
cancer cell migration is regulated by p53 mutation status. Cancers
(Basel). 13:5312021. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Sun Y and Li H: Functional
characterization of SAG/RBX2/ROC2/RNF7, an antioxidant protein and
an E3 ubiquitin ligase. Protein Cell. 4:103–116. 2013. View Article : Google Scholar
|
|
112
|
Xu Q, Lin G, Xu H, Hu L, Wang Y, Du S,
Deng W, Hu W, Cheng W and Jiang K: MLN4924 neddylation inhibitor
promotes cell death in paclitaxel-resistant human lung
adenocarcinoma cells. Oncol Lett. 15:515–521. 2018.PubMed/NCBI
|
|
113
|
Zhang Q, Karnak D, Tan M, Lawrence TS,
Morgan MA and Sun Y: FBXW7 facilitates nonhomologous End-Joining
via K63-Linked Polyubiquitylation of XRCC4. Mol Cell. 61:419–433.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Zhou X, Tan M, Nyati MK, Zhao Y, Wang G
and Sun Y: Blockage of neddylation modification stimulates tumor
sphere formation in vitro and stem cell differentiation and wound
healing in vivo. Proc Natl Acad Sci USA. 113:E2935–2944. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Takahashi K and Yamanaka S: Induction of
pluripotent stem cells from mouse embryonic and adult fibroblast
cultures by defined factors. Cell. 126:663–676. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Park JB, Seo J, Park JW and Chun YS:
Neddylation blockade induces HIF-1α driven cancer cell migration
via upregulation of ZEB1. Sci Rep. 10:182102020. View Article : Google Scholar
|
|
117
|
Korrodi-Gregório L, Soto-Cerrato V,
Vitorino R, Fardilha M and Pérez-Tomás R: From proteomic analysis
to potential therapeutic targets: Functional profile of two lung
cancer cell lines, A549 and SW900, widely studied in pre-clinical
research. PLoS One. 11:e01659732016. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Ni S, Chen X, Yu Q, Xu Y, Hu Z, Zhang J,
Zhang W, Li B, Yang X, Mao F, et al: Discovery of candesartan
cilexetic as a novel neddylation inhibitor for suppressing tumor
growth. Eur J Med Chem. 185:1118482020. View Article : Google Scholar
|
|
119
|
Chen X, Yang X, Mao F, Wei J, Xu Y, Li B,
Zhu J, Ni S, Jia L and Li J: Development of novel
benzimidazole-derived neddylation inhibitors for suppressing tumor
growth invitro and invivo. Eur J Med Chem. 210:1129642021.
View Article : Google Scholar
|
|
120
|
An H and Statsyuk AV: An inhibitor of
ubiquitin conjugation and aggresome formation. Chem Sci.
6:5235–5245. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Milhollen MA, Thomas MP, Narayanan U,
Traore T, Riceberg J, Amidon BS, Bence NF, Bolen JB, Brownell J,
Dick LR, et al: Treatment-emergent mutations in NAEβ confer
resistance to the NEDD8-activating enzyme inhibitor MLN4924. Cancer
Cell. 21:388–401. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Toth JI, Yang L, Dahl R and Petroski MD: A
gatekeeper residue for NEDD8-activating enzyme inhibition by
MLN4924. Cell Rep. 1:309–316. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Verma S, Singh A and Mishra A: Molecular
dynamics investigation on the poor sensitivity of A171T mutant
NEDD8-activating enzyme (NAE) for MLN4924. J Biomol Struct Dyn.
32:1064–1073. 2014. View Article : Google Scholar
|
|
124
|
Hammill JT, Scott DC, Min J, Connelly MC,
Holbrook G, Zhu F, Matheny A, Yang L, Singh B, Schulman BA and Guy
RK: Piperidinyl ureas chemically control defective in cullin
neddylation 1 (DCN1)-Mediated cullin neddylation. J Med Chem.
61:2680–2693. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Hammill JT, Bhasin D, Scott DC, Min J,
Chen Y, Lu Y, Yang L, Kim HS, Connelly MC, Hammill C, et al:
Discovery of an orally bioavailable inhibitor of defective in
cullin neddylation 1 (DCN1)-Mediated cullin neddylation. J Med
Chem. 61:2694–2706. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Zhou H, Lu J, Liu L, Bernard D, Yang CY,
Fernandez-Salas E, Chinnaswamy K, Layton S, Stuckey J, Yu Q, et al:
A potent small-molecule inhibitor of the DCN1-UBC12 interaction
that selectively blocks cullin 3 neddylation. Nat Commun.
8:11502017. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Zhou H, Lu J, Chinnaswamy K, Stuckey JA,
Liu L, McEachern D, Yang CY, Bernard D, Shen H, Rui L, et al:
Selective inhibition of cullin 3 neddylation through covalent
targeting DCN1 protects mice from acetaminophen-induced liver
toxicity. Nat Commun. 12:26212021. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Li Y, Bao Q, Yang S, Yang M and Mao C:
Bionanoparticles in cancer imaging, diagnosis, and treatment. View.
3:202000272022. View Article : Google Scholar
|
|
129
|
Zhang Z, Zhang J, Tian J and Li H: A
polydopamine nanomedicine used in photothermal therapy for liver
cancer knocks down the anti-cancer target NEDD8-E3 ligase ROC1
(RBX1). J Nanobiotechnology. 19:3232021. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Deng L, Meng T, Chen L, Wei W and Wang P:
The role of ubiquitination in tumorigenesis and targeted drug
discovery. Signal Transduct Target Ther. 5:112020. View Article : Google Scholar : PubMed/NCBI
|



















