Introduction
Osteosarcoma is one of the most common primary
malignant tumour observed in children and adolescents, with a
current annual incidence rate of 3/1,000,000 (1). At present, combined neoadjuvant
chemotherapy and surgery are mainly used to treat osteosarcoma,
however, patient survival rates remain low (2). Studies of potential gene therapy
candidates for the treatment of osteosarcoma have increased with
the development of molecular biology and genomic sciences (3,4). In
previous studies (5,6), we utilised a limiting dilution to
obtain an osteosarcoma MG63 monoclonal cell substrain. Cell
electrophoresis and invasion assays were then used to identify MG63
monoclonal cell substrains with variations in transfer and
malignant characteristics. A cell substrain with high transfer and
malignant characteristics (M8) and another with low transfer and
malignant characteristics (M6) were verified by tumour transfer
experiments in vitro. A gene chip was then used to scan and
compare M8 with M6. Cyclin-dependent kinase 2 (Cdc2) expression in
M8 was high at 2.67. Following this, quantitative
reverse-transcription polymerase chain reaction (qRT-PCR) was
utilised to verify the increase in gene expression of Cdc2 in M8
compared with M6 cells (5,6) and Cdc2 was identified as a key gene
for the proliferation and apoptosis of osteosarcoma MG63 cells. In
addition, silencing of Cdc2 expression was demonstrated to inhibit
proliferation and promote apoptosis of osteosarcoma MG63 cells. To
verify these findings, a small interfering RNA (siRNA) expression
plasmid targeting the Cdc2 gene was constructed in the current
study and the effect of siRNA interference of Cdc2 expression on
the proliferation and apoptosis of osteosarcoma MG63 cells was
investigated. The results obtained may serve as an experimental
basis for gene therapy of osteosarcoma.
Materials and methods
Cell culture
MG63 cells were grown in RPMI-1640 medium
supplemented with 10% calf serum, 100 μg/ml streptomycin and 100
U/ml penicillin in a humidified 5% CO2 and 95% air
incubator at 37°C.
Plasmid construction
The genetic code sequence for Cdc2 was obtained from
GenBank (NM_001170406) in accordance with the principles of siRNA
design. One siRNA-2 interference sequence was designed by
Invitrogen Life Technologies (Carlsbad, CA, USA). An additional
siRNA-1 interference sequence that was previously reported to be
more effective (7) than that in
the human genome was identified only as a Cdc2 gene interference
sequence. A non-specific siRNA-scrambled (siRNA-Scramble) sequence,
designed in a similar manner to serve as the negative control, was
obtained from the Institute of Molecular Biology of (Three Gorges
University; Table I). The three
synthesised siRNA sequences were annealed using sticky ends and
BamHI and HindIII digestion and then connected to a
psilencer 2.1-U6 that had been cut by the same enzyme. The three
restructured psilencer 2.1/siRNA plasmids were then transformed
into Escherichia coli DH5, screened for ampicillin
resistance on an agar plate and then amplified in culture.
Following this, plasmids were selected and sequenced.
 | Table ITemplate sequence of Cdc2 siRNA. |
Table I
Template sequence of Cdc2 siRNA.
| siRNA | Sequence (5′-3′) |
|---|
| Negative |
| siRNA-Scramble |
| Forward |
GATCCACTACCGTTGTTATAGGTGTTCAAGAGACACCTATAACAACGGTAGTTTTTTGGAAA |
| Reverse |
AGCTTTTCCAAAAAACTACCGTTGTTATAGGTGTCTCTTGAACACCTATAACAACGGTAGG |
| Cdc2 siRNA-1 |
| Forward |
GATCCGGGGTTCCTAGTACTGCAATTCAAGAGATTGCAGTACTAGGAACCCCTTTTTTGGAAA |
| Reverse |
AGCTTTTCCAAAAAAGGGGTTCCTAGTACTGCAATCTCTTGAATTGCAGTACTAGGAACCCCG |
| Cdc2 siRNA-2 |
| Forward |
GATCCGTTGATCAACTCTTCAGGATTTCAAGAGAATCCTGAAGAGTTGATCAATTTTTTGGAAA |
| Reverse |
AGCTTTTCCAAAAAATTGATCAACTCTTCAGGATTCTCTTGAAATCCTGAAGAGTTGATCAACG |
Transient transfection
One day prior to transient transfection, the cells
were plated in six-well plates at a concentration of
2×105 cells/well. When the cell density reached ~80%,
transfection was performed using Lipofectamine™ 2000 in accordance
with the manufacturer’s instructions. Following transfection (~24
h), the proteins were extracted from the cells. Western blot
analysis was then performed to determine the siRNA silencing
efficiency.
Stable transfection
MG63 cells were plated in six-well plates at a
concentration of 2×105 cells/well and then cultured in a
conventional culture medium, as previously described. Cells were
transfected the following day according to the manufacturer’s
instructions. In brief, diluted DNA and Lipofectamine™ 2000
complexes (total volume, 25 ml) were prepared and added to each
MG63 cell-containing well. Cells were then incubated for 4–6 h.
Post-transfection (~48 h), cells were incubated with 100 μg/ml
hygromycin in RPMI-1640 medium with 10% calf serum for three weeks.
Single clones with low Cdc2 expression were then identified via
RT-PCR, western blot analysis and cell immunofluorescence.
Quantitative PCR
RNA from the cell lines was extracted using TRIzol
reagent. cDNA was synthesised using SuperScript™ II Reverse
Transcriptase according to the manufacturer’s instructions. The
primers used for amplification were as follows: Cdc2,
5′-TACCTATGGAGTTGTGTATAA-3′ and 5′-ATTCCA CTTCTGGCCACACTT-3′;
β-actin, 5′-CCACAGCTGAG AGGGAAATC-3′ and
5′-ATCCTCTTCCTCCCTGGAGA-3′. PCR cycling conditions were as follows:
94°C for 5 min, 25 cycles at 94°C for 30 sec, 55°C for 30 sec, 72°C
for 30 sec and 78°C for 1 sec for plate reading and 72°C for 5 min.
PCR products were separated via electrophoresis at 100 V for 30 min
on a 1% agarose gel and then detected using ethidium bromide
staining. The expected sizes of specific PCR products were verified
using a 1-kb DNA reference ladder.
Western blot analysis
Cell lysates were prepared from six cloning strains.
Normal MG63 and negative control cells were cultured for 24 h.
Protein samples were subjected to 12% sodium dodecyl
sulphate-polyacrylamide gel electrophoresis and then transferred
onto polyvinylidene fluoride membranes. Following blocking with 5%
non-fat dry milk in a Tris-buffered saline Tween-20 buffer for 1 h,
the samples were probed with a primary antibody against Cdc2 and a
horseradish peroxidase-conjugated secondary antibody (Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA). Protein bands were
detected using an enhanced chemiluminescence detection system and
X-ray film exposure (Kodak, Rochester, NY, USA). β-actin was used
as a loading control.
Cell immunofluorescence
MG63 and MG63-siRNA-Cdc2 cells were seeded into
24-well culture plates. When the growth density reached 60%, cells
were fixed with ice-cold 4% paraformaldehyde and incubated with 10%
(v/v) goat serum as a blocking background. Following overnight
incubation with the primary antibody against Cdc2, the cells were
washed three times with phosphate-buffered saline (PBS) and then
incubated with rhodamine-123-labeled anti-rabbit IgG for 1 h at
37°C. The cells were washed three times with PBS and then examined
via fluorescence/phase-contrast microscopy (TE2000S; Nikon, Tokyo,
Japan).
Assay of cellular proliferation
MG63, MG63-siRNA/Scramble and MG63-siRNA/Cdc2 cells
were seeded into 96-well culture plates (2,000 cells/well in 100 μl
RPMI-1640 medium). Culture medium was removed following culture for
0, 24, 48, 72 and 96 h. A 200 μl MTT solution (0.25 g/l in
RPMI-1640 medium) was added to each well and the cells were
incubated at 37°C for 4 h. The medium was removed and 200 μl
dimethyl sulfoxide was added to each well. Absorbance (A) at 570 nm
was recorded following 30 min incubation at room temperature. The
cell survival rate was calculated as follows: Cell survival rate =
Adrug/Acontrol × 100.
Apoptosis protein detection via western
blot analysis
Experiments were conducted as described for Cdc2.
Primary monoclonal antibodies against B-cell lymphoma 2 (Bcl-2) and
Bcl-2-associated X (Bax) were utilised. Secondary antibodies were
mouse and goat anti-rabbit IgG (both 1:2,000).
Flow cytometry
MG63, MG63-siRNA/Scramble and MG63-siRNA/Cdc2 were
collected, washed twice with ice-cold PBS and then fixed in 75%
ethanol overnight at 4°C. Following centrifugation at 2,000 rpm for
5 min, cell pellets were resuspended in PBS containing 0.1% Triton
X-100 and 50 μg/ml RNase A. The cell samples were then stained with
50 mg/l propidium iodide for 30 min at 37°C in the dark prior to
flow cytometry.
Statistical analysis
Data are expressed as mean ± SD. Differences between
two groups were analysed using the Student’s t-test, whereas
differences between three or more groups were analysed using ANOVA.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Plasmid construction
The siRNA-restructured plasmid was connected to
psilencer 2.1-U6. The identity of the restructured plasmid
psilencer 2.1/siRNA was confirmed via DNA sequencing (Fig. 1).
Cdc2 low-expression cell line
siRNA was transiently transfected into MG63 cells.
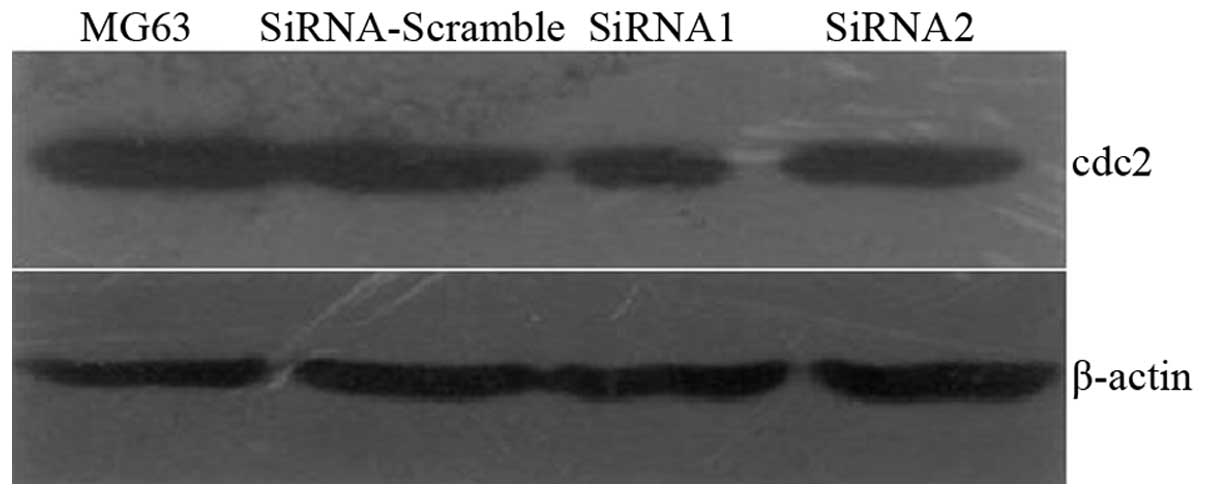
Western blot analysis was then performed to determine the siRNA
silencing efficiency (Fig. 2).
siRNA-Scramble had no effect on Cdc2 expression and siRNA-1
exhibited higher silencing efficiency than siRNA-2. Therefore,
siRNA-1 was used in the stable transfection. Western blot analysis
and RT-PCR detected Cdc2 expression in six clones (Figs. 3 and 4). The siRNA1-5 clone exhibited the
highest silencing efficiencies: 89 and 86% at the protein and mRNA
levels, respectively. Immunofluorescence results demonstrated that,
compared with normal MG63, MG63-siRNA/Cdc2 cells shrank and their
structures changed into short, irregular, oblate spheroids. In
addition, a reduced number of MG63-siRNA/Cdc2 cells were identified
to express Cdc2 protein. No Cdc2 expression was observed in
MG63-siRNA/Cdc2 cells.
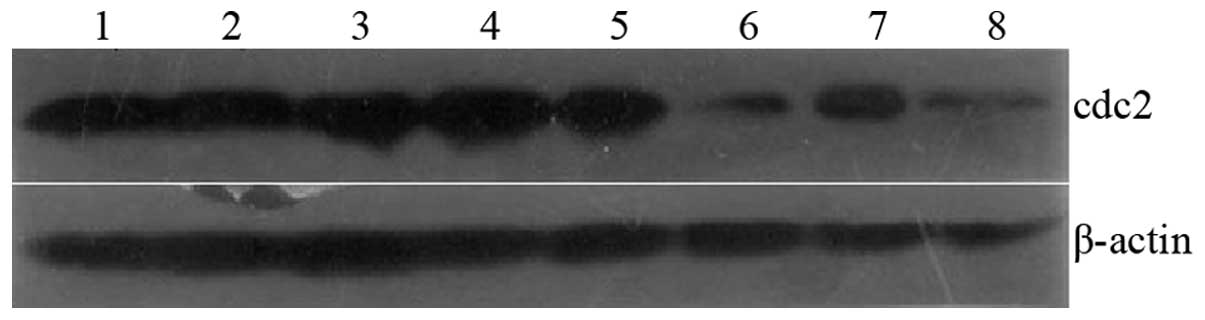 | Figure 3Cdc2 expression in six cloned strains
was detected by western blot analysis. siRNA1-5 demonstrated the
best effect in Cdc2 silencing. Lane 1, MG63; 2, siRNA-Scramble; 3,
siRNA1-1; 4, siRNA1-2; 5, siRNA1-3; 6, siRNA1-5; 7, siRNA1-7; 8,
siRNA1-8. Cdc2, cyclin-dependent kinase 2; siRNA, small interfering
RNA. |
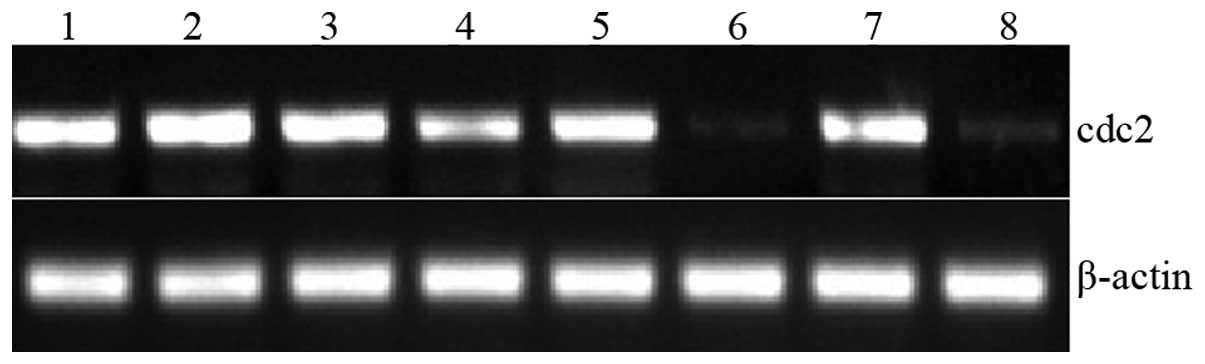 | Figure 4Cdc2 expression in six cloned strains
was analysed by RT-PCR.siRNA1-5 was identified to have the best
silencing effect on Cdc2. Lane 1, MG63; 2, siRNA-Scramble; 3,
siRNA1-1; 4, siRNA1-2; 5, siRNA1-3; 6, siRNA1-5; 7, siRNA1-7; 8,
siRNA1-8. Cdc2, cyclin-dependent kinase 2; siRNA, small interfering
RNA; RT-PCR, reverse-transcription polymerase chain reaction. |
MTT assay
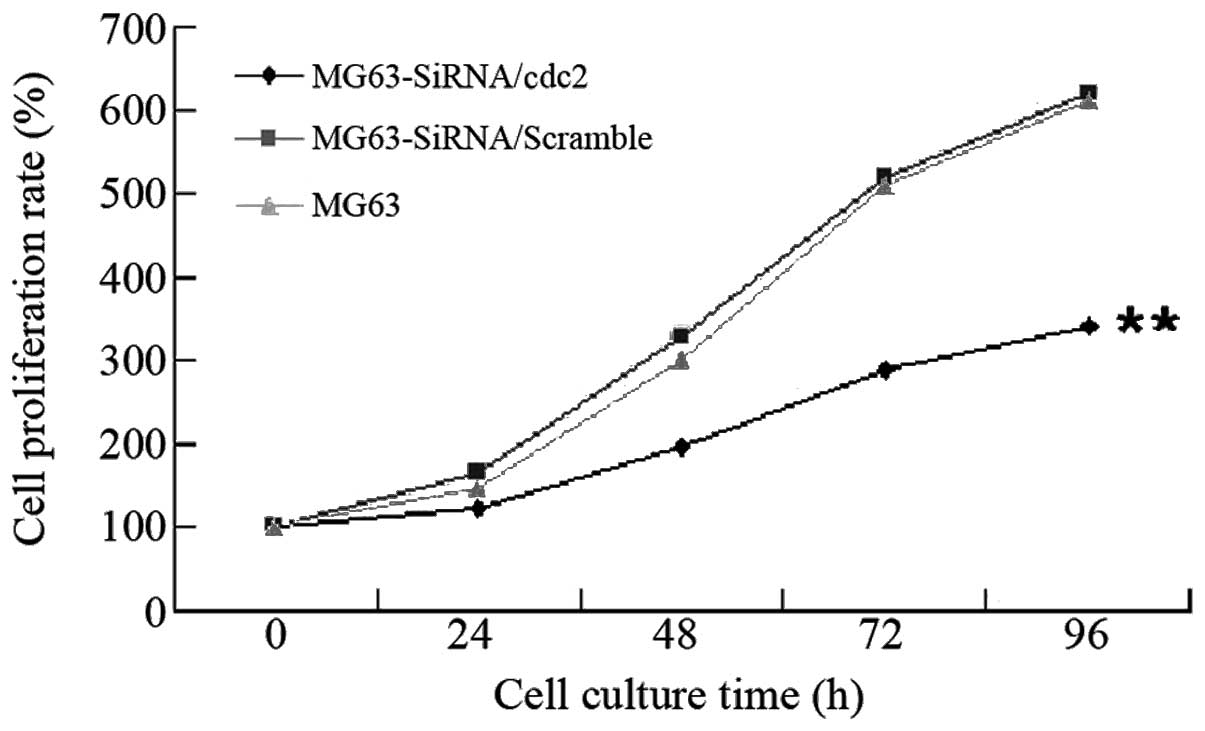
The MTT assay detected MG63 cell proliferation
following Cdc2 expression silencing (Fig. 5). Following culture for 24 h, the
growth rate of the MG63-siRNA/Cdc2 was identified as significantly
lower than those of the MG63-siRNA/scramble and MG6 cells
(P<0.01). These observations indicate that Cdc2 expression
silencing inhibits cell proliferation.
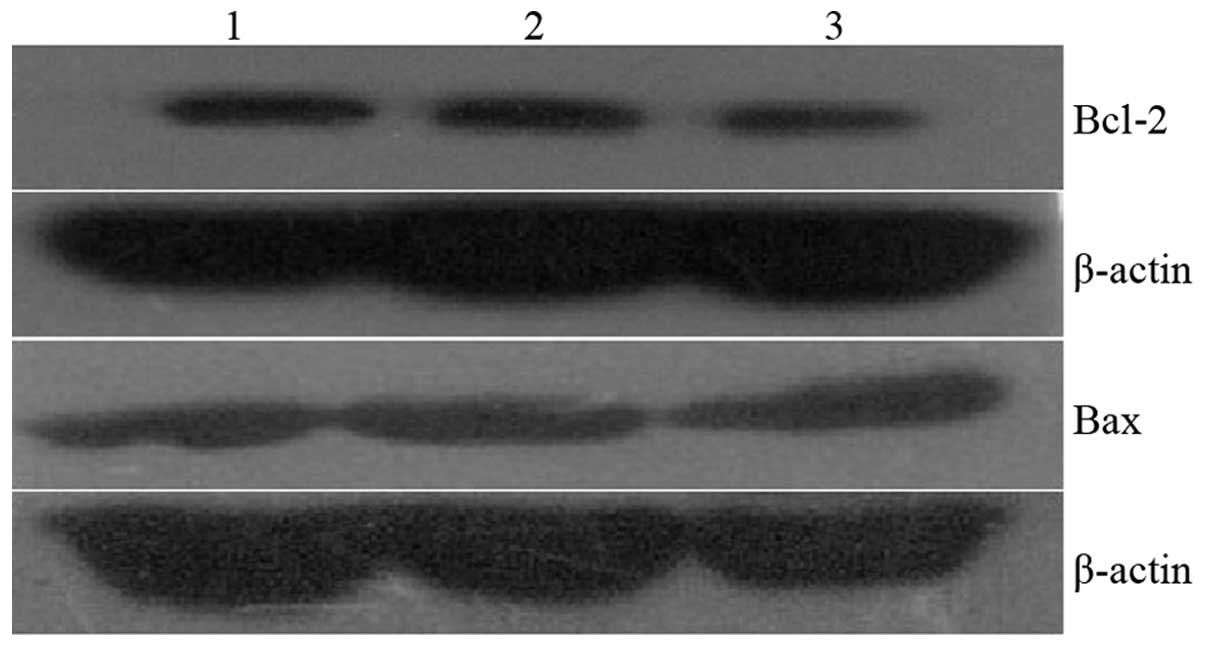
Bcl-2, Bax gene expression
Bcl-2 expression in the MG63-siRNA/Cdc2 was not
revealed to decrease significantly compared with the
MG63-siRNA/scramble and MG63 cells (126±16 vs. 148±18, 151±13,
respectively; P>0.05). In addition, Bcl-2 expression in the
MG63-siRNA/Cdc2 cells was not identified to increase significantly
(198±21 vs. 186±26, 178±25, respectively; P>0.05; Fig. 6).
Cell cycle
Following Cdc2 expression silencing, MG63-siRNA/Cdc2
accumulated at the G2/M and G0/G1
phases compared with MG63-siRNA/scramble and MG63 cells. However,
the number of MG63-siRNA/Cdc2 cells in the S phase was observed to
be significantly reduced (P<0.05; Table II).
 | Table IIEffect of Cdc2-siRNA on the cell cycle
in MG63 cells (n=3). |
Table II
Effect of Cdc2-siRNA on the cell cycle
in MG63 cells (n=3).
|
G0/G1 | S | G2/M |
|---|
| MG63 | 63.1±0.8 | 25.0±0.7 | 11.9±0.2 |
|
MG63-siRNA/Scramble | 62.8±0.9 | 26.7±0.6 | 10.5±0.2 |
| MG63-siRNA/Cdc2 |
66.8±0.7a |
11.8±0.4b |
21.4±0.3b |
Discussion
Osteosarcoma is the most common highly malignant
tumour of the bone, often manifesting during the second or third
decade of life. It accounts for ~60% of all malignant bone tumours
that occur during the first 20 years of life (8). The development of neoadjuvant
chemotherapy has increased long-term survival rates from 10–20 to
60–70%. However, although adjuvant chemotherapy effectively
improves patient survival, as well as the treatment of primary
tumours (9), patients who present
with metastatic disease and/or tumour relapse remain associated
with a poor prognosis. The estimated five-year survival rate
following relapse is only 25% (10). To date, gene therapy studies of
osteosarcoma pathogenesis and progression, have investigated p53,
ezrin and surivin (11–16). However, few studies have focused on
the molecular mechanism of Cdc2 in osteosarcoma cells.
In the present study, siRNA interference technology
was used to obtain a low-Cdc2 expression cell line. The cell line
was then used to study the effects of siRNA-induced interference of
Cdc2 expression on the proliferation and apoptosis of osteosarcoma
MG63 cells. Normal MG63 cells exhibit long shuttle-like or
triangular structures. Following Cdc2 expression silencing, cells
exhibited short, irregular, oblate shapes which accumulated easily.
MTT results demonstrate that Cdc2 expression silencing inhibits
cell proliferation by blocking the cell cycle at the
G2/M phase. Expression of the apoptosis genes, Bcl-2 and
Bax, was also analysed to determine the possible effects of Cdc2
expression silencing on cell apoptosis. Cdc2 expression silencing
had no effect on Bcl-2 and Bax, indicating that Cdc2 silencing does
not promote MG63 cell apoptosis. Results of the present study
indicate that Cdc2 expression silencing in MG63 cells inhibits cell
proliferation but does not promote cell apoptosis. However, we
previously hypothesised that disruption of Cdc2 expression inhibits
osteosarcoma MG63 cell proliferation and promotes apoptosis. The
absence of an observable effect of Cdc2 expression silencing on the
apoptosis of MG63 cells may be due to the large number of genes
associated with the regulation of apoptosis of MG63 cells. Although
Cdc2 expression in MG63 cells, itself a regulation mechanism, is
silenced, additional genes may be activated to offset low Cdc2
expression. Therefore, cells do not undergo apoptosis when Cdc2
alone is silenced. Inhibition of cell proliferation by Cdc2
expression silencing is attributed to the association between Cdc2
and the eukaryotic cell division cycle gene CDC2, which codes for a
serine/threonine protein kinase with a molecular weight of 3,400
Da. Cdc2 enzyme activation is an indicator of cell division. Cdc2
is vital to cell cycle regulation, inducing DNA replication and
cell mitosis (17–19). Overexpression of Cdc2 is associated
with cell cycle progression disorders, including abnormal growth
and cell differentiation, as well as malignant cell proliferation
and tumour formation (20). A
number of previous studies have reported overexpression of Cdc2 in
breast cancer (21,22), B-cell lymphoma (23), colon carcinoma (24), ovarian cancer (25) and glioma (7) cells, as well as a marked correlation
with tumour stage, grade, relapse and prognosis. Results of the
current study are consistent with these observations.
In the present study, the expression levels of Bax
and Bcl-2 only were analysed to detect cell apoptosis. Therefore,
additional data are required to verify these results. Future
studies must include analyses using an anticancer drug. The effect
of Cdc2 expression silencing on the strength of the anticancer drug
activity may then be determined. Three cell lines will be implanted
in mice to investigate the effect of Cdc2 expression silencing on
tumour growth, transfer and prognosis in vivo.
Acknowledgements
The present study was supported by the Natural
Science Foundation of Hubei Province (D20101206).
References
|
1
|
Picci P: Osteosarcoma (osteogenic
sarcoma). Orphanet J Rare Dis. 2:62007. View Article : Google Scholar
|
|
2
|
Chou AJ, Geller DS and Gorlick R: Therapy
for osteosarcoma: where do we go from here? Paediatr Drugs.
10:315–327. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tan ML, Choong PF and Dass CR:
Osteosarcoma: Conventional treatment vs. gene therapy. Cancer Biol
Ther. 8:106–117. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Broadhead ML, Clark JC, Choong PF and Dass
CR: Making gene therapy for osteosarcoma a reality. Expert Rev
Anticancer Ther. 10:477–480. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li XZ, Meng L, Chen AM, Guo FJ, Luo ZQ and
Zeng H: Screening of differentially expressed genes in osteosarcoma
cell lines with various metastatic potentialities. Zhonghua Zhong
Liu Za Zhi. 1:71–76. 2010.(In Chinese).
|
|
6
|
Li XZ, Meng L, Chen AM, Guo FJ, Luo ZQ and
Zeng H: Differentially expressed gene in osteosarcoma cell lines
with different metastatic potentials. J Nanjing Med Univ.
23:352–358. 2009. View Article : Google Scholar
|
|
7
|
Chen H, Huang Q, Dong J, Zhai DZ, Wang AD
and Lan Q: Overexpression of CDC2/CyclinB1 in gliomas, and CDC2
depletion inhibits proliferation of human glioma cells in vitro and
in vivo. BMC Cancer. 8:292008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Winkler K, Beron G, Kotz R, et al:
Neoadjuvant chemotherapy for osteogenic sarcoma: results of a
Cooperative German/Austrian study. J Clin Oncol. 2:617–624.
1984.PubMed/NCBI
|
|
9
|
Marina NM, Pratt CB, Rao BN, Shema SJ and
Meyer WH: Improved prognosis of children with osteosarcoma
metastatic to the lung(s) at the time of diagnosis. Cancer.
70:2722–2727. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kempf-Bielack B, Bielack SS, Jürgens H, et
al: Osteosarcoma relapse after combined modality therapy: an
analysis of unselected patients in the Cooperative Osteosarcoma
Study Group (COSS). J Clin Oncol. 23:559–568. 2005. View Article : Google Scholar
|
|
11
|
Oshima Y, Sasaki Y, Negishi H, et al:
Antitumor effect of adenovirus-mediated p53 family gene transfer on
osteosarcoma cell lines. Cancer Biol Ther. 6:1058–1066. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Walkley CR, Qudsi R, Sankaran VG, et al:
Conditional mouse osteosarcoma, dependent on p53 loss and
potentiated by loss of Rb, mimics the human disease. Genes.
22:1662–1676. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liang X, Da M, Zhuang Z, et al: Effects of
Survivin on cell proliferation and apoptosis in MG-63 cells in
vitro. Cell Biol Int. 33:119–124. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu YF, Liang XJ, Liu YY, et al: +Antisense
oligonucleotide targeting survivin inhibits growth by inducing
apoptosis in human osteosarcoma cells MG-63. Neoplasma. 57:501–506.
2010.
|
|
15
|
Kim C, Shin E, Hong S, et al: Clinical
value of ezrin expression in primary osteosarcoma. Cancer Res
Treat. 41:138–144. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang YF, Shen JN, Xie XB, Wang J and Huang
G: Expression change of ezrin as a prognostic factor in primary
osteosarcoma. Med Oncol. 28(Suppl 1): S636–S643. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xu N and Chang DC: Different thresholds of
MPF inactivation are responsible for controlling different mitotic
events in mammalian cell division. Cell Cycle. 6:1639–1645. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kim DH: Prognostic implications of
cyclinB1, p34cdc2, p27(Kip1) and p53 expression in gastric cancer.
Yonsei Med J. 48:694–700. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chang JT, Wang HM, Chang KW, et al:
Identification of differentially expressed genes in oral squamous
cell carcinoma (OSCC): overexpression of NPM, CDK1 and NDRG1 and
underexpression of CHES1. Int J Cancer. 114:942–949. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Diril MK, Ratnacaram CK, Padmakumar VC, et
al: Cyclin-dependent kinase 1 (Cdk1) is essential for cell division
and suppression of DNA re-replication but not for liver
regeneration. Proc Natl Acad Sci USA. 109:3826–3831. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim SJ, Nakayama S, Miyoshi Y, et al:
Determination of the specific activity of CDK1 and CDK2 as a novel
prognostic indicator for early breast cancer. Ann Onco. 19:68–72.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Choi HJ and Zhu BT: Critical role of
cyclin B1/Cdc2 up-regulation in the induction of mitotic
prometaphase arrest in human breast cancer cells treated with
2-methoxyestradiol. Biochim Biophys Acta. 1823:1306–1315. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao MY, Auerbach A, D’Costa AM, et al:
Phospho-p70S6K/p85S6K and cdc2/cdk1 are novel targets for diffuse
large B-cell lymphoma combination therapy. Clin Cancer Res.
15:1708–1720. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Meyer A, Merkel S, Brückl W, et al: Cdc2
as prognostic marker in stage UICC II colon carcinomas. Eur J
Cancer. 45:1466–1473. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shi HR and Zhang RT: Expression and
significance of P53, P21WAF1 and CDK1 proteins in epithelial
ovarian cancer. Ai Zheng. 28:882–885. 2009.(In Chinese).
|