Introduction
Acupuncture is one of the major therapies that has
been used in traditional Chinese medicine for at least 2,500 years,
and it remains an effective, safe and convenient intervention for
patients. Acupuncture practitioners insert thin, solid, metallic
needles into specific acupuncture points on the skin known as
acupoints, which are special nodes (or outlets) on the meridians.
Acupoints are utilized to correct the imbalances of the flow of qi
in the body to treat various diseases. The World Health
Organization (WHO) listed 43 indications for acupuncture in 1980
(1) and classified the diseases
treated by acupuncture into 4 categories, including 107 illnesses,
in 2002 (2). The National
Institutes of Health (NIH) recommended acupuncture as an adjunctive
treatment in comprehensive management programs, such as those for
addiction, stroke rehabilitation and asthma (3,4). In
recent years, acupuncture has also emerged as an alternative and
satisfactory treatment for symptom management in cancer (5), functional gastrointestinal disorders
(6) and rheumatic conditions
(7).
Acupuncture causes multiple biological alterations
in humans (4). In recent years,
researchers have attempted to identify changes in active substances
after acupuncture and the correlation between the changes in active
substances and the effect of acupuncture. It is thought that the
therapeutic effect of acupuncture includes 2 parts: the
psychological and physiological effects (8). The physiological effect of
acupuncture is composed of point-specific, treatment-specific and
non-invasive skin contact physiological effects (9). Due to the significant effect on
healthy volunteers (10,11), the condition-specific physiological
effect is thought to be one of the most important physiological
effects of acupuncture. Although a large number of studies have
been carried out to examine the effects of acupuncture under
different morbid conditions, the regulatory mechanism of
acupuncture remains unclear, and the evidence that has been
acquired is not compelling. We will only be able to distinguish the
effects of acupuncture on specific active substances under morbid
conditions once we are able to accurately analyze the general
effects of acupuncture under normal conditions, and clarify the
regulatory mechanisms involved.
Previous studies have shown that acupuncture in
healthy volunteers has given rise to biological responses that
regulate important physiological processes. Using functional
magnetic resonance imaging (fMRI) of the brain, a previous
randomized, controlled trial showed that acupuncture improves motor
and sensory functions in healthy volunteers (12). There was a statistically
significant increase in the number of CD4+ and
CD8+ cells, and in interleukin (IL)-4, IL-1β and
interferon-γ (IFN-γ) levels in the cells after stimulation of
meridian points by acupuncture (13). Acupuncture also modifies the
mediation of the autonomic innervations of the heart. Acupuncture
at the PC6 (wrists) and ST36 (lower legs) points has been shown to
help athletes significantly decrease their maximum heart rate,
oxygen consumption and blood lactic acid production 30 min after
exercise (14). It has been
reported that both specific and non-specific factors may play a
role in acupuncture therapy for pain; however, only real
acupuncture (non-placebo) has shown specific physiological effects
(15). Another study demonstrated
that the physiological effects of acupuncture may be influenced by
anxiety (16). Additionally,
acupuncture treatment regulates autonomic nervous system functions,
such as blood pressure regulation, sphincter of Oddi relaxation,
immune modulation (17) and
electrodermal activity (18).
However, gene expression profile studies on the physiological
effects of acupuncture under normal conditions are still
lacking.
High-throughput technologies, such as microarray and
serial analyses of gene expression (SAGE), may help to reveal the
background gene expression in response to acupuncture and highlight
the regulatory mechanism of acupuncture under morbid conditions.
SAGE, a powerful expression profiling method that is very useful in
dissecting this complex system, has been applied to qualitatively
and quantitatively evaluate the transcription of genes by length
without the prerequisite of a hybridization probe for each
transcript (19).
In this study, two SAGE tag libraries from the lung
tissues of normal and acupuncture-treated rats were constructed.
The aim of this study was to determine the influence of acupuncture
on the gene expression profiles of lung tissue from normal rats.
The results from this analysis may elucidate the regulatory
mechanism of acupuncture under normal conditions and help to
specify its effects under morbid conditions in the future.
Materials and methods
Animal and acupuncture treatment
Pathogen-free, male, Sprague-Dawley (SD) rats (4
weeks of age, 110–130 g; SLAC Laboratory Animal Co. Ltd., Shanghai,
China) were raised in a pathogen-free, rodent facility and were
provided with food and water ad libitum. The rats were
randomly divided into 2 groups (each group contained 8 rats):
normal rats (control) and normal rats treated with acupuncture. The
rats were kept in animal facilities that had been approved by the
Shanghai Committee for Accreditation of Laboratory Animals, and the
animal experiments conformed to the regulations of the State
Science and Technology Commission.
Three common acupoints for treating lung-related
diseases were selected and manipulated at the same time in the
acupuncture group, namely dazhui (GV14, located between the C7 and
T1 vertebrae), bilateral fengmen (BL12, foveola, located laterally
between the T2 and T3 vertebrae) and bilateral feishu (BL13,
foveola, located laterally between the T3 and T4 vertebrae). Manual
acupuncture was performed once every other day for 2 weeks by a
well-trained acupuncturist (7 times in total). The protocol of
acupuncture treatment has been described previously (20). Briefly, disposable, stainless
needles (0.30×13 mm) were inserted ~5 mm deep into the skin, and
the needles were twisted evenly, ~360°, at the rate of 60 times/min
for 20 sec. The needles were manipulated every 5 min, and were then
withdrawn after 20 min. Each rat was placed on a suspended shelf
(50×45 mm, ~50 cm above the ground) in order to allow the animal to
stand still without anesthesia. Rats in the control group were
handled in the same manner as the animals in the acupuncture group,
with the exception of acupuncture treatments. At the end of 2
weeks, all animals were sacrificed. Total RNA was extracted from
the lungs of the rats and frozen immediately in liquid
nitrogen.
Construction, annotation and confirmation
of the SAGE libraries
The construction and annotation of the SAGE
libraries have been described previously (21). The confirmation of the 2 SAGE
libraries was performed by quantitative real-time PCR (qRT-PCR) on
an Applied Biosystems 7300 Real-Time PCR System using the Toyobo
Real-time PCR Master Mix (Toyobo, Osaka, Japan). Primer sequences
are listed in Table I. The
expression ratio was calculated according to the 2−ΔΔCt
method (22). Transcripts with a
2-fold increase in expression were considered upregulated, and
those with a 0.5-fold decrease in expression were considered
downregulated.
 | Table IPrimer sequences of the real-time
PCR. |
Table I
Primer sequences of the real-time
PCR.
| Genes | Sequences
(5′-3′) |
|---|
| Sftpa1 | Forward
AGCCAGTTTCGCATTCCCT |
| Reverse
ATGTGAAGGCCCATGAGCA |
| Col6a2 | Forward
ATGGAAGCCAGAACCAGCAAC |
| Reverse
CCACGTGCGAGAAAGAATTGA |
| Col4a1 | Forward
GCAATGCTGAATCGTCCCA |
| Reverse
TGGAGATGCCAGATGGTTAGG |
| GAPDH | Forward
TCCTGCACCACCAACTGCTTAG |
| Reverse
AGTGGCAGTGATGGCATGGACT |
Bioinformatic analysis of SAGE tags
Genes that were differentially expressed (P<0.05)
between the 2 SAGE libraries were functionally annotated and
classified using the Database for Annotation, Visualization and
Integrated Discovery (DAVID) Functional Annotation Tool (http://david.abcc.ncifcrf.gov/), a web-based tool
that provides integrated solutions for the annotation and analyses
of genome-scale datasets that are derived from high-throughput
technologies, such as microarray or SAGE (23).
To understand the key regulatory processes in
acupuncture treatment, the Gene Ontology (GO) Tree Machine
(http://www.genereg.ornl.gov/gotm/)
was applied for bioinformatic analyses of the acupuncture-regulated
expression data. The GO Tree Machine generates a tree-like
structure that is used to navigate the GO categories for the input
gene sets (24). Statistical
analyses of the enrichments were performed to identify the most
significant GO categories of the input gene sets and to suggest
their potential biological importance in the categories.
Statistical analyses
Statistical analyses used to determine the
significance of each of the 2 SAGE libraries were performed using
Monte Carlo analysis. The enrichments of the GO Tree Machine were
determined to be statistically significant by the hypergeometric
test (24).
Results
General analysis of SAGE libraries
The 2 SAGE libraries of the rat lungs were deposited
into the SAGEmap database at the National Center for Biotechnology
Information (NCBI; http://www.ncbi.nlm.nih.gov/geo), and the accession
numbers given to these libraries are GSM45195 and GSM279945. The
genes that were matched to the expressed sequence tags (ESTs) of
the 2 libraries are listed in Table
II. By comparing the SAGE data of the control and acupuncture
libraries, 144 differentially expressed tags (P<0.05, Table III) were identified. Among these
tags, 78 were upregulated and 66 were downregulated.
 | Table IISummary of serial analysis of gene
expression (SAGE) analysis of the 2 libraries. |
Table II
Summary of serial analysis of gene
expression (SAGE) analysis of the 2 libraries.
| SAGE tag | Control | Acupuncture |
|---|
| Total tags | 28,284 | 29,284 |
| Unique tags | 12,857 | 12,412 |
| Genes matched | 54.1% | 50.5% |
| ESTs matched | 38.5% | 17.0% |
| Not matched | 7.4% | 32.5% |
 | Table IIIOne hundred and forty-four
differentially expressed tags between the libraries of control and
acupuncture. |
Table III
One hundred and forty-four
differentially expressed tags between the libraries of control and
acupuncture.
| Tag sequences | Control | Acupuncture | Fold | UniGene | Annotation |
|---|
| Upregulated (78) |
| CACACTAACC | 288 | 499 | 1.7 | 2283 | Lyz, lysozyme |
| ATGAAATCAA | 197 | 267 | 1.4 | 40171 | Rps4x, ribosomal
protein S4, X-linked |
| TGGGTTGTCT | 193 | 283 | 1.5 | 36610 | Tpt1, tumor protein,
translationally-controlled 1 |
| ATACGAACTG | 93 | 134 | 1.4 | 11343 | Sftpa1, surfactant,
pulmonary-associated protein A1 |
| AGGAGGCTAC | 38 | 67 | 1.8 | 108039 | Rpl14, ribosomal
protein L14 |
| GGCAAGCCCC | 35 | 59 | 1.7 | 2262 | Rpl10a, ribosomal
protein L10A |
| AACCGCTTTT | 35 | 58 | 1.7 | 2989 | GIIg15b, global
ischemia induced protein GIIG15B |
| AAAATGCACA | 32 | 55 | 1.7 | 2267 | Defb2, defensin
β-2 |
| CGGAAGGCGG | 25 | 51 | 2.0 | 1439 | Rpl36, ribosomal
protein L36 |
| GTGAAGGCGG | 24 | 46 | 1.9 | 94935 | Rps3a, ribosomal
protein S3a |
| GGTAGCCACT | 18 | 38 | 2.1 | 106034 | Rps27a, ribosomal
protein S27a |
| TTTGCACCTT | 13 | 29 | 2.2 | 17145 | Ctgf, connective
tissue growth factor |
| GATGTGGCTG | 12 | 28 | 2.3 | 3910 | LOC363241, similar to
eukaryotic translation elongation factor 1 β2 |
| TGGACCTAGA | 9 | 24 | 2.7 | 1997 | Ctsh, cathepsin
H |
| CTGTGTGATC | 9 | 22 | 2.4 | 22047 | Transcribed sequence
with weak similarity to protein pir:T46271 (H. sapiens)
T46271 hypothetical protein DKFZp564P1263.1 - human |
| TTGATTTTTT | 8 | 21 | 2.6 | 59630 | Transmembrane and
coiled-coil domains 3 |
| GGCTCAGCCT | 8 | 21 | 2.6 | 84435 | Gpi, glucose
phosphate isomerase |
| TATGTCAAGC | 7 | 23 | 3.3 | 8400 | Rps12, ribosomal
protein S12 |
| CCCTGAGTCC | 7 | 19 | 2.7 | 94978 | Actb, actin, β |
| TGTACTCAAT | 6 | 15 | 2.5 | 25771 | Hnrpa1, heterogeneous
nuclear ribonucleoprotein A1 |
| TACATTTTCA | 6 | 16 | 2.7 | 8527 | Transcribed
sequence with strong similarity to protein pir:S55054 (H.
sapiens) S55054 Sm protein G - human |
| AAGACAGCTG | 6 | 16 | 2.7 | 83667 | RT1-Aw2, RT1 class
Ib, locus Aw2 |
| TTCAAAAAAA | 4 | 12 | 3.0 | 19267 | Pecr, perosisomal
2-enoyl-CoA reductase |
| TGCTGCGAAA | 4 | 16 | 4.0 | 22087 | LOC293618 |
| TGTAATGTGT | 3 | 14 | 4.7 | 2589 | Cdo1, cytosolic
cysteine dioxygenase 1 |
| GAGGGAGAGG | 3 | 11 | 3.7 | 105953 | LOC315326 similar
to Tenc1 protein |
| GAATATCGGA | 3 | 13 | 4.3 | 10696 | Pspla1,
phosphatidylserine-specific phospholipase A1 |
| AGAAAAAAAA | 3 | 11 | 3.7 | 55036 | Ciliary
neurotrophic factor receptor |
| GTTCTTCCGT | 2 | 10 | 5.0 | 29258 | Atp5g2 ATP
synthase, H transporting, mitochondrial F0 complex, subunit c
(subunit 9), isoform 2 |
| GTCCTGAGAG | 2 | 9 | 4.5 | 82672 | Vamp8,
vesicle-associated membrane protein 8 |
| GGTGGGACAC | 2 | 9 | 4.5 | 8509 | Tmp21, integral
membrane protein Tmp21-I (p23) |
| GCCACTTAGG | 2 | 12 | 6.0 | 12102 | LOC311545, similar
to histocompatibility 13; presenilin-like protein 3 |
| GATTGTCTTG | 2 | 15 | 7.5 | 1464 | LOC299104, similar
to 25 kDa FK506-binding protein |
| CTGTCATTTG | 2 | 11 | 5.5 | 9002 | LOC361814, similar
to splicing factor, arginine/serine-rich 3 (Pre-mRNA splicing
factor SRP20) (X16 protein) |
| CTCTCTGAAT | 2 | 9 | 4.5 | 5106 | Hmgcs1,
3-hydroxy-3-methylglutaryl-coenzyme A synthase 1 |
| CCCGTGTGCT | 2 | 9 | 4.5 | 109735 | Rps9, ribosomal
protein S9 |
| TTTCAGCAGT | 1 | 7 | 7.0 | 47 | Hprt, hypoxanthine
guanine phosphoribosyl transferase |
| TTGAGCGACA | 1 | 8 | 8.0 | 4223 | Dkc1, dyskeratosis
congenita 1, dyskerin |
| TGGTCTGAAA | 1 | 9 | 9.0 | 2722 | LOC315642, similar
to 60S ribosomal protein L27A |
| TCCTTGTTTA | 1 | 7 | 7.0 | 14866 | Transcribed
sequences |
| TATGAAATTT | 1 | 8 | 8.0 | 29782 | Fh1, fumarate
hydratase 1 |
| TATAGAGAAA | 1 | 9 | 9.0 | 36797 | HECT domain
containing 1 |
| GGAAAAGAAG | 1 | 7 | 7.0 | 32080 | Aif1, allograft
inflammatory factor 1 |
| GCTCTGATAT | 1 | 8 | 8.0 | 3285 | Dbi, diazepam
binding inhibitor |
| CAACCGTCAT | 1 | 9 | 9.0 | 108012 | Laptm4a,
lysosomal-associated protein transmembrane 4α |
| AGGACACCGC | 1 | 9 | 9.0 | 2759 | LOC315707, similar
to Tyrosine-protein kinase CSK (C-SRC kinase) |
| TTTTAAAAGC | 0 | 5 | 5.0 | 13322 | LOC306542, similar
to RNA polymerase III transcription initiation factor BRF2 |
| TTCAATGGTG | 0 | 5 | 5.0 | 98380 | Pgpep1,
pyroglutamyl-peptidase I |
| TTAAGCACTT | 0 | 6 | 6.0 | 44465 | Cmklr1,
chemokine-like receptor 1 |
| TGGAAGCTGA | 0 | 15 | 15.0 | 55487 | Gnb2l1, guanine
nucleotide binding protein, β polypeptide 2-like 1 |
| TGAGCTCTGG | 0 | 10 | 10.0 | 2694 | Mcam l-gicerin |
| TCGCTGTGTA | 0 | 5 | 5.0 | 23906 | LOC361305, similar
to T-cell activation WD repeat protein |
| TAAGATTCTT | 0 | 6 | 6.0 | 33807 | LOC288620, similar
to CCT (chaperonin containing TCP-1) ζ subunit |
| GGAAAAAATA | 0 | 5 | 5.0 | 9406 | LOC309259, similar
to mage-g1 |
| GCGTCTGCTC | 0 | 5 | 5.0 | 1677 | Gpr56, G
protein-coupled receptor 56 |
| GCAGGGTTTT | 0 | 5 | 5.0 | 10293 | Lrpap1, low density
lipoprotein receptor-related protein associated protein 1 |
| GATCTTTCCC | 0 | 5 | 5.0 | 13589 | LOC287710, similar
to polymerase I-transcript release factor; PTRF |
| GACTGAACCC | 0 | 5 | 5.0 | 25727 | Transcribed
sequence with moderate similarity to protein sp:P00722 (E.
coli) BGAL_ECOLI β-galactosidase |
| CTGTCCTTTC | 0 | 5 | 5.0 | 2776 | Becn1, beclin
1 |
| CTGCAGCCTG | 0 | 5 | 5.0 | 5782 | Stx5a, syntaxin
5a |
| CTGAGTAAAC | 0 | 8 | 8.0 | 9829 | Ager advanced
glycosylation end product-specific receptor |
| CAACTACACA | 0 | 10 | 10.0 | 2564 | Transcribed
sequences |
| ATTTGATATT | 0 | 5 | 5.0 | 18892 | Transcribed
sequence with moderate similarity to protein sp:P00722 (E.
coli) BGAL_ECOLI β-galactosidase |
| AGCCTGGAAA | 0 | 6 | 6.0 | 6606 | LOC300036, similar
to MHC class I tum-transplantation antigen |
| GATGCCCCCC | 85 | 126 | 1.5 | | No match |
| GTGACCACGG | 41 | 83 | 2.0 | | No match |
| ACCCGCCGGG | 7 | 17 | 2.4 | | No match |
| CTAACTAGTT | 4 | 20 | 5.0 | | No match |
| CTGGCCTGAG | 4 | 17 | 4.3 | | No match |
| CCGACGGGCG | 3 | 11 | 3.7 | | No match |
| TACACTAACC | 1 | 9 | 9.0 | | No match |
| CAGAGGTCCT | 1 | 7 | 7.0 | | No match |
| GTTCAGGGTC | 0 | 6 | 6.0 | | No match |
| CACCAGGGTG | 0 | 6 | 6.0 | | No match |
| CTTTTATAAG | 0 | 5 | 5.0 | | No match |
| CGAAGTGAAA | 0 | 5 | 5.0 | | No match |
| AAAAATCATC | 0 | 5 | 5.0 | | No match |
| TAAATTCGAT | 0 | 5 | 5.0 | | No match |
| Downregulated
(66) |
| ATAACACATA | 544 | 401 | 1.4 | 3658 | LOC287805, similar
to retinoic acid inducible protein 3 |
| CACGCCTCTC | 276 | 186 | 1.5 | 107334 | Hba1, hemoglobin,
α1 |
| TATGGCTTTA | 46 | 21 | 2.2 | 3793 | Similar to
tensin |
| AGCCATCCCT | 44 | 30 | 1.5 | 98846 | Fga, fibrinogen, α
polypeptide |
| CCAACAAGAA | 37 | 24 | 1.5 | 13685 | Tetraspanin 7 |
| TCTTCTAGAA | 33 | 15 | 2.2 | 1952 | Sftpb, surfactant,
pulmonary-associated protein B |
| ATTTGAAATA | 30 | 18 | 1.7 | 3036 | Gnai2, GTP-binding
protein (G-α-i2) |
| TGCGAATGAT | 18 | 7 | 2.6 | 93479 | LOC301563, similar
to RIKEN cDNA 5230400G24 |
| GGCTTTACCC | 18 | 6 | 3.0 | 104607 | LOC287444, similar
to Eukaryotic translation initiation factor 5A (eIF-5A) (eIF-4D)
(Rev-binding factor) |
| CCCAATGGCC | 17 | 8 | 2.1 | 11889 | Procollagen, type
VI, α2 |
| TTGCATTCCC | 16 | 5 | 3.2 | 3321 | Transcribed
sequence with weak similarity to protein sp |
| TTGAAAAAAA | 14 | 5 | 2.8 | 11330 | Uox, urate
oxidase |
| CCTCTCAAGG | 14 | 6 | 2.3 | 40119 | Ly6c, Ly6-C antigen
gene |
| GCTGAATGTC | 13 | 5 | 2.6 | 102005 | LOC287212, similar
to hypothetical protein FLJ31951 |
| ACAACTTCCT | 12 | 4 | 3.0 | 98783 | Gm2a, GM2
ganglioside activator protein |
| TATTCAAATA | 11 | 3 | 3.7 | 9954 | Tgfbr2,
transforming growth factor, β receptor II |
| TACAATAAAC | 11 | 3 | 3.7 | 7685 | LOC361940, similar
to 4631434O19Rik protein |
| TCTGGCTCCT | 10 | 2 | 5.0 | 17033 | LOC300996, similar
to RNA binding motif protein 5 |
| GACTCGAGCC | 10 | 3 | 3.3 | 54541 | Scn6a, sodium
channel, voltage-gated, type 6, α polypeptide |
| GAAATAACGG | 10 | 1 | 10.0 | 108127 | Pgk1,
phosphoglycerate kinase 1 |
| GAAAATAAAA | 10 | 3 | 3.3 | 100627 | Fibronectin type
III domain containing 3a |
| CCTTTGAATA | 10 | 3 | 3.3 | 1838 | Clic5, chloride
intracellular channel 5 |
| CCCTGATTTT | 10 | 3 | 3.3 | 103276 | Eif4g2, eukaryotic
translation initiation factor 4γ, 2 |
| TTCAGGTGGT | 9 | 2 | 4.5 | 114499 | Transcribed
sequences |
| TTCAATATTA | 9 | 2 | 4.5 | 6387 | Transcribed
sequences |
| TGCTGGACAT | 9 | 2 | 4.5 | 41063 | SREBP-2, sterol
regulatory element binding protein 2 |
| TCTACAAGAA | 8 | 1 | 8.0 | 98667 | Heat shock protein
90 kDa α (cytosolic), class B member 1 |
| GTGCTACTCC | 8 | 1 | 8.0 | 53801 | Col4a1,
procollagen, type IV, α 1 |
| CCTTCTCAGA | 8 | 1 | 8.0 | 61687 | Ppap2a,
phosphatidate phosphohydrolase type 2a |
| CAGAAAGATA | 8 | 1 | 8.0 | 50677 | LOC314336, similar
to DEAD-box protein abstrakt homolog |
| TTTGTGGGAT | 7 | 1 | 7.0 | 12550 | Nfkbia, nuclear
factor of κ light chain gene enhancer in B-cells inhibitor, α |
| TTTGATTAAA | 7 | 1 | 7.0 | 25124 | Rat insulin-like
growth factor I mRNA, 3′ end of mRNA |
| TGGTCCTTCC | 7 | 1 | 7.0 | 8180 | Neuronal
regeneration related protein |
| GTGACGTCCT | 7 | 1 | 7.0 | 65477 | Lfng, lunatic
fringe gene homolog |
| GTCCCAAGGA | 7 | 1 | 7.0 | 80835 | Dci,
dodecenoyl-coenzyme A δ isomerase |
| GCACCTCTTA | 7 | 1 | 7.0 | 95170 | LOC361522, similar
to EIB-55 kDa associated protein 5 |
| GAGTGATCCT | 7 | 1 | 7.0 | 104649 | LOC290923 |
| GAAACAACCC | 7 | 0 | 7.0 | 123755 | LRRGT00170 mRNA,
complete cds |
| ACCTTAAACC | 7 | 1 | 7.0 | 6660 | LOC361037, similar
to mitochondrial ribosomal protein L52 CG1577-PA |
| TTTTTTTGTG | 6 | 0 | 6.0 | 4108 | Ppp2r1a, protein
phosphatase 2 (formerly 2A), regulatory subunit A (PR 65), α
isoform |
| TTGAGCCAGC | 6 | 0 | 6.0 | 92643 | Marta1, MAP2 RNA
trans-acting protein MARTA1 |
| TGGGGCAGGC | 6 | 0 | 6.0 | 11887 | Transcribed
sequence with strong similarity to protein ref:NP_080275.1 (M.
musculus) RIKEN cDNA 2610110L04 |
| TACTATTTAT | 6 | 0 | 6.0 | 71377 | LOC298573, similar
to eukaryotic translation initiation factor 4 γ, 3 |
| GTGAGTAGTG | 6 | 0 | 6.0 | 98685 | Tdg, thymine-DNA
glycosylase |
| GCTCCGTGGC | 6 | 0 | 6.0 | 15842 | LOC362456, similar
to GDP-dissociation inhibitor |
| GATCAGATGG | 6 | 0 | 6.0 | 11540 | LOC289144, similar
to calcyclin binding protein |
| TTAATTTGTT | 5 | 0 | 5.0 | 11763 | Smc1l1, SMC-like 1
(yeast) |
| TGAGGAACAA | 5 | 0 | 5.0 | 40233 | LOC287063, similar
to RIKEN cDNA 1110025F24 |
| TCTAAATAAA | 5 | 0 | 5.0 | 37427 | Tceb3,
transcription elongation factor B (SIII), polypeptide 3 |
| TATTCATCAG | 5 | 0 | 5.0 | 17321 | Pank4, pantothenate
kinase 4 |
| TAAGGTTTTT | 5 | 0 | 5.0 | 8299 | Transcribed
sequences |
| GATTAAATAA | 5 | 0 | 5.0 | 40420 | LOC292766, similar
to proteasome 26S non-ATPase subunit 8 |
| GAGTCCTTCC | 5 | 0 | 5.0 | 33218 | Stx4a, syntaxin
4 |
| GAATCCAACT | 5 | 0 | 5.0 | 3377 | LOC299310, similar
to neuronal protein 15.6 |
| GAAACCGTTA | 5 | 0 | 5.0 | 3510 | Transcribed
sequences |
| CTGGTAAAAT | 5 | 0 | 5.0 | 3264 | Ssr3, TRAP-complex
γ subunit |
| CCCTTGGAAT | 5 | 0 | 5.0 | 25717 | RT1-Ba, RT1 class
II, locus Ba |
| ACCAGCTTCC | 5 | 0 | 5.0 | 101762 | LOC292949, similar
to p53 apoptosis-associated target |
| ATACTGACAC | 252 | 226 | 1.1 | | No match |
| GACTGACCCT | 23 | 8 | 2.9 | | No match |
| CTACTCGAAT | 14 | 6 | 2.3 | | No match |
| GGATGCATTT | 12 | 4 | 3.0 | | No match |
| GTGAATTCGG | 8 | 0 | 8.0 | | No match |
| AAAGAAAAAA | 5 | 0 | 5.0 | | No match |
| GGATTCGAGC | 5 | 0 | 5.0 | | No match |
| TAATAAGCTT | 5 | 0 | 5.0 | | No match |
Confirmation of SAGE results by
qRT-PCR
To confirm the expression profiles of the 2 SAGE
libraries, 3 genes of interest that were differentially expressed
before and after acupuncture were chosen, and their expression
levels were evaluated by qRT-PCR. The first gene encodes
surfactant, pulmonary-associated protein A1 (Sftpa1), which binds
to surfactant phospholipids and aids in decreasing the surface
tension at the air-liquid interface in the alveoli of the mammalian
lung. This process occurs in the presence of calcium ions and is
essential for normal respiration. The second gene encodes
procollagen, the α2 chain of collagen VI (Col6a2), which is a major
constituent of microfibrils that are found in different organs and
tissues and may play an important role in cell migration and
differentiation. The third gene encodes procollagen, the α1 chain
of collagen IV (Col4a1), which is the major structural component of
basement membranes. The expression profiles of the representative
genes by qRT-PCR analysis corresponded to the SAGE profiles
(Fig. 1), thus validating our data
and indicating that our data sets could be used for further
bioinformatic analyses.
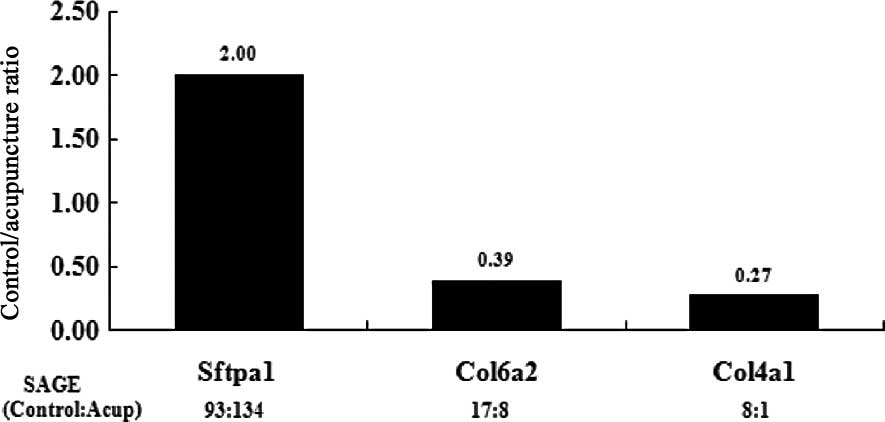 | Figure 1Quantitative real-time PCR (qRT-PCR)
confirmation of differentially regulated genes of interest, as
predicted by serial analysis of gene expression (SAGE). The
expression levels of Sftpa1 in normal rats treated with acupuncture
(Acup) were upregulated by 2-fold, when compared to the control.
The expression levels of Col6a2 and Col4a1 in the Acup group were
downregulated by >0.5-fold, when compared to the control.
Sftpa1, surfactant, pulmonary-associated protein A1; Col6a2,
procollagen, type VI, α2; Col4a1, procollagen, type IV, α1. |
DAVID gene functional classification
For the functional annotation of the differentially
expressed genes, 110 known genes and 12 ESTs with a UniGene ID were
assigned by applying the DAVID functional annotation tool to the
data sets. Of the 122 UniGenes, 105 were functionally classified
into 18 groups using the default settings (medium classification
stringency) and all GO terms for biological processes. Among these
18 groups, 6 had enrichment scores (ES) that were ≥1. These groups
included ‘cellular biosynthetic processes’ (51 genes, ES=2.04),
‘cellular metabolic processes’ (78 genes, ES=1.78), ‘cell
proliferation’ (37 genes, ES=1.58), ‘cellular lipid metabolic
processes’ (10 genes, ES=1.34), ‘defense response’ (10 genes,
ES=1.3) and ‘transport’ (25 genes, ES=1.11). The gene lists of each
group are shown in Table IV.
 | Table IVList of the Database for Annotation,
Visualization and Integrated Discovery (DAVID) gene functional
classification groups. |
Table IV
List of the Database for Annotation,
Visualization and Integrated Discovery (DAVID) gene functional
classification groups.
| Classification | Enrichment
score | UniGene | Annotation |
|---|
| Cellular
biosynthetic process | 2.04 | Rn.9954 | Transforming growth
factor, β receptor ii |
| | Rn.1997 | Cathepsin h |
| | Rn.2589 | Cysteine
dioxygenase 1, cytosolic |
| | Rn.98380 |
Pyroglutamyl-peptidase i |
| | Rn.29258 | Similar to atp
synthase, h+ transporting, mitochondrial f0 complex, subunit c
(subunit 9), isoform 2 |
| | Rn.108039 | Ribosomal protein
l14 |
| | Rn.17145 | Connective tissue
growth factor |
| | Rn.84435 | Glucose phosphate
isomerase |
| | Rn.36797 | Similar to hect
domain containing 1 |
| | Rn.82672 | Vesicle-associated
membrane protein 8 |
| | Rn.13589 | Polymerase i and
transcript release factor |
| | Rn.37427 | Transcription
elongation factor b (siii), polypeptide 3 |
| | Rn.98846 | Fibrinogen, α
polypeptide |
| | Rn.17321 | Pantothenate kinase
4 |
| | Rn.3510 | Similar to bb128963
protein |
| | Rn.2722 | Ribosomal protein
l27a |
| | Rn.103276 | Eukaryotic
translation initiation factor 4γ, 2 |
| | Rn.17033 | Rna binding motif
protein 5 |
| | Rn.61687 | Phosphatidic acid
phosphatase 2a |
| | Rn.11763 | Structural
maintenance of chromosomes 1 like 1 (S. cerevisiae) |
| | Rn.47 | Hypoxanthine
guanine phosphoribosyl transferase |
| | Rn.13322 | Brf2, subunit of
rna polymerase iii transcription initiation factor, brf1-like |
| | Rn.19267 | Peroxisomal
trans-2-enoyl-CoA reductase |
| | Rn.94935 | Ribosomal protein
s3a |
| | Rn.2759 | C-src tyrosine
kinase |
| | Rn.92643 | KH-type splicing
regulatory protein |
| | Rn.3285 | Diazepam binding
inhibitor |
| | Rn.25771 | Heterogeneous
nuclear ribonucleoprotein a1 |
| | Rn.98667 | Heat shock 90 kda
protein 1, β |
| | Rn.9406 | Necdin-like 2 |
| | Rn.8400 | Ribosomal protein
s12 |
| | Rn.33807 | Chaperonin subunit
6a (ζ) |
| | Rn.106034 | Ribosomal protein
s27a |
| | Rn.98783 | Gm2 ganglioside
activator protein |
| | Rn.29782 | Fumarate hydratase
1 |
| | Rn.4223 | Dyskeratosis
congenita 1, dyskerin |
| | Rn.71377 | Eukaryotic
translation initiation factor 4γ, 3 |
| | Rn.5106 |
3-Hydroxy-3-methylglutaryl-coenzyme a
synthase 1 |
| | Rn.3910 | Eukaryotic
translation elongation factor 1, β2 |
| | Rn.2989 | Homeobox only
domain |
| | Rn.11540 | Similar to
calcyclin binding protein |
| | Rn.94978 | Actin, β |
| | Rn.41063 | Sterol regulatory
element binding factor 2 |
| | Rn.104649 |
Aspartylglucosaminidase |
| | Rn.6606 | Tissue specific
transplantation antigen p35b |
| | Rn.1439 | Ribosomal protein
l36 |
| | Rn.98685 | Thymine-dna
glycosylase |
| | Rn.107334 | Hemoglobin α, adult
chain 1 |
| | Rn.109735 | Ribosomal protein
s9 |
| | Rn.2262 | Ribosomal protein
l10a |
| | Rn.108127 | Phosphoglycerate
kinase 1 |
| Cellular metabolic
process | 1.78 | Rn.9954 | Transforming growth
factor, β receptor ii |
| | Rn.1997 | Cathepsin h |
| | Rn.2589 | Cysteine
dioxygenase 1, cytosolic |
| | Rn.29258 | Similar to atp
synthase, h+ transporting, mitochondrial f0 complex, subunit c
(subunit 9), isoform 2 |
| | Rn.98380 |
Pyroglutamyl-peptidase i |
| | Rn.55036 | Ciliary
neurotrophic factor receptor |
| | Rn.108039 | Ribosomal protein
l14 |
| | Rn.17145 | Connective tissue
growth factor |
| | Rn.3793 | Similar to
tensin |
| | Rn.84435 | Glucose phosphate
isomerase |
| | Rn.8509 | Transmembrane
trafficking protein 21 |
| | Rn.36797 | Similar to hect
domain containing 1 |
| | Rn.82672 | Vesicle-associated
membrane protein 8 |
| | Rn.13589 | Polymerase i and
transcript release factor |
| | Rn.37427 | Transcription
elongation factor b (siii), polypeptide 3 |
| | Rn.98846 | Fibrinogen, α
polypeptide |
| | Rn.108012 |
Lysosomal-associated protein transmembrane
4a |
| | Rn.17321 | Pantothenate kinase
4 |
| | Rn.3510 | Similar to bb128963
protein |
| | Rn.103276 | Eukaryotic
translation initiation factor 4γ, 2 |
| | Rn.2722 | Ribosomal protein
l27a |
| | Rn.17033 | Rna binding motif
protein 5 |
| | Rn.3264 | Signal sequence
receptor, γ |
| | Rn.11763 | Structural
maintenance of chromosomes 1 like 1 (S. cerevisiae) |
| | Rn.61687 | Phosphatidic acid
phosphatase 2a |
| | Rn.47 | Hypoxanthine
guanine phosphoribosyl transferase |
| | Rn.13322 | Brf2, subunit of
rna polymerase iii transcription initiation factor, brf1-like |
| | Rn.19267 | Peroxisomal
trans-2-enoyl-CoA reductase |
| | Rn.2759 | C-src tyrosine
kinase |
| | Rn.94935 | Ribosomal protein
s3a |
| | Rn.105953 | Tensin like c1
domain containing phosphatase |
| | Rn.101762 | Perp, tp53
apoptosis effector |
| | Rn.92643 | KH-type splicing
regulatory protein |
| | Rn.3285 | Diazepam binding
inhibitor |
| | Rn.3036 | Guanine nucleotide
binding protein, α inhibiting 2 |
| | Rn.32080 | Allograft
inflammatory factor 1 |
| | Rn.44465 | Chemokine-like
receptor 1 |
| | Rn.1677 | G protein-coupled
receptor 56 |
| | Rn.25771 | Heterogeneous
nuclear ribonucleoprotein a1 |
| | Rn.98667 | Heat shock 90 kda
protein 1, β |
| | Rn.9406 | Necdin-like 2 |
| | Rn.8400 | Ribosomal protein
s12 |
| | Rn.36610 | Tumor protein,
translationally-controlled 1 |
| | Rn.22087 | Interferon induced
transmembrane protein 1 |
| | Rn.9829 | Advanced
glycosylation end product-specific receptor |
| | Rn.2776 | Beclin 1
(coiled-coil, myosin-like bcl2-interacting protein) |
| | Rn.33807 | Chaperonin subunit
6a (ζ) |
| | Rn.106034 | Ribosomal protein
s27a |
| | Rn.98783 | Gm2 ganglioside
activator protein |
| | Rn.29782 | Fumarate hydratase
1 |
| | Rn.1952 | Surfactant
associated protein b |
| | Rn.4223 | Dyskeratosis
congenita 1, dyskerin |
| | Rn.71377 | Eukaryotic
translation initiation factor 4γ, 3 |
| | Rn.80835 | Dodecenoyl-coenzyme
a δ isomerase |
| | Rn.5106 |
3-Hydroxy-3-methylglutaryl-coenzyme a
synthase 1 |
| | Rn.3910 | Eukaryotic
translation elongation factor 1 β2 |
| | Rn.2989 | Homeobox only
domain |
| | Rn.11540 | Similar to
calcyclin binding protein |
| | Rn.94978 | Actin, β |
| | Rn.55487 | Guanine nucleotide
binding protein (g protein) |
| | Rn.2283 | Lysozyme |
| | Rn.41063 | Sterol regulatory
element binding factor 2 |
| | Rn.104649 |
Aspartylglucosaminidase |
| | Rn.12550 | Nuclear factor of κ
light chain gene enhancer in b-cells inhibitor, α |
| | Rn.6606 | Tissue specific
transplantation antigen p35b |
| | Rn.1439 | Ribosomal protein
l36 |
| | Rn.11330 | Urate oxidase |
| | Rn.98685 | Thymine-dna
glycosylase |
| | Rn.107334 | Hemoglobin α, adult
chain 1 |
| | Rn.10696 |
Phosphatidylserine-specific phospholipase
a1 |
| | Rn.2694 | Melanoma cell
adhesion molecule |
| | Rn.109735 | Ribosomal protein
s9 |
| | Rn.15842 | Rho, gdp
dissociation inhibitor (gdi) β |
| | Rn.2262 | Ribosomal protein
l10a |
| | Rn.108127 | Phosphoglycerate
kinase 1 |
| | Rn.5782 | Syntaxin 5a |
| | Rn.3658 | G protein-coupled
receptor, family c, group 5, member c |
| | Rn.33218 | Syntaxin 4a
(placental) |
| Cell
proliferation | 1.58 | Rn.22087 | Interferon induced
transmembrane protein 1 |
| | Rn.9829 | Advanced
glycosylation end product-specific receptor |
| | Rn.9954 | Transforming growth
factor, β receptor ii |
| | Rn.2776 | Beclin 1
(coiled-coil, myosin-like bcl2-interacting protein) |
| | Rn.59630 | Similar to riken
cdna b230339h12 |
| | Rn.106034 | Ribosomal protein
s27a |
| | Rn.55036 | Ciliary
neurotrophic factor receptor |
| | Rn.98783 | Gm2 ganglioside
activator protein |
| | Rn.1952 | Surfactant
associated protein b |
| | Rn.17145 | Connective tissue
growth factor |
| | Rn.4223 | Dyskeratosis
congenita 1, dyskerin |
| | Rn.84435 | Glucose phosphate
isomerase |
| | Rn.71377 | Eukaryotic
translation initiation factor 4γ, 3 |
| | Rn.11343 | Surfactant,
pulmonary-associated protein a1 |
| | Rn.13589 | Polymerase i and
transcript release factor |
| | Rn.2989 | Homeobox only
domain |
| | Rn.37427 | Transcription
elongation factor b (siii), polypeptide 3 |
| | Rn.98846 | Fibrinogen, α
polypeptide |
| | Rn.55487 | Guanine nucleotide
binding protein (g protein) |
| | Rn.41063 | Sterol regulatory
element binding factor 2 |
| | Rn.11763 | Structural
maintenance of chromosomes 1 like 1 (S. cerevisiae) |
| | Rn.61687 | Phosphatidic acid
phosphatase 2a |
| | Rn.47 | Hypoxanthine
guanine phosphoribosyl transferase |
| | Rn.13322 | Brf2, subunit of
rna polymerase iii transcription initiation factor, brf1-like |
| | Rn.12550 | Nuclear factor of κ
light chain gene enhancer in b-cells inhibitor, α |
| | Rn.94935 | Ribosomal protein
s3a |
| | Rn.2759 | C-src tyrosine
kinase |
| | Rn.92643 | KH-type splicing
regulatory protein |
| | Rn.101762 | Perp, tp53
apoptosis effector |
| | Rn.98685 | Thymine-dna
glycosylase |
| | Rn.107334 | Hemoglobin α, adult
chain 1 |
| | Rn.3036 | Guanine nucleotide
binding protein, α inhibiting 2 |
| | Rn.32080 | Allograft
inflammatory factor 1 |
| | Rn.15842 | Rho, gdp
dissociation inhibitor (gdi) β |
| | Rn.9406 | Necdin-like 2 |
| | Rn.33218 | Syntaxin 4a
(placental) |
| | Rn.36610 | Tumor protein,
translationally-controlled 1 |
| Cellular lipid
metabolic process | 1.34 | Rn.3285 | Diazepam binding
inhibitor |
| | Rn.10696 |
Phosphatidylserine-specific phospholipase
a1 |
| | Rn.1952 | Surfactant
associated protein b |
| | Rn.3510 | Similar to bb128963
protein |
| | Rn.41063 | Sterol regulatory
element binding factor 2 |
| | Rn.61687 | Phosphatidic acid
phosphatase 2a |
| | Rn.5106 |
3-Hydroxy-3-methylglutaryl-coenzyme a
synthase 1 |
| | Rn.80835 | Dodecenoyl-coenzyme
a δ isomerase |
| | Rn.19267 | Peroxisomal
trans-2-enoyl-CoA reductase |
| | Rn.98783 | Gm2 ganglioside
activator protein |
| Defense
response | 1.3 | Rn.9829 | Advanced
glycosylation end product-specific receptor |
| | Rn.22087 | Interferon induced
transmembrane protein 1 |
| | Rn.9954 | Transforming growth
factor, β receptor ii |
| | Rn.32080 | Allograft
inflammatory factor 1 |
| | Rn.2589 | Cysteine
dioxygenase 1, cytosolic |
| | Rn.2283 | Lysozyme |
| | Rn.98667 | Heat shock 90 kda
protein 1, β |
| | Rn.2776 | Beclin 1
(coiled-coil, myosin-like bcl2-interacting protein) |
| | Rn.12550 | Nuclear factor of κ
light chain gene enhancer in b-cells inhibitor, α |
| | Rn.2267 | Defensin β3 |
| Transport | 1.11 | Rn.9829 | Advanced
glycosylation end product-specific receptor |
| | Rn.9954 | Transforming growth
factor, β receptor ii |
| | Rn.54541 | Sodium channel,
voltage-gated, type 6, α polypeptide |
| | Rn.29258 | Similar to atp
synthase, h+ transporting, mitochondrial f0 complex, subunit c
(subunit 9), isoform 2 |
| | Rn.25717 | Butyrophilin-like 2
(mhc class ii associated) |
| | Rn.98783 | Gm2 ganglioside
activator protein |
| | Rn.17145 | Connective tissue
growth factor |
| | Rn.3793 | Similar to
tensin |
| | Rn.8509 | Transmembrane
trafficking protein 21 |
| | Rn.1838 | Chloride
intracellular channel 5 |
| | Rn.82672 | Vesicle-associated
membrane protein 8 |
| | Rn.11343 | Surfactant,
pulmonary-associated protein a1 |
| | Rn.108012 |
Lysosomal-associated protein transmembrane
4a |
| | Rn.3264 | Signal sequence
receptor, γ |
| | Rn.12550 | Nuclear factor of κ
light chain gene enhancer in b-cells inhibitor, α |
| | Rn.92643 | KH-type splicing
regulatory protein |
| | Rn.3285 | Diazepam binding
inhibitor |
| | Rn.3036 | Guanine nucleotide
binding protein, α inhibiting 2 |
| | Rn.32080 | Allograft
inflammatory factor 1 |
| | Rn.107334 | Hemoglobin α, adult
chain 1 |
| | Rn.25771 | Heterogeneous
nuclear ribonucleoprotein a1 |
| | Rn.15842 | Rho, gdp
dissociation inhibitor (gdi) β |
| | Rn.5782 | Syntaxin 5a |
| | Rn.33218 | Syntaxin 4a
(placental) |
| | Rn.36610 | Tumor protein,
translationally-controlled 1 |
Analysis of gene categories by GO Tree
Machine
In total, 10 GO categories were enriched after
acupuncture, and these categories included ‘biosynthesis’ (16
genes), ‘cellular biosynthesis’ (14 genes), ‘macromolecule
biosynthesis’ (11 genes), ‘protein biosynthesis’ (11 genes),
‘protein kinase C activation’ (2 genes), ‘cytolysis’ (2 genes),
‘vesicle targeting’ (2 genes), ‘regulation of liquid surface
tension’ (2 genes), ‘mRNA transport’ (2 genes) and ‘mRNA export
from the nucleus’ (2 genes). The gene lists of each category are
shown in the Table V.
 | Table VGene lists of categories by the Gene
Ontology (GO) Tree Machine. |
Table V
Gene lists of categories by the Gene
Ontology (GO) Tree Machine.
| Enriched GO
categories | Gene nos. | UniGene | Annotation |
|---|
| Biosynthesis | 16 | Rn.92643 | KH-type splicing
regulatory protein |
| | Rn.47 | Hypoxanthine
guanine phosphoribosyl transferase |
| | Rn.3285 | Diazepam binding
inhibitor |
| | Rn.2722 | Ribosomal protein
L27a |
| | Rn.40171 | Ribosomal protein
S4, X-linked |
| | Rn.5106 |
3-Hydroxy-3-methylglutaryl-Coenzyme A
synthase 1 |
| | Rn.71377 | Eukaryotic
translation initiation factor 4γ, 3 |
| | Rn.6606 | Tissue specific
transplantation antigen P35B |
| | Rn.103276 | Eukaryotic
translation initiation factor 4, γ2 |
| | Rn.3910 | Eukaryotic
translation elongation factor 1 β2 |
| | Rn.61687 | Phosphatidic acid
phosphatase 2a |
| | Rn.108039 | Ribosomal protein
L14 |
| | Rn.8400 | Ribosomal protein
S12 |
| | Rn.2589 | Cysteine
dioxygenase 1, cytosolic |
| | Rn.109735 | Ribosomal protein
S9 |
| | Rn.106034 | Ribosomal protein
S27a |
| Cellular
biosynthesis | 14 | Rn.92643 | KH-type splicing
regulatory protein |
| | Rn.47 | Hypoxanthine
guanine phosphoribosyl transferase |
| | Rn.2722 | Ribosomal protein
L27a |
| | Rn.40171 | Ribosomal protein
S4, X-linked |
| | Rn.71377 | Eukaryotic
translation initiation factor 4 gamma, 3 |
| | Rn.6606 | Tissue specific
transplantation antigen P35B |
| | Rn.103276 | Eukaryotic
translation initiation factor 4, γ2 |
| | Rn.3910 | Eukaryotic
translation elongation factor 1 β2 |
| | Rn.61687 | Phosphatidic acid
phosphatase 2a |
| | Rn.108039 | Ribosomal protein
L14 |
| | Rn.8400 | Ribosomal protein
S12 |
| | Rn.2589 | Cysteine
dioxygenase 1, cytosolic |
| | Rn.109735 | Ribosomal protein
S9 |
| | Rn.106034 | Ribosomal protein
S27a |
| Macromolecule
biosynthesis | 11 | Rn.106034 | KH-type splicing
regulatory protein |
| | Rn.2722 | Ribosomal protein
L27a |
| | Rn.40171 | Ribosomal protein
S4, X-linked |
| | Rn.71377 | Eukaryotic
translation initiation factor 4γ, 3 |
| | Rn.6606 | Tissue specific
transplantation antigen P35B |
| | Rn.103276 | Eukaryotic
translation initiation factor 4, γ2 |
| | Rn.3910 | Eukaryotic
translation elongation factor 1 β2 |
| | Rn.108039 | Ribosomal protein
L14 |
| | Rn.8400 | Ribosomal protein
S12 |
| | Rn.109735 | Ribosomal protein
S9 |
| | Rn.106034 | Ribosomal protein
S27a |
| Protein
biosynthesis | 11 | Rn.92643 | KH-type splicing
regulatory protein |
| | Rn.2722 | Ribosomal protein
L27a |
| | Rn.40171 | Ribosomal protein
S4, X-linked |
| | Rn.71377 | Eukaryotic
translation initiation factor 4γ, 3 |
| | Rn.6606 | Tissue specific
transplantation antigen P35B |
| | Rn.103276 | Eukaryotic
translation initiation factor 4, γ2 |
| | Rn.3910 | Eukaryotic
translation elongation factor 1 β2 |
| | Rn.108039 | Ribosomal protein
L14 |
| | Rn.8400 | Ribosomal protein
S12 |
| | Rn.109735 | Ribosomal protein
S9 |
| | Rn.106034 | Ribosomal protein
S27a |
| Protein kinase C
activation | 2 | Rn.61687 | Phosphatidic acid
phosphatase 2a |
| | Rn.55487 | Guanine nucleotide
binding protein (G protein, β polypeptide 2 like 1) |
| Cytolysis | 2 | Rn.47 | Hypoxanthine
guanine phosphoribosyl transferase |
| | Rn.2283 | Lysozyme |
| Vesicle
targeting | 2 | Rn.5782 | Syntaxin 5a |
| | Rn.8509 | Transmembrane
emp24-like trafficking protein 10 (yeast) |
| Regulation of
liquid surface tension | 2 | Rn.1952 | Surfactant
associated protein B |
| | Rn.11343 | Surfactant,
pulmonary-associated protein A1 |
| mRNA transport | 2 | Rn.92643 | KH-type splicing
regulatory protein |
| | Rn.25771 | Heterogeneous
nuclear ribonucleoprotein A1 |
| mRNA export from
nucleus | 2 | Rn.92643 | KH-type splicing
regulatory protein |
| | Rn.25771 | Heterogeneous
nuclear ribonucleoprotein A1 |
Finding KEGG pathways
To understand the functional roles of the
differentially expressed genes, KEGG pathway analysis was assigned
by applying the DAVID annotation tool. Fifteen KEGG pathways
matched the differentially expressed tags of the 2 libraries
(Table VI). Two of these
pathways, ‘soluble (N-ethylmaleimide-sensitive fusion) NSF
attachment protein receptor (SNARE) interactions in vesicular
transport’ and ‘tight junction’ pathways, were matched to 3
UniGenes. SNAREs are small, abundant, plasma membrane-bound
proteins; the cyclic assembly and disassembly of the SNARE complex
is required for regulated secretory vesicle fusion with the plasma
membrane. Tight junctions are the closely associated areas between
2 cells whose membranes join to form a virtually impermeable
barrier to fluid.
 | Table VIKyoto Encyclopedia of Genes and
Genomes (KEGG) pathways of differentially expressed tags. |
Table VI
Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathways of differentially expressed tags.
| Pathway name | Gene nos. | UniGene | Annotation |
|---|
| SNARE interactions
in vesicular transport | 3 | Rn.82672 | Vesicle-associated
membrane protein 8 |
| | Rn.5782 | Syntaxin 5a |
| | Rn.33218 | Syntaxin 4a |
| Tight junction | 3 | Rn.3036 | Guanine nucleotide
binding protein, α inhibiting 2 |
| | Rn.94978 | Actin, β |
| | Rn.4108 | Protein phosphatase
2 (formerly 2a), regulatory subunit a (pr 65), α isoform |
| ECM-receptor
interaction | 2 | Rn.53801 | Procollagen, type
iv, α1 |
| | Rn.100627 | Fibronectin type
iii domain containing 3 |
| Adherens
junction | 2 | Rn.9954 | Transforming growth
factor, β receptor ii |
| | Rn.94978 | Actin, β |
| Long-term
depression | 2 | Rn.3036 | Guanine nucleotide
binding protein, α inhibiting 2 |
| | Rn.4108 | Protein phosphatase
2 (formerly 2a), regulatory subunit a (pr 65), α isoform |
| Small cell lung
cancer | 2 | Rn.53801 | Procollagen, type
iv, α1 |
| | Rn.12550 | Nuclear factor of κ
light chain gene enhancer in b-cells inhibitor, α |
| TGF-β signaling
pathway | 2 | Rn.9954 | Transforming growth
factor, β receptor ii |
| | Rn.4108 | Protein phosphatase
2 (formerly 2a), regulatory subunit a (pr 65), α isoform |
| Chronic myeloid
leukemia | 2 | Rn.9954 | Transforming growth
factor, β receptor ii |
| | Rn.12550 | Nuclear factor of κ
light chain gene enhancer in b-cells inhibitor, α |
| Antigen processing
and presentation | 2 | Rn.98667 | Heat shock 90 kda
protein 1, β |
| | Rn.25717 | Butyrophilin-like 2
(mhc class ii associated) |
| Purine
metabolism | 2 | Rn.11330 | Urate oxidase |
| | Rn.47 | Hypoxanthine
guanine phosphoribosyl transferase |
| Leukocyte
transendothelial migration | 2 | Rn.3036 | Guanine nucleotide
binding protein, α inhibiting 2 |
| | Rn.94978 | Actin, β |
| Cell
communication | 2 | Rn.94978 | Actin, β |
| | Rn.53801 | Procollagen, type
iv, α1 |
| Cytokine-cytokine
receptor interaction | 2 | Rn.9954 | Transforming growth
factor, β receptor ii |
| | Rn.55036 | Ciliary
neurotrophic factor receptor |
| Focal adhesion | 2 | Rn.94978 | Actin, β |
| | Rn.53801 | Procollagen, type
iv, α1 |
| Regulation of actin
cytoskeleton | 2 | Rn.94978 | Actin, β |
| | Rn.2759 | C-src tyrosine
kinase |
Discussion
A number of experiments and theories have been used
to explain the possible mechanisms of acupuncture, which are
believed to have a strong biological basis (4). In contrast to target-specific and
one-way adjustment chemical reagents, acupuncture upregulates
various organ systems that are hypofunctional, or downregulates
hyperfunctional systems. Acupuncture is considered as a mechanical
activation and signaling process (25). The observation and identification
of the global alterations in gene expression in response to
acupuncture that are involved in specific physiological processes
will help to integrate the knowledge of the underlying signaling
and metabolic pathways, biosynthetic processes and the functional
interactions of cells, tissues and organs (26). The present study provides abundant
experimental data and reveals the background gene expression
profile in rats before and after acupuncture treatment. This
information highlights the active gene alterations in a particular
disease.
DAVID functional classification suggested that the
genes involved in metabolism were regulated after acupuncture
intervention. These genes included those with roles in ‘cellular
biosynthetic processes’, ‘cellular lipid metabolic processes’ and
‘transport’. The corresponding regulated genes included the
following: glucose phosphate isomerase, β-actin and lysozyme.
Glucose phosphate isomerase is an enzyme that catalyzes the
conversion of glucose-6-phosphate into fructose-6-phosphate in the
second step of glycolysis. β-actin is one of 6 different actin
isoforms that have been identified in humans and is one of most
important non-muscle, cytoskeletal actins that have highly
conserved roles in cell motility, structure and integrity. Lysozyme
is an enzyme that damages the cell walls of bacteria, which are
abundant in a number of secretions, such as tears, saliva, human
milk and mucus. Finding these genes in our study indicates that
dynamic changes occur involving various biological processes and
that protein, lipid and sugar synthesis are adjusted separately
after acupuncture. In this way, acupuncture completes the
regulation of nerve and body fluids.
The GO Tree Machine analysis of the gene categories
suggested that acupuncture influences the processes of RNA
transcription and protein translation. In the SAGE libraries, genes
known as ‘KH-type splicing regulatory protein (KHSRP)’ and
‘heterogeneous nuclear ribonucleoprotein A1’ belong to the gene
categories of ‘mRNA transport’ and ‘mRNA export from the nucleus’.
This finding indicates that acupuncture may influence the
expression of these 2 genes to regulate the movement of mRNA from
the nucleus to the cytoplasm. By doing so, acupuncture controls
mRNA expression and protein translation at its source. The category
termed ‘regulation of liquid surface tension’ included 2 genes,
‘Sftpa1’ and ‘surfactant associated protein C’. After inserting the
needle into the point of feishu (BL13), the expression of these
proteins was increased. This indicated that the change in
expression may be a specific effect of the selected acupuncture
point.
Fifteen pathways were identified as regulated by
acupuncture treatment using KEGG pathway analysis. One of these
pathways was ‘SNARE interactions in vesicular transport’. SNARE
proteins are a large superfamily of proteins that consists of more
than 60 members in yeast and mammalian cells, and play an integral
part in membrane fusion events in the secretory and endocytic
pathways (27). Syntaxin 5a was
found in our analysis to be regulated in response to acupuncture.
This protein is a member of the integrated SNARE proteins that
participate in exocytosis, which mediates endoplasmic reticulum to
Golgi transport. These results suggest that acupuncture may
activate the processes of membrane fusion and intercellular signal
transduction, which regulate the transport of substances into or
out of cells.
KEGG pathway selection provides a platform for
integrating and elucidating useful data. The matching of pathways,
such as ‘tight junction’, ‘adherens junction’, ‘focal adhesion’,
‘cell communication’, ‘cytokine-cytokine receptor interaction’ and
‘regulation of actin cytoskeleton’, in our research suggests that
the process of acupuncture activates intercellular signal
transduction. Tight and adherens junctions are 2 types of
intercellular junctions in vertebrates (28). The local electrical activity of the
cell membrane, interactions with the cytoskeleton or the activation
of certain receptors may play a part in activating these signaling
pathways. Based on our data, we suggest that interactions between
cells and re-arrangement of the cytoskeleton occur after
acupuncture, and signals are transferred from one cell to another.
The process of cell communication may last for a certain time, and
then cells relay the message and certain processes commence, such
as new mRNA transcription, synthesizing macromolecular proteins and
enzymes, and conducting purine metabolism. β-actin and the c-src
tyrosine kinase were 2 genes that were regulated by acupuncture.
β-actin is an important component of the cytoskeleton, and it
participates in the local signal exchange between cells, their
surroundings or with other cells. c-src tyrosine kinase plays a
role in phosphorylation and acidification in cell-cell interactions
(29), which also controls the
dynamic actin cytoskeleton (30).
The results from our study suggest that acupuncture may regulate
intercellular signal transduction in normal rat lung tissue through
specific genes, such as β-actin and c-src tyrosine kinase.
We found that the gene expression profile in
response to acupuncture under normal conditions shares similar
DAVID gene functional classifications and GO categories with the
gene expression profiles of asthmatic rats in response to
acupuncture (20). In the DAVID
gene functional classifications, the categories entitled ‘cellular
biosynthetic process’, ‘cellular lipid metabolic process’ and
‘cellular process’ were found to change in response to acupuncture
in normal and asthmatic rats. Furthermore, in the GO Tree Machine
analysis, the gene categories entitled ‘regulation of liquid
surface tension’ and ‘biosynthesis’ were found to be regulated by
acupuncture in both types of rats. However, the genes were
different under the same functional classification and enriched GO
category of biological process. Therefore, this suggests that
acupuncture initiates different branches of the same biological
processes under normal and morbid conditions. This comparison
demonstrates that the cellular biosynthesis and cellular lipid
metabolism are the common regulations of biological processes in
response to acupuncture in normal and asthmatic rats.
In this study, we present the gene expression
profiles of lung tissues derived from normal and
acupuncture-treated, normal rats by SAGE analysis. A series of
physiological alterations occurs after acupuncture treatment, and
the essential effects of acupuncture include the regulation of
biosynthesis, transportation and metabolism. Acupuncture
orchestrates the activity of an organism by regulating the
expression of specific genes. Cellular biosynthesis and cellular
lipid metabolism are the common regulations of biological processes
in response to acupuncture under normal and morbid conditions,
which may be the general physiological effects of acupuncture.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (no. 30873299, 90409014, 81001548,
81173341, 81173332), the ‘Chen Guang’ Project supported by the
Shanghai Municipal Education Commission and Shanghai Education
Development Foundation (10CG45), the Shanghai Leading Academic
Discipline Project (S30304), and the Key Program of the State
Administration of Traditional Chinese Medicine of China.
Abbreviations:
|
DAVID
|
Database for Annotation, Visualization
and Integrated Discovery
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
References
|
1
|
Use of acupuncture in modern health care.
WHO Chron. 34:294–301. 1980.
|
|
2
|
World Health Organization. Acupuncture:
Review and Analysis of Reports on Controlled Clinical Trials.
2002
|
|
3
|
Acupuncture. NIH Consens Statement.
15:1–34. 1997.
|
|
4
|
NIH Consensus Development Panel on
Acupuncture: Acupuncture. JAMA. 280:1518–1524. 1998.
|
|
5
|
Dean-Clower E, Doherty-Gilman AM,
Keshaviah A, et al: Acupuncture as palliative therapy for physical
symptoms and quality of life for advanced cancer patients. Integr
Cancer Ther. 9:158–167. 2010.
|
|
6
|
Ouyang H and Chen JD: Review article:
therapeutic roles of acupuncture in functional gastrointestinal
disorders. Aliment Pharmacol Ther. 20:831–841. 2004.
|
|
7
|
Ernst E and Lee MS: Acupuncture for
rheumatic conditions: an overview of systematic reviews.
Rheumatology (Oxford). 49:1957–1961. 2010.
|
|
8
|
Han JS: Acupuncture analgesia: areas of
consensus and controversy. Pain. 152:S41–S48. 2011.
|
|
9
|
Irnich D, Salih N, Offenbacher M and
Fleckenstein J: Is sham laser a valid control for acupuncture
trials. Evid Based Complement Alternat Med. 2011:4859452011.
|
|
10
|
Hammerschlag R: The physiology of
acupuncture. Contemporary Chinese Medicine and Acupuncture. Cassidy
CM: Churchill Livingstone; 2001
|
|
11
|
Hammerschlag R, Langevin HM, Lao L and
Lewith G: Physiological dynamics of acupuncture: correlations and
mechanisms. Acupuncture Research: Strategies for Establishing an
Evidence Base. MacPherson H: Elsevier; Philadelphia, PA: 2007
|
|
12
|
Huang Y, Li TL, Lai XS, et al: Functional
brain magnetic resonance imaging in healthy people receiving
acupuncture at Waiguan versus Waiguan plus Yanglingquan points: a
randomized controlled trial. Zhong Xi Yi Jie He Xue Bao. 7:527–531.
2009.
|
|
13
|
Yamaguchi N, Takahashi T, Sakuma M, et al:
Acupuncture regulates leukocyte subpopulations in human peripheral
blood. Evid Based Complement Alternat Med. 4:447–453. 2007.
|
|
14
|
Lin ZP, Lan LW, He TY, et al: Effects of
acupuncture stimulation on recovery ability of male elite
basketball athletes. Am J Chin Med. 37:471–481. 2009.
|
|
15
|
Pariente J, White P, Frackowiak RS and
Lewith G: Expectancy and belief modulate the neuronal substrates of
pain treated by acupuncture. Neuroimage. 25:1161–1167. 2005.
|
|
16
|
Vickland V, Rogers C, Craig A and Tran Y:
Anxiety as a factor influencing physiological effects of
acupuncture. Complement Ther Clin Pract. 15:124–128. 2009.
|
|
17
|
Kim SK and Bae H: Acupuncture and immune
modulation. Auton Neurosci. 157:38–41. 2010.
|
|
18
|
Vickland V, Rogers C, Craig A and Tran Y:
Electrodermal activity as a possible physiological marker for
acupuncture. Complement Ther Clin Pract. 14:83–89. 2008.
|
|
19
|
Velculescu VE, Zhang L, Zhou W, et al:
Characterization of the yeast transcriptome. Cell. 88:243–251.
1997.
|
|
20
|
Yin LM, Jiang GH, Wang Y, et al: Use of
serial analysis of gene expression to reveal the specific
regulation of gene expression profile in asthmatic rats treated by
acupuncture. J Biomed Sci. 16:462009.
|
|
21
|
Yin LM, Jiang GH, Wang Y, et al: Serial
analysis of gene expression in a rat lung model of asthma.
Respirology. 13:972–982. 2008.
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
|
|
23
|
Dennis G Jr, Sherman BT, Hosack DA, et al:
DAVID: Database for annotation, visualization, and integrated
discovery. Genome Biol. 4:P32003.
|
|
24
|
Zhang B, Schmoyer D, Kirov S and Snoddy J:
GO Tree Machine (GOTM): a web-based platform for interpreting sets
of interesting genes using Gene Ontology hierarchies. BMC
Bioinformatics. 5:16–23. 2004.
|
|
25
|
Yang ES, Li PW, Nilius B and Li G: Ancient
Chinese medicine and mechanistic evidence of acupuncture
physiology. Pflugers Arch. 462:645–653. 2011.
|
|
26
|
Strange K: From genes to integrative
physiology: ion channel and transporter biology in
Caenorhabditis elegans. Physiol Rev. 83:377–415. 2003.
|
|
27
|
Lundeberg T and Stener-Victorin E: Is
there a physiological basis for the use of acupuncture in pain?
International Congress Series. 1238:3–10. 2002.
|
|
28
|
Bazzoni G and Dejana E: Endothelial
cell-to-cell junctions: molecular organization and role in vascular
homeostasis. Physiol Rev. 84:869–901. 2004.
|
|
29
|
Lau AF: c-Src: bridging the gap between
phosphorylation- and acidification-induced gap junction channel
closure. Sci STKE. 2005:pe332005.
|
|
30
|
Avizienyte E, Keppler M, Sandilands E, et
al: An active Src kinase-beta-actin association is linked to actin
dynamics at the periphery of colon cancer cells. Exp Cell Res.
313:3175–3188. 2007.
|















