Introduction
There is increasing epidemiological evidence
implicating vitamin B6 as a protective factor against colon cancer
(1–4). Consistent with these previous
results, the current study observed that dietary vitamin B6 intake,
from a supplemental vitamin B6 diet to a low vitamin B6 diet,
caused a marked reduction in colon tumorigenesis in mice exposed to
azoxymethane (5). Our animal
studies suggest that the anti-colon tumor effect of dietary vitamin
B6 is partially ascribed to lowering colon cell proliferation,
oxidative stress, inflammation and epithelium cell damage (5–8).
Furthermore, we observed that vitamin B6 inhibited
lipopolysaccharide (LPS)-induced expression of inducible nitric
oxide synthase (iNOS) and cyclooxygenase-2 (COX-2) in mouse
macrophage RAW264.7 cells via suppression of nuclear factor-κB
(NF-κB) activation (9). In
addition, dietary vitamin B6 inhibited iNOS activity in the liver
of rats exposed to LPS (9).
Notably, vitamin B6 has been observed to inhibit DNA and RNA
polymerase (10–12), topoisomerase-IB and angiogenesis
(13,14). However, the molecular mechanisms
involved in the antitumor effect of vitamin B6 are not yet clearly
understood.
According to preliminary experimental results in the
current study, which used DNA microarray analysis, a number of
genes were upregulated by pyridoxal (PL; 500 μM) in human colon
cancer cells (HT29). Insulin-like growth factor-binding protein 1
(IGFBP1) was one of these upregulated genes in HT29 cells,
confirmed by quantitative PCR. IGFBP1 is primarily produced and
secreted from the liver (15) and
binds to insulin growth factors (IGFs), modulating their actions
(15). Previous studies have
suggested that IGFBP1 may be a tumor suppressor (16–19).
Low serum IGFBP1 levels are associated with a number of chronic
diseases, including colon cancer, cardiovascular disease and
diabetes (20–22). A previous study showed that
increased circulating IGFBP1 levels improve insulin sensitivity,
lower blood pressure and protect against atherosclerosis (22). IGFBP1 is rapidly induced during
liver regeneration and is implicated in the maintenance of
hepatocyte differentiation and metabolism (23,24).
The liver is a central organ involved in regulating vitamin B6
metabolism (25). In addition,
Nakari et al(26)
demonstrated that a high dose of pyridoxine (PN; 10 mM) induced the
expression of the insulin-like growth factor-binding protein 3
(IGFBP3) in human breast adenocarcinoma MCF-7 cells. Therefore, the
objective of the current study was to explore the effect of vitamin
B6 on the expression of IGFBP1 in human hepatoma HepG2 cells.
Materials and methods
Materials
PL hydrochloride, PN hydrochloride and pyridoxal
5′-phosphate (PLP) were obtained from Nacalai Tesque (Kyoto, Japan)
and pyridoxamine (PM) dihydrochloride was obtained from Calbiochem
(La Jolla, CA, USA). Human colorectal cancer HT29 cells and human
hepatoma HepG2 cells were purchased from the Health Science
Research Resources Bank (Osaka, Japan) and the Japan Health Science
Foundation (Tokyo, Japan), respectively. Dulbecco’s modified
Eagle’s medium (DMEM) was purchased from Sigma-Aldrich (St. Louis,
MO, USA). Antibodies specific for IGFBP1 and p-c-Jun were products
of Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). An
antibody against p-ERK1/2 was obtained from Cell Signaling
Technology Inc. (Danvers, MA, USA). Tubulin antibody was obtained
from Harlan Sera-Lab (Leicestershire, UK).
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
was purchased from Sigma-Aldrich. Dimethyl sufoxide (DMSO) was
obtained from Nacalai Tesque. PD98059 (ERK inhibitor) was purchased
from Wako Pure Chemical Industries (Osaka, Japan). Cycloheximide
(protein synthesis inhibitor) was also purchased from Wako Pure
Chemical Industries.
Cell culture and treatment
HT29 and HepG2 cells were maintained in DMEM
supplemented with 10% fetal calf serum, 100 U/ml penicillin and 100
mg/ml streptomycin at 37°C in 5% CO2. PL, PN, PM or PLP
were dissolved directly in culture medium and filtered through a
Millex-HV 0.45-μm syringe filter (Millipore, Billerica, MA, USA).
PD98059 was dissolved in DMSO and added directly to the culture
medium. DMSO, instead of the inhibitor, was added to the other
groups.
LDH assay
Cytotoxicity of PL in HepG2 cells was determined
using a lactate dehydrogenase (LDH) assay kit (Promega, Madison,
WI, USA). Cells (1.5×104 cells/well) were seeded in a
96-well plate with 120 μl culture medium. Following 24 h, cells
were treated with or without PL at a concentration of 500 μM for 6,
12 or 24 h. A total amount of 100 μl of supernatant for each well
was transferred to a new 96-well plate and 100 μl of reconstituted
substrate mix was added to each well and the plates were maintained
at room temperature for 60 min. Absorbance was recorded at 490 nm
with an ELNX 96 reader (TFB, Tokyo, Japan). Each experiment was
repeated eight times.
MTT assay
Cell culture medium suspensions (3,000 cells/100 μl)
were plated into 96-well plates. Following 24 h incubation, 100 μl
culture medium with or without PL (500 μM) was added to the wells
and incubated for 6, 12, 24 or 48 h. At the end of the incubation,
20 μl 0.5% MTT solution was added to each well. Plates were
returned to the incubator for a period of 4 h. Absorbance was read
on a spectrophotometer with an ELNX 96 reader at 550 nm. Each
experiment was repeated eight times.
mRNA analysis
Total RNA from HT29 or HepG2 cells was isolated
using TRIzol™ (Invitrogen Life Technologies, Carlsbad, CA, USA).
Total RNA (1 μg) was reverse transcribed using the First Strand
cDNA Synthesis kit (Toyobo, Osaka, Japan) according to the
manufacturer’s instructions. Quantitative PCR was performed with a
StepOne™ Real-Time PCR System (Applied Biosystems, Carlsbad, CA,
USA) using Thunderbird SYBR qPCR Mix (Toyobo). The primer sets for
IGFBP1 and GADPH were purchased from Greiner Bio-One (Tokyo, Japan)
and were as follows: IGFBP1, 5′-GCCAAACTGCAA CAAGAATG-3′ and
5′-ATCCTCTTCCCATTCCAAG-3′; and GADPH, 5′-CAATGACCCCTTCATTGACC-3′
and 5′-TGG AAGATGGTGATGGGATT-3′. The cycling parameters were as
follows: Initial step at 90°C for 10 sec, followed by 40 cycles of
90°C for 5 sec, 60°C for 10 sec and 72°C for 10 sec. Relative gene
expression levels were calculated using the 2−ΔΔCt
method normalizing to GAPDH expression levels and fold differences
in expression were calculated relative to control samples.
Western blot analysis
Western blot analysis experiments for IGFBP1
detection were performed using HepG2 cell lysate and culture
medium. Cells were grown to 70% confluency in a 6-well plate.
Following PL treatment, cells were washed twice with PBS and lysed
in RIPA buffer [20 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM
MgCl2 and 1 mM CaCl2] with 1% Triton X-100.
Cell lysate was centrifuged at 12,000 × g for 10 min to pellet
debris. Total protein samples were removed and assayed for protein
content using a Bio-Rad Protein Assay kit (Bio-Rad, Hertfordshire,
UK). Sodium dodecyl sulphate-polyacrylamide gel electrophoresis
(SDS-PAGE) sample buffer was added to the protein pellets and
samples were boiled for 3 min at 95°C. Samples were loaded (10 μg
of total protein for cell lysate) and electrophoretically separated
on 10% polyacrylamide gels and transferred to polyvinylidene
difluoride (PVDF) membranes. Western blot analysis was performed to
standard instructions and proteins were visualized using the
following primary antibodies: IGFBP1 (rabbit polyclonal antibody;
1:1,000), p-ERK1/2 (rabbit polyclonal antibody; 1:1,000), p-c-Jun
(mouse monoclonal antibody; 1:1,000) and tubulin (rat monoclonal
antibody; 1:1,000).
To detect IGFBP1 in the culture medium, culture
medium (1 ml) was collected and centrifuged at 12,000 × g for 10
min. A total of 100 μl of supernatant was added to 30 μl SDS-PAGE
sample buffer and boiled for 3 min at 95°C. Each sample (10 μl) was
loaded for western blot analysis.
Statistical analysis
Data are presented as the mean ± SE. Differences
among the average means of treatment groups were analyzed using a
one-way ANOVA and P<0.05 was considered to indicate a
statistically significant difference, as determined by Scheffe’s
multiple-range test. For experiments which included only two
groups, a Student’s t-test was used and the statistical difference
was set at P<0.05.
Results
PL stimulates expression of IGFBP1
mRNA
According to DNA microarray analysis, IGFBP1 was
upregulated in HT29 cells exposed to PL (500 μM). Stimulation of
IGFBP1 by PL in HT29 cells was confirmed by quantitative PCR
(Fig. 1A). HepG2 cells were
incubated in the presence or absence of PL (500 μM) for 24 h and
IGFBP1 mRNA levels were determined by quantitative PCR. As shown in
Fig. 1B, IGFBP1 mRNA levels were
markedly elevated by PL in HepG2 cells (P<0.01). The effects of
B6 vitamers (500 μM), including PL, PM, PN and PLP, on the
expression of IGFBP1 mRNA levels were analyzed (Fig. 1C). The results show that PL has a
marked stimulation effect (P<0.05); however, other B6 vitamers
did not exhibit the same effect.
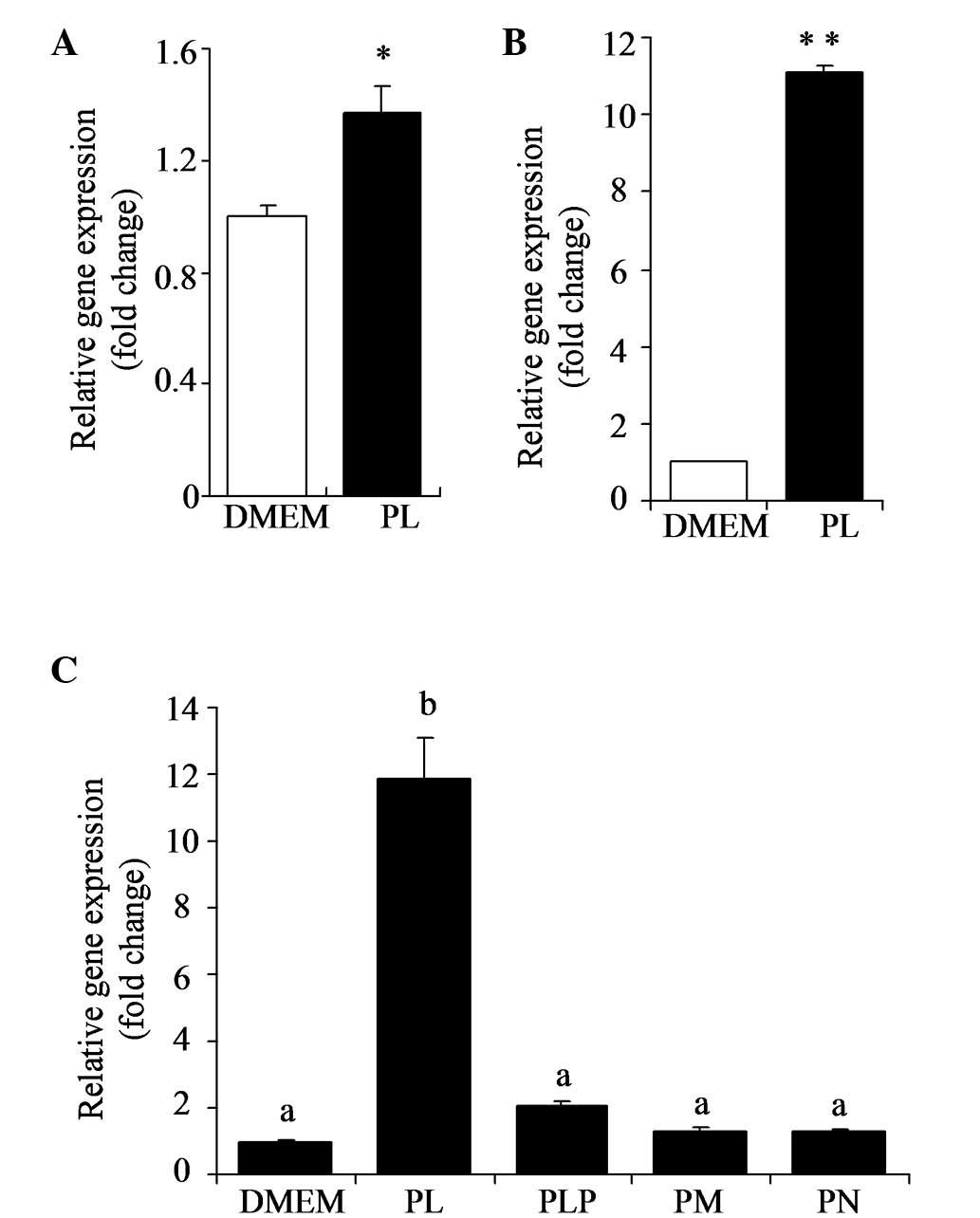 | Figure 1Stimulation of expression of IGFBP1
mRNA by PL in HT29 and HepG2 cells. (A) HT29 or (B) HepG2 cells
were incubated in presence or absence of PL (500 μM) over 24 h. (C)
HepG2 cells were incubated with various vitamers (PL, PLP, PM or
PN) at a concentration of 500 μM over 24 h. IGFBP1 mRNA levels were
determined by quantitative PCR. Cells cultured with medium (DMEM
with 10% fetal calf serum, 100 units/ml penicillin and 100 mg/ml
streptomycin) were used as control. Values are presented as the
mean ± SE (n=4). *P<0.05 and **P<0.01,
vs. relative control. Groups with different letters are
significantly different from each other (P<0.05). IGFBP1,
insulin-like growth factor-binding protein 1; PL, pyridoxal; PLP,
pyridoxal 5′-phosphate; PM, pyridoxamine; PN, pyridoxine; DMEM,
Dulbecco’s modified Eagle’s medium. |
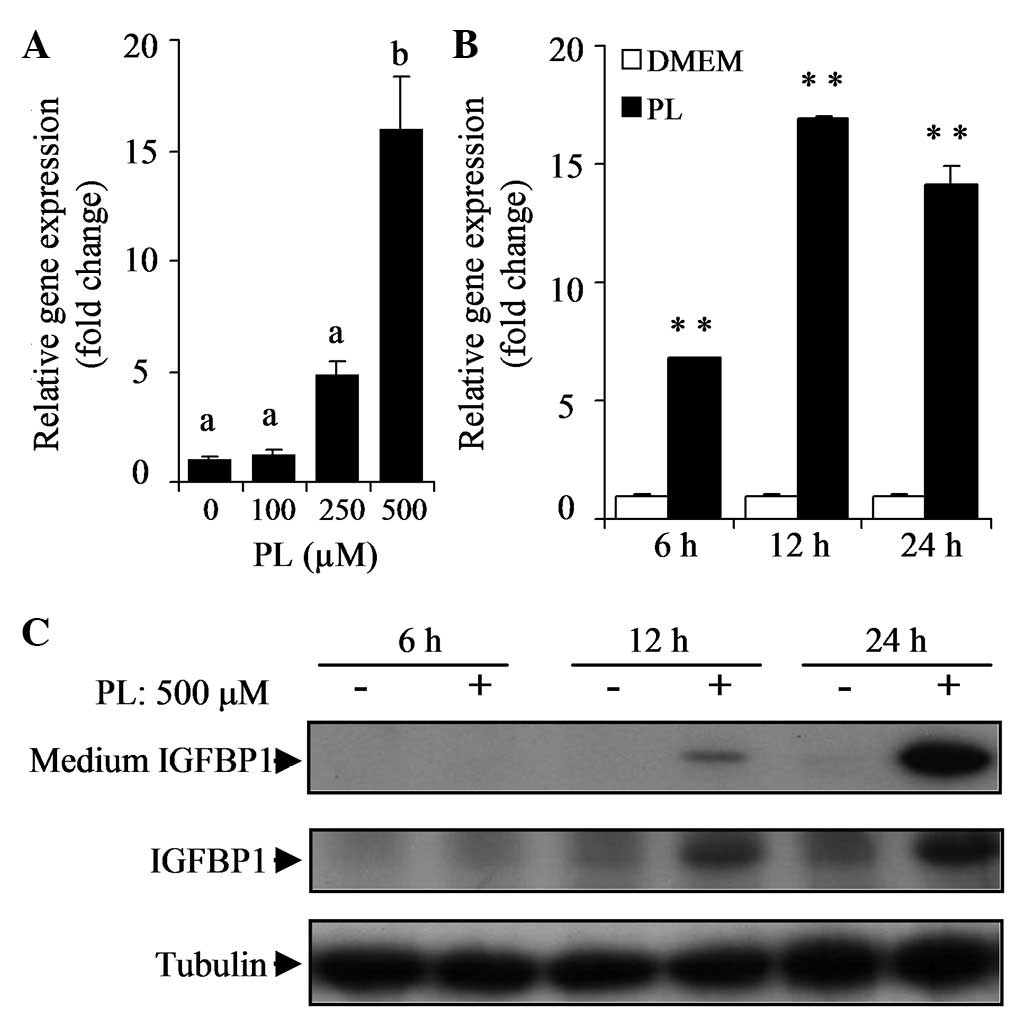
PL stimulates IGFBP1 expression in a
time- and dose-dependent manner
The effects of various concentrations of PL on the
expression of IGFBP1 mRNA were investigated. Following 24 h
incubation of HepG2 cells with varying concentrations of PL (100,
250 or 500 μM), IGFBP1 mRNA levels were elevated in a
dose-dependent manner (Fig. 2A).
At a concentration of 500 μM, PL markedly stimulated the expression
of IGFBP1 mRNA levels in HepG2 cells (P<0.05). To examine the
time-dependent effect of PL, HepG2 cells were cultured with or
without PL (500 mM) for 6, 12 or 24 h. The results showed that
treatment with PL increased IGFBP1 mRNA levels in a time-dependent
manner (P<0.01; Fig. 2B). The
expression of IGFBP1 protein in the cell lysate and culture medium
increased in a time-dependent manner (Fig. 2C).
PL affects cell growth of HepG2 without
showing cytotoxicity
The current cell culture studies were conducted with
a supraphysiological dose of PL (500 mM), thus the cytotoxicity of
PL was examined at this dose. An LDH assay was performed to examine
the cytotoxicity of PL (500 mM) for various incubation times (6, 12
or 24 h). The results show that PL, at a concentration of 500 mM,
caused no cytotoxicity to HepG2 cells over 24 h (Fig. 3A). In addition, the effect of PL on
cell growth was examined with an MTT assay. HepG2 cells were
incubated with PL (500 mM) for 6, 12, 24 or 48 h. PL significantly
elevated cell growth over 12 h (P<0.01; Fig. 3B), while PL significantly
suppressed cell growth over 24 and 48 h (P<0.01). Thus, the
induction of IGFBP1 by PL appears to begin prior to alteration in
cell growth.
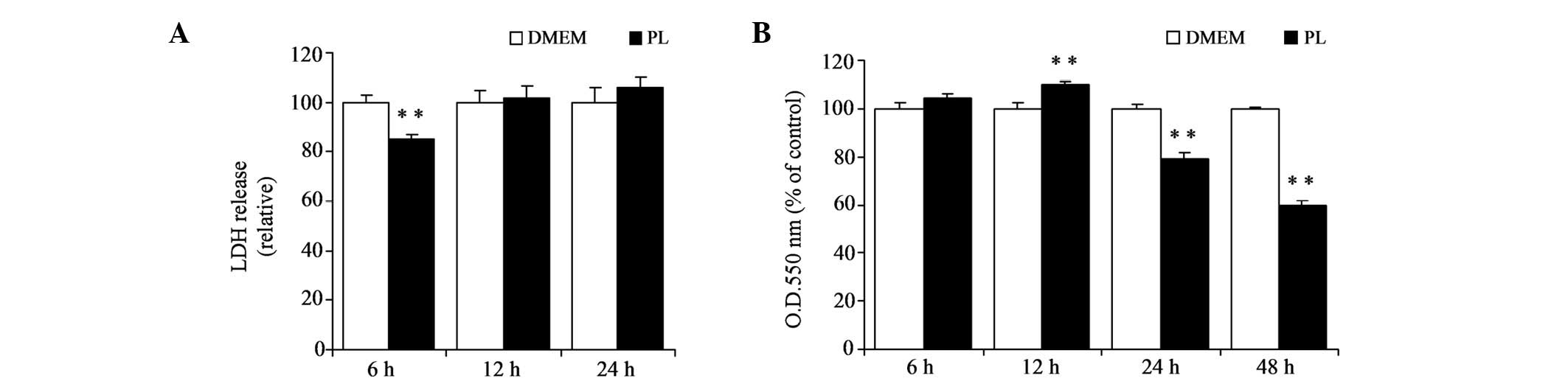 | Figure 3LDH and MTT assays in HepG2 cells
exposed to PL. HepG2 cells were exposed to PL (500 μM) for various
times (6, 12 or 24 h). (A) Cytotoxicity was determined by measuring
the amount of LDH released from the cells into the culture medium.
Cultured cells (3×103 cells/well) were exposed to the
medium with or without PL (500 μM) over 6, 12, 24 or 48 h. (B) Cell
growth was determined by the MTT assay. Cells cultured with medium
(DMEM with 10% fetal calf serum, 100 U/ml penicillin and 100 mg/ml
streptomycin) were used as control. The data are presented as the
mean ± SE (n=8). **P<0.01, vs. relative control. LDH,
lactate dehydrogenase; MTT,
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; PL,
pyridoxal. |
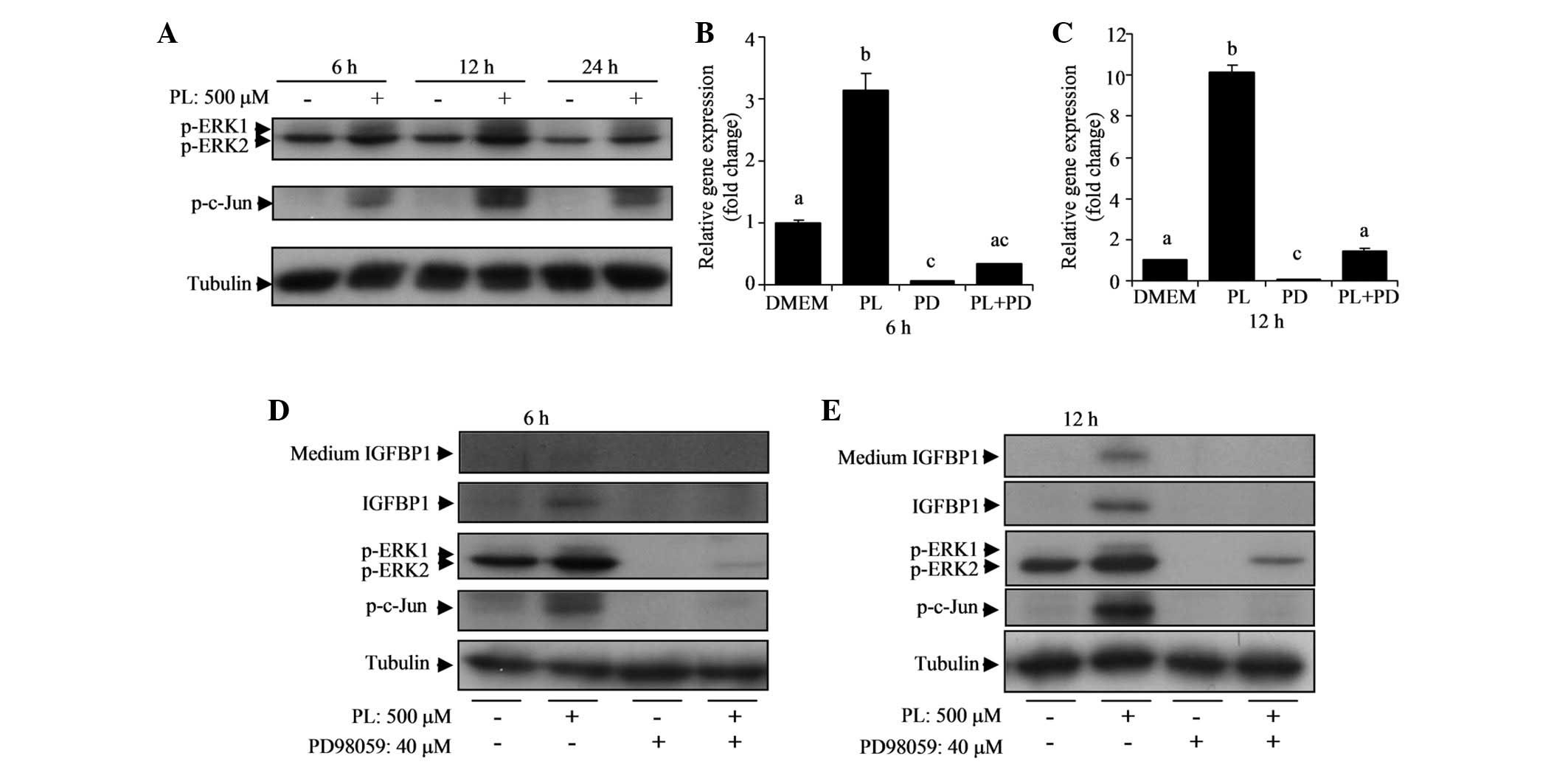
PL activates the ERK pathway responsible
for the stimulation of IGFBP1
Multiple mitogen-activated protein kinase (MAPK)
pathways are reportedly involved in the stimulation of IGFBP1
(27–29). Therefore, in the current study, the
protein expression of ERK, p-ERK, p-JNK and p-c-Jun (downstream
factor of ERK and JNK) was examined in HepG2 cells treated with or
without PL over 6, 12 or 24 h. The results indicate that PL
elevated p-ERK1 and p-c-Jun proteins significantly at 6 and 12 h
(Fig. 4A). Total ERK and p-JNK
expression were not affected by PL (data not shown).
Activation of the ERK pathway by PL led to the
hypothesis that the ERK pathway is involved in the stimulation of
IGFBP1 by PL in HepG2 cells. PD98059, an ERK inhibitor, was used to
test this assumption. The ERK inhibitor (40 μM) effectively reduced
IGFBP1 mRNA levels in the control and PL-treated cells (Fig. 4B and C). Notably, in the presence
of the ERK inhibitor, there was no significant difference in IGFBP1
mRNA levels between cells with or without PL at 6 h (Fig. 4B). In addition, the elevated IGFBP1
protein expression by PL in the cell lysate and culture medium was
entirely eradicated by the ERK inhibitor (Fig. 4D and E). Elevations in p-ERK1 and
p-c-Jun protein expression by PL were eliminated by PD98059
(Fig. 4D and E). These results
indicate that the ERK pathway is important in the stimulation of
IGFBP1 by PL.
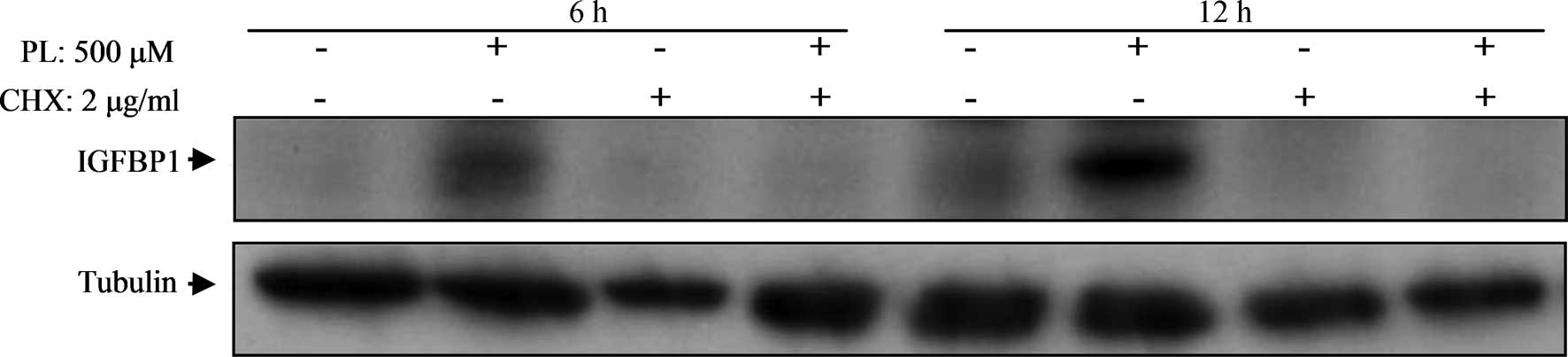
Inhibition of protein synthesis inhibits
elevation in the expression of IGFBP1 protein by PL
To understand the role of protein synthesis in the
stimulation of IGFBP1 by PL, HepG2 cells were treated with
cycloheximide for 6 or 12 h. Cycloheximide (2 μg/ml) completely
inhibited the elevation of IGFBP1 protein expression induced by PL
(Fig. 5). These results suggest
that higher expression of IGFBP1 protein by PL is dependent on
protein synthesis.
Discussion
The present study demonstrated that PL elevates the
expression of IGFBP1 mRNA and protein levels in HepG2 cells.
Notably, IGFBP1 protein observed in the cell culture medium was
also elevated by PL, indicating that a high concentration of IGFBP1
was released into the culture medium. PL was observed to cause a
marked elevation in IGFBP1 mRNA, but for other B6-vitamers,
including PLP, PN and PM, this effect was not observed on IGFBP1.
Our previous study showed that the inhibitory effect of PL on the
LPS-induced expression of iNOS and COX-2 in RAW264.7 cells was
stronger compared with PLP (9).
Kanouchi et al(30) showed
that when RAW264.7 cells were cultured in a culture medium treated
with the B6 vitamers PL, PM, PN or PLP, only PL interacted with the
cell surface. PL is known as a primary form of transport in blood
and is capable of freely passing through the cell membrane, while
PLP cannot easily cross the cell membrane and must be hydrolyzed by
alkaline phosphatase into PL (31–33).
Therefore, PL appears to affect IGFBP1 expression in HepG2 cells as
PL crosses cell membranes from a cultured medium.
Higher expression of IGFBP1 mRNA by PL was inhibited
by PD98059, an ERK inhibitor, suggesting that the ERK pathway may
be important in PL-induced IGFBP1 gene expression. Previous studies
have observed that the expression of IGFBP1 may be regulated via
the MAPK pathway (JNK and ERK). Inhibitors of the MAPK pathway were
able to entirely eliminate IGFBP1 expression by various inducers
(27,29). The current results show that
PL-induced IGFBP1 expression was inhibited by the ERK inhibitor.
The phosphorylated c-Jun protein was also reduced by the ERK
inhibitor. A previous study showed that c-Jun is required to
promote the maximal expression of the IGFBP1 promoter in HepG2
cells in the presence of IL-6 (28). Phosphorylation of c-Jun by ERK in
PC12 cell differentiation has been demonstrated (34). According to the present results and
previous studies (28,34), PL may stimulate the expression of
IGFBP1 in HepG2 cells by elevating the ERK/c-Jun pathway. Since the
JNK pathway is hypothesized to be involved in the stimulation of
IGFBP1 gene expression in HepG2 cells (29), PL was investigated to determine
whether it activates JNK in HepG2 cells. The activation of JNK by
PL was not observed. Thus, the JNK pathway does not appear to be
involved in PL-induced IGFBP1 expression. The current study shows
that PL at 500 mM significantly affects the growth of HepG2 cells
between 12 and 48 h. It is unclear whether the alteration in cell
growth is associated with the modulation of IGFBP1 and ERK.
In summary, the present study provides evidence for
PL stimulating IGFBP1 expression in HepG2 cells. The ERK/c-Jun
pathway may be involved in the induction of IGFBP1 by PL. Vitamin
B6 and IGFBP1 are hypothesized as protective factors against
cancers and cardiovascular disease (7,15,22,35).
Thus, it is of interest to examine whether vitamin B6 intake causes
a higher production of IGFBP1 in the liver, resulting in higher
circulating IGFBP1, which in turn leads to lower development of
these diseases.
Acknowledgements
This study was supported in part by a Grant-in-Aid
for Scientific Research from the Ministry of Education, Culture,
Sports, Science and Technology of Japan.
Abbreviations:
|
iNOS
|
inducible nitric oxide synthase
|
|
COX-2
|
cyclooxygenase-2
|
|
IGFBP1
|
insulin-like growth factor-binding
protein 1
|
|
NF-κB
|
nuclear factor-κB
|
|
LPS
|
lipopolysaccharide
|
|
IGFs
|
insulin growth factors
|
|
IGFBP3
|
insulin-like growth factor-binding
protein 3
|
|
PL
|
pyridoxal
|
|
PN
|
pyridoxine
|
|
PM
|
pyridoxamine
|
|
PLP
|
pyridoxal 5′-phosphate
|
|
DMEM
|
Dulbecco modified Eagle’s medium
|
|
MTT
|
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
|
|
LDH
|
lactate dehydrogenase
|
|
DMSO
|
dimethyl sulfoxide
|
|
SDS-PAGE
|
sodium dodecyl sulphate-polyacrylamide
gel electrophoresis
|
|
MAPK
|
mitogen-activated protein kinase
|
|
PVDF
|
polyvinylidene difluoride
|
|
PD
|
PD98059
|
|
CHX
|
cycloheximide
|
References
|
1
|
Ishihara J, Otani T, Inoue M, Iwasaki M,
Sasazuki S and Tsugane S; Japan Public Health Center-based
Prospective Study Group. Low intake of vitamin B-6 is associated
with increased risk of colorectal cancer in Japanese men. J Nutr.
137:1808–1814. 2007.PubMed/NCBI
|
|
2
|
Theodoratou E, Farrington SM, Tenesa A,
McNeill G, Cetnarskyj R, Barnetson RA, Porteous ME, Dunlop MG and
Campbell H: Dietary vitamin B6 intake and the risk of colorectal
cancer. Cancer Epidemiol Biomarkers Prev. 17:171–182. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Larsson SC, Orsini N and Wolk A: Vitamin
B6 and risk of colorectal cancer: a meta-analysis of prospective
studies. JAMA. 303:1077–1083. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang XH, Ma J, Smith-Warner SA, Lee JE
and Giovannucci E: Vitamin B6 and colorectal cancer: current
evidence and future directions. World J Gastroenterol.
19:1005–1010. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Komatsu SI, Watanabe H, Oka T, Tsuge H,
Nii H and Kato N: Vitamin B-6-supplemented diets compared with a
low vitamin B-6 diet suppress azoxymethane-induced colon
tumorigenesis in mice by reducing cell proliferation. J Nutr.
131:2204–2207. 2001.PubMed/NCBI
|
|
6
|
Komatsu S, Watanabe H, Oka T, Tsuge H and
Kat N: Dietary vitamin B6 suppresses colon tumorigenesis,
8-hydroxyguanosine, 4-hydroxynonenal, and inducible nitric oxide
synthase protein in azoxymethane-treated mice. J Nutr Sci Vitaminol
(Tokyo). 48:65–68. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Komatsu S, Yanaka N, Matsubara K and Kato
N: Antitumor effect of vitamin B6 and its mechanisms. Biochim
Biophys Acta. 1647:127–130. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kayashima T, Tanaka K, Okazaki Y,
Matsubara K, Yanaka N and Kato N: Consumption of vitamin B6 reduces
colonic damage and protein expression of HSP70 and HO-1, the
anti-tumor targets, in rats exposed to 1,2-dimethylhydrazine. Oncol
Lett. 2:1243–1246. 2011.PubMed/NCBI
|
|
9
|
Yanaka N, Koyama TA, Komatsu S, Nakamura
E, Kanda M and Kato N: Vitamin B6 suppresses NF-kappaB activation
in LPS-stimulated mouse macrophages. Int J Mol Med. 16:1071–1075.
2005.PubMed/NCBI
|
|
10
|
Diffley JF: Affinity labeling the DNA
polymerase alpha complex. I. Pyridoxal 5′-phosphate inhibition of
DNA polymerase and DNA primase activities of the DNA polymerase
alpha complex from Drosophila melanogaster embryos. J Biol
Chem. 263:14669–14677. 1988.
|
|
11
|
Matsubara K, Matsumoto H, Mizushina Y, Lee
JS and Kato N: Inhibitory effect of pyridoxal 5′-phosphate on
endothelial cell proliferation, replicative DNA polymerase and DNA
topoisomerase. Int J Mol Med. 12:51–55. 2003.
|
|
12
|
Martial J, Zaldivar J, Bull P, Venegas A
and Valenzuela P: Inactivation of rat liver RNA polymerases I and
II and yeast RNA polymerase I by pyridoxal 5′-phosphate. Evidence
for the participation of lysyl residues at the active site.
Biochemistry. 14:4907–4911. 1975.PubMed/NCBI
|
|
13
|
Vermeersch JJ, Christmann-Franck S,
Karabashyan LV, Fermandjian S, Mirambeau G and Der Garabedian PA:
Pyridoxal 5′-phosphate inactivates DNA topoisomerase IB by
modifying the lysine general acid. Nucleic Acids Res. 32:5649–5657.
2004.
|
|
14
|
Matsubara K, Mori M, Matsuura Y and Kato
N: Pyridoxal 5′-phosphate and pyridoxal inhibit angiogenesis in
serum-free rat aortic ring assay. Int J Mol Med. 8:505–508.
2001.
|
|
15
|
Lee PD, Giudice LC, Conover CA and Powell
DR: Insulin-like growth factor binding protein-1: recent findings
and new directions. Proc Soc Exp Biol Med. 216:319–357. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yee D, Jackson JG, Kozelsky TW and
Figueroa JA: Insulin-like growth factor binding protein 1
expression inhibits insulin-like growth factor I action in MCF-7
breast cancer cells. Cell Growth Differ. 5:73–77. 1994.PubMed/NCBI
|
|
17
|
Van den Berg CL, Cox GN, Stroh CA,
Hilsenbeck SG, Weng CN, McDermott MJ, Pratt D, Osborne CK,
Coronado-Heinsohn EB and Yee D: Polyethylene glycol conjugated
insulin-like growth factor binding protein-1 (IGFBP-1) inhibits
growth of breast cancer in athymic mice. Eur J Cancer.
33:1108–1113. 1997.PubMed/NCBI
|
|
18
|
Zhang X and Yee D: Insulin-like growth
factor binding protein-1 (IGFBP-1) inhibits breast cancer cell
motility. Cancer Res. 62:4369–4375. 2002.PubMed/NCBI
|
|
19
|
Ngo TH, Barnard RJ, Leung PS, Cohen P and
Aronson WJ: Insulin-like growth factor I (IGF-I) and IGF binding
protein-1 modulate prostate cancer cell growth and apoptosis:
possible mediators for the effects of diet and exercise on cancer
cell survival. Endocrinology. 144:2319–2324. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wei EK, Ma J, Pollak MN, Rifai N, Fuchs
CS, Hankinson SE and Giovannucci E: A prospective study of
C-peptide, insulin-like growth factor-I, insulin-like growth factor
binding protein-1, and the risk of colorectal cancer in women.
Cancer Epidemiol Biomarkers Prev. 14:850–855. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Le Marchand L, Wang H, Rinaldi S, Kaaks R,
Vogt TM, Yokochi L and Decker R: Associations of plasma C-peptide
and IGFBP1 levels with risk of colorectal adenoma in a multiethnic
population. Cancer Epidemiol Biomarkers Prev. 19:1471–1477.
2010.PubMed/NCBI
|
|
22
|
Rajwani A, Ezzat V, Smith J, Yuldasheva
NY, Duncan ER, Gage M, Cubbon RM, Kahn MB, Imrie H, Abbas A,
Viswambharan H, Aziz A, Sukumar P, Vidal-Puig A, Sethi JK, Xuan S,
Shah AM, Grant PJ, Porter KE, Kearney MT and Wheatcroft SB:
Increasing circulating IGFBP1 levels improves insulin sensitivity,
promotes nitric oxide production, lowers blood pressure, and
protects against atherosclerosis. Diabetes. 61:915–924. 2012.
View Article : Google Scholar
|
|
23
|
Taub R: Liver regeneration 4:
transcriptional control of liver regeneration. FASEB J. 10:413–427.
1996.PubMed/NCBI
|
|
24
|
Leu JI, Crissey MA, Craig LE and Taub R:
Impaired hepatocyte DNA synthetic response posthepatectomy in
insulin-like growth factor binding protein 1-deficient mice with
defects in C/EBP beta and mitogen-activated protein
kinase/extracellular signal-regulated kinase regulation. Mol Cell
Biol. 23:1251–1259. 2003. View Article : Google Scholar
|
|
25
|
Ink SL and Henderson LM: Vitamin B6
metabolism. Ann Rev Nutr. 4:455–470. 1984. View Article : Google Scholar
|
|
26
|
Nakari M, Kanouchi H and Oka T: High dose
of pyridoxine induces IGFBP-3 mRNA expression in MCF-7 cells and
its induction is inhibited by the p53-specific inhibitor
pifithrin-α. J Nutr Sci Vitaminol (Tokyo). 57:280–284.
2011.PubMed/NCBI
|
|
27
|
Frost RA, Nystrom GJ and Lang CH:
Stimulation of insulin-like growth factor binding protein-1
synthesis by interleukin-1beta: requirement of the
mitogen-activated protein kinase pathway. Endocrinology.
141:3156–3164. 2000.PubMed/NCBI
|
|
28
|
Leu JI, Crissey MA, Leu JP, Ciliberto G
and Taub R: Interleukin-6-induced STAT3 and AP-1 amplify hepatocyte
nuclear factor 1-mediated transactivation of hepatic genes, an
adaptive response to liver injury. Mol Cell Biol. 21:414–424. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Magne L, Blanc E, Marchand A, Fafournoux
P, Barouki R, Rouach H and Garlatti M: Stabilization of IGFBP1 mRNA
by ethanol in hepatoma cells involves the JNK pathway. J Hepatol.
47:691–698. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kanouchi H, Shibuya M, Tsukamoto S,
Fujimura Y, Tachibana H, Yamada K and Oka T: Comparisons of uptake
and cell surface binding among pyridoxal, pyridoxine, and
pyridoxamine in RAW264.7 cells. Nutrition. 26:648–652. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lumeng L, Brashear RE and Li TK: Pyridoxal
5′-phosphate in plasma: source, protein-binding, and cellular
transport. J Lab Clin Med. 84:334–343. 1974.
|
|
32
|
Anderson BB, O’Brien H, Griffin GE and
Mollin DL: Hydrolysis of pyridoxal-5′-phosphate in plasma in
conditions with raised alkaline phosphate. Gut. 21:192–194.
1980.
|
|
33
|
Sakurai T, Asakura T, Mizuno A and Matsuda
M: Absorption and metabolism of pyridoxamine in mice. I. Pyridoxal
as the only form of transport in blood. J Nutr Sci Vitaminol
(Tokyo). 37:341–348. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Leppä S, Saffrich R, Ansorge W and Bohmann
D: Differential regulation of c-Jun by ERK and JNK during PC12 cell
differentiation. EMBO J. 17:4404–4413. 1998.PubMed/NCBI
|
|
35
|
Shen J, Lai CQ, Mattei J, Ordovas JM and
Tucker KL: Association of vitamin B-6 status with inflammation,
oxidative stress, and chronic inflammatory conditions: the Boston
Puerto Rican Health Study. Am J Clin Nutr. 91:337–342. 2010.
View Article : Google Scholar : PubMed/NCBI
|