Introduction
Tuberculosis (TB) is a chronic infectious disease
caused by the bacterium Mycobacterium tuberculosis and it
remains a significant public health risk worldwide. The pestilence
of TB in the Cosmopolitan population has been eased with the
introduction of anti-TB and anti-HIV drugs during the 1980’s
(1,2). Pneumoconiosis is an important
occupational disease in employees of the Huainan coal mine. Once
pneumoconiosis is associated with TB, not only is it accelerated,
but it also worsens. The prognosis of patients with the disease is
usually poor and the curative effect of current treatments is not
as good as expected. At present, the therapeutic regimen includes
curing pneumoconiosis and TB; however, anti-TB drugs are key for
curing the disease since pneumoconiosis lacks a radically curative
drug. As the first-elected anti-TB drug, the clinical therapeutic
efficacy of isoniazid (INH) has markedly decreased due to the
emergence of multi-drug-resistant and mycobacterial cell
wall-deficient strains. At present, MDR-TB is a major drawback for
the complete elimination of TB (3). According to the statistics, mutations
in the katG gene in MDR-TB account for 40–90% of
INH-resistant strains (4),
however, no studies investigating the resistance of the L-form have
been conducted. The present study applied a DNA sequence analysis
technique for analyzing the mutational characteristics of the
katG gene in MDR-TB L-form isolates in order to gain further
information on the mechanisms underlying drug resistance in
MDR-TB.
Materials and methods
Experimental subjects
The subjects included male patients aged 42–71 years
old (mean age, 55.15±8.75 years) diagnosed with stage II and III
pneumoconiosis complicated with TB in the active stage (n=114),
treated in The Affiliated Hospital of Anhui University of Science
and Technology (Huainan, China) between May 2011 and May 2012. All
the subjects were instructed to spit phlegm from the bottom of the
trachea into a sterile wide-mouthed bottle following gargling
several times. The strain used for quality control was
H37Rv, which was provided by the National Institute for
the Control of Pharmaceutical and Biological products (Beijing,
China). Patient consent was obtained from all patients. Approval
was obtained from the Ethics Committee of the School of Medicine,
Anhui University of Science and Technology (Huainan, China).
Experimental methods
MDR-TB L-form identification
In accordance with the book of the Chinese Medical
Laboratory, an indirect absolute concentration method was adopted
(1,5). INH and rifampin (RFP), used to detect
drug sensitivity of MDR-TB L-forms, were purchased from
Becton-Dickinson (Franklin Lakes, NJ, USA). Firstly, the medicine
(INH and RFP) was added to 92-3 TB-L liquid culture medium (Bengbu
Medical College, Bengbu, China), respectively, to prepare the
antimicrobial susceptibility medium. The concentrations used were
10 μg/ml (high concentration) and 1 μg/ml (low concentration) for
INH, and 250 μg/ml (high concentration) and 50 μg/ml (low
concentration) for RFP. Under bacteria-free conditions, 0.1 ml of
specimen was removed and inoculated to the medium. Simultaneously,
the blank control used medium without medicine and 0.1 ml of
specimen was added. Subsequently, the medium was mixed thoroughly
using a dropper and placed at 37°C for 3 weeks. The medium was
observed three days later and three times every week for 4 weeks to
determine whether or not there was growth on the surface or bottom
of the medium. If there was growth, a deposit was obtained to
produce a smear, and was identified by intensified Kinyoun acid
fast staining. All the specimens were resistant to INH and RFP at
low concentrations, and the specimens resistant to high
concentrations were identified as the MDR-TB L-form.
DNA extraction
The sputum specimens of patients were inactivated
with autoclave. Genomic DNA was extracted using DNA extraction kits
(Takara Biotech, Dalian, China). DNA lysate (200 μl) was added to
the inactivated specimen, following a water bath at 55°C for 1–3 h
and at 95°C for 5 min. The lysate was extracted twice with
phenol:chloroform:isoamyl alcohol (25:24:1). DNA was precipitated
by adding ammonium acetate. Two volumes of pure ethanol were added,
followed by incubation at −20°C overnight. DNA was pelleted by
centrifugation at 12,000 × g for 15 min, washed with 70% ethanol,
air-dried and dissolved in the TE buffer (6).
Primer synthesis
The primers were synthesized by Sangon Biotech Co.
(Shanghai, China) according to the MDR-TB conserved genomic DNA
sequence (NM_X68081). The primer sequences for katG, used in
the present study were as follows: Forward: 5′-CGCGATGAGCGTTACAG-3′
and reverse: 5′-CGTCCTTGGCGGTGTATTG-3, with a product size of 458
bp.
Polymerase chain reaction (PCR) and
sequence analysis
The total PCR reaction volume was 50 μl and included
5.0 μl of 10X buffer (15 mmol/l MgCl2), 4.0 μl dNTP (200
μmol/l), 1.0 μl of each primer (25 pmol/μl), 0.5 μl Taq DNA
polymerase (5 U/μl) and 4 μl of the DNA template. Deionized water
was added to produce a total volume of 50 μl. The reaction
conditions were: 95°C for 5 min of denaturation, 94°C for 30 sec,
55°C for 90 sec and 72°C for 30 sec, for 33 cycles in total,
followed by maintenance at 72°C for 10 min. The amplified product
(5 μl) was separated by electrophoresis with 2% agarose gel
electrophoresis (containing 0.5 mg/ml ethidium bromide), then
images were captured and analyzed using the Image Master Totallab
software (Nonlinear Dynamics Ltd., Durham, NC, USA). A 458 bp
specific band indicated that a mutation in katG existed,
otherwise it was negative. The DNA sequences of the PCR products
were detected using an ABI PRISM 7700 sequencer (Takara
Biotech).
Statistical analysis
Statistical analyses were performed using SPSS 12.0
software (SPSS, Inc., Chicago, IL, USA).
Results
Antimicrobial susceptibility test (AST)
analysis
In 114 cases of MTB L-form isolated from sputum
samples, 31 cases of MDR-TB L-form isolates were detected, and the
resistance rate was 27.19% (31/114). In total, 42 strains were
detected to tolerate INH, including 20 strains with high-level
resistance and 42 strains with low-level resistance. In addition,
101 strains were detected to tolerate RFP, including 53 strains
with high-level resistance and 101 strains with low-level
resistance (Table I).
 | Table IResults of drug resistance using the
antimicrobial susceptibility test analysis in 114 cases of
Mycobacterium tuberculosis L-forms. |
Table I
Results of drug resistance using the
antimicrobial susceptibility test analysis in 114 cases of
Mycobacterium tuberculosis L-forms.
| Group | Drug concentration
(μg/ml) and results | Total (no. of
cases) |
|---|
|
|---|
| INH | RFP |
|---|
|
|
|---|
| 10 | 1 | 250 | 50 |
|---|
| 1 | + | + | − | − | 4 |
| 2 | − | − | − | + | 39 |
| 3 | − | − | + | + | 31 |
| 4 | + | + | + | + | 10 |
| 5 | − | + | + | + | 12 |
| 6 | − | + | − | − | 7 |
| 7 | − | + | − | + | 3 |
| 8 | + | + | − | + | 6 |
| 9 | − | − | − | − | 2 |
| Total | 20 | 42 | 53 | 101 | 114 |
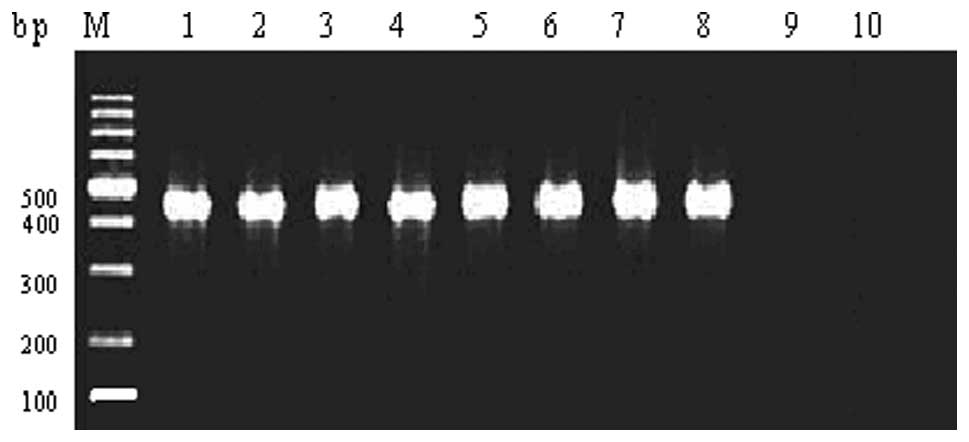
PCR analysis
In 31 cases of MDR-TB L-form strains isolated from
sputum samples, 19 INH-resistant strains and 12 INH-sensitive
strains were identified (Fig.
1).
Sequence analysis of DNA of the MDR-TB
L-form
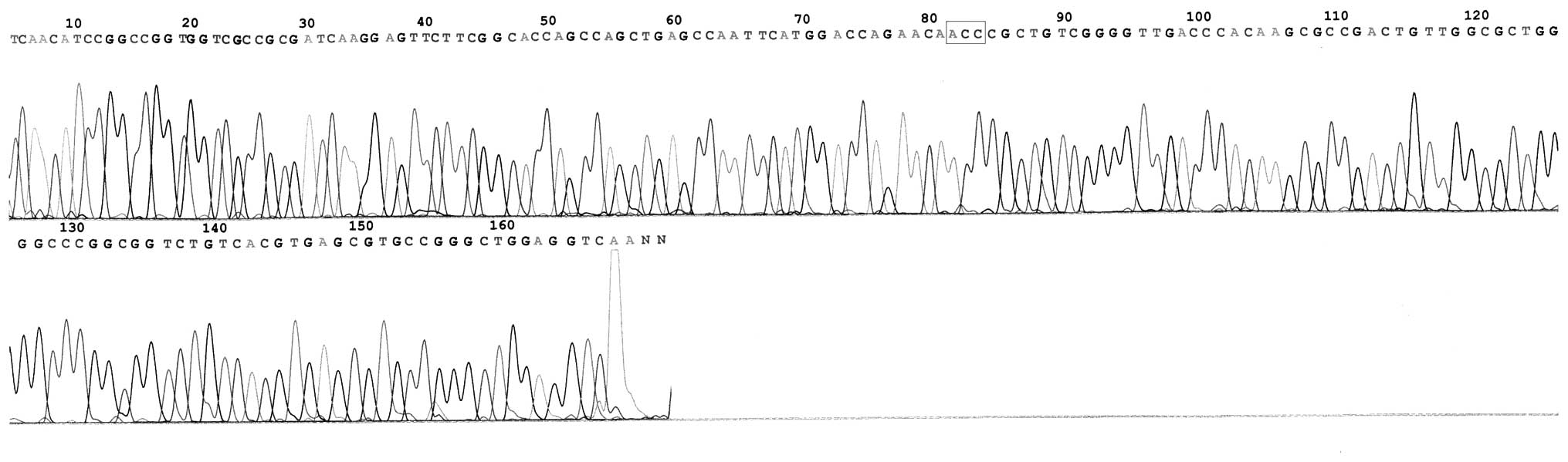
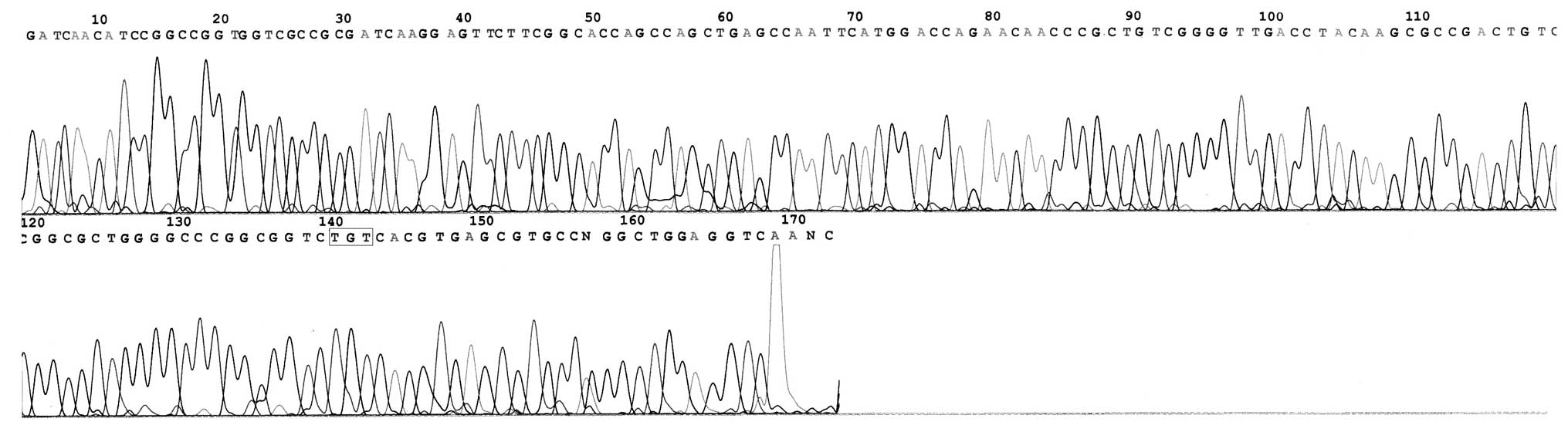
In total, 19 mutational strains of the katG
gene were identified in 31 INH-resistant strains. The mutation rate
was 61.29% (19/31; Table II) and
mutations were mainly concentrated in codon 315 (58.06%; 18/31) and
431 (3.23%; 1/31). Base substitutions were detected, however, no
multisite mutations were identified (Figs. 2–4). The former included 15 strains of
Ser→Thr (AGC→ACC; 48.39%) and 3 strains of Ser→Asn (AGC→AAC;
9.68%), and the latter was chiefly Ala→Val (GCG→GTG). No mutation
in katG was identified in 10 randomly selected INH-sensitive
strains.
 | Table IIMutational characteristics of the
katG gene in 31 clinical drug-resistant strains of
multi-drug resistant Mycobacterium tuberculosis L-form. |
Table II
Mutational characteristics of the
katG gene in 31 clinical drug-resistant strains of
multi-drug resistant Mycobacterium tuberculosis L-form.
| Strain (no. of
cases) | Location of amino
acid | Codon change | Amino acid
change | Percentage (%) |
|---|
| 15 | 315 | AGC→ACC | Ser→Thr | 48.39 |
| 3 | 315 | AGC→AAC | Ser→Asn | 9.68 |
| 1 | 431 | GCG→GTG | Ala→Val | 3.23 |
| 12 | − | − | − | 38.17 |
Discussion
The World Health Organization and the International
Union Against Tuberculosis and Lung Disease identified that MDR-TB
refers to at least two types of drug resistance, including
resistance to RFP and INH (2). The
emergence of resistant Mycobacterium tuberculosis,
particularly the generation and propagation of MDR-TB, is an
important cause of the high global TB incidence. Compared with
other types of TB, MDR-TB is more severe and leads to a higher
incidence of mortality (7). Due to
a deficiency in the cell wall of bacteria, the biological
characteristics and drug sensitivity of the MDR-TB L-form is able
to be altered. Several previous studies have demonstrated that
common clinical therapeutic concentrations of RFP, INH and
streptomycin, used at the same time in order to inhibit or kill
MDR-TB, are also able to induce the formation of MTB L-form
bacteria, causing difficulty in the diagnosis and treatment of TB
(8,9). Therefore, there is an urgent
requirement to investigate the drug resistance mechanisms
underlying the MDR-TB L-form to aid in the development of anti-TB
drugs and novel diagnostic methods.
As the main anti-TB drug, INH is the basis for a
variety of drug and chemotherapy combined treatments for TB. The
molecular mechanisms underlying the resistance of MDR-TB to INH was
associated with the katG gene mutation that encodes
catalase-peroxidase. INH is a hydrazine chemical synthetic drug,
which is able to be oxidized to isonicotinic acid by the
catalase-peroxidase encoded by the katG gene that
participates in the synthesis of coenzyme I (NAD) to inhibit the
biosynthesis of mycolic acid of the cell wall in Mycobacterium
tuberculosis, so as to damage the MDR-TB’s barricade of
resisting antioxygen and invasion. Due to deletion or mutation in
the katG gene, resistance is able to be generated as the
enzymatic activity is lost or degraded, thus, inhibiting the
activation of INH (10–13).
The Mycobacterium tuberculosis L-form was also
termed the mycobacterium cell wall-deficient form. In 1960,
Mattmand (14) examined the
biological characteristics in detail and revealed that alterations
in the biological characteristics, drug sensitivity and DNA of the
L-form were owed to the partial or complete absence of the cell
wall. The L-form is a type of mutation. Several mechanisms are able
to induce the occurrence of this phenomenon, for instance,
chemotherapeutics, lysozymes and bacteriophages. The
Mycobacterium tuberculosis L-form possesses pathogenicity
and previously caused chronic transformation of the disease
process, a worse prognosis and lacked typical tubercles. This has
led to difficulty in diagnosing and treating TB (15).
In the present study, 31 cases of MDR-TB L-form
isolates were detected by AST analysis in 114 cases of MTB L-forms
isolated from sputum samples, and the resistance rate was 27.19%
(31/114). The present study demonstrated that the situation of
multidrug resistance is currently severe in MDR-TB L-form isolates
among patients with pneumoconiosis complicated with TB in the
Huainan coal mine, and that the commonly used concentrations of
anti-TB drugs in the clinic are less effective.
The results of PCR and gene sequence analysis
demonstrated that there were 19 mutational strains of katG
in 31 INH-resistant strains, the mutation rate was 61.29% (19/31),
mainly concentrated in codon 315 (Ser315Thr, AGC→ACC, 48.39%;
Ser315Asn, AGC→AAC, 9.68%) and 431 (Ala431Val, GCG→GTG, 3.23%), and
involved base substitutions. The results indicated that point
mutations in katG of MDR-TB L-forms concentrated in codon
315 (94.74%; 18/19), were greater than the mutation rate of
bacteric MDR-TB, which was reported to be 50–60% in previous
studies (16,17).
Furthermore, the present study also demonstrated
that katG mutations in 12 INH-resistant isolates (38.17%;
12/31) were not detected. This verified that other mechanisms
leading to INH resistance in the MDR-TB L-form exist. In addition,
no point mutations, multisite mutations, large fragment deletions
and insertion mutations were identified at codons 327, 144 and 143
(4). The probable cause may be
associated with novel characteristics of mutations of the MDR-TB
L-form and the effects of sampling error, thus, it must be verified
using larger sample sizes.
In conclusion, sequence analysis of DNA is the most
reliable method for detecting gene mutations, not only can it be
used for mutational screening, but it is also able to determine the
mutation site. The present study investigated the mutational
characteristics of the drug-resistant gene of katG in MDR-TB
L-form among patients with pneumoconiosis complicated with TB, and
provided an experimental basis for the clinical diagnosis and
treatment of the disease.
Acknowledgements
This study was supported by a grant from the
National Natural Science Foundation of China (grant no. 81172778)
and a grant from the Natural Science Foundation of Anhui Province
(grant nos. KJ2010A087 and KJ2012A081).
References
|
1
|
Li YL: The Book of Chinese Medical
Laboratory Science. 1st edition. People’s Medical Publishing House
Co Ltd; Beijing: pp. 1119–1120. 2000, (In Chinese).
|
|
2
|
du Toit LC, Pillay V and Danckwerts MP:
Tuberculosis chemotherapy: current drug delivery approaches. Respir
Res. 7:1182006.PubMed/NCBI
|
|
3
|
Telenti A and Iseman M: Drug-resistant
tuberculosis: what do we do now? Drugs. 59:171–179. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gui J, Wang F, Li JL, et al: Genetic and
phenotypic characterization of drug-resistant Mycobacterium
tuberculosis isolates in Shenzhen of China. Chin J Microbiol
Immunol. 30:466–471. 2010.(In Chinese).
|
|
5
|
Zhu M, Xia P, Zhang Y, et al: Detecting
mycobacteria and their L-forms in peripheral blood from pulmonary
tuberculosis patients by cultivation with hemolyzed-centrifugated
blood in liquid medium. Zhonghua Jie He He Hu Xi Za Zhi.
23:556–558. 2000.(In Chinese).
|
|
6
|
Cheng XD, Yu WB, Bie LF, et al: The PCR in
detecting INH drug resistant genetic mutation of Mycobacterium
tuberculosis. Di 4 Jun Yi Da Xue Xue Bao. 24:849–851. 2003.(In
Chinese).
|
|
7
|
Raviglione MC, Dye C, Schmidt S and Kochi
A: Assessment of worldwide tuberculosis control. Lancet.
350:624–629. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lu J, Ye S, Li CP, et al: Sequence
analysis on drug-resistant gene of rpoB in MDR-TB among
pneumoconiosis patients complicated with tuberculosis in Huainan
mining district. Zhonghua Lao Dong Wei Sheng Zhi Ye Bing Za Zhi.
30:579–581. 2012.(In Chinese).
|
|
9
|
Wang H and Chen Z: Observations of
properties of the L-form of M. tuberculosis induced by the
antituberculosis drugs. Zhonghua Jie He He Hu Xi Za Zhi. 24:52–55.
2001.(In Chinese).
|
|
10
|
Ferguson LA and Rhoads J:
Multidrug-resistant and extensively drug-resistant tuberculosis:
the new face of an old disease. J Am Acad Nurse Pract. 21:603–609.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Drobniewski FA and Wilson SM: The rapid
diagnosis of isoniazid and rifampicin resistance in
Mycobacterium tuberculosis - a molecular story. J Med
Microbiol. 47:189–196. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shim TS, Yoo CG, Han SK, Shim YS and Kim
YW: Isoniazid resistance and the point mutation of codon 463 of
katG gene of Mycobacterium tuberculosis. J Korean Med Sci.
12:92–98. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lee H, Cho SN, Bang HE, et al: Exclusive
mutations related to isoniazid and elihionamide resistance among
Mycobacterium tuberculosis isolates from Korea. Int J Tuberc
Lung Dis. 4:441–443. 2000.PubMed/NCBI
|
|
14
|
Mattmand LH: Variation in mycobacteria. Am
Rev Respir Dis. 202:82–85. 1960.
|
|
15
|
Lu J, Ye S, Li CP, et al: Drug resistance
of tuberculosis mycobacteria L forms and related gene mutation of
tuberculosis patients with pneumoconiosis in Huainan mine area.
Zhonghua Lao Dong Wei Sheng Zhi Ye Bing Za Zhi. 25:369–370.
2007.(In Chinese).
|
|
16
|
Abe C, Kobayashi I, Mitarai S, et al:
Biological and molecular characteristics of Mycobacterium
tuberculosis clinical isolates with low-level resistance to
isoniazid in Japan. J Clin Microbiol. 46:2263–2268. 2008.PubMed/NCBI
|
|
17
|
Zhu M, Fan YM and Sheng GP: Study on the
characteristics of mutations on mycobacterium tuberculosis
katG, katG, embB gene in Zhejiang province. Yi Xue Yan Jin Za Zhi.
37:26–29. 2008.(In Chinese).
|