Introduction
Hepatocellular carcinoma (HCC) is the most common
type of malignant tumor and the leading cause of cancer-related
mortality in the Guangxi Zhuang Autonomous Region of China
(1). Epidemiological observations
have shown that hepatitis B virus (HBV) infection and exposure to
aflatoxin B1 are two major etiological risk factors for the high
prevalence of HCC in this area (2–3).
However, only a fraction of individuals who are hepatitis B surface
antigen (HBsAg) carriers and have chronic aflatoxin exposure
develop HCC later in life (4). The
exact mechanism of hepatocarcinogenesis is not well understood and
the risk factors for HCC require further investigation.
Interleukin-23 (IL-23) is an inflammatory
hemopoietic cytokine comprised of a heterodimer of p40/p19, which
shares a common p40 subunit with IL-12 (5). IL-23 is secreted predominantly by
activated dendritic cells and phagocytic cells. Its main function
is to induce T cells or dendritic cells to produce IL-17, IL-6 and
IFN-γ. The IL-23 receptor (IL-23R) complex is comprised of IL-23R
and IL-12Rβ1, which is also a component of the IL-12 receptor.
IL-23R is highly expressed on the surface of T cells, monocyte
cells and natural killer T cells, and has been identified to be
involved in several chronic inflammatory diseases (6–7). The
IL-23R gene is located on chromosome 1p31. Polymorphisms of
the IL-23R gene have been reported to be associated with
susceptibility to chronic inflammatory diseases, including
inflammtory bowel disease (IBD) (8–9),
psoriasis (10) and multiple
sclerosis (11). IL-23R
gene polymorphisms have been found to be associated with gastric
cancer (12–13), esophageal cancer (14), colorectal carcinoma (15) and oral cancer (16), indicating that IL-23R gene
polymorphisms may represent one of the genetic factors contributing
to the regulation of the IL-23 signaling pathway and thus
potentially modulating susceptibility to inflammatory cancers. HCC
is a typical inflammation-associated malignancy and the
pathological stage from hepatitis to HCC is a chronic inflammatory
process (17). In the present
study, a hospital-based case-control study was conducted to
investigate whether IL-23R gene polymorphisms contribute to
susceptibility to HBV-infected diseases in a Guangxi population in
China.
Materials and methods
Study populations
The current study was designed as a retrospective
study. The study consisted of 84 patients with HBV-related HCC (73
males and 11 females; mean age, 49.5 years old), 57 patients with
HBV-induced liver cirrhosis (LC; 44 males and 13 females; mean age,
46.0 years old) and 87 patients with chronic hepatitis B (CHB; 74
males and 13 females; mean age, 40.3 years old). Patients with
HBV-related HCC had not undergone previous radiotherapy or
chemotherapy. All patients with HBV-infected disease were
consecutively selected from the First Affiliated Hospital of
Guangxi Medical University (Guangxi, China) between January and
July 2011. Control subjects were matched to the patients on the
basis of age and were selected accordingly from a group of 97
control subjects (71 males and 23 females; mean age, 42.0 years
old). Control subjects underwent a routine medical check-up in the
outpatient clinic of the Department of Internal Medicine (First
Affiliated Hospital of Guangxi Medical University, Guangxi, China)
between January and July 2011. According to thorough clinical and
laboratory evaluation, there was no personal or family history of
cancer or other serious diseases observed.
Following a period of at least 6 months with
elevated alanine aminotransferase or aspartate aminotransferase
(>40 IU/ml), CHB was defined as positive for HBsAg. LC was
diagnosed based on pathological examinations or typical
morphological observations from computed tomography (CT) or
ultrasonography and laboratory features. In the HCC group, newly
diagnosed HCC patients were included and patients with a medical
history of HCC or other cancers were excluded. Diagnosis of
HBV-related HCC was based on histological or cytological findings,
or on elevated serum α-fetoprotein levels >400 ng/ml combined
with at least one positive liver image on CT, magnetic resonance
imaging or ultrasonography. All patients included were further
confirmed as HBsAg-positive, HBV core antibody-positive and
hepatitis B e antigen or hepatitis B e antibody-positive for at
least six months. All subjects were Chinese from the Guangxi
District and provided written informed consent. The study was
approved by the ethics committee of the First Affiliated Hospital
of Guangxi Medical University, Nanning, Guangxi, China.
Single nucleotide polymorphism (SNP)
selection and genotyping
rs10889677, rs1884444 and rs11465817 were selected
as candidate SNPs for determining the association between
IL-23R gene polymorphisms and HBV-infected patients, as
earlier studies reported positive associations of these particular
SNPs with specific inflammatory diseases and types of cancer
(12). Venous blood from each
patient was collected into vacutainer tubes containing EDTA and
stored at 4°C. Amplification of the target DNA was performed by
polymerase chain reaction (PCR). Primer sequences and reaction
conditions are presented in Table
I. The SNPs studied were genotyped by PCR-restriction fragment
length polymorphism analysis. PCR products were visualized on a
2.0% agarose gel and stained with ethidium bromide (Promega
Corporation, Madison, WI, USA) to detect the quality and the
quantity of the amplified products. Digestion products were
visualized on a 3.0% agarose gel and stained with ethidium bromide.
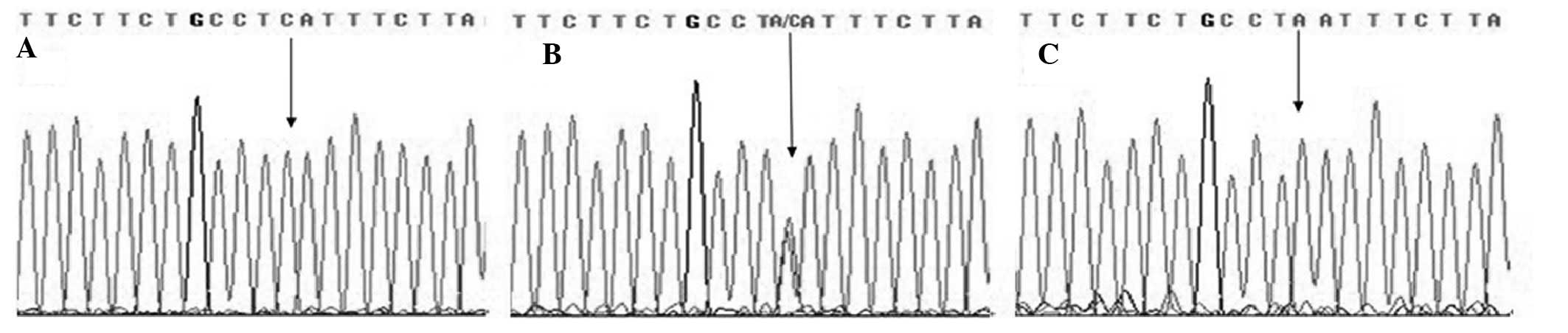
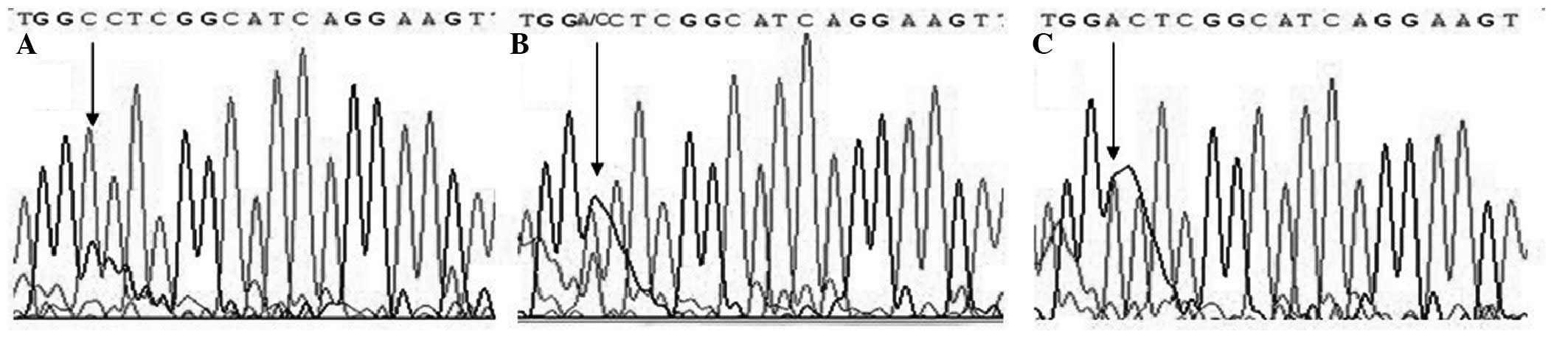
Direct sequencing was also performed by Sangon Biotech Company
(Shanghai, China) using randomly selected subjects (10% of all
samples) to validate the method used in this study and the results
were 100% concordant (Figs.
1–3).
 | Table IPrimer sequences and reaction
conditions for genotyping IL-23 gene polymorphisms. |
Table I
Primer sequences and reaction
conditions for genotyping IL-23 gene polymorphisms.
| Polymorphism | Primer sequence | Ta,
°C | Restriction
enzyme | Product size,
bp |
|---|
| rs10889677 | F:
5′-AGGGGATTGCTGGGCCATAT-3′ | 61.0 | MnII | A: 216 |
| R:
5′-TGTGCCTGTATGTGGACCA-3′ | | | C: 154+62 |
| rs1884444 | F:
5′-TCTTAGGGAAAAATGTTATGCTTTTT-3′ | 56.0 | HaeIII | T: 132 |
| R:
5′-GCATCCCATTGAATAGTGGC-3′ | | | G: 106+26 |
| rs11465817 | F:
5′-CATTAAGTAAGAGATGAAAACTTTGG-3′ | 54.0 | HaeIII | A: 136 |
| R:
5′-CTGTAGTGAGCTGTGACCATG-3′ | | | C: 117+19 |
Statistical analysis
Demographic and clinical data among groups were
compared using a χ2 test for categorical variables and
Student’s t-test for continuous variables. Hardy-Weinberg
equilibrium was tested with a goodness of fit χ2 test
with one degree of freedom to compare the observed genotype
frequencies among the subjects with the expected genotype
frequencies. Genotype and allele frequencies of IL-23R gene
polymorphisms were compared among various groups using the
χ2 test and Fisher’s exact test when appropriate, and
odds ratios (OR) and 95% confidence intervals (CIs) were calculated
by binary logistic regression and adjusted for age and gender to
assess the relative risk conferred by a particular allele and
genotype. The linkage disequilibrium (LD) between polymorphisms was
quantified using Shi’s standardized coefficient D′ (18). Haplotypes and their frequencies
were estimated based on a Bayesian algorithm using the Phase
program (19). The SPSS 13.0
software package (SPSS, Inc., Chicago, IL, USA) was used for all
statistical analysis. All tests were two-tailed. P<0.05 was
considered to indicate a statistically significant difference.
Results
Clinical characteristics of the study
participants
Results showed that the three candidate
IL-23R genetic variants were in Hardy-Weinberg equilibrium
in the patients and the control group. Demographic data of all
subjects are presented in Table
II. There was no statistically significant difference in age
between the case and control groups. However, a significant
difference was observed in gender distribution between the HCC
patients and controls.
 | Table IIClinical characteristics of the study
population. |
Table II
Clinical characteristics of the study
population.
| Variable | Controls | CHB patients | LC patients | HCC patients |
|---|
| Overall | 94 | 87 | 57 | 84 |
| Gender, n (%) |
| Male | 71 (75.5) | 74 (85.1) | 44 (77.2) | 73 (86.9) |
| Female | 23 (24.5) | 13 (14.9) | 13 (22.8) | 11 (13.1) |
| P-valuea | | 0.109 | 0.816 | 0.054 |
| Age (years)b | 42.0±10.8 | 40.3±11.9 | 46.0±12.6 | 49.5±10.3 |
| P-valuea | | 0.269 | 0.152 | 0.471 |
Correlation between IL-23R polymorphisms
and risk of CHB
Genotype and allele frequencies of the IL-23R
rs10889677, rs1884444 and rs11465817 polymorphisms between CHB
patients and healthy controls are presented in Table III. The genotype distributions of
three polymorphisms between the CHB cases and healthy controls were
in Hardy-Weinberg equilibrium. No significant differences were
identified between the genotype and allele frequencies of the
IL-23R rs10889677, rs1884444 and rs11465817 polymorphisms
and CHB risk. Similarily, binary logistic regression analyses
adjusted for age and gender did not reveal any significant
difference between IL-23R rs10889677, rs1884444 and
rs11465817 polymorphisms and CHB risk.
 | Table IIIDistribution of alleles and genotypes
of three SNPs in the IL-23R gene and their association with
CHB risk. |
Table III
Distribution of alleles and genotypes
of three SNPs in the IL-23R gene and their association with
CHB risk.
| Polymorphism | CHB patients, n=87
(%) | Control, n=94
(%) | Crude OR (95%
CI) | Adjusted OR (95%
CI)a | Adjusted
P-valuea |
|---|
| rs10889677 |
| Genotype |
| AA | 57 (65.5) | 55 (58.5) | 1.00 (Ref) | 1.00 (Ref) | |
| AC | 25 (28.7) | 34 (36.2) | 0.71
(0.38–1.34) | 0.74
(0.39–1.40) | 0.35 |
| CC | 5 (5.7) | 5 (5.3) | 0.97
(0.27–3.52) | 0.93
(0.25–3.40) | 0.91 |
| Allele |
| A | 139 (79.9) | 144 (76.6) | 1.00 (Ref) | 1.00 (Ref) | |
| C | 35 (20.1) | 44 (23.4) | 0.82
(0.50–1.36) | 0.83
(0.5–1.37) | 0.46 |
| rs1884444 |
| Genotype |
| TT | 45 (51. 7) | 60 (63.8) | 1.00 (Ref) | 1.00 (Ref) | |
| TG | 33 (37.9) | 24 (25.5) | 1.83
(0.95–3.52) | 1.82
(0.93–3.53) | 0.08 |
| GG | 9 (10.3) | 10 (10.6) | 1.20
(0.45–3.20) | 1.21
(0.45–3.24) | 0.71 |
| Allele |
| T | 123 (70.7) | 144 (76.6) | 1.00 (Ref) | 1.00 (Ref) | |
| G | 51 (29.3) | 44 (23.4) | 1.36
(0.85–2.17) | 1.34
(0.83–2.14) | 0.23 |
| rs11465817 |
| Genotype |
| AA | 34 (39.1) | 41 (43.6) | 1.00 (Ref) | 1.00 (Ref) | |
| AC | 41 (47.1) | 34 (36.2) | 1.45
(0.77–2.77) | 1.39
(0.73–2.67) | 0.32 |
| CC | 12 (13.8) | 19 (20.2) | 0.76
(0.32–1.79) | 0.73
(0.31–1.75) | 0.48 |
| Allele |
| A | 109 (62.4) | 116 (61.7) | 1.00 (Ref) | 1.00 (Ref) | |
| C | 65 (37.4) | 72 (38.3) | 0.96
(0.63–1.47) | 0.97
(0.63–1.49) | 0.89 |
Correlation between IL-23R polymorphisms
and risk of LC patients
Genotype and allele frequencies of the IL-23R
rs10889677, rs1884444 and rs11465817 polymorphisms between LC
patients and healthy controls are presented in Table IV. Genotype distributions of three
polymorphisms in the LC cases and healthy controls were in
Hardy-Weinberg equilibrium. No significant differences were
identified between the genotype and allele frequencies of the
IL-23R rs10889677, rs1884444 and rs11465817 polymorphisms
and LC risk. Similarily, binary logistic regression analyses
adjusted for age and gender did not show any significant difference
between IL-23R rs10889677, rs1884444 and rs11465817
polymorphisms and LC risk.
 | Table IVDistribution of alleles and genotypes
of three SNPs in the IL-23R gene and their association with
LC risk. |
Table IV
Distribution of alleles and genotypes
of three SNPs in the IL-23R gene and their association with
LC risk.
| Polymorphism | LC patients, n=57
(%) | Control, n=94
(%) | Crude OR (95%
CI) | Adjusted OR (95%
CI)a | Adjusted
P-valuea |
|---|
| rs10889677 |
| Genotype |
| AA | 40 (70.2) | 55 (58.5) | 1.00 (Ref) | 1.00 (Ref) | |
| AC + CC | 17 (29.8) | 39 (41.5) | 0.60
(0.30–1.21) | 0.57
(0.28–1.16) | 0.12 |
| Allele |
| A | 97 (85.1) | 144 (76.6) | 1.00 (Ref) | 1.00 (Ref) | |
| C | 17 (14.9) | 44 (23.4) | 0.57
(0.31–1.06) | 0.55
(0.30–1.03) | 0.06 |
| rs1884444 |
| Genotype |
| TT | 34 (59.6) | 60 (63.8) | 1.00 (Ref) | 1.00 (Ref) | |
| TG | 15 (26.3) | 24 (25.5) | 1.10
(0.51–2.38) | 1.10
(0.50–2.43) | 0.81 |
| GG | 8 (14.0) | 10 (10.6) | 1.41
(0.51–3.92) | 1.45
(0.51–4.12) | 0.49 |
| Allele |
| T | 83 (72.8) | 144 (76.6) | 1.00 (Ref) | 1.00 (Ref) | |
| G | 31 (27.5) | 44 (23.4) | 1.22
(0.72–2.08) | 1.21
(0.70–2.08) | 0.50 |
| rs11465817 |
| Genotype |
| AA | 16 (28.1) | 41 (43.6) | 1.00 (Ref) | 1.00 (Ref) | |
| AC | 28 (49.1) | 34 (36.2) | 2.11
(0.98–4.53) | 1.96
(0.90–4.26) | 0.09 |
| CC | 13 (22.8) | 19 (20.2) | 1.75
(0.70–4.36) | 1.64
(0.65–4.15) | 0.30 |
| Allele |
| A | 60 (52.6) | 116 (61.7) | 1.00 (Ref) | 1.00 (Ref) | |
| C | 54 (47.4) | 72 (38.3) | 1.45
(0.91–2.32) | 1.36
(0.84–2.20) | 0.21 |
Correlation between IL-23R polymorphisms
and risk of HCC
Genotype and allele distributions of the
IL-23R rs10889677, rs1884444 and rs11465817 polymorphisms
between HCC patients and healthy controls are presented in Table V. The observed genotype frequencies
for three variants were all in Hardy-Weinberg equilibrium in the
case group and controls. Results showed no significant differences
between the genotype and allele frequencies of the IL-23R
rs10889677 and rs11465817 polymorphisms and HCC risk. However, in
the rs1884444 polymorphism, the frequencies of the TT, TG and GG
genotypes were 63.8, 25.5 and 10.6% in controls and were 45.2, 42.9
and 11.9% in case groups, respectively. The frequencies of the T
and G alleles were 76.6 and 23.4% in controls, respectively and
were 66.7 and 33.3% in case groups, respectively. The results
indicated that there were significant differences in the genotype
and allele frequencies of the IL-23R rs1884444 polymorphism
between HCC patients and healthy controls. The rs1884444 TG
genotype was associated with a significantly increased risk of HCC
as compared with the TT genotype (OR, 2.37; 95% CI, 1.23–4.57;
P=0.009). The rs1884444 G allele was associated with a
significantly increased risk of HCC as compared with the T allele
(OR, 1.64, 95% CI, 1.03–2.61; P=0.037). In the binary logistic
regression analysis adjusted for age and gender, the results showed
that the rs1884444 TG genotype was significantly associated with
increased risk of HCC when compared with the TT genotype (adjusted
OR, 2.86; 95% CI, 1.39–5.85; P=0.00). Similarily, the rs1884444 G
allele was associated with a trend towards increased risk of HCC
when compared with the T allele (adjusted OR, 1.58; 95% CI,
0.96–2.60; P=0.07); however, this difference was not
significant.
 | Table VDistribution of alleles and genotypes
of three SNPs in the IL-23R gene and their association with
HCC risk. |
Table V
Distribution of alleles and genotypes
of three SNPs in the IL-23R gene and their association with
HCC risk.
| Polymorphism | HCC patients, n=84
(%) | Controls, n=94
(%) | Crude OR (95%
CI) | Adjusted OR (95%
CI)a | Adjusted
P-valuea |
|---|
| rs10889677 |
| Genotype |
| AA | 55 (65.5) | 55 (58.5) | 1.00 (Ref) | 1.00 (Ref) | |
| AC | 26 (31.0) | 34 (36.2) | 0.77
(0.41–1.44) | 0.85
(0.43–1.67) | 0.63 |
| CC | 3 (3.6) | 5 (5.3) | 0.60
(0.14–2.63) | 0.66
(0.14–3.06) | 0.60 |
| Allele |
| A | 136 (81.0) | 144 (76.6) | 1.00 (Ref) | 1.00 (Ref) | |
| C | 32 (19.0) | 44 (23.4) | 0.77
(0.46–1.29) | 0.83
(0.48–1.42) | 0.50 |
| rs1884444 |
| Genotype |
| TT | 38 (45.2) | 60 (63.8) | 1.00 (Ref) | 1.00 (Ref) | |
| TG | 36 (42.9) | 24 (25.5) | 2.37
(1.23–4.57) | 2.86
(1.39–5.85) | 0.00 |
| GG | 10 (11.9) | 10 (10.6) | 1.58
(0.60–4.15) | 1.24
(0.43–3.51) | 0.69 |
| Allele |
| T | 112 (66.7) | 144 (76.6) | 1.00 (Ref) | 1.00 (Ref) | |
| G | 56 (33.3) | 44 (23.4) | 1.64
(1.03–2.61) | 1.58
(0.96–2.60) | 0.07 |
| rs11465817 |
| Genotype |
| AA | 42 (50.0) | 41 (43.6) | 1.00 (Ref) | 1.00 (Ref) | |
| AC | 26 (31.0) | 34 (36.2) | 0.75
(0.38–1.46) | 0.72
(0.34–1.50) | 0.38 |
| CC | 16 (19.0) | 19 (20.2) | 0.82
(0.37–1.82) | 0.99
(0.42–2.31) | 0.97 |
| Allele |
| A | 110 (65.5) | 116 (61.7) | 1.00 (Ref) | 1.00 (Ref) | |
| C | 58 (34.5) | 72 (38.3) | 0.85
(0.55–1.31) | 0.92
(0.58–1.46) | 0.72 |
Haplotype analysis of IL-23R gene
polymorphisms and HCC risk
Haplotype analyses were performed in HCC patients
and healthy controls using the SHEsis software (18) and the possible eight haplotype
frequencies are presented in Table
VI. Moderate to minor LD was observed between the alleles of
SNPs rs1884444 and rs11465817 (D′=0.164), rs1884444 and rs10889677
(D′=0.145) and rs11465817 and rs10889677 (D′=0.230). The results
showed that the major ATA haplotype accounted for 41.1 and 41.0% of
these eight haplotypes in the cases and the controls, respectively.
Using haplotype analyses, the AGC haplotype was observed to be
associated with a significantly increased risk of HCC as compared
with the ATA haplotype (OR, 2.71; 95% CI, 1.06–6.93; P=0.03).
 | Table VIFrequencies of the haplotypes formed
by rs10889677, rs1884444 and rs11465817 SNPs in HCC patients and
controls. |
Table VI
Frequencies of the haplotypes formed
by rs10889677, rs1884444 and rs11465817 SNPs in HCC patients and
controls.
| Haplotype | HCC cases, 2n=168
(%) | Controls, 2n=188
(%) | OR (95% CI) | P-value |
|---|
| ATA | 69 (41.1) | 77 (41.0) | 1.00 (Ref) | |
| ATC | 24 (14.3) | 39 (20.7) | 0.69
(0.38–1.26) | 0.22 |
| AGA | 25 (14.9) | 20 (10.6) | 1.40
(0.71–2.73) | 0.33 |
| AGC | 17 (10.1) | 7 (3.7) | 2.71
(1.06–6.93) | 0.03a |
| CTA | 10 (6.0) | 13 (6.9) | 0.86
(0.35–2.08) | 0.74 |
| CTC | 8 (4.8) | 15 (8.0) | 0.60
(0.24–1.49) | 0.26 |
| CGA | 7 (4.2) | 6 (3.2) | 1.30
(0.42–4.06) | 0.65 |
| CGC | 8 (4.8) | 11 (5.9) | 0.81
(0.31–2.13) | 0.67 |
Discussion
To the best of our knowledge, this is the first
study to evaluate the association between the IL-23R gene
polymorphisms and HCC risk in a Guangxi population in China.
IL-23R rs1884444 T>G polymorphism was observed to be
significantly associated with HCC. The rs1884444 TG genotype was
associated with a significantly increased risk of HCC compared with
the rs1884444 TT genotypes (adjusted OR, 2.37; 95%CI, 1.23–4.57;
P=0.00). The rs1884444 G allele was associated with a trend towards
an increased risk effect of HCC as compared with the T allele;
however, this difference was not significant (adjusted OR, 1.58;
95% CI, 0.96–2.60; P=0.07). The haplotype AGC was associated with a
significantly increased risk of HCC as compared with the major ATA
haplotype (OR, 2.71; 95% CI, 1.06–6.93; P=0.03). These results
indicate that the IL-23R rs1884444 T>G polymorphism
increases HCC risk and may be used as genetic susceptibility
markers for HCC.
Several epidemiological studies in diverse ethnic
populations have previously been conducted and results consistent
with the current study have not been observed. In the current
study, the frequencies of the rs10889677 C, rs1884444 G and
rs11465817 C alleles among the healthy controls were 0.234, 0.234
and 0.383, respectively, and these were similar to those
frequencies observed in healthy Korean and Japanese populations
(20,21); however, the frequencies were
significantly lower than those of European Caucasians (0.689, 0.618
and 0.725, respectively) (22). In
addition, the rs10889677 A>C, rs1884444 T>G and rs11465817
A>C polymorphisms were observed in moderate-to-minor LD. Major
haplotype frequency of the ATA among the controls in the present
study was 0.410, which was significantly higher than those of a
study performed in Caucasians (0.213) (23), indicating that the distribution of
IL-23R gene frequencies and haplotypes may vary among ethnic
groups.
HCC is a typical inflammation-related malignancy
(24). In addition, increasing
evidence indicates that inflammation is important in the process of
hepatocarcinogenesis (25–26). IL-23 is a member of the IL-12
cytokine family that stimulates memory CD4+ T cells to
produce interferon-γ and to promote the development of distinctive
Th17 cells (27). Studies have
indicated that IL-23 may enhance the proliferation of memory T
cells and the production of interferon-γ by activated T cells
(5), indicating a role for IL-23
in cell-mediated antitumor immunity. IL-23R is the initial sensor
of the IL-23 signal. It is also the determinant of Th17 cell
expansion and in turn, serves as an important gate for the
Th17-cell-mediated autoimmune responses. Due to the biological
functions of IL-23R, this molecule has been hypothesized to be
involved in the development of cancer and the associated
complications. In agreement with this hypothesis, mice lacking
IL-23 p19 display resistance to endogenous tumor formation
when challenged in a chemical carcinogenesis protocol (28).
The IL-23R gene is located on chromosome
1p31. A number of studies have been conducted to investigate the
associations between IL-23R gene polymorphisms and cancer
risk (12,14,29).
Chen et al(13) observed
that the nonsynonymous SNP rs1884444 of the IL-23R gene may
modify the risk of gastric cancer, particularly in intestinal types
of gastric cancer. In addition, Zhang et al(29) previously indicated that
IL-23R polymorphisms may be important in the susceptibility
and prognosis of ovarian cancer and Chu et al(14) hypothesized that the IL-23R
rs6682925 and rs1884444 variants affect esophageal cancer
susceptibility through modulation of IL-23R biological activity.
These results are consistent with the current data. In the current
study, the rs1884444 TG genotype was associated with a
significantly increased risk of HCC as compared with the TT
genotype (adjusted OR, 2.37; 95%CI, 1.23–4.57; P=0.00). In the
binary logistic regression analysis, rs1884444 G allele levels were
insignificant when compared with the T allele (adjusted OR, 1.58;
95% CI, 0.96–2.60; P=0.07), which may be attributed to the fact
that the limited sample size prevented the analysis from reaching a
significant P-value. The rs1884444 T>G polymorphism is located
at exon 2 of the IL-23R, which is responsible for the signal
peptide of IL-23R(30).
Thus far, the exact biological mechanism of the variant on HCC
carcinogenesis has not been elucidated. Notably, according to the
web-based SNP analysis tool, PupaSuite3, the T to G base change of
rs1884444 may disrupt an exonic splicing enhancer, resulting in
exon skipping, malformation or transcript alternative splicing.
Another possible explanation is that this polymorphism results in
an amino acid change in codon 3 (His>Gln) and may affect the
ligand receptor binding specificity and affinity, modulating the
pro-inflammatory effect by Th17 cells and may be involved in the
development of HCC. However, these hypotheses are based on
postulations or predictions and require confirmation by biological
assays in future studies.
Haplotypes are a set of closely linked genetic
markers present on one chromosome which tend to be inherited
together and appear frequently in a block pattern owing to the
presence of LD (31). In the
current study, the AGC haplotype was observed to be significantly
higher in the HCC patients than in controls when compared with the
ATA haplotype (P=0.03). It is possible that a higher prevalence of
the AGC haplotype in the IL-23R gene may modulate the
expression of IL-23R, resulting in downregulation of the
Th17 cell-mediated immune response and escape of tumor cells from
immune surveillance. The current results indicate that the GCC
haplotype of IL-23R gene may play a facilitative role in the
development of HCC.
In summary, the rs1884444 polymorphism of the
IL-23R gene was observed to be significantly correlated with
the risk of HCC. These results indicate that the IL-23R gene
may contribute to an inherited predisposition to HCC. Additional
studies with larger sample sizes must be performed to confirm the
current observations in diverse ethnic populations.
Abbreviations:
|
HBV
|
hepatitis B virus
|
|
CHB
|
chronic hepatitis B
|
|
LC
|
liver cirrhosis
|
|
HCC
|
hepatocellular carcinoma
|
|
IL-23R
|
interleukin-23 receptor
|
|
OR
|
odds ratio
|
|
95% CI
|
95% confidence interval
|
|
SNP
|
single nucleotide polymorphism
|
|
PCR
|
polymerase chain reaction
|
References
|
1
|
Wang JS, Huang T, Su J, et al:
Hepatocellular carcinoma and aflatoxin exposure in Zhuqing Village,
Fusui County, People’s Republic of China. Cancer Epidemiol
Biomarkers Prev. 10:143–146. 2001.PubMed/NCBI
|
|
2
|
Yeh FS, Yu MC, Mo CC, Luo S, Tong MJ and
Henderson BE: Hepatitis B virus, aflatoxins and hepatocellular
carcinoma in southern Guangxi, China. Cancer Res. 49:2506–2509.
1989.PubMed/NCBI
|
|
3
|
Tao P, Zhi-Ming L, Tang-Wei L, et al:
Associated factors in modulating aflatoxin B1-albumin adduct level
in three Chinese populations. Dig Dis Sci. 50:525–532. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zemel R, Issachar A and Tur-Kaspa R: The
role of oncogenic viruses in the pathogenesis of hepatocellular
carcinoma. Clin Liver Dis. 15:261–279. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Oppmann B, Lesley R, Blom B, et al: Novel
p19 protein engages IL-12p40 to form a cytokine, IL-23, with
biological activities similar as well as distinct from IL-12.
Immunity. 13:715–725. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Abraham C and Cho J: Interleukin-23/Th17
pathways and inflammatory bowel disease. Inflamm Bowel Dis.
15:1090–1100. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jiang Z, Yang P, Hou S, et al: IL-23R gene
confers susceptibility to Behcet’s disease in a Chinese Han
population. Ann Rheum Dis. 69:1325–1328. 2010.PubMed/NCBI
|
|
8
|
Duerr RH, Taylor KD, Brant SR, et al: A
genome-wide association study identifies IL-23R as an inflammatory
bowel disease gene. Science. 314:1461–1463. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Venegas M, Beltrán CJ, Alvarez L, et al:
IL-23R Arg381Gln polymorphism in Chilean patients with inflammatory
bowel disease. Eur Cytokine Netw. 19:190–195. 2008.PubMed/NCBI
|
|
10
|
Wu Y, Lu Z, Chen Y, Xue F, Chen X and
Zheng J: Replication of association between interleukin-23 receptor
(IL-23R) and its ligand (IL-12B) polymorphisms and psoriasis in the
Chinese Han population. Hum Immunol. 71:1255–1258. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Illes Z, Safrany E, Peterfalvi A, et al:
3′UTR C2370A allele of the IL-23 receptor gene is associated with
relapsing-remitting multiple sclerosis. Neurosci Lett. 431:36–38.
2008.
|
|
12
|
Chen J, Lu Y, Zhang H, et al: A
nonsynonymous polymorphism in IL23R gene is associated with risk of
gastric cancer in a Chinese population. Mol Carcinog. 49:862–868.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen B, Zeng Z, Xu L, et al: IL23R
+2199A/C polymorphism is associated with decreased risk of certain
subtypes of gastric cancer in Chinese: a case-control study. Cancer
Epidemiol. 35:165–169. 2011.
|
|
14
|
Chu H, Cao W, Chen W, et al: Potentially
functional polymorphisms in IL-23 receptor and risk of esophageal
cancer in a Chinese population. Int J Cancer. 130:1093–1097. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Poole EM, Curtin K, Hsu L, et al: Genetic
variability in IL23R and risk of colorectal adenoma and colorectal
cancer. Cancer Epidemiol. 36:e104–e110. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chien MH, Hsin CH, Chou LS, et al:
Interleukin-23 receptor polymorphism as a risk factor for oral
cancer susceptibility. Head Neck. 34:551–556. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Martin M and Herceg Z: From hepatitis to
hepatocellular carcinoma: a proposed model for cross-talk between
inflammation and epigenetic mechanisms. Genome Med. 4:82012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shi YY and He L: SHEsis, a powerful
software platform for analyses of linkage disequilibrium, haplotype
construction and genetic association at polymorphism loci. Cell
Res. 15:97–98. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Stephens M, Smith NJ and Donnelly P: A new
statistical method for haplotype reconstruction from population
data. Am J Hum Genet. 68:978–989. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ban Y, Tozaki T, Taniyama M, Nakano Y,
Yoneyama K, Ban Y and Hirano T: Association studies of the IL-23R
gene in autoimmune thyroid disease in the Japanese population.
Autoimmunity. 42:126–130. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim ES, Kim SW, Moon CM, et al:
Interactions between IL17A, IL23R and STAT4 polymorphisms confer
susceptibility to intestinal Behcet’s disease in Korean population.
Life Sci. 90:740–746. 2012.PubMed/NCBI
|
|
22
|
Faragó B, Magyari L, Sáfrány E, et al:
Functional variants of interleukin-23 receptor gene confer risk for
rheumatoid arthritis but not for systemic sclerosis. Ann Rheum Dis.
67:248–250. 2008.PubMed/NCBI
|
|
23
|
Ferguson LR, Han DY, Fraser AG, Huebner C,
Lam WJ and Morgan AR: IL23R and IL12B SNPs and haplotypes strongly
associate with Crohn’s disease risk in a New Zealand population.
Gastroenterol Res Pract. 2010:5394612010.PubMed/NCBI
|
|
24
|
Liao R, Sun TW, Yi Y, et al: Expression of
TREM-1 in hepatic stellate cells and prognostic value in hepatitis
B-related hepatocellular carcinoma. Cancer Sci. 103:984–992. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Szabo G and Lippai D: Molecular hepatic
carcinogenesis: impact of inflammation. Dig Dis. 30:243–248. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Qin LX: Inflammatory immune responses in
tumor microenvironment and metastasis of hepatocellular carcinoma.
Cancer Microenviron. 5:203–209. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
McGeachy MJ, Chen Y, Tato CM, et al: The
interleukin 23 receptor is essential for the terminal
differentiation of interleukin 17-producing effector T helper cells
in vivo. Nat Immunol. 10:314–324. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Langowski JL, Zhang X, Wu L, et al: IL-23
promotes tumour incidence and growth. Nature. 442:461–465. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang Z, Zhou B, Zhang J, et al:
Association of interleukin-23 receptor gene polymorphisms with risk
of ovarian cancer. Cancer Genet Cytogenet. 196:146–152. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mancini G, Kan SH and Gallagher G: A novel
insertion variant of the human IL-23 receptor-alpha chain
transcript. Genes Immun. 9:566–569. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Seng KC and Seng CK: The success of the
genome-wide association approach: a brief story of a long struggle.
Eur J Hum Genet. 16:554–564. 2008. View Article : Google Scholar : PubMed/NCBI
|