Macroautophagy, hereafter referred to as autophagy,
is a catabolic process where cytoplasmic materials are sequestered
by double-membrane-bound vesicles, called autophagosomes, to be
degraded via fusion with lysosomes. Autophagy, from the Greek
autóphagos, meaning 'self-eating', is a process that is
highly conserved from yeast to humans and is necessary for
maintenance of cellular homeostasis under nutrient deprivation and
other stress conditions (1). It
is also involved in physiological processes such as defense against
intracellular pathogens, organelle quality control and removal of
misfolded or aggregated proteins (2).
Numerous studies demonstrate that autophagy acts as
a double-edged sword in cancer. Namely, it can inhibit tumor
initiation by removing damaged proteins and organelles and
preventing genome instability (3-6).
Tissue-specific deletion of key autophagic molecules (such as
autophagy related genes ATG7 and ATG5) restricts cancer development
in several mouse models of inducible cancer such as melanoma and
pancreatic cancer (7-12). On the other hand, when a tumor has
already developed, autophagy can support cancer cell survival under
stressful conditions such as hypoxia and metabolic stress,
promoting the persistence of tumor cells in hostile environment
(13-16). Autophagy affects anchorage to the
extracellular matrix, cytoskeletal remodeling and
epithelial-to-mesenchymal transition (EMT), further supporting its
involvement in cancer progression (17).
The autophagy machinery consists of a number of
molecules, involved in the different steps of autophagy:
Initiation, nucleation, elongation, autophagosome-lysosome fusion
and degradation of substrates (18). A group of ~20 molecules, called
autophagy-related genes, was initially discovered through genetic
studies in yeast and found to be necessary for the control of
several key phases of the autophagy process (19,20). The group of autophagic molecules
has been studied in humans and gradually expanded to include other
proteins; The Autophagy Project of the BioGRID repository contains
information on ~200 human proteins involved in autophagy
(thebiogrid.org/project/6/autophagy.html; accessed on
October 2023) (21) (Table SI).
Numerous autophagy machinery molecules (AMMs) have
been reported to extend their functions beyond autophagy.
Non-canonical autophagy-related processes have been described
(22-25). For example, molecules involved in
autophagic vesicle elongation are also involved in a form of
exocytosis termed secretory autophagy (22). In addition, there is increasing
evidence that individual AMMs exert autophagy-independent functions
in various types of diseases, including cancer (23-25). The present review aimed to discuss
the autophagy-independent functions of AMMs with a particular focus
on cancer progression.
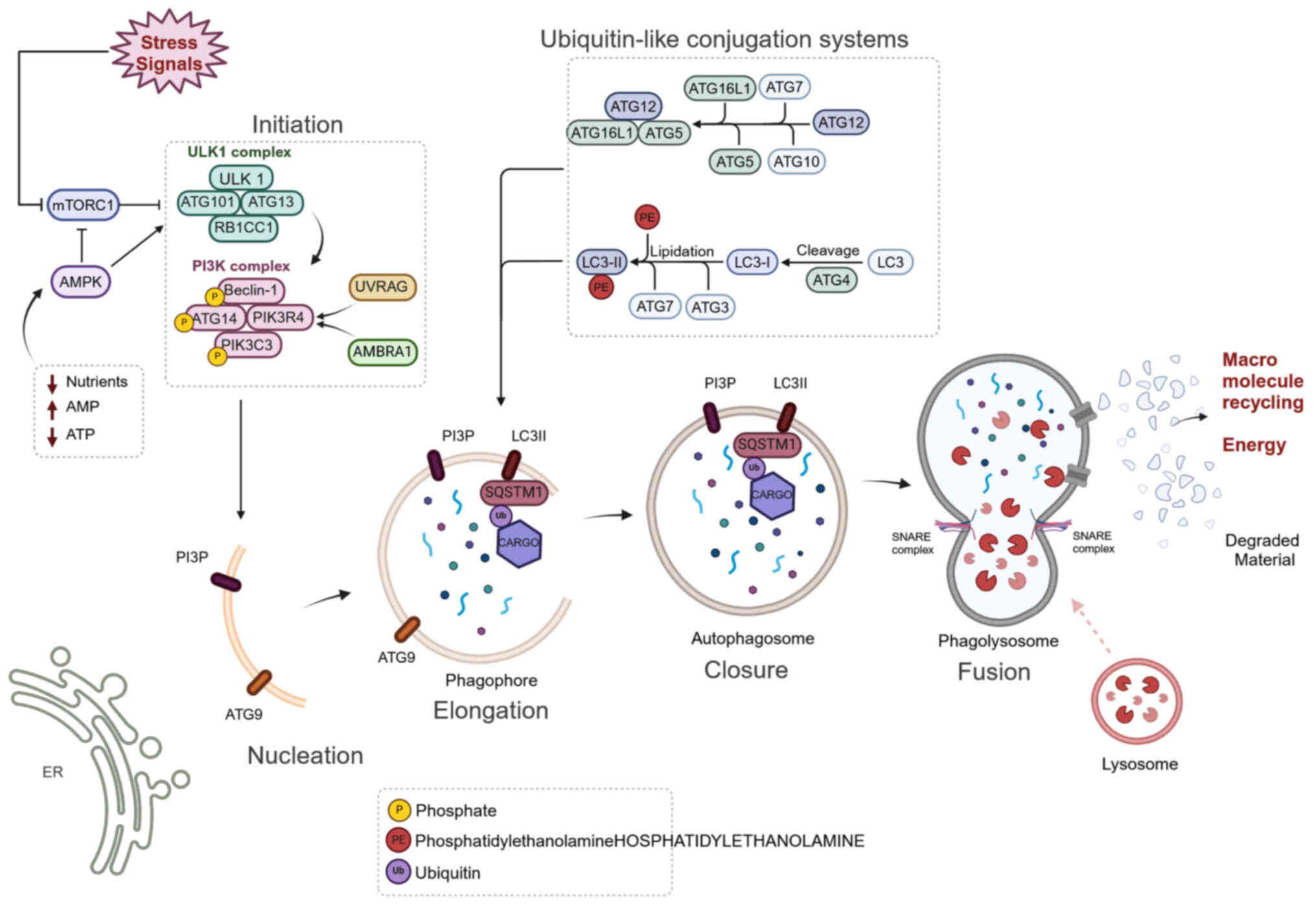
Nutrient shortage and stress conditions are the main
triggers for the autophagy process (Fig. 1). One of the key sensors of
energy, nutrient and redox status is the mTORC1 protein complex,
which consists of the Ser/Thr kinase mTOR and other regulatory
components (27). Under
nutrient-limited conditions, the amount of ATP decreases and
increased AMP/ATP ratio triggers the activation of AMPK, which in
turn restrains mTORC1 and its inhibitory activity towards
Unc-51-like autophagy-activating kinase 1 (ULK1). ULK1 can form a
complex with autophagy related proteins ATG101, ATG13 and RB1CC1
(family kinase-interacting protein 200/RB1 Inducible Coiled-Coil
1), which initiates the autophagy cascade (28) by phosphorylating components of
PI3K complex I (PIK3C3, Beclin-1, ATG14 and PIK3R4) (29). This complex is essential for the
nucleation phase of autophagy. Activated PI3K complex I
phosphorylates phosphatidylinositol (PI) to form PI-3-phosphate
(PI3P) (29), which binds to the
nascent phagophore membrane (30). PI3K complex I is positively
regulated by the ultraviolet radiation resistance-associated gene
(UVRAG) (31) and autophagy and
Beclin-1 regulator 1 (AMBRA1) (32). The process is supported by ATG9, a
lipid scramblase that is incorporated into vesicles involved in the
nucleation of phagophores and subsequently assists the elongation
process (33,34). Two ubiquitin-like conjugation
systems are then activated: The phagophore elongation complexes
ATG5-ATG12-ATG16L1 and the LC3 system. The ATG5-ATG12-ATG16L1
complex is formed by a reaction cascade involving ATG7 (E1-like
enzyme) and ATG10 (E2-like enzyme), which mediate covalent binding
between ATG5 and ATG12. Subsequently, the ATG5-ATG12 conjugate
binds ATG16L1 and forms a ternary complex located at the
autophagosomal membrane (35).
Studies propose an alternative model for the formation of the
ATG5-ATG12-ATG16L1 complex, which first requires an interaction
between ATG5 and ATG16L1. Then, the transient ATG5-ATG16L1 duplet
allows recruitment of ATG12 and the formation of a stable trimeric
structure via formation of a covalent bond between ATG12 and ATG5
(36-38). The ATG5-ATG12-ATG16L1 complex
serves as a scaffold and promotes LC3 lipidation (35). Microtubule-associated protein
1-light chain 3 (MAP1LC3) is the ortholog of Atg8 in yeast. LC3 is
first cleaved at its carboxy terminus by ATG4 to form LC3 I.
Following ATG4-mediated cleavage, LC3 is activated by ATG7 (E1-like
enzyme) and ATG3 (E2-like enzyme) and finally conjugated to
phosphatidylethanolamine (PE) to form the active LC3 (LC3-II)
(39). Lipidated LC3, together
with the ATG5-ATG12-ATG16L1 complex, enables elongation of the
autophagic phagophore membrane (40).
The nascent phagophore sequesters specific cargo
material via simultaneous interaction between LC3-II molecules and
cargo receptors such as sequestrosome-1 (SQSTM1 or p62),
toll-interacting Protein), and neighbor Of BRCA1 Gene 1) (41). SQSTM1 oligomerizes via its PB1
domain and forms filaments that interact with polyubiquitinated
cargoes and LC3-II via LC3-interacting regions. These interactions
enable autophagy-mediated degradation of specific cargo material
(42).
In the late stages of the autophagic process, the
phagophore closes and fuses with the lysosome to form the
autophagolysosome. This phase relies on soluble
N-ethylmaleimide-sensitive factor attachment protein receptor
(SNARE) proteins, which are found in both membranes (43). The activity of the two known SNARE
complexes (TX17-SNAP29-VAMP7/VAMP812 and STX7-SNAP29-YKT6
complexes) is facilitated by the tethering factors such as the
homotypic fusion and protein sorting) complex, pleckstrin Homology
And RUN Domain Containing M1) and EPG5 (Ectopic P-Granules 5
Autophagy Tethering Factor), which promote close interaction
between the membranes. In the autophagolysosome, acidic hydrolases
degrade sequestered material and generate metabolites that are
released into the cytoplasm (44). The nutrients obtained via the
autophagy pathway stimulate mTOR activation. A negative feedback
mechanism stops autophagy when availability of nutrients is
restored (44).
During tumor transformation and progression, several
biological functions are altered (45). Cancer cells acquire genome
instability, which gives them selective advantages. The
aggressiveness of the transformed clones is characterized by
sustained proliferation and death resistance. Cancer cells have
invasive behavior determined by the activation of molecular
invasion programs and the ability to control the tumor
microenvironment by sending signals to surrounding cells, including
immune cells (45). Increasing
evidence suggests that AMMs may serve functions that are not
exclusive to lysosomal degradation of autophagy substrates
(22-24). AMMs are involved in the processes
of cancer initiation and progression (Table I).
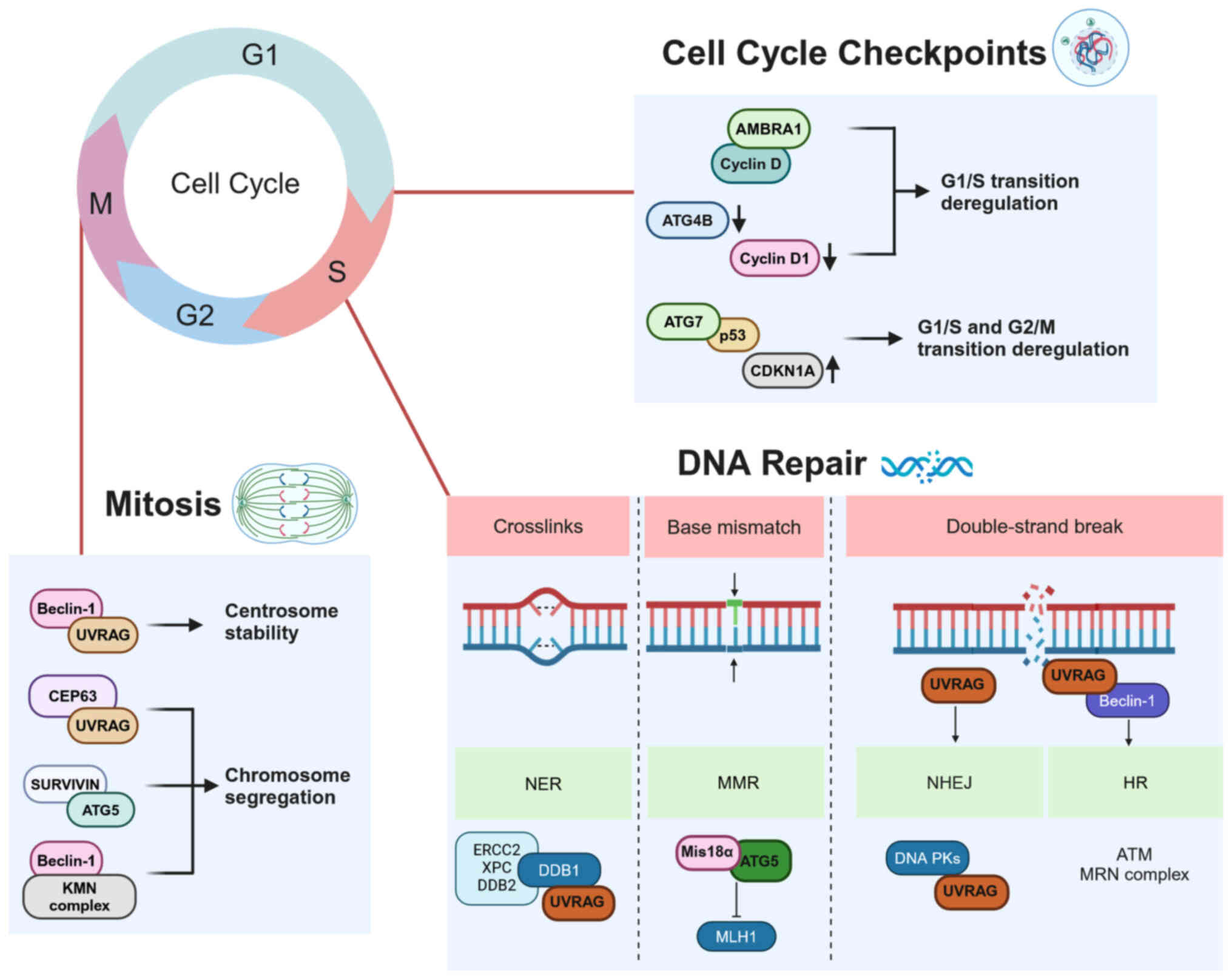
Normal cells control DNA integrity to ensure genome
stability. Following DNA damage, cell cycle progression is delayed
or blocked by a number of cell cycle control mechanisms to allow
repair of DNA damage and prevent abnormal cell division (45).
Following treatment with 5-fluorouracil (5-FU), ATG5
translocates to the nucleus independently of its autophagic
function and interacts with Mis18α (MIS18 kinetochore Protein A), a
protein localized in the centromere and involved in methylation of
the underlying chromatin. This binding increases the levels of
promoter methylation of MLH1 (MutL Homolog 1) gene (a component of
DNA mismatch repair), thereby downregulating MLH1 expression and
enhancing mismatch repair defects and resistance to 5-FU (53). Similarly, following DNA-damaging
treatment, ATG5 interacts with survivin in the nucleus, disrupting
chromosome segregation and triggering an abnormal mitotic process
known as mitotic catastrophe (54). This suggests control of cell cycle
progression by ATG5 independent of autophagy.
AMBRA1 is another AMM that controls cell cycle
progression by mediating degradation of cyclin D, which regulates
the G1/S phase transition. A defective AMBRA1/cyclin D axis leads
to premature entry into S phase, resulting in replication stress
and genome instability (55). In
addition, ATG4B downregulation in colorectal cancer has been shown
to decrease expression and activity of cyclin D1 (56). The aforementioned study
demonstrated an inhibition of mTOR and induction of autophagy in
ATG4B-silenced cells; however it is unclear how this fits with the
hypothesis that ATG4B can prime LC3B for lipidation and autophagy
induction (56). ATG7 promotes
CDKN1A (p21) expression and cell cycle arrest by directly binding
to p53, a master keeper of cell cycle, apoptosis and CDKN1A (p21)
expression. The effect of ATG7 on this process is enhanced by
nutrient starvation, which is known to stimulate autophagy
(57). Nevertheless, the E1-like
enzymatic activity of ATG7, which is central to its autophagic
involvement, is not required for this cell cycle arrest (57).
The component of the PI3K complex Beclin-1 is
crucial for mitotic progression as it interacts with the
KNL-1/Mis12/Ndc80 complex involved in the precise anchoring of the
kinetochore to the mitotic spindle (58). Moreover, a variant of the PI3K
complex, which includes Bax-Interacting Factor 1), is involved in
cytokinesis, and thus exerts a tumor suppressor function distinct
from its role in the early steps of autophagy (59). PI3P generated by the activated
PI3K complex mediates contact with proteins of the centrosome and
regulates completion of cytokinesis (60).
Autophagy is a multifaceted process that promotes
either cell survival or death, depending on the physiological state
of the cell and environmental conditions. Autophagy and apoptosis
are closely associated processes in which AMMs can be activated by
apoptotic factors and vice versa (61). For example, the autophagosome
membrane and its associated autophagy machinery serve as a platform
for recruitment and activation of the apoptotic caspase cascade
(62). On the other hand,
proapoptotic caspase-9 has been shown to interact with ATG7 and
contribute to autophagy in various human cancer cell lines
(63).
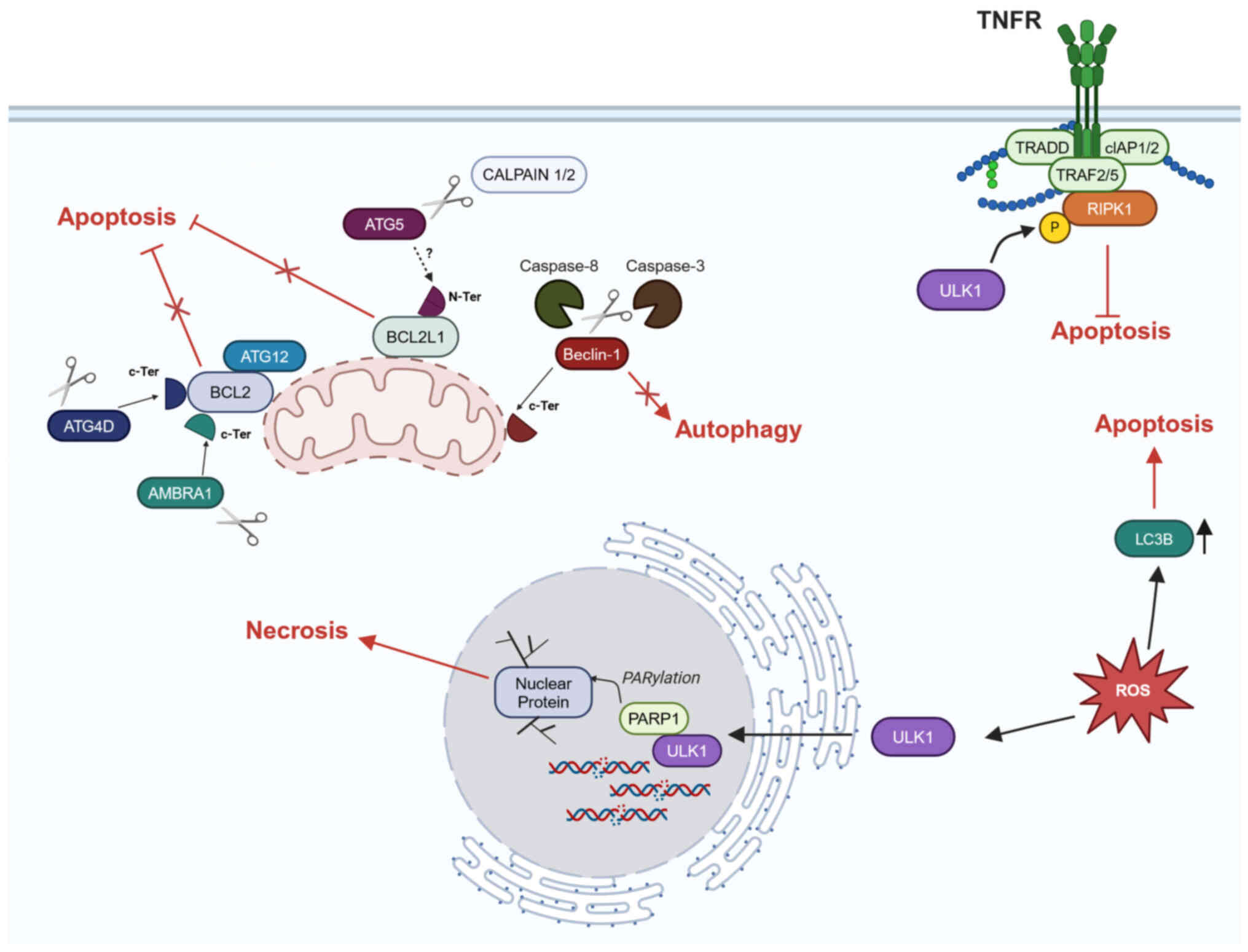
Under certain circumstances, autophagy and apoptosis
appear to be mutually exclusive processes mediated by common
players (Fig. 3).
Protease-mediated cleavage of certain AMMs inhibits autophagy and
promotes apoptosis. For example, under unfavorable conditions, such
as cell starvation and drug treatment, cleavage of Beclin-1 by
caspase-3 or caspase-8 results in formation of an
autophagy-impaired Beclin-1 fragment that localizes to mitochondria
and promotes apoptosis (61,64,65).
AMBRA1 carries a BH3 motif that, after being
released by caspase-mediated cleavage, binds and blocks BCL2, one
of the key inhibitors of apoptosis (66). Overexpression of ATG4D and a form
of ATG4D cleaved by caspase-3 leads to their recruitment to
mitochondria, where they contribute to apoptosis. Again, this
pro-apoptotic function of ATG4D relies on its C-terminal BH3
domain, which specifically interacts with members of the BCL2
family (67). Similarly, ATG5 is
specifically cleaved by calpains 1/2 during apoptosis,
independently of cell type and apoptotic stimulus. Truncated ATG5
translocates to the mitochondria, associates with BCL2-like protein
1 (BCL2L1) and triggers caspase activation via an
autophagy-independent mechanism (68). The ATG5 partner ATG12 can control
apoptosis independently of the other AMMs by interacting with BCL2
protein family via the BH3 motif (69). ATG12 is an unstable protein that,
when not conjugated to ATG5, is subject to proteasomal degradation
(36). Due to aberrant
proteasomal blockade, ATG12 accumulates in osteosarcoma cells,
antagonizes BCL2 and activates apoptosis (38). In colorectal cancer cell lines,
oncogenic Ras promotes cancer cell survival by decreasing ATG12
levels and thus ATG12-mediated inhibition of apoptosis (70). ATG12 also serves a role in the
control of mitochondrial homeostasis. It interacts with ATG3 in a
complex that does not affect autophagy, but is critical for
controlling mitochondrial fission and fusion and regulating the
function of mitochondria-mediated cell death pathways (71).
Not only apoptosis, but also other forms of cell
death are influenced by AMMs. Lung and breast cancer cell lines in
which ATG12 expression is reduced undergo oncosis, a
caspase-independent cell death triggered by energy deficiency
(72). In this condition, the
imbalance of mitochondrial ions and impaired metabolism cause
changes in osmotic pressure, leading to organelle swelling and
cytoplasmic blebs that disrupt cellular function. Similarly, Ni
et al (73) identified a
novel phosphorylation site in ATG4B that, once phosphorylated,
allows binding with the soluble catalytic core F1, and the
membrane-spanning component, Fo, subunits of ATP synthase. This
interaction results in impaired mitochondrial function, which leads
to an increase in mitochondrial reactive oxygen species (ROS) and
metabolic reprogramming of hepatocellular carcinoma cells towards
the Warburg effect (73).
Following autophagy-mediated activation, ULK1 can phosphorylate
RIPK1, a component of the TNF receptor (TNFR)-mediated cell death
complex, thereby improving survival of mouse embryonic fibroblasts
(MEFs) (74). However, stress
conditions, such as increased ROS species, relocate ULK1 to the
nucleus, limiting the autophagic response and allowing nuclear ULK1
to promote PARP1-dependent necrosis (75). Similarly, pharmacological
induction of ROS increases LC3B production without activating
autophagic flux and enhances both expression of apoptotic molecules
and induction of anoikis (76).
The majority of cancer deaths are caused by
metastasis. The metastatic process begins when cancer cells leave
the primary neoplasm, invade the surrounding matrix and colonize
other tissue via the bloodstream and lymphatic system. These
processes, combined with the uncontrolled proliferative capacity of
cancer cells, lead to the destruction of the physiological
functions of distant organs (77). At the molecular level, invasive
and metastatic behavior is supported by features of EMT, which is
an embryonic molecular program that is abnormally activated in
tumor cells (78). Several
studies have demonstrated the role of autophagy in EMT (79-82). Nevertheless, AMMs have also been
reported to have non-autophagic functions in EMT (25,83-85),(Fig.
4).
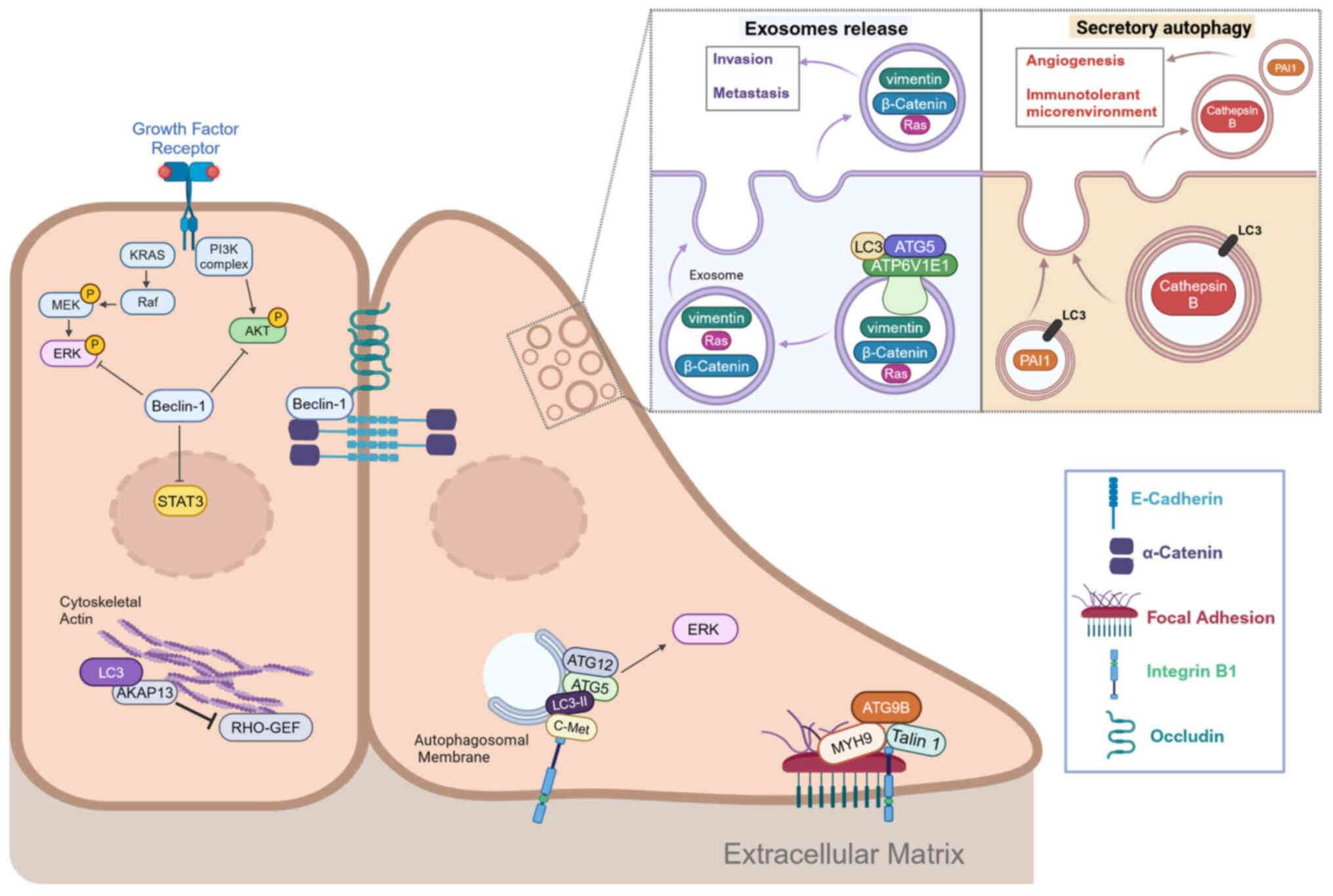
The metastatic potential of cancer cells is ensured
by intracellular processes that enable cells to survive under
stress conditions. Autophagic membranes carrying LC3-II and
ATG5-ATG12 conjugates serve as scaffolds for ERK pathway activation
(83). In colorectal cancer, B1
integrin can promote c-Met internalization and ERK1/2 activation,
allowing cancer cells to survive to anoikis (84). c-Met and B1 integrin colocalize on
autophagic membranes for their pro-survival signals and require
LC3-II, Beclin-1 and ATG5 for this purpose, but not other canonical
autophagy mediators. Loss of Beclin-1 impairs endosomal signaling
and results in prolonged ERK and AKT activation, leading to
migratory and invasive behavior in breast cancer (84). Similarly Beclin-1 suppresses cell
migration in colorectal cancer cells by interacting with
transcription factor STAT3 (which is abnormally activated in
numerous types of cancer) and blocking its phosphorylation by JAK2
(25).
In addition, Beclin-1 and several other AMMs
influence cytoskeletal dynamics and cell-cell adhesion. Beclin-1
promotes membrane localization of the adhesion molecules E-cadherin
and α-catenin in breast cancer (85). The aforementioned study also
suggested a contribution of UVRAG to control of membrane
localization of E-cadherin, but the molecular mechanism needs
further investigation. Similarly, Beclin-1 localizes to the cell
membrane surface and mediates endocytosis of tight junction protein
occludin. However, it is not clear whether the downregulation of
occludin mediated by Beclin-1 is dependent on autophagy (86). Autophagy mediates the degradation
of E-cadherin (87).
LC3 has been reported to regulate cytoskeletal
dynamics by interacting with the selective Rho-A exchange factor
AKP13 and regulating Rho family of GTPases-dependent reorganization
of the actin cytoskeleton (88).
ATG9B is involved in regulation of cell-matrix contacts and
invasiveness. In colorectal cancer, it serves a non-autophagic
function that contributes to the formation of focal adhesions and
promotes metastasis (89). In
this context, the interaction between ATG9B and myosin heavy chain
gene (MYH9) increases the stability of both proteins by preventing
their degradative ubiquitination. This favors the interaction
between ATG9B, integrin B1 and talin 1, two key molecules of focal
adhesions. Immunohistochemical data have confirmed that high
expression of ATG9B and MYH9 is associated with poor prognosis in
colorectal carcinoma (89).
Endocytosis and exocytosis can be used by tumor
cells to create the favorable microenvironment they need for
aberrant behavior (90). The
ATG5-ATG12 complex is involved in the clathrin membrane trafficking
system that affects endocytosis under both normal and starvation
conditions in MEFs (37). ATG16L1
has been proposed as a key regulator of several steps of the
secretory machinery (especially vesicular release). Its interaction
with small GTPase Rab33A has been shown to be key in the process of
hormone secretion and may be an hallmark of neuroendocrine tissue
(91).
Exosome release and secretory autophagy (SA) are two
pathways that mediate secretion and require a number of AMMs.
However, the molecular details of these two processes are not yet
fully clarified. In the breast cancer cell line MDA-MB-231, ATG5
acts independently of autophagy to sort LC3 into multivesicular
bodies, where it binds a component of the vacuolar ATPase
H+ Transporting V1 Subunit E1 and causes a decrease in
vesicular acidification (92).
The increase in pH promotes the fusion of vesicles with plasma
membrane and their release as exosomes. These exosomes have been
shown to contain invasion mediators (RAS, β-catenin and vimentin)
and thus promote invasion and metastasis of breast cancer cells in
mice (93). SA involves
unconventional release of molecules into the extracellular space to
affect the tumor microenvironment and is used for molecules that
cannot enter the conventional endoplasmic reticulum-Golgi secretion
system because they lack a signal peptide (93). In bladder cancer, cathepsin-B is
released into the tumor microenvironment via SA and stimulates
endothelial cells to undergo angiogenesis (94). Elevated levels of cathepsin-B are
associated with invasiveness, metastasis and poor prognosis in
bladder cancer (94).
In melanoma, SA is activated by pharmacological
stimuli and mediates secretion of plasminogen activator inhibitor
(PAI-1), which is involved in the formation of a pro-tumor immune
microenvironment (95). Moreover,
cancer-associated fibroblasts in head and neck cancer use part of
the autophagic machinery to secrete tumor-promoting cytokines (IL-6
and IL-8) (96). Whether this
mechanism of secretion is SA is not clear.
LC3-associated phagocytosis (LAP) is a process that
generates anti-inflammatory and immunosuppressive signals that lead
to immune tolerance. Studies show that LAP is involved in M2
macrophage polarization and helps to promote an immunosuppressive
environment that favors tumor growth (97,98).
Conversely, in patients with colorectal cancer,
expression of an ATG16L1 variant (T600A) is responsible for an
increase in IFN-I levels (99).
Via the mitochondrial antiviral signaling pathway, cancer cells
produce IFN-I, which promotes host antitumor immunity and inhibits
the proliferation and metastasis of cancer cells (99).
Expression of critical AMMs is regulated by miRNAs,
which are also involved in carcinogenesis (100). miRNAs are a class of small
non-coding RNAs (20-24 nucleotides) that control gene expression
primarily by either inhibiting the translation or promoting decay
of target mRNAs (101).
Downregulation of several miRNAs has been shown to promote both
tumor progression and autophagy by targeting AMMs (102,103). This is the case for a number of
miRNAs that directly target core autophagy molecules such as ATG5
(miR-137, miR-153-3p), ATG12 (miR-30a-3p and miR-214), ATG7
(miR-138-5p and miR-375) and Beclin-1 (miR-17-5p, miR-26a, miR-30a,
miR-124-3p, miR-216a and miR-409-3p) (104-114). The downregulation of these
miRNAs relieves both oncogenic signaling pathways and autophagy
that usually are inhibited by them. This suggests a link between
autophagy and cancer progression.
As aforementioned, AMMs play a crucial role in
cancer, both dependent on autophagy and independent of it.
Therefore, changes in the expression of AMMs can be associated with
the prognosis of patients with cancer. Molecular AMM signatures
with potential diagnostic and prognostic value have been defined in
triple-negative breast cancer and sarcoma (115,116).
Moreover, several studies have investigated the
prognostic role of the core LC3 family nuclear proteins and shown
that their expression is associated with poor prognosis in various
cancers such as lung, breast, gastric and other types of carcinomas
(117-122).
Similarly, a signature based on high SQSTM1 and LC3
levels has been considered a negative prognostic factor in squamous
cell carcinoma (123). It has
been frequently observed that SQSTM1 exerts a pro-tumorigenic
function in cancer (124,125).
In a meta-analysis, SQSTM1 was shown to serve a negative prognostic
role in a number of solid tumors (126). However, it is worth noting that
these AMMs are typically degraded by active autophagy and their
expression is used to monitor autophagic flux. Thus, these studies
may reveal the prognostic role of autophagy rather than that of
AMMs. At the same time, SQSTM1 functions as a scaffold protein for
multiple signaling pathways and its prognostic role may therefore
be independent of autophagy (127).
Beclin-1 is considered a haploinsufficient tumor
suppressor in a number of cancers. Monoallelic deletion of the
BECN1 gene is frequently observed in breast and ovarian cancer
(128). However, the BECN1 gene
is located in proximity to the known tumor suppressor BRCA1 in both
humans (chromosome 17) and mice (chromosome 11) and the two loci
are simultaneously deleted in breast and ovarian cancer; therefore,
the actual role of Beclin-1 in tumorigenesis has been questioned
(129-131). The observation that low
expression of the BECN1 transcript, but not BRCA1, is associated
with poor prognosis in breast cancer supports the role of Beclin-1
as a tumor suppressor (132). In
addition, Beclin-1 enhances the efficacy of chemotherapeutic agents
in cervical and gastric cancer cells (133).
AMMs influence tumorigenesis via canonical and
non-canonical autophagy functions; however, the dominance of these
functions is unclear. Certain AMMs, such as ATG12, Beclin-1 and
AMBRA1, have a domain that is activated by caspase and typically
prevents activity of anti-apoptotic proteins (65,66,69). These AMMs therefore may link
autophagy and apoptosis. Under certain stress conditions, their
levels determine the fate of cells towards survival (autophagy) or
death (apoptosis). Similarly, high levels of ROS promote
autophagy-independent involvement of ULK1 in triggering necrosis
(74). Thus, it would be useful
to clarify how AMMs are regulated in cancer. For example, the LC3
and GABA type A Receptor-Associated Protein) family consists of at
least 7 proteins that have different functions in membrane
management for autophagic and non-autophagic purposes (142). Furthermore, they may be
regulated by different transcription factors in different tissue,
which could explain the activation of tissue-specific molecular
programs beyond autophagy (142). As aforementioned, expression of
LC3B and LC3A is associated with poor prognosis in gastric, breast
and other types of cancer (117-122).
The existence of alternative variants has been
demonstrated for several AMMs and may account for novel
autophagy-independent functions (143). The autophagy-incompetent ATG7
p.Arg659*, has been proposed as a cholangiocarcinoma-associated
gene (144). In addition, an
ATG7 splice variant has been described that is unable to lipidate
LC3 and is incompetent for autophagy (145), but, it remains to be clarified
whether this variant has a function in cancer. As aforementioned,
an ATG16L1 variant (T600A) stimulates an anti-tumor immune response
and is associated with a good prognosis (99). SQSTM1 is expressed in several
variants: N-Ter truncated isoform lacking the domain responsible
for SQSTM1 oligomerization and autophagic cargo sorting ability
(146); splice variant affecting
the p62/Keap1/NRF2 axis (147)
and SQSTM1 3' untranslated region-truncated variant associated with
aggressiveness and resistance to therapy in patients with breast
cancer (148). Therefore, it
would be of interest to determine whether these different AMMs
isoforms exhibit autophagy-independent functions.
Determining the autophagy-independent function of a
single AMM in cancer is challenging because autophagy is a
redundant signaling pathway that can find alternative routes to
function and influence other cellular processes (5). Therefore, modulation of multiple
autophagy markers should be considered before claiming that AMM
activity is independent of autophagy. Alternatively, it is
advisable to investigate the autophagy-independent role of AMMs
in vitro by using autophagy-incompetent mutants, such as the
ATG5 variant that cannot bind its autophagic partner ATG5K130R
(149).
In summary, the role of AMMs is not limited to
canonical autophagy but also involves autophagy-independent
functions in various biological processes. Nevertheless, further
studies that elucidate the link between autophagy-dependent and
-independent pathways will help to clarify the activity of AMMs in
cancer progression and response to therapies as well as in the
identification of novel therapeutic targets.
Not applicable.
GT and MS conceived the review, analyzed the
literature and wrote the manuscript. GT collected and reviewed the
literature and produced the figures. RM critically revised the
manuscript.
All authors have read and approved the final
manuscript. Data authentication is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
The present study was supported by the Italian Ministry of
Health-Ricerca Corrente.
|
1
|
Suzuki K, Kubota Y, Sekito T and Ohsumi Y:
Hierarchy of Atg proteins in pre-autophagosomal structure
organization. Genes Cells. 12:209–218. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Reggiori F and Klionsky DJ: Autophagic
processes in yeast: Mechanism, machinery and regulation. Genetics.
194:341–361. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pankiv S, Clausen TH, Lamark T, Brech A,
Bruun JA, Outzen H, Øvervatn A, Bjørkøy G and Johansen T:
p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of
ubiquitinated protein aggregates by autophagy. J Biol Chem.
282:24131–24145. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Elmore SP, Qian T, Grissom SF and
Lemasters JJ: The mitochondrial permeability transition initiates
autophagy in rat hepatocytes. FASEB J. 15:2286–2287. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mizushima N and Levine B: Autophagy in
human diseases. N Engl J Med. 383:1564–1576. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bustos SO, Antunes F, Rangel MC and
Chammas R: Emerging autophagy functions shape the tumor
microenvironment and play a role in cancer progression-implications
for cancer therapy. Front Oncol. 10:6064362020. View Article : Google Scholar
|
|
7
|
Xie X, Koh JY, Price S, White E and
Mehnert JM: Atg7 overcomes senescence and promotes growth of
BrafV600E-Driven Melanoma. Cancer Discov. 5:410–423. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang A, Herter-Sprie G, Zhang H, Lin EY,
Biancur D, Wang X, Deng J, Hai J, Yang S, Wong KK and Kimmelman AC:
Autophagy sustains pancreatic cancer growth through both
cell-autonomous and nonautonomous mechanisms. Cancer Discov.
8:276–287. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shchors K, Massaras A and Hanahan D: Dual
targeting of the autophagic regulatory circuitry in gliomas with
repurposed drugs elicits cell-lethal autophagy and therapeutic
benefit. Cancer Cell. 28:456–471. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Santanam U, Banach-Petrosky W, Abate-Shen
C, Shen MM, White E and DiPaola RS: Atg7 cooperates with Pten loss
to drive prostate cancer tumor growth. Genes Dev. 30:399–407. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Karsli-Uzunbas G, Guo JY, Price S, Teng X,
Laddha SV, Khor S, Kalaany NY, Jacks T, Chan CS, Rabinowitz JD and
White E: Autophagy is required for glucose homeostasis and lung
tumor maintenance. Cancer Discov. 4:914–927. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huo Y, Cai H, Teplova I, Bowman-Colin C,
Chen G, Price S, Barnard N, Ganesan S, Karantza V, White E and Xia
B: Autophagy opposes p53-mediated tumor barrier to facilitate
tumorigenesis in a model of PALB2-associated hereditary breast
cancer. Cancer Discov. 3:894–907. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Degenhardt K, Mathew R, Beaudoin B, Bray
K, Anderson D, Chen G, Mukherjee C, Shi Y, Gélinas C, Fan Y, et al:
Autophagy promotes tumor cell survival and restricts necrosis,
inflammation, and tumorigenesis. Cancer Cell. 10:51–64. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Karantza-Wadsworth V, Patel S, Kravchuk O,
Chen G, Mathew R, Jin S and White E: Autophagy mitigates metabolic
stress and genome damage in mammary tumorigenesis. Genes Dev.
21:1621–1635. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Moussay E, Kaoma T, Baginska J, Muller A,
Van Moer K, Nicot N, Nazarov PV, Vallar L, Chouaib S, Berchem G and
Janji B: The acquisition of resistance to TNFα in breast cancer
cells is associated with constitutive activation of autophagy as
revealed by a transcriptome analysis using a custom microarray.
Autophagy. 7:760–770. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bildik G, Liang X, Sutton MN, Bast RC Jr
and Lu Z: DIRAS3: An Imprinted tumor suppressor gene that regulates
RAS and PI3K-driven cancer growth, motility, autophagy and tumor
dormancy. Mol Cancer Ther. 21:25–37. 2021. View Article : Google Scholar
|
|
17
|
Dower CM, Wills CA, Frisch SM and Wang HG:
Mechanisms and context underlying the role of autophagy in cancer
metastasis. Autophagy. 14:1110–1128. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mizushima N, Yoshimori T and Ohsumi Y: The
role of Atg proteins in autophagosome formation. Annu Rev Cell Dev
Biol. 27:107–132. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kuma A, Hatano M, Matsui M, Yamamoto A,
Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T and Mizushima N: The
role of autophagy during the early neonatal starvation period.
Nature. 432:1032–1036. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Levine B and Kroemer G: Biological
functions of autophagy genes: A disease perspective. Cell.
176:11–42. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Oughtred R, Rust J, Chang C, Breitkreutz
BJ, Stark C, Willems A, Boucher L, Leung G, Kolas N, Zhang F, et
al: The BioGRID database: A comprehensive biomedical resource of
curated protein, genetic, and chemical interactions. Protein Sci.
30:187–200. 2021. View Article : Google Scholar
|
|
22
|
Leidal AM and Debnath J: Emerging roles
for the autophagy machinery in extracellular vesicle biogenesis and
secretion. FASEB Bioadv. 3:377–386. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Okamoto T, Yeo SK, Hao M, Copley MR, Haas
MA, Chen S and Guan JL: FIP200 suppresses immune checkpoint therapy
responses in breast cancers by limiting AZI2/TBK1/IRF signaling
independent of its canonical autophagy function. Cancer Res.
80:3580–3592. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Guo H, Sadoul R and Gibbings D:
Autophagy-independent effects of autophagy-related-5 (Atg5) on
exosome production and metastasis. Mol Cell Oncol. 5:e14459412018.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hu F, Li G, Huang C, Hou Z, Yang X, Luo X,
Feng Y, Wang G, Hu J and Cao Z: The autophagy-independent role of
BECN1 in colorectal cancer metastasis through regulating STAT3
signaling pathway activation. Cell Death Dis. 11:3042020.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yamamoto H, Zhang S and Mizushima N:
Autophagy genes in biology and disease. Nat Rev Genet. 24:382–400.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Agarwal S, Bell CM, Rothbart SB and Moran
RG: AMP-activated Protein Kinase (AMPK) Control of mTORC1 Is
p53-and TSC2-independent in pemetrexed-treated carcinoma cells. J
Biol Chem. 290:27473–27486. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hosokawa N, Hara T, Kaizuka T, Kishi C,
Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et
al: Nutrient-dependent mTORC1 association with the
ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell.
20:1981–1991. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mercer TJ, Gubas A and Tooze SA: A
molecular perspective of mammalian autophagosome biogenesis. J Biol
Chem. 293:5386–5395. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zachari M and Ganley IG: The mammalian
ULK1 complex and autophagy initiation. Essays Biochem. 61:585–596.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liang C, Feng P, Ku B, Dotan I, Canaani D,
Oh BH and Jung JU: Autophagic and tumour suppressor activity of a
novel Beclin1-binding protein UVRAG. Nat Cell Biol. 8:688–699.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fimia GM, Stoykova A, Romagnoli A, Giunta
L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A,
Schwartz P, et al: Ambra1 regulates autophagy and development of
the nervous system. Nature. 447:1121–1125. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sawa-Makarska J, Baumann V, Coudevylle N,
von Bülow S, Nogellova V, Abert C, Schuschnig M, Graef M, Hummer G
and Martens S: Reconstitution of autophagosome nucleation defines
Atg9 vesicles as seeds for membrane formation. Science.
369:eaaz77142020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Matoba K, Kotani T, Tsutsumi A, Tsuji T,
Mori T, Noshiro D, Sugita Y, Nomura N, Iwata S, Ohsumi Y, et al:
Atg9 is a lipid scramblase that mediates autophagosomal membrane
expansion. Nat Struct Mol Biol. 27:1185–1193. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dooley HC, Razi M, Polson HE, Girardin SE,
Wilson MI and Tooze SA: WIPI2 links LC3 conjugation with PI3P,
autophagosome formation, and pathogen clearance by recruiting
Atg12-5-16L1. Mol Cell. 55:238–252. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wible DJ, Chao HP, Tang DG and Bratton SB:
ATG5 cancer mutations and alternative mRNA splicing reveal a
conjugation switch that regulates ATG12-ATG5-ATG16L1 complex
assembly and autophagy. Cell Discov. 5:422019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Baines K, Yoshioka K, Takuwa Y and Lane
JD: The ATG5 interactome links clathrin-mediated vesicular
trafficking with the autophagosome assembly machinery. Autophagy
Rep. 1:88–118. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Haller M, Hock AK, Giampazolias E, Oberst
A, Green DR, Debnath J, Ryan KM, Vousden KH and Tait SW:
Ubiquitination and proteasomal degradation of ATG12 regulates its
proapoptotic activity. Autophagy. 10:2269–2278. 2014. View Article : Google Scholar
|
|
39
|
Tanida I, Ueno T and Kominami E: LC3
conjugation system in mammalian autophagy. Int J Biochem Cell Biol.
36:2503–2518. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Melia TJ, Lystad AH and Simonsen A:
Autophagosome biogenesis: From membrane growth to closure. J Cell
Biol. 219:e2020020852020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gatica D, Lahiri V and Klionsky DJ: Cargo
recognition and degradation by selective autophagy. Nat Cell Biol.
20:233–242. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Liu WJ, Ye L, Huang WF, Guo LJ, Xu ZG, Wu
HL, Yang C and Liu HF: p62 links the autophagy pathway and the
ubiqutin-proteasome system upon ubiquitinated protein degradation.
Cell Mol Biol Lett. 21:292016. View Article : Google Scholar
|
|
43
|
Yim WW and Mizushima N: Lysosome biology
in autophagy. Cell Discov. 6:62020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Dikic I and Elazar Z: Mechanism and
medical implications of mammalian autophagy. Nat Rev Mol Cell Biol.
19:349–364. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hanahan D: Hallmarks of cancer: New
Dimensions. Cancer Discov. 12:31–46. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Mathew R, Kongara S, Beaudoin B, Karp CM,
Bray K, Degenhardt K, Chen G, Jin S and White E: Autophagy
suppresses tumor progression by limiting chromosomal instability.
Genes Dev. 21:1367–1381. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Marteijn JA, Lans H, Vermeulen W and
Hoeijmakers JH: Understanding nucleotide excision repair and its
roles in cancer and ageing. Nat Rev Mol Cell Biol. 15:465–481.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yang Y, He S, Wang Q, Li F, Kwak MJ, Chen
S, O'Connell D, Zhang T, Pirooz SD, Jeon YH, et al: Autophagic
UVRAG Promotes UV-Induced Photolesion Repair by Activation of the
CRL4(DDB2) E3 Ligase. Mol Cell. 62:507–519. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhao Z, Oh S, Li D, Ni D, Pirooz SD, Lee
JH, Yang S, Lee JY, Ghozalli I, Costanzo V, et al: A dual role for
UVRAG in maintaining chromosomal stability independent of
autophagy. Dev Cell. 22:1001–1016. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Park JM, Tougeron D, Huang S, Okamoto K
and Sinicrope FA: Beclin 1 and UVRAG confer protection from
radiation-induced DNA damage and maintain centrosome stability in
colorectal cancer cells. PLoS One. 9:e1008192014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Knævelsrud H, Ahlquist T, Merok MA,
Nesbakken A, Stenmark H, Lothe RA and Simonsen A: UVRAG mutations
associated with microsatellite unstable colon cancer do not affect
autophagy. Autophagy. 6:863–870. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
He S, Zhao Z, Yang Y, O'Connell D, Zhang
X, Oh S, Ma B, Lee JH, Zhang T, Varghese B, et al: Truncating
mutation in the autophagy gene UVRAG confers oncogenic properties
and chemosensitivity in colorectal cancers. Nat Commun. 6:78392015.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Sun SY, Hu XT, Yu XF, Zhang YY, Liu XH,
Liu YH, Wu SH, Li YY, Cui SX and Qu XJ: Nuclear translocation of
ATG5 induces DNA mismatch repair deficiency (MMR-D)/microsatellite
instability (MSI) via interacting with Mis18α in colorectal cancer.
Br J Pharmacol. 178:2351–2369. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Maskey D, Yousefi S, Schmid I, Zlobec I,
Perren A, Friis R and Simon HU: ATG5 is induced by DNA-damaging
agents and promotes mitotic catastrophe independent of autophagy.
Nat Commun. 4:21302013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Maiani E, Milletti G, Nazio F, Holdgaard
SG, Bartkova J, Rizza S, Cianfanelli V, Lorente M, Simoneschi D, Di
Marco M, et al: AMBRA1 regulates cyclin D to guard S-phase entry
and genomic integrity. Nature. 592:799–803. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Liu PF, Leung CM, Chang YH, Cheng JS, Chen
JJ, Weng CJ, Tsai KW, Hsu CJ, Liu YC, Hsu PC, et al: ATG4B promotes
colorectal cancer growth independent of autophagic flux. Autophagy.
10:1454–1465. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Lee IH, Kawai Y, Fergusson MM, Rovira II,
Bishop AJ, Motoyama N, Cao L and Finkel T: Atg7 modulates p53
activity to regulate cell cycle and survival during metabolic
stress. Science. 336:225–228. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Frémont S, Gérard A, Galloux M, Janvier K,
Karess RE and Berlioz-Torrent C: Beclin-1 is required for
chromosome congression and proper outer kinetochore assembly. EMBO
Rep. 14:364–372. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Thoresen SB, Pedersen NM, Liestøl K and
Stenmark H: A phosphatidylinositol 3-kinase class III sub-complex
containing VPS15, VPS34, Beclin 1, UVRAG and BIF-1 regulates
cytokinesis and degradative endocytic traffic. Exp Cell Res.
316:3368–3378. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Sagona AP, Nezis IP, Pedersen NM, Liestøl
K, Poulton J, Rusten TE, Skotheim RI, Raiborg C and Stenmark H:
PtdIns(3) P controls cytokinesis through KIF13A-mediated
recruitment of FYVE-CENT to the midbody. Nat Cell Biol. 12:362–371.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zhu Y, Zhao L, Liu L, Gao P, Tian W, Wang
X, Jin H, Xu H and Chen Q: Beclin 1 cleavage by caspase-3
inactivates autophagy and promotes apoptosis. Protein Cell.
1:468–477. 2010. View Article : Google Scholar
|
|
62
|
Young MM, Takahashi Y, Khan O, Park S,
Hori T, Yun J, Sharma AK, Amin S, Hu CD, Zhang J, et al:
Autophagosomal membrane serves as platform for intracellular
death-inducing signaling complex (iDISC)-mediated caspase-8
activation and apoptosis. J Biol Chem. 287:12455–12468. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Han J, Hou W, Goldstein LA, Stolz DB,
Watkins SC and Rabinowich H: A Complex between Atg7 and Caspase-9:
A novel mechanism of cross-regulation between autophagy and
apoptosis. J Biol Chem. 289:6485–6497. 2014. View Article : Google Scholar :
|
|
64
|
Wirawan E, Vande Walle L, Kersse K,
Cornelis S, Claerhout S, Vanoverberghe I, Roelandt R, De Rycke R,
Verspurten J, Declercq W, et al: Caspase-mediated cleavage of
Beclin-1 inactivates Beclin-1-induced autophagy and enhances
apoptosis by promoting the release of proapoptotic factors from
mitochondria. Cell Death Dis. 1:e182010. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Li X, Su J, Xia M, Li H, Xu Y, Ma C, Ma L,
Kang J, Yu H, Zhang Z and Sun L: Caspase-mediated cleavage of
Beclin1 inhibits autophagy and promotes apoptosis induced by S1 in
human ovarian cancer SKOV3 cells. Apoptosis. 21:225–238. 2016.
View Article : Google Scholar
|
|
66
|
Strappazzon F, Di Rita A, Cianfanelli V,
D'Orazio M, Nazio F, Fimia GM and Cecconi F: Prosurvival AMBRA1
turns into a proapoptotic BH3-like protein during mitochondrial
apoptosis. Autophagy. 12:963–975. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Betin VM and Lane JD: Caspase cleavage of
Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial
targeting and apoptosis. J Cell Sci. 122(Pt 14): 2554–2566. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Yousefi S, Perozzo R, Schmid I, Ziemiecki
A, Schaffner T, Scapozza L, Brunner T and Simon HU:
Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis.
Nat Cell Biol. 8:1124–1132. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Rubinstein AD, Eisenstein M, Ber Y, Bialik
S and Kimchi A: The autophagy protein Atg12 associates with
antiapoptotic Bcl-2 family members to promote mitochondrial
apoptosis. Mol Cell. 44:698–709. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Yoo BH, Khan IA, Koomson A, Gowda P,
Sasazuki T, Shirasawa S, Gujar S and Rosen KV: Oncogenic
RAS-induced downregulation of ATG12 is required for survival of
malignant intestinal epithelial cells. Autophagy. 14:134–151. 2018.
View Article : Google Scholar :
|
|
71
|
Radoshevich L, Murrow L, Chen N, Fernandez
E, Roy S, Fung C and Debnath J: ATG12 conjugation to ATG3 regulates
mitochondrial homeostasis and cell death. Cell. 142:590–600. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Liu H, He Z, Germič N, Ademi H, Frangež Ž,
Felser A, Peng S, Riether C, Djonov V, Nuoffer JM, et al: ATG12
deficiency leads to tumor cell oncosis owing to diminished
mitochondrial biogenesis and reduced cellular bioenergetics. Cell
Death Differ. 27:1965–1980. 2020. View Article : Google Scholar :
|
|
73
|
Ni Z, He J, Wu Y, Hu C, Dai X, Yan X, Li
B, Li X, Xiong H, Li Y, et al: AKT-mediated phosphorylation of
ATG4B impairs mitochondrial activity and enhances the Warburg
effect in hepatocellular carcinoma cells. Autophagy. 14:685–701.
2018. View Article : Google Scholar :
|
|
74
|
Wu W, Wang X, Berleth N, Deitersen J,
Wallot-Hieke N, Böhler P, Schlütermann D, Stuhldreier F, Cox J,
Schmitz K, et al: The autophagy-initiating kinase ULK1 Controls
RIPK1-mediated cell death. Cell Rep. 31:1075472020. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Joshi A, Iyengar R, Joo JH, Li-Harms XJ,
Wright C, Marino R, Winborn BJ, Phillips A, Temirov J, Sciarretta
S, et al: Nuclear ULK1 promotes cell death in response to oxidative
stress through PARP1. Cell Death Differ. 23:216–230. 2016.
View Article : Google Scholar :
|
|
76
|
Satyavarapu EM, Das R and Mandal C,
Mukhopadhyay A and Mandal C: Autophagy-independent induction of
LC3B through oxidative stress reveals its non-canonical role in
anoikis of ovarian cancer cells. Cell Death Dis. 9:9342018.
View Article : Google Scholar
|
|
77
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: the next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Aiello NM, Maddipati R, Norgard RJ, Balli
D, Li J, Yuan S, Yamazoe T, Black T, Sahmoud A, Furth EE, et al:
EMT subtype influences epithelial plasticity and mode of cell
migration. Dev Cell. 45:681–695.e4. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Zada S, Hwang JS, Ahmed M, Lai TH, Pham TM
and Kim DR: Control of the epithelial-to-mesenchymal transition and
cancer metastasis by autophagy-dependent SNAI1 degradation. Cells.
8:1292019. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Han JH, Kim YK, Kim H, Lee J, Oh MJ, Kim
SB, Kim M, Kim KH, Yoon HJ, Lee MS, et al: Snail acetylation by
autophagy-derived acetyl-coenzyme A promotes invasion and
metastasis of KRAS-LKB1 co-mutated lung cancer cells. Cancer Commun
(Lond). 42:716–749. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Sharifi MN, Mowers EE, Drake LE, Collier
C, Chen H, Zamora M, Mui S and Macleod KF: Autophagy promotes focal
adhesion disassembly and cell motility of metastatic tumor cells
through the direct interaction of paxillin with LC3. Cell Rep.
15:1660–1672. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Santarosa M and Maestro R: The autophagic
route of E-Cadherin and cell adhesion molecules in cancer
progression. Cancers (Basel). 13:63282021. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Martinez-Lopez N, Athonvarangkul D,
Mishall P, Sahu S and Singh R: Autophagy proteins regulate ERK
phosphorylation. Nat Commun. 4:27992013. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Rohatgi RA, Janusis J, Leonard D, Bellvé
KD, Fogarty KE, Baehrecke EH, Corvera S and Shaw LM: Beclin 1
regulates growth factor receptor signaling in breast cancer.
Oncogene. 34:5352–5362. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Wijshake T, Zou Z, Chen B, Zhong L, Xiao
G, Xie Y, Doench JG, Bennett L and Levine B: Tumor-suppressor
function of Beclin 1 in breast cancer cells requires E-cadherin.
Proc Natl Acad Sci USA. 118:e20204781182021. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Wong M, Ganapathy AS, Suchanec E, Laidler
L, Ma T and Nighot P: Intestinal epithelial tight junction barrier
regulation by autophagy-related protein ATG6/beclin 1. Am J
Physiol, Cell Physiol. 316:C753–C765. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Damiano V, Spessotto P, Vanin G, Perin T,
Maestro R and Santarosa M: The autophagy machinery contributes to
E-cadherin turnover in breast cancer. Front Cell Dev Biol.
8:5452020. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Baisamy L, Cavin S, Jurisch N and Diviani
D: The ubiquitin-like protein LC3 regulates the Rho-GEF activity of
AKAP-Lbc. J Biol Chem. 284:28232–28242. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Zhong Y, Long T, Gu CS, Tang JY, Gao LF,
Zhu JX, Hu ZY, Wang X, Ma YD, Ding YQ, et al: MYH9-dependent
polarization of ATG9B promotes colorectal cancer metastasis by
accelerating focal adhesion assembly. Cell Death Differ.
28:3251–3269. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Galluzzi L and Green DR:
Autophagy-Independent functions of the autophagy machinery. Cell.
177:1682–1699. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Ishibashi K, Uemura T, Waguri S and Fukuda
M: Atg16L1, an essential factor for canonical autophagy,
participates in hormone secretion from PC12 cells independently of
autophagic activity. Mol Biol Cell. 23:3193–3202. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Guo H, Chitiprolu M, Roncevic L, Javalet
C, Hemming FJ, Trung MT, Meng L, Latreille E, Tanese de Souza C,
McCulloch D, et al: Atg5 Disassociates the V1V0-ATPase to promote
exosome production and tumor metastasis independent of canonical
macroautophagy. Dev Cell. 43:716–730.e7. 2017. View Article : Google Scholar
|
|
93
|
Ponpuak M, Mandell MA, Kimura T, Chauhan
S, Cleyrat C and Deretic V: Secretory autophagy. Curr Opin Cell
Biol. 35:106–116. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Li X, Wei Z, Yu H, Xu Y, He W, Zhou X and
Gou X: Secretory autophagy-induced bladder tumour-derived
extracellular vesicle secretion promotes angiogenesis by activating
the TPX2-mediated phosphorylation of the AURKA-PI3K-AKT axis.
Cancer Lett. 523:10–28. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Tzeng HT, Yang JL, Tseng YJ, Lee CH, Chen
WJ and Chyuan IT: Plasminogen activator inhibitor-1 secretion by
autophagy contributes to melanoma resistance to chemotherapy
through tumor microenvironment modulation. Cancers (Basel).
13:12532021. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
New J, Arnold L, Ananth M, Alvi S,
Thornton M, Werner L, Tawfik O, Dai H, Shnayder Y, Kakarala K, et
al: Secretory autophagy in cancer-associated fibroblasts promotes
head and neck cancer progression and offers a novel therapeutic
target. Cancer Res. 77:6679–6691. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Cunha LD, Yang M, Carter R, Guy C, Harris
L, Crawford JC, Quarato G, Boada-Romero E, Kalkavan H, Johnson MDL,
et al: LC3-Associated phagocytosis in myeloid cells promotes tumor
immune tolerance. Cell. 175:429–441.e16. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Liu X, Zhang W, Xu Y, Xu X, Jiang Q, Ruan
J, Wu Y, Zhou Y, Saw PE and Luo B: Targeting PI3Kγ/AKT Pathway
Remodels LC3-Associated phagocytosis induced immunosuppression
after radiofrequency ablation. Adv Sci (Weinh). 9:e21021822022.
View Article : Google Scholar
|
|
99
|
Grimm WA, Messer JS, Murphy SF, Nero T,
Lodolce JP, Weber CR, Logsdon MF, Bartulis S, Sylvester BE,
Springer A, et al: The Thr300Ala variant in ATG16L1 is associated
with improved survival in human colorectal cancer and enhanced
production of type I interferon. Gut. 65:456–464. 2016. View Article : Google Scholar
|
|
100
|
Peng Y and Croce CM: The role of MicroRNAs
in human cancer. Signal Transduct Target Ther. 1:150042016.
View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Chipman LB and Pasquinelli AE: miRNA
Targeting: Growing beyond the Seed. Trends Genet. 35:215–222. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
de la Cruz-Ojeda P, Flores-Campos R,
Navarro-Villarán E and Muntané J: The role of non-coding RNAs in
autophagy during carcinogenesis. Front Cell Dev Biol.
10:7993922022. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Shan C, Chen X, Cai H, Hao X, Li J, Zhang
Y, Gao J, Zhou Z, Li X, Liu C, et al: The emerging roles of
autophagy-related MicroRNAs in cancer. Int J Biol Sci. 17:134–150.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Zhu H, Wu H, Liu X, Li B, Chen Y, Ren X,
Liu CG and Yang JM: Regulation of autophagy by a beclin 1-targeted
microRNA, miR-30a, in cancer cells. Autophagy. 5:816–823. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Wang ZC, Huang FZ, Xu HB, Sun JC and Wang
CF: MicroRNA-137 inhibits autophagy and chemosensitizes pancreatic
cancer cells by targeting ATG5. Int J Biochem Cell Biol. 111:63–71.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Chen Y, Zhou J, Wu X, Huang J, Chen W, Liu
D, Zhang J, Huang Y and Xue W: miR-30a-3p inhibits renal cancer
cell invasion and metastasis through targeting ATG12. Transl Androl
Urol. 9:646–653. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Hu JL, He GY, Lan XL, Zeng ZC, Guan J,
Ding Y, Qian XL, Liao WT, Ding YQ and Liang L: Inhibition of
ATG12-mediated autophagy by miR-214 enhances radiosensitivity in
colorectal cancer. Oncogenesis. 7:162018. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Pan X, Chen Y, Shen Y and Tantai J:
Knockdown of TRIM65 inhibits autophagy and cisplatin resistance in
A549/DDP cells by regulating miR-138-5p/ATG7. Cell Death Dis.
10:4292019. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Chang Y, Yan W, He X, Zhang L, Li C, Huang
H, Nace G, Geller DA, Lin J and Tsung A: miR-375 inhibits autophagy
and reduces viability of hepatocellular carcinoma cells under
hypoxic conditions. Gastroenterology. 143:177–187.e8. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Zhang X, Shi H, Lin S, Ba M and Cui S:
MicroRNA-216a enhances the radiosensitivity of pancreatic cancer
cells by inhibiting beclin-1-mediated autophagy. Oncol Rep.
34:1557–1564. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Li M, Chen XM, Wang DM, Gan L and Qiao Y:
Effects of miR-26a on the expression of Beclin 1 in retinoblastoma
cells. Genet Mol Res. 15:2016.
|
|
112
|
Hou W, Song L, Zhao Y, Liu Q and Zhang S:
Inhibition of beclin-1-mediated autophagy by MicroRNA-17-5p
enhanced the radiosensitivity of glioma cells. Oncol Res. 25:43–53.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Zhang F, Wang B, Long H, Yu J, Li F, Hou H
and Yang Q: Decreased miR-124-3p expression prompted breast cancer
cell progression mainly by targeting Beclin-1. Clin Lab.
62:1139–1145. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Tan S, Shi H, Ba M, Lin S, Tang H, Zeng X
and Zhang X: miR-409-3p sensitizes colon cancer cells to
oxaliplatin by inhibiting Beclin-1-mediated autophagy. Int J Mol
Med. 37:1030–1038. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Wang Y, Li J, Shao C, Tang X, Du Y, Xu T,
Zhao Z, Hu H, Sheng Y, Hu C and Xi Y: Systematic profiling of
diagnostic and prognostic value of autophagy-related genes for
sarcoma patients. BMC Cancer. 21:582021. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Yang Q, Sun K, Xia W, Li Y, Zhong M and
Lei K: Autophagy-related prognostic signature for survival
prediction of triple negative breast cancer. PeerJ. 10:e128782022.
View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Sivridis E, Koukourakis MI, Zois CE,
Ledaki I, Ferguson DJ, Harris AL, Gatter KC and Giatromanolaki A:
LC3A-positive light microscopy detected patterns of autophagy and
prognosis in operable breast carcinomas. Am J Pathol.
176:2477–2489. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Gachechiladze M, Uberall I, Skanderova D,
Matchavariani J, Ibrahim M, Shani I, Smickova P, Kolek V, Cierna L,
Klein J, et al: LC3A positive 'stone like structures' are
differentially associated with survival outcomes and CD68
macrophage infiltration in patients with lung adenocarcinoma and
squamous cell carcinoma. Lung Cancer. 156:129–135. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Terabe T, Uchida F, Nagai H, Omori S,
Ishibashi-Kanno N, Hasegawa S, Yamagata K, Gosho M, Yanagawa T and
Bukawa H: Expression of autophagy-related markers at the surgical
margin of oral squamous cell carcinoma correlates with poor
prognosis and tumor recurrence. Hum Pathol. 73:156–163. 2018.
View Article : Google Scholar
|
|
120
|
Giatromanolaki A, Koukourakis MI, Georgiou
I, Kouroupi M and Sivridis E: LC3A, LC3B and Beclin-1 Expression in
gastric cancer. Anticancer Res. 38:6827–6833. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Bortnik S, Tessier-Cloutier B, Leung S, Xu
J, Asleh K, Burugu S, Magrill J, Greening K, Derakhshan F, Yip S,
et al: Differential expression and prognostic relevance of
autophagy-related markers ATG4B, GABARAP, and LC3B in breast
cancer. Breast Cancer Res Treat. 183:525–547. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Kim JW, Jun SY, Kim JM, Oh YH, Yoon G,
Hong SM and Chung JY: Prognostic value of LC3B and p62 expression
in small intestinal adenocarcinoma. J Clin Med. 10:53982021.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Langer R, Neppl C, Keller MD, Schmid RA,
Tschan MP and Berezowska S: Expression analysis of autophagy
related markers LC3B, p62 and HMGB1 indicate an
autophagy-independent negative prognostic impact of High p62
expression in pulmonary squamous cell carcinomas. Cancers (Basel).
10:2812018. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Mathew R, Karp CM, Beaudoin B, Vuong N,
Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al:
Autophagy suppresses tumorigenesis through elimination of p62.
Cell. 137:1062–1075. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Islam MA, Sooro MA and Zhang P: Autophagic
regulation of p62 is critical for cancer therapy. Int J Mol Sci.
19:14052018. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Ruan H, Xu J, Wang L, Zhao Z, Kong L, Lan
B and Li X: The prognostic value of p62 in solid tumor patients: A
meta-analysis. Oncotarget. 9:4258–4266. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Sánchez-Martín P, Saito T and Komatsu M:
p62/SQSTM1: 'Jack of all trades' in health and cancer. FEBS J.
286:8–23. 2019. View Article : Google Scholar
|
|
128
|
Laddha SV, Ganesan S, Chan CS and White E:
Mutational landscape of the essential autophagy gene BECN1 in human
cancers. Mol Cancer Res. 12:485–490. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Delaney JR, Patel CB, Bapat J, Jones CM,
Ramos-Zapatero M, Ortell KK, Tanios R, Haghighiabyaneh M, Axelrod
J, DeStefano JW, et al: Autophagy gene haploinsufficiency drives
chromosome instability, increases migration, and promotes early
ovarian tumors. PLoS Genet. 16:e10085582020. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh
H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, et al:
Promotion of tumorigenesis by heterozygous disruption of the beclin
1 autophagy gene. J Clin Invest. 112:1809–1820. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Ajazi A and Foiani M: Vps30/Atg6/BECN1 at
the crossroads between cell metabolism and DNA damage response.
Autophagy. 18:1202–1204. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Tang H, Sebti S, Titone R, Zhou Y, Isidoro
C, Ross TS, Hibshoosh H, Xiao G, Packer M, Xie Y and Levine B:
Decreased BECN1 mRNA expression in human breast cancer is
associated with estrogen receptor-negative subtypes and poor
prognosis. EBioMedicine. 2:255–263. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Liu C, Yan X, Wang HQ, Gao YY, Liu J, Hu
Z, Liu D, Gao J and Lin B: Autophagy-independent enhancing effects
of Beclin 1 on cytotoxicity of ovarian cancer cells mediated by
proteasome inhibitors. BMC Cancer. 12:6222012. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Xu C, Zang Y, Zhao Y, Cui W, Zhang H, Zhu
Y and Xu M: comprehensive pan-cancer analysis confirmed that ATG5
Promoted the maintenance of tumor metabolism and the occurrence of
tumor immune escape. Front Oncol. 11:6522112021. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Zhou S, Wang X, Ding J, Yang H and Xie Y:
Increased ATG5 expression predicts poor prognosis and promotes EMT
in cervical carcinoma. Front Cell Dev Biol. 9:7571842021.
View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Qin YQ, Liu SY, Lv ML and Sun WL: Ambra1
in cancer: Implications for clinical oncology. Apoptosis.
27:720–729. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Nitta T, Sato Y, Ren XS, Harada K, Sasaki
M, Hirano S and Nakanuma Y: Autophagy may promote carcinoma cell
invasion and correlate with poor prognosis in cholangiocarcinoma.
Int J Clin Exp Pathol. 7:4913–4921. 2014.PubMed/NCBI
|
|
138
|
Ko YH, Cho YS, Won HS, Jeon EK, An HJ,
Hong SU, Park JH and Lee MA: Prognostic significance of
autophagy-related protein expression in resected pancreatic ductal
adenocarcinoma. Pancreas. 42:829–835. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Falasca L, Torino F, Marconi M, Costantini
M, Pompeo V, Sentinelli S, De Salvo L, Patrizio M, Padula C,
Gallucci M, et al: AMBRA1 and SQSTM1 expression pattern in prostate
cancer. Apoptosis. 20:1577–1586. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Ieni A, Cardia R, Giuffrè G, Rigoli L,
Caruso RA and Tuccari G: Immunohistochemical expression of
autophagy-related proteins in advanced tubular gastric
adenocarcinomas and its implications. Cancers (Basel). 11:3892019.
View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Tang DY, Ellis RA and Lovat PE: Prognostic
impact of autophagy biomarkers for cutaneous melanoma. Front Oncol.
6:2362016. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Schaaf MB, Keulers TG, Vooijs MA and
Rouschop KM: LC3/GABARAP family proteins: Autophagy-(un)related
functions. FASEB J. 30:3961–3978. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
González-Rodríguez P, Klionsky DJ and
Joseph B: Autophagy regulation by RNA alternative splicing and
implications in human diseases. Nat Commun. 13:27352022. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Greer SU, Ogmundsdottir MH, Chen J, Lau
BT, Delacruz RGC, Sandoval IT, Kristjansdottir S, Jones DA, Haslem
DS, Romero R, et al: Genetic risk of cholangiocarcinoma is linked
to the autophagy gene ATG7. BioRxiv. 2019.
|
|
145
|
Ogmundsdottir MH, Fock V, Sooman L,
Pogenberg V, Dilshat R, Bindesbøll C, Ogmundsdottir HM, Simonsen A,
Wilmanns M and Steingrimsson E: A short isoform of ATG7 fails to
lipidate LC3/GABARAP. Sci Rep. 8:143912018. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Somlapura M, Gottschalk B, Lahiri P,
Kufferath I, Pabst D, Rülicke T, Graier WF, Denk H and Zatloukal K:
Different Roles of p62 (SQSTM1) isoforms in keratin-related protein
aggregation. Int J Mol Sci. 22:62272021. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Kageyama S, Saito T, Obata M, Koide RH,
Ichimura Y and Komatsu M: Negative Regulation of the Keap1-Nrf2
Pathway by a p62/Sqstm1 Splicing Variant. Mol Cell Biol.
38:e006422018. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Guo Q, Wang H, Duan J, Luo W, Zhao R, Shen
Y, Wang B, Tao S, Sun Y, Ye Q, et al: An alternatively spliced p62
isoform confers resistance to chemotherapy in breast cancer. Cancer
Res. 82:4001–4015. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Otomo C, Metlagel Z, Takaesu G and Otomo
T: Structure of the human ATG12~ATG5 conjugate required for LC3
lipidation in autophagy. Nat Struct Mol Biol. 20:59–66. 2013.
View Article : Google Scholar :
|