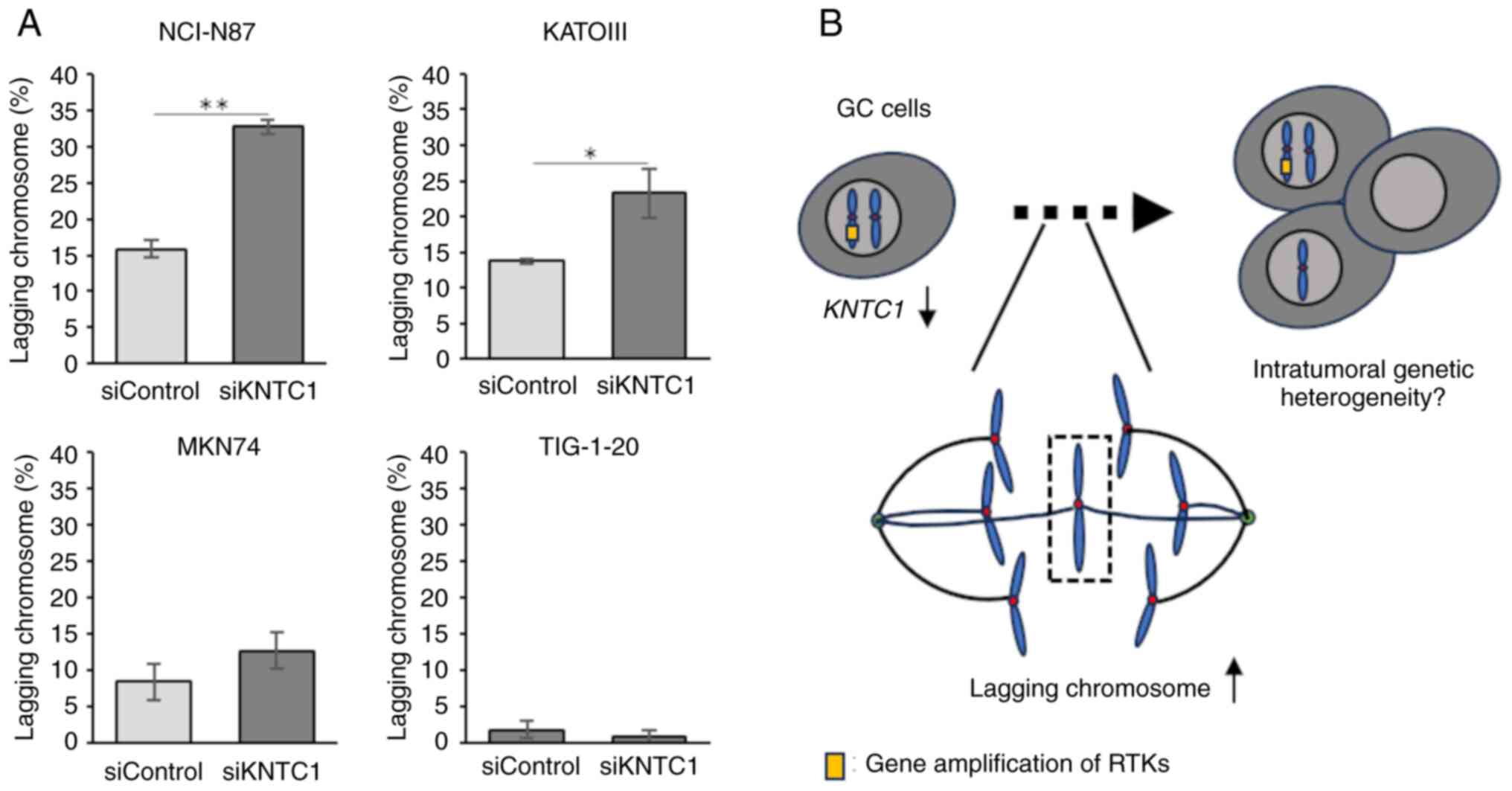
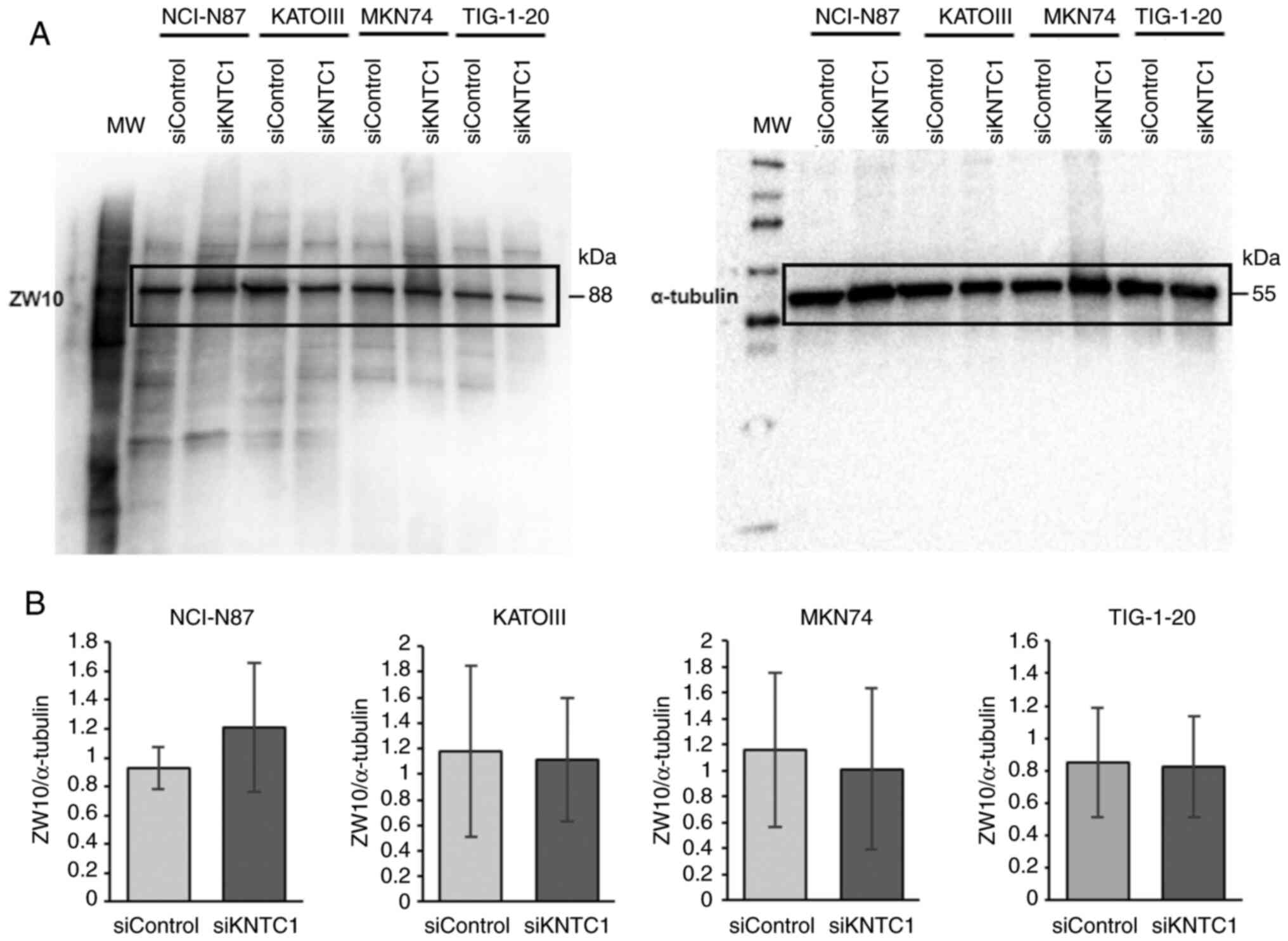
|
1
|
Sung H, Ferlay J, Siegel RL, Sung H,
Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A and
Bray F: Global cancer statistics 2020: GLOBOCAN estimates of
incidence and mortality worldwide for 36 cancers in 185 countries.
CA Cancer J Clin. 71:209–249. 2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Sato Y, Okamoto K, Kawano Y, Kasai A,
Kawaguchi T, Sagawa T, Sogabe M, Miyamoto H and Takayama T: Novel
biomarkers of gastric cancer: Current research and future
perspectives. J Clin Med. 12(4646)2023.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Cancer Genome Atlas Research Network.
Comprehensive molecular characterization of gastric adenocarcinoma.
Nature. 513:202–209. 2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Ignatova EO, Kozlov E, Ivanov M, Mileyko
V, Menshikova S, Sun H, Fedyanin M, Tryakin A and Stilidi I:
Clinical significance of molecular subtypes of gastrointestinal
tract adenocarcinoma. World J Gastrointest Oncol. 14:628–645.
2022.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Sohn BH, Hwang JE, Jang HJ, Lee HS, Oh SC,
Shim JJ, Lee KW, Kim EH, Yim SY, Lee SH, et al: Clinical
significance of four molecular subtypes of gastric cancer
identified by the cancer genome atlas project. Clin Cancer Res.
23:4441–4449. 2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Nemtsova MV, Kuznetsova EB and Bure IV:
Chromosomal instability in gastric cancer: Role in tumor
development, progression, and therapy. Int J Mol Sci.
24(16961)2023.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Grillo F, Fassan M, Sarocchi F, Fiocca R
and Mastracci L: HER2 heterogeneity in gastric/gastroesophageal
cancers: From benchside to practice. World J Gastroenterol.
22:5879–5887. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Wakatsuki T, Yamamoto N, Sano T, Chin K,
Kawachi H, Takahari D, Ogura M, Ichimura T, Nakayama I, Osumi H, et
al: Clinical impact of intratumoral HER2 heterogeneity on
trastuzumab efficacy in patients with HER2-positive gastric cancer.
J Gastroenterol. 53:1186–1195. 2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Gassmann R, Essex A, Hu JS, Maddox PS,
Motegi F, Sugimoto A, O'Rourke SM, Bowerman B, McLeod I, Yates III
JR, et al: A new mechanism controlling kinetochore–microtubule
interactions revealed by comparison of two dynein-targeting
components: SPDL-1 and the Rod/Zwilch/Zw10 complex. Genes Dev.
22:2385–2399. 2008.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Lara-Gonzalez P, Westhorpe FG and Taylor
SS: The spindle assembly checkpoint. Curr Biol. 22:R966–R980.
2012.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Vallee RB, Varma D and Dujardin DL: ZW10
function in mitotic checkpoint control, dynein targeting, and
membrane trafficking: Is dynein the unifying theme? Cell Cycle.
5:2447–2451. 2006.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Cmentowski V, Ciossani G, d'Amico E,
Wohlgemuth S, Owa M, Dynlacht B and Musacchio A: RZZ-Spindly and
CENP-E form an integrated platform to recruit dynein to the
kinetochore corona. EMBO J. 42(e114838)2023.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Pereira C, Reis RM, Gama JB, Celestino R,
Cheerambathur DK, Carvalho AX and Gassmann R: Self-assembly of the
RZZ complex into filaments drives kinetochore expansion in the
absence of microtubule attachment. Curr Biol. 28:3408–3421.e8.
2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Barbosa J, Conde C and Sunkel C:
RZZ-SPINDLY-DYNEIN: You got to keep 'em separated. Cell Cycle.
19:1716–1726. 2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Zhang G, Lischetti T, Hayward DG and
Nilsson J: Distinct domains in Bub1 localize RZZ and BubR1 to
kinetochores to regulate the checkpoint. Nat Commun.
6(7162)2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Lara-Gonzalez P, Pines J and Desai A:
Spindle assembly checkpoint activation and silencing at
kinetochores. Semin Cell Dev Biol. 117:86–98. 2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Scaërou F, Aguilera I, Saunders R, Kane N,
Blottière L and Karess R: The rough deal protein is a new
kinetochore component required for accurate chromosome segregation
in Drosophila. J Cell Sci. 112:3757–3768. 1999.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Williams BC, Li Z, Liu S, Williams EV,
Leung G, Yen TJ and Goldberg ML: Zwilch, a new component of the
ZW10/ROD complex required for kinetochore functions. Mol Biol Cell.
14:1379–1391. 2003.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Hirose H, Arasaki K, Dohmae N, Takio K,
Hatsuzawa K, Nagahama M, Tani K, Yamamoto A, Tohyama M and Tagaya
M: Implication of ZW10 in membrane trafficking between the
endoplasmic reticulum and Golgi. EMBO J. 23:1267–1278.
2004.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Défachelles L, Raich N, Terracol R, Baudin
X, Williams B, Goldberg M and Karess RE: RZZ and Mad1 dynamics in
Drosophila mitosis. Chrom Res. 23:333–342. 2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Williams BC and Goldberg ML: Determinants
of drosophila zw10 protein localization and function. J Cell Sci.
107:785–798. 1994.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Zhengxiang Z, Yunxiang T, Zhiping L and
Zhimin Y: KNTC1 knockdown suppresses cell proliferation of colon
cancer. 3 Biotech. 11(262)2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Liu L, Chen H, Chen X, Yao C, Shen W and
Jia C: KNTC1 as a putative tumor oncogene in pancreatic cancer. J
Cancer Res Clin Oncol. 149:3023–3031. 2023.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Liu CT, Min L, Wang YJ, Li P, Wu YD and
Zhang ST: shRNA-mediated knockdown of KNTC1 suppresses cell
viability and induces apoptosis in esophageal squamous cell
carcinoma. Int J Oncol. 54:1053–1060. 2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Bakhoum SF and Cantley LC: The
multifaceted role of chromosomal instability in cancer and its
microenvironment. Cell. 174:1347–1360. 2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Bakhoum SF, Ngo B, Laughney AM, Cavallo
JA, Murphy CJ, Ly P, Shah P, Sriram RK, Watkins TBK, Taunk NK, et
al: Chromosomal instability drives metastasis through a cytosolic
DNA response. Nature. 553:467–472. 2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Sansregret L, Vanhaesebroeck B and Swanton
C: Determinants and clinical implications of chromosomal
instability in cancer. Nat Rev Clin Oncol. 15:139–150.
2018.PubMed/NCBI View Article : Google Scholar
|