Introduction
Mature microRNAs (miRNAs) are single-stranded RNA
molecules, between 20- and 23-nucleotides in length, that control
gene expression in cellular processes. These molecules typically
decrease the stability of mRNAs, including those of genes that
mediate processes in tumorigenesis, including cell cycle
regulation, differentiation, apoptosis, invasion and stress
responses (1). miRNAs were
initially linked to tumorigenesis due to their proximity to
chromosomal breakpoints and their unregulated expression levels in
a number of malignancies. Given their integral role in development,
miRNAs were observed as being significant in tumorigenesis. miRNA
targeting is achieved through specific base-pairing interactions
between the ‘seed’ region of the miRNA and sites within coding and
untranslated regions of mRNAs. Target sites in the 3′ untranslated
region (UTR) lead to more effective mRNA destabilization.
As the most abundant liver-specific miRNA, miR-122,
was identified as constituting 70% of total hepatic microRNA, while
cloning small RNAs from different tissues of the body (2,3). Its
expression was activated gradually with the growth of embryonic
development. miR-122 is implicated in fatty acid and cholesterol
metabolism, and depletion of miR-122 by an antisense oligopeptide
lowers the cholesterol level in plasma (4). miR-122 is also required by the
hepatitis C virus and is frequently suppressed in primary
hepatocellular carcinomas (HCCs) (5).
To determine the physiological roles of miR-122 and
the biotransformation of hepatoma carcinoma cells highly expressing
miR-122, the Hep3B cell line, which stably overexpressed miR-122,
was generated by gene transfection methods and drug screening.
Materials and methods
Cell cultures
Hep3B was obtained from the Chinese Academy of
Sciences Cell Bank (Shanghai, China) and the cell line was
maintained in Dulbecco’s modified Eagle’s medium supplemented with
10% fetal bovine serum. A humidified incubator was set at 37°C, 5%
CO2.
Plasmid construction
The oligonucleotide chains Pre-miR-122-1 and
Pre-miR-122-2, which contained SmaI and EcoRI
endonucleases, were designed by precursor sequences of has-miR-122
from miRBASE (http://www.mirbase.org/cgi-bin/mirna_entry.pl?acc=MI0000442)
and were as follows: PreMIR-122-1, 5′-TCCCCCGGGGGACCGGGA
TCCGCCTTAGCAGAGCTGTGGAGTGTGACAATGGTG
TTTGTGTCTAAACTATCAAACG-3′; and PreMIR-122-2, 5′-TCCCCCGGGGGACGGAATTCAAAAAAGGCCTAGCAG
TAGCTATTTAGTGTGATAATGGCGTTTGATAGTTTAG CA-3′ (restriction sites
indicated by underlining). The 3′UTR of miR-122-Luc, containing the
miR-122 response element (miR-122-Luc-1, 5′-CTAGAACAAA
CACCATTGTCACACTCCAT-3′; miR-122-Luc-2, 5′-CTA
GATGGAGTGTGACAATGGTGTTTGTT-3′), was cloned into the
pGL-control vector plasmid (Promega Corporation, Madison, WI, USA),
which contained the XbaI endonuclease. The double-strand was
cloned into pGL-control vector to acquire the recombinant plasmid
pGL3-miR122-Luc.
Stable transfection of miR-122
pSIREN-miR-122 was transfected into Hep3B cells
using Effectene® Transfection Reagent (Qiagen, Hilden,
Germany) according to the manufacturer’s instructions. After 24 h,
the fresh culture medium was replaced with 2 μg/ml puromycin to
obtain the positive monoclonal cells which expressed stably
(Hep3B-122 cells). Hep3B-pSIREN-RetroQ cells were generated by
transfecting Hep3B cells with RNAi-Ready pSIREN-RetroQ vector as
the plasmid control group (Hep3B-Q).
Dual-luciferase reporter assay
The 3′UTR fragments of the candidate target genes
were subcloned into the XbaI site downstream of the
luciferase gene in the pGL3-control vector (Promega Corporation).
Under the control of the SV40 promoter and enhancer, pGL3-control
vector expresses marked luciferase activity in numerous types of
mammalian cells. To verify pSIREN-miR-122’s activity in cells,
pGL3-miR122-Luc, pSIREN-miR-122 and pRL-SV40 were transfected in
Hep3B at the same time using a Dual-Luciferase® Reporter
assay System kit (Promega Corporation) according to the
manufacturer’s instructions. The luciferase activity of Firefly and
Renilla was obtained from pGL3-miR122-Luc and pRL-SV40 by Infinite
200 Pro multimode plate readers (Tecan Systems Inc., San Jose, CA,
USA).
qPCR
RNA was harvested using TRIzol (Invitrogen, Hong
Kong, China) according to the manufacturer’s instructions.
Quantification of total RNA was performed using a NanoDrop ND-1000
spectrophotometer (NanoDrop, Thermo Scientific, Wilmington, DE,
USA). To analyze miRNA expression, PCR master mix (2×; Superarray,
Valencia, CA, USA) was used to quantify levels of mature miRNAs
according to the manufacturer’s instructions. The PCR reaction was
as follows: enzyme activation for 10 min at 95°C followed by 40
cycles of 95°C for 15 sec and 60°C for 60 sec. Primers are shown in
Table I. Data was analyzed using
the 2-ΔΔCT method.
 | Table IQuantitative polymerase chane reaction
primers. |
Table I
Quantitative polymerase chane reaction
primers.
| Gene | Primer | Sequence |
|---|
| miR-122 | Reverse
transcription |
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCAAACAC-3′ |
| Forward |
5′-ACACTCCAGCTGGGTGGAGTGTGACAATGGTGTTTG-3′ |
| Reverse |
5′-CTCAACTGGTGTCGTGGA-3′ |
| U6 | Forward |
5′-CTCGCTTCGGCAGCACA-3′ |
| Reverse |
5′-AACGCTTCACGAATTTGCGT-3′ |
Results
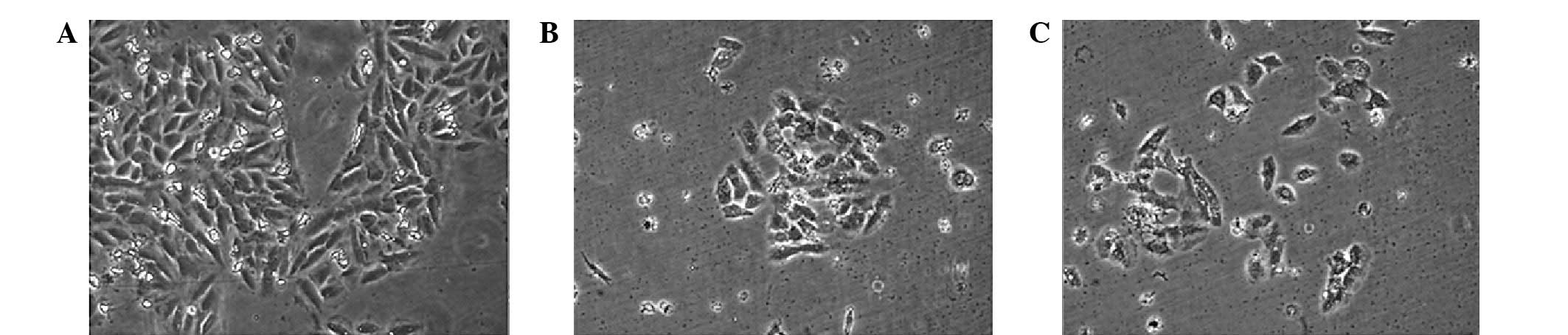
Cell reproduction rate was decreased when visualized
following 48 h transfection; however cellular morphology did not
exhibit significant differences (Fig.
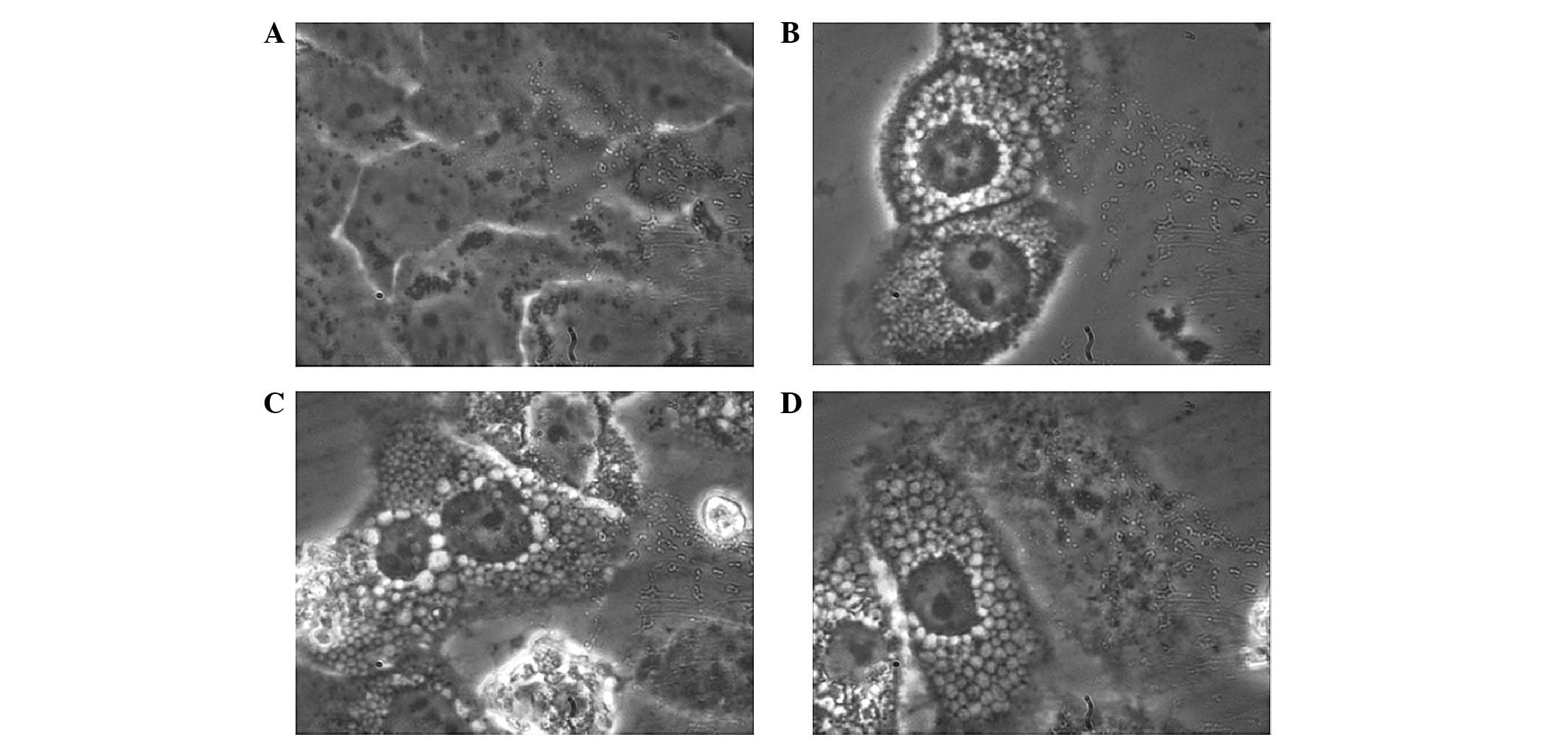
1). Ten hours later (~60 h following transfection), there was a
significant morphological change observed at high magnification
(Fig. 2). A significant boundary
between the cytoplasm and nucleus was observed with the increase in
cell volume. In addition, the nucleolus and nuclear matrix/skeleton
were enlarged and easy to recognize. The nucleolus was more easily
distinguishable compared with in the control group. Notably, the
cavitating structures of varying sizes appeared throughout the
cytoplasm and increased in size with increased proximity to the
nucleus.
To utilise the puromycin resistance the RNAi-Ready
pSIREN-RetroQ vector possesses, the Hep3B cell lines were obtained
with stable high expression of miR-122 by high intensity-drug
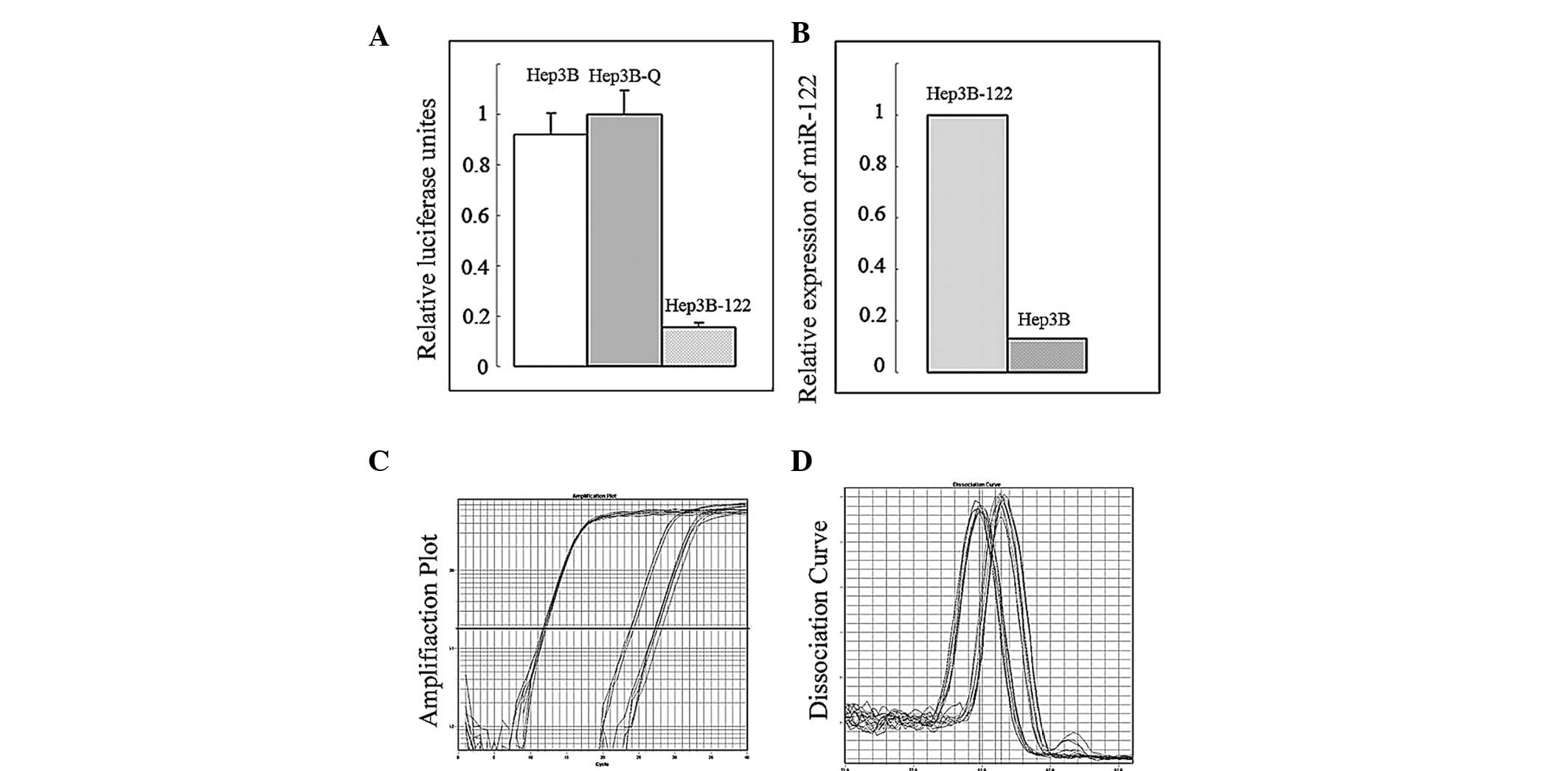
screening. To verify the content and activity expressive of miR-122
in cells, a Dual-Luciferase® Reporter assay and
quantitative PCR were performed (Fig.
3). Compared with the control group, the pSIREN-miR-122 plasmid
was capable of expressing the mature series of miR-122 abundantly
and miR-122 efficiently performed biological functions that
inhibited the expression of the target gene.
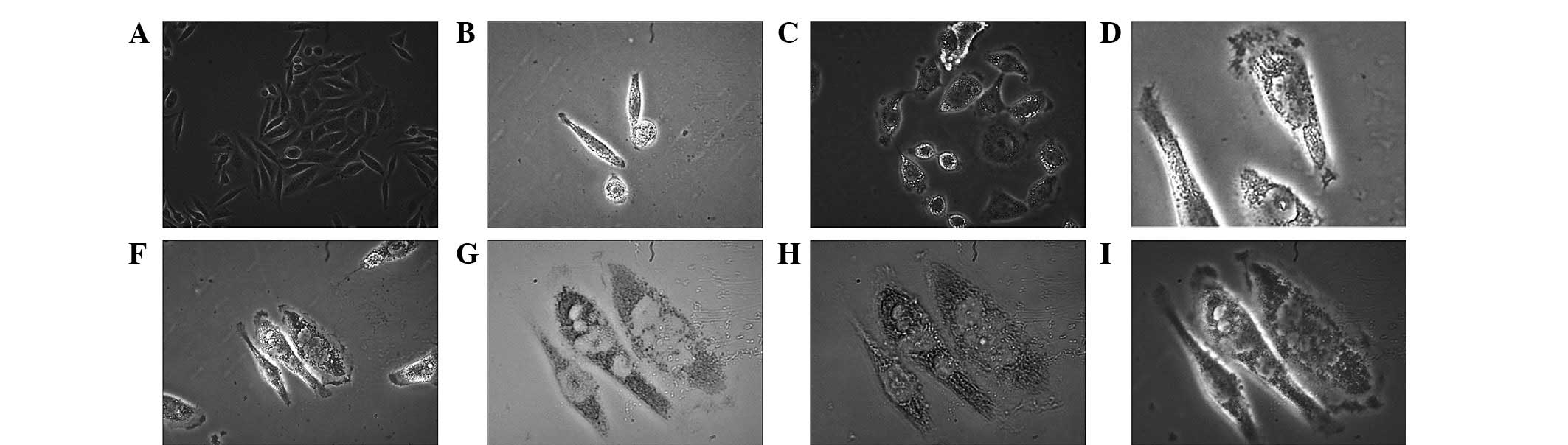
In the stage of stable high expression of miR-122
following drug screening and miR-122 detection, Hep3B-122 cells
showed further characteristic features (Fig. 4). Simultaneous to the increase in
volume, cells swelled and expanded and the edges became smooth and
bright. In addition, similar to a lipid granule, a transparent
droplet gathered at the edge or two poles of the cells and a
vacuole appeared in the transfection phase (~60 h after
transfection) and gradually integrated into larger vacuoles in the
center.
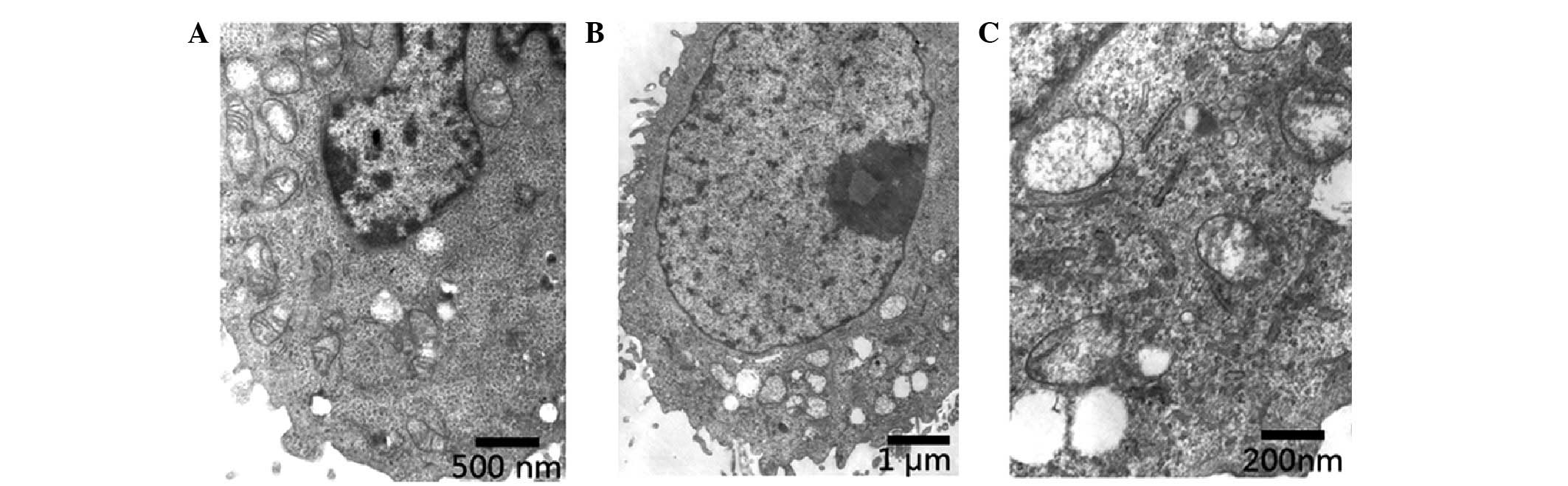
To further identify this transformation, the
Hep3B-122 group and the control group were scanned by transmission
electron microscopy and compared with the control groups (Fig. 5). The nucleus was observed to be
enlarged, the nucleus was located at the edge of the nucleus and
the quantity of heterochromatin decreased. In addition to the
swelling of mitochondria, the double membrane structure was not
clear as the mitochondria exhibited cavities. Furthermore, the
cytoplasm was vacuolated and the microvilli on the cell membrane
were damaged.
Discussion
Biomolecules are generated by healthy livers for
export to consumer tissues of the body, including proteins, fats,
carbohydrates, vitamins and minerals. Protein and fat are
decomposed into amino acids and fatty acids, respectively, in the
liver which contains a large number of mitochondria. Mitochondria
are known as the ‘energy factory’ as they are the primary producer
of adenosine triphosphate in cells. The balance of liver energy and
material metabolism is destroyed in hepatocarcinoma and hepatocytes
metabolize glucose into lactate, even in the presence of sufficient
oxygen to support mitochondrial oxidative phosphorylation, a
phenomenon referred as the ‘Warburg Effect’ (6). The liver-specific miR-122 is
frequently suppressed in primary HCCs. Thus, it may be concluded
that miR-122 is required to maintain and promote the aforementioned
functions and metabolism. In the current study, a number of
cavities of varying sizes appeared throughout the cytoplasm 60 h
after transfection. Following the integration of the recombinant
plasmid, there was high expression of miR-122. miR-122 maintains
and promotes the hepatocyte’s functions and metabolism. Notably,
the cavitating structure was observed to be endoplasmic reticulum
(ER); miRNA mediates ER more sensitively and earlier than the
thousands of genes in the synthetic process of ER. (7). Cavities located closer to the nucleus
were larger. A thick ER was observed with ER-associated ribosomes
connected to the nucleus by nuclear pore complexes that contain
central transporter linked nucleoplasmic rings with a cytoplasmic
ring. Bcl-2 resides in the nuclear envelope, ER and outer
mitochondrial membrane in a nonuniform distribution and is
suggested to have a role in protein complexes that may be involved
in certain aspects of transport (8). miR-122 targets the Bcl family
directly, which is repressed by elevated levels of miR-122, and is
an endogenous apoptosis regulator in these HCC-derived cell lines
(9). It may be inferred that in
Hep3B-122 cells, the ER may cause changes in structural
absorbance.
The morphology of a cancer cell may be modulated by
miRNA activity (10,11) and a number of miRNA-overexpressing
cells exhibited markedly different morphologies (12). miR-122 is significantly
downregulated in liver cancers and restoration of miR-122
significantly decreased in vitro migration, invasion and
anchorage-independent growth, as well as in vivo
tumorigenesis, angiogenesis and intrahepatic metastasis in an
orthotopic liver cancer model (13). The results of the present study
showed that cell morphology and cell viability were altered.
Following drug screening, Hep3B-122-treated cells appeared more
translucent compared with the control group, with the appearance of
a lipid granule-like vacuoles. miR-122 has previously been linked
to the regulation of cholesterol and lipid metabolism (14) and the knockdown of miR-122
expression has previously been recognized to result in the
downregulation of cholesterol and lipid-metabolizing enzymes
(15). Conversely, in the present
study the overexpression of miR-122 in Hep3B-122 cells resulted in
the accumulation of lipid granule-like transparent droplets at the
cell periphery or the two poles of the cells suggesting that
miR-122 upregulates cholesterol and lipid metabolism. miR-122 is
essential for hepatitis C virus (HCV) RNA accumulation in cultured
liver cells and it stimulates the translation of HCV RNA.
Therefore, virus infection and lipodystrophy separately promote the
elevated expression of miR-122 in the liver although how they
interact is unclear.
Although miR-122 regulates mitochondrial metabolism
and its loss may be detrimental to sustaining critical liver
function in a number of studies (5,16),
high expression of miR-122 also stimulates the translation of HCV
RNA in the liver. The HCV core protein alters mitochondrial
function and results directly in an increase in the cellular
abundance of reactive oxygen species, with consequent increases in
cellular lipid peroxidation (16).
Retrospective analysis provided evidence that silencing of miR-122
downregulates the replication of HCV for therapeutic purpose. By
contrast, miR-122, as a tumor suppressor, targets anti-apoptotic
genes to inhibit the tumorigenic properties of HCC (10). Thus, miR-122 performs specific
functions at various expression levels. The present study; however,
did not provide an explanation for the regulation of mitochondrial
metabolism. Transmission electron microscopy images provide novel
evidence that elevated expression of miR-122 may not be beneficial
to mitochondria. miR-122 caused an increase in the size of the
nucleus and relocated the nucleus to the edge of the nucleus and in
addition to the swelling of mitochondria, the double membrane
structure was not as clear due to mitochondrial cavities.
In conclusion, miR-122 was observed to markedly
alter the cell morphology and negatively regulate mitochondria.
miR-122 function was observed to correlate with its expression
level in HCC cells.
Acknowledgements
This study was financially supported by grants from
the National Natural Science Foundation of China (grant no.
81160529) and the Jilin Province Science and Technology Development
Project (grant no. 200905207).
References
|
1
|
Farazi TA, Spitzer JI, Morozov P and
Tuschl T: miRNAs in human cancer. J Pathol. 223:102–115. 2011.
View Article : Google Scholar
|
|
2
|
Chang J, Guo JT, Jiang D, et al:
Liver-specific microRNA miR-122 enhances the replication of
hepatitis C virus in nonhepatic cells. J Virol. 82:8215–8223. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lagos-Quintana M, Rauhut R, Yalcin A, et
al: Identification of tissue-specific microRNAs from mouse. Curr
Biol. 12:735–739. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chang JH, Nicolas E, Marks D, et al:
miR-122, a mammalian liver-specific microRNA, is processed from hcr
mRNA and may downregulate the high affinity cationic amino acid
transporter CAT-1. RNA Biol. 1:106–113. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lanford RE, Hildebrandt-Eriksen ES, Petri
A, et al: Therapeutic silencing of microRNA-122 in primates with
chronic hepatitis C virus infection. Science. 327:198–201. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Skárka L and Ostádal B: Mitochondrial
membrane potential in cardiac myocytes. Physiol Res. 51:425–434.
2002.
|
|
7
|
Mato JM and Lu SC: The hepatocarcinogenic
effect of methionine and choline deficient diets: an adaptation to
the Warburg effect? Alcohol Clin Exp Res. 35:811–814. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Selbach M, Schwanhäusser B, Thierfelder N,
Fang Z, Khanin R and Rajewsky N: Widespread changes in protein
synthesis induced by microRNAs. Nature. 455:58–63. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Krajewski S, Tanaka S, Takayama S,
Schibler MJ, Fenton W and Reed JC: Investigation of the subcellular
distribution of the bcl-2 oncoprotein: residence in the nuclear
envelope, endoplasmic reticulum, and outer mitochondrial membranes.
Cancer Res. 53:4701–4714. 1993.
|
|
10
|
Lin CJ, Gong HY, Tseng HC, Wang WL and Wu
JL: miR-122 targets an anti-apoptotic gene, Bcl-w, in human
hepatocellular carcinoma cell lines. Biochem Biophys Res Commun.
375:315–320. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tavazoie SF, Alarcón C, Oskarsson T, et
al: Endogenous human microRNAs that suppress breast cancer
metastasis. Nature. 451:147–152. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sossey-Alaoui K, Bialkowska K and Plow EF:
The miR200 family of microRNAs regulates WAVE3-dependent cancer
cell invasion. J Biol Chem. 284:33019–33029. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Elson-Schwab I, Lorentzen A and Marshall
CJ: MicroRNA-200 family members differentially regulate
morphological plasticity and mode of melanoma cell invasion. PLoS
One. 5:e131762010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tsai WC, Hsu PW, Lai TC, et al:
MicroRNA-122, a tumor suppressor microRNA that regulates
intrahepatic metastasis of hepatocellular carcinoma. Hepatology.
49:1571–1582. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Krützfeldt J, Rajewsky N, Braich R, Rajeev
KG, Tuschl T, Manoharan M and Stoffel M: Silencing of microRNAs in
vivo with ‘antagomirs’. Nature. 438:685–689. 2005.
|
|
16
|
Burchard J, Zhang CS, Liu AM, et al:
microRNA-122 as a regulator of mitochondrial metabolic gene network
in hepatocellular carcinoma. Mol Syst Biol. 6:4022010. View Article : Google Scholar : PubMed/NCBI
|