Introduction
The leaves of Perilla frutescens are commonly
used as decorative elements in Asia, including China, Japan and
Taiwan. Dried red Perilla leaves are also used as ‘soyou’ in
Chinese herbal medicine and are components of ‘saiboku-to’, which
is a Japanese herbal formula that is commonly used to treat asthma.
Previous studies have reported that Perilla frutescens leaf
extracts (PLE) display a range of biological activities, including
inhibition of tumor necrosis factor (TNF)-α (1), suppression of IgA nephropathy
(2), and anti-inflammatory and
anti-allergic activity (3,4). However, the mechanisms underlying the
anti-inflammatory properties of PLE are not well understood.
Inflammation is a response of organisms to pathogens
and chemical or mechanical injury. Inflammatory response and tissue
damage are induced by inflammatory mediators generated through
upregulation of a number of inducible genes, including inducible
nitric oxide (iNOS), cyclooxygenase (COX)-2,
interleukin (IL)-6 and IL-8. Nitric oxide (NO)
is a messenger molecule that has critical functions in vascular
regulation, host immune defense, neuronal signal transduction, and
other pathways (5–7). NO is produced from the conversion of
L-arginine to L-citrulline by the nitric oxide synthase (NOS) in
the presence of oxygen and NADPH (8). NOS is inducible in macrophages and
hepatocytes, and is activated following infection (9). Therefore, inducible (i)NOS-derived NO
is an ubiquitous mediator of a wide range of inflammatory
conditions, and its expression level reflects the degree of
inflammation, thus providing a measure of the inflammatory response
(10,11). COX-2 also participates in immune
system modulation and is involved in pathophysiological events
(12). COX-2 is not expressed or
is slightly detectable in most tissues under normal conditions;
high expression of COX-2 following induction by pro-inflammatory
mediators such as bacterial lipopolysaccharide (LPS), is involved
in the pathogenesis of sepsis and inflammation (13,14).
In addition, compounds interfering with both iNOS and COX-2
generally act as inhibitors of a transcription factor required for
iNOS and COX-2 expression, nuclear factor (NF)-κB (15).
Macrophages play a crucial role in eliciting
NF-κB-related cascades at the acute phase of the inflammatory
response. LPS stimulation of mouse macrophages leads to increased
phosphorylation and activation of mitogen-activated protein kinases
(MAPKs), such as extracellular-signal-regulated kinase (ERK)1/2,
and c-Jun N-terminal kinase (JNK) (16). Therefore, the development of
specific drugs to inhibit the production of these inflammatory
modulators may be effective in the treatment of inflammatory
diseases. In the present study, we investigated the
anti-inflammatory effects and the underlying mechanism of a P.
frutescens leaf extract (PLE). Specifically, we studied the
regulation of gene expression of iNOS, COX-2, and
pro-inflammatory cytokines in RAW264.7 cells stimulated with
LPS.
Materials and methods
Chemicals and reagents
Aprotinin, leupeptin, LPS,
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide
(MTT), penicillin, streptomycin and other chemicals used in this
study were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum (FBS)
and supplements for cell culture were purchased from Gibco-BRL
(Gaithersburg, MD, USA). Antibodies targeting phosphorylated Erk1/2
(p-Erk1/2), total Erk1/2 (t-Erk1/2), phosphorylated JNK (p-JNK),
total JNK (t-JNK), phosphorylated p38 (p-p38), total p38 (t-p38),
IκBα and NF-κB (p65 subunit) were purchased from Cell Signaling
Technologies (Beverly, MA, USA). The antibody targeting the
glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was purchased from
Sigma-Aldrich. HRP-conjugated secondary antibodies targeting mouse
and rabbit IgGs were purchased from Abcam (Cambridge, UK). The
murine macrophage RAW264.7 cell line was obtained from the American
Type Culture Collection (ATCC; Rockville, MD, USA).
Preparation of P. frutescens leaf extract
(PLE)
Perilla frutescens plants were purchased from
a certified herbal pharmacy (Chung-Yi Chinese Herbal Medicine
Pharmacy, Taichung, Taiwan). After dehydration, 100 g of the leaves
were homogenized, and the powder was passed through a mesh (0.05
mm). The filtered powder was resuspended into 1 liter of 100%
methanol and stirred at room temperature for 24 h. Following
filtration on a Whatman no. 1 filter paper, the solution was
lyophilized. Stock solution (20 mg/ml) of the extract was prepared
in dimethylsulfoxide, and stored at −20°C until further use.
Cell cultures and treatment
The RAW264.7 cells were incubated in DMEM
supplemented with 0.1% sodium bicarbonate, 2 mM glutamine,
penicillin G (100 U/ml), streptomycin (100 μg/ml) and 10% FBS, and
were maintained at 37°C in a humidified incubator containing 5%
CO2. Following pre-incubation with different
concentrations of PLE for 4 h, 1 μg/ml LPS was added and then
incubated for 2 h (NO assay), 3 h (RT-PCR and qRT-PCR analysis), or
24 h (cell viability assay and cell morphology analyses).
Cell viability assay
Cell viability was determined by an assay based on
the mitochondrial-dependent reduction of MTT to formazan. Briefly,
10 μl of MTT solution (5 mg/ml in DMEM) were added to the cell
supernatant and incubated for 4 h at 37°C. After removal of the
medium, 2-propanol was added to lyse the cells and to solubilize
the formazan. The optical density of formazan was measured at 570
nm using a microplate reader (Benchmark; Bio-Rad Laboratories,
Hercules, CA, USA). The optical density of formazan generated by
untreated cells was used to determine the 100% viability.
RNA extraction, reverse transcription
polymerase chain reaction (RT-PCR) and quantitative (q)RT-PCR
Total RNA was isolated from individual samples,
according to the manufacturer’s instructions, using the RNeasy kit
(Qiagen, Valencia, CA, USA). The purified RNA was used as a
template to generate cDNA using the RevertAid™ First Strand cDNA
Synthesis kit (Fermentas Life Sciences, St. Leon-Rot, Germany). The
primer sequences used for RT-PCR and qRT-PCR are listed in Table I. RT-PCR experiments were performed
in triplicates for each sample. Amplifications were performed using
the ABI PRISM 7700 sequence detection system (Applied Biosystems,
Foster City, CA, USA). For mRNA quantification, the FastStart
Universal SYBR-Green Master mix (Roche Applied Science, Mannheim,
Germany) was used. The cycle threshold (Ct) values were calculated
using the ΔΔCT method, and relative expression values were
expressed by normalizing to the expression of GAPDH. qRT-PCR
experiments were performed in duplicates for each sample. The size
of the PCR products was confirmed by agarose gel
electrophoresis.
 | Table IPrimer sequences used for reverse
transcription polymerase chain reaction (RT-PCR) and quantitative
(q)RT-PCR. |
Table I
Primer sequences used for reverse
transcription polymerase chain reaction (RT-PCR) and quantitative
(q)RT-PCR.
| Gene name | RT-PCR | qRT-PCR |
|---|
| IL-6 | F:
5′-ATGAACTCCTTCTCCACAAGCGC-3′ | F:
5′-GTAGTGAGGAACAAGCCAGAGC-3′ |
| R:
5′-GAAGAGCCCTCAGGCTGGACTG-3′ | R:
5′-GGCATTTGTGGTTGGGTCA-3′ |
| IL-8 | F:
5′-AGATATTGCACGGGAGAA-3′ | F:
5′-CTCTTGGCAGCCTTCCTGATTT-3′ |
| R:
5′-GAAATAAAGGAGAAACCA-3′ | R:
5′-CGCAGTGTGGTCCACTCTCAAT-3′ |
| iNOS | F:
5′-CTGAGGGCTCTGTTGAGGTC-3′ | F:
5′-GGCAGCCTGTGAGACCTTTG-3′ |
| R:
5′-CCTTGTTCAGCTACGCCTTC-3′ | R:
5′-GCATTGGAAGTGAAGCGTTTC-3′ |
| COX2 | F:
5′-GGAGAGACTATCAAGATAGTGATC-3′ | F:
5′-CAGAACCGCATTGCCTCTG-3′ |
| R:
5′-ATGGTCAGTAGACCTTTACAGCTC-3′ | R:
5′-TTGTAACTTCTGGTCCTCATGTCGA-3′ |
| TNF-α | F:
5′-GCGACGTGGAACTGGCAGAAG-3′ | F:
5′-GACCCTCACACTCAGATCATCTTCT-3′ |
| R:
5′-TCCATGCCGTTGGCCAGGAGG-3′ | R:
5′-CCTCCACTTGGTGGTTTGCT-3′ |
| GAPDH | F:
5′-ACCACAGTCCATGCCATCAC-3′ | F:
5′-ATGCCTCCTGCACCACCA-3′ |
| R:
5′-TCCACCACCCTGTTGCTGTA-3′ | R:
5′-CCATCACGCCACAGTTTCC-3′ |
NO assay
The NO level in cell culture supernatants was
determined using the Griess test. Briefly, cells were treated with
1, 5, or 15 μg/ml PLE for 1 h, followed by incubation with 1 μg/ml
LPS for 24 h. Nitrite in the culture supernatants was mixed with
the same volume of Griess reagent [1% sulfanilamide and 0.1%
N-(1-naphthyl)ethylenediamine dihydrochloride in 5% phosphoric
acid]. Absorbance was measured at 540 nm, and the nitrite
concentration was determined using sodium nitrite
(NaNO2) as a standard (17).
Enzyme-linked immunosorbent assay
(ELISA)
To analyze the production of pro-inflammatory
mediators, cells were seeded in 6-well plates at an initial density
of 5×105 cells/ml, starved in serum-free medium for 16
h, pre-incubated with different concentrations of PLE (5, 10 and 20
μg/ml) for 1 h, then treated with 1 μg/ml LPS for 24 h. The
supernatants were collected, and the concentrations of IL-6, IL-8
and TNF-α, as well as of prostaglandin E2
(PGE2) were determined using DuoSet ELISA kits and the
Prostaglandin E2 Parameter Assay kit (R&D Systems,
Abingdon, UK) respectively, following the manufacturer’s
instructions.
Subcellular fractionation
Cells were washed with physiological saline and
incubated with lysis buffer [10 mM HEPES, pH 7.6, containing 15 mM
KCl, 2 mM MgCl2, 0.1 mM EDTA, 1 mM dithiothreitol, 0.05%
v/v IGEPAL® CA-630, 1 mM phenylmethanesulfonyl fluoride
(PMSF), 1 mM sodium orthovanadate, 50 mM sodium fluoride, 10 μg/ml
leupeptin, and 10 μg/ml aprotinin] for 10 min. Following
centrifugation at 2,500 × g for 10 min at 4°C, the supernatant was
transferred into a new Eppendorf tube, further centrifuged at
20,000 × g for 15 min at 4°C, and the new supernatant was collected
as the cytosolic fraction. The pellets containing the nuclei were
washed with physiological saline, incubated with a nucleus lysis
buffer (25 mM HEPES, pH 7.6, 0.1% v/v IGEPAL® CA-630, 1
M KCl, 0.1 mM EDTA, 1 mM PMSF, 1 mM sodium orthovanadate, 2 mM
sodium fluoride, 10 μg/ml leupeptin, and 10 μg/ml aprotinin), and
then centrifuged at 10,000 × g for 15 min at 4°C. The resulting
supernatants were collected as the nuclear fraction.
Immunoblotting
Cells (5×105 cells/ml) were harvested,
washed twice with ice-cold phosphate-buffered saline (PBS), and
lysed in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% v/v
IGEPAL® CA-630, 1 mM PMSF, 1 mM sodium fluoride, and 10
μg/ml aprotinin and leupeptin). The cell lysates were centrifuged
at 14,000 × g for 10 min at 4°C to remove the debris. The
supernatants were collected and crude protein concentrations were
determined using the BCA™ protein assay kit (Pierce, Rockford, IL,
USA). Crude proteins (30 μg/lane) were electrophoresed and
transferred onto a nitrocellulose membrane (Millipore, Bedford, MA,
USA). After blocking with 5% w/v skimmed milk in PBS, the membrane
was incubated for 2 h with a 1/1,000 dilution of the specific
primary antibodies. Bound antibodies were detected using a 1/2,000
dilution of peroxidase-conjugated secondary antibodies (Abcam) and
ECL chemiluminescence reagent (Millipore) as the substrate. Three
independent experiments were performed, and results were quantified
by densitometry.
Statistical analysis
Data were expressed as means ± standard deviation
(SD) of the mean from three independent experiments. Statistical
comparisons were performed by a one-way analysis of variance
(ANOVA), followed by a Duncan multiple comparison test. Differences
were considered statistically significant at P<0.05.
Results
Effects of PLE on viability of murine
macrophage RAW264.7 cells
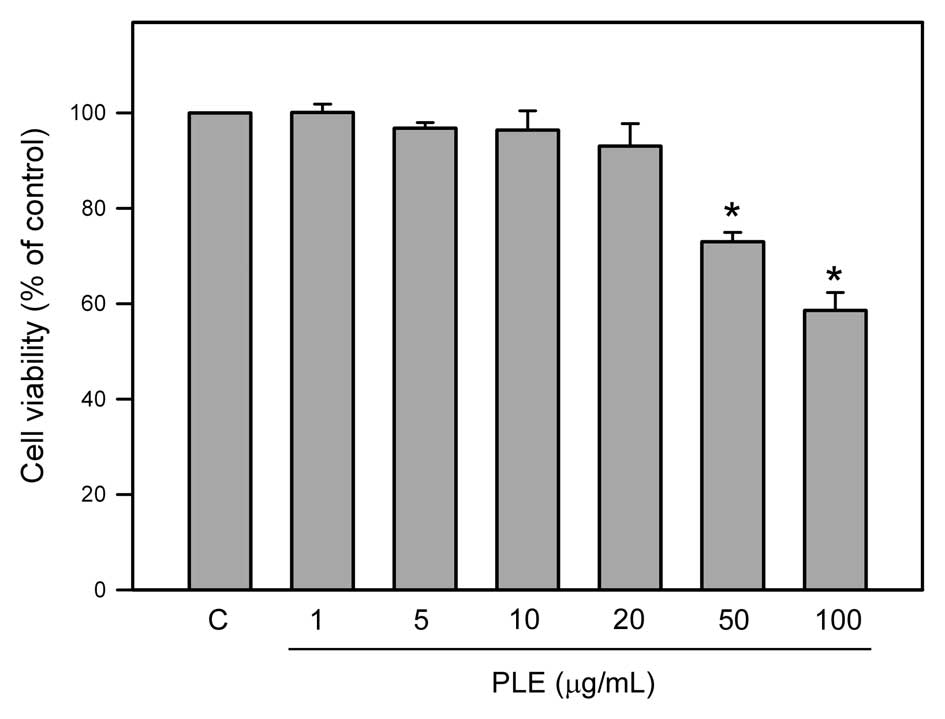
The cytotoxic effect of PLE on viability of RAW264.7
cells was investigated by the MTT assay. As shown in Fig. 1, the viability of RAW264.7 cells
was determined following incubation with different concentrations
(1, 5, 15, 30, 50 and 100 μg/ml) of PLE for 24 h. The results
revealed that treatment with concentrations of 50 and 100 μg/ml PLE
significantly decreased the viability of RAW264.7 cells (P<0.05)
compared to control (untreated) cells, while lower concentrations
(1, 5, 15 and 30 μg/ml) of PLE had no significant effect on
viability of RAW264.7 cells.
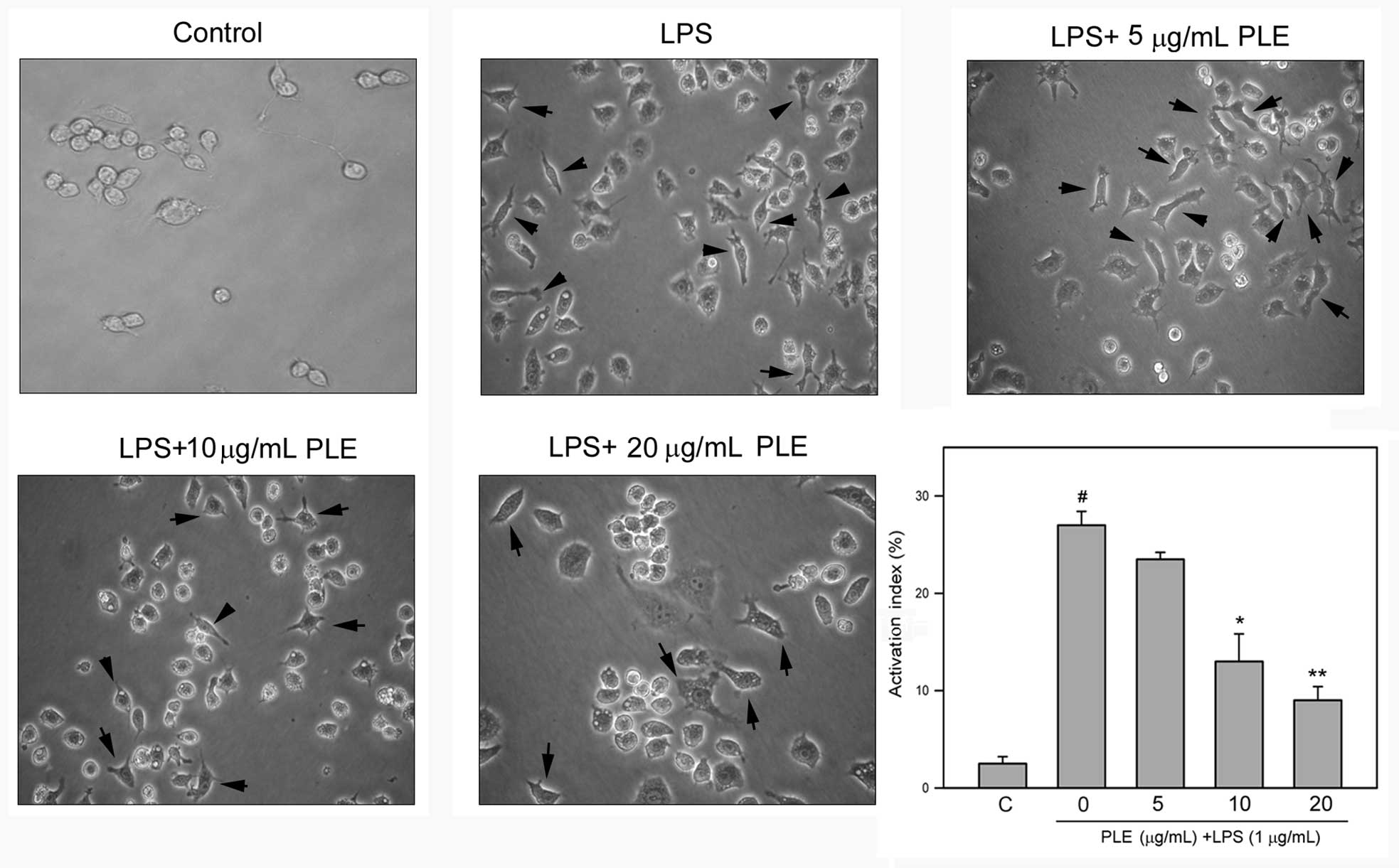
PLE inhibits LPS-induced dendritic
transformation of RAW264.7 cells
LPS is known to induce morphological transformation
of macrophage RAW264.7 cells (18). Therefore, whether PLE affects the
LPS-induced morphological changes of RAW264.7 cells was
investigated. As shown in Fig. 2,
LPS alone significantly induced the morphological transformation of
RAW264.7 cells to dendritic cells, and this transformation was
attenuated by PLE pretreatment in a dose-dependent manner. Further
quantitative analysis revealed that LPS significantly increases the
number of cells having a dendritic morphology to 27.0±0.5%
(P<0.05 as compared to the control). When cells were pretreated
with 10 or 15 μg/ml PLE and then treated with LPS, the number of
cells with dendritic morphology was decreased to 13.6±1.6 and
9.4±0.6%, respectively (P<0.05 as compared to LPS treatment
alone).
PLE inhibits LPS-induced mRNA expression
and production of IL-6, IL-8 and TNF-α in RAW264.7 cells
To investigate whether PLE affects the expression of
pro-inflammatory cytokines in LPS-stimulated macrophages, the mRNA
levels of IL-6, IL-8 and TNF-α were determined
in LPS-treated RAW264.7 cells. RT-PCR analysis revealed that LPS
alone enhances the expression of IL-6, IL-8 and
TNF-α, and this increase is reverted by pretreatment with
PLE in a dose-dependent manner (Fig.
3A). Further quantitative analysis using qRT-PCR showed that
treatment with LPS alone significantly increases the mRNA levels of
IL-6, IL-8 and TNF-α, 3.9±0.4-, 13.8±1.5- and
5.4±0.3-fold respectively (Fig.
3B), as compared to the control (untreated cells). In RAW264.7
cells pretreated with 20 μg/ml PLE and then treated with 1 μg/ml
LPS, the mRNA levels of IL-6, IL-8 and TNF-α
were reduced 1.4±0.2-, 5.1±0.3- and 3.4±0.2-fold as compared to the
control, respectively (P<0.005 as compared to LPS alone).
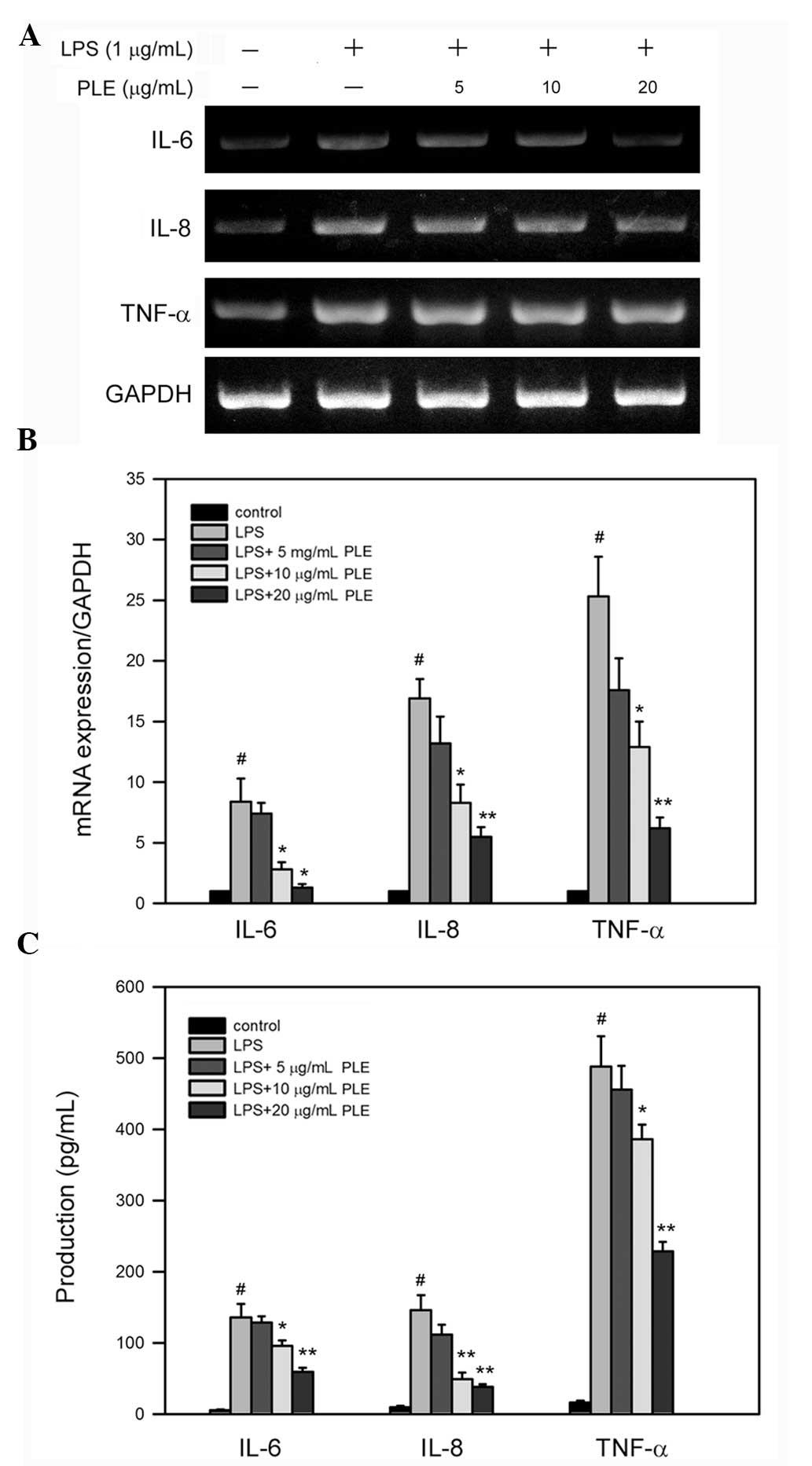 | Figure 3PLE reduces the mRNA level and the
protein production of pro-inflammatory cytokines IL-6, IL-8 and
TNF-α in LPS-stimulated RAW264.7 cells. Cells were pre-incubated
with the indicated concentrations of PLE in the DMEM culture medium
for 4 h, and then stimulated with 1 μg/ml LPS for 6 or 24 h.
Following treatment, the cells were lysed for mRNA extraction, and
the gene expression level was analyzed by (A) RT-PCR and quantified
by (B) qRT-PCR, while the culture medium was collected to quantify
the production of corresponding proteins by (C) ELISA. Bars denote
standard deviation (SD) of the mean from three independent
experiments. #P <0.05 as compared to the control
(untreated cells); *P<0.05 as compared to LPS alone.
PLE, Perilla frutescens leaf extract; IL, interleukin;
TNF-α, tumor necrosis factor-α; LPS, lipopolysaccharide; RT-PCR,
reverse transcription-polymerase chain reaction; q,
quantitative. |
In addition to mRNA expression, LPS alone increased
the protein production of IL-6, IL-8, and TNF-α in RAW264.7 cells
(Fig. 3C) up to 135.7±18.9,
145.9±21.3 and 488.2±33.2 pg/ml/104 cells, respectively
(P<0.05 as compared to the control). PLE pretreatment
dose-dependently reduced the protein production of the tested
cytokines (Fig. 3C) up to 59.3±5.6
(IL-6), 38.2±3.9 (IL-8) and 228.7±13.3 pg/ml/104 cells
(TNF-α), respectively (P<0.05 as compared to LPS alone).
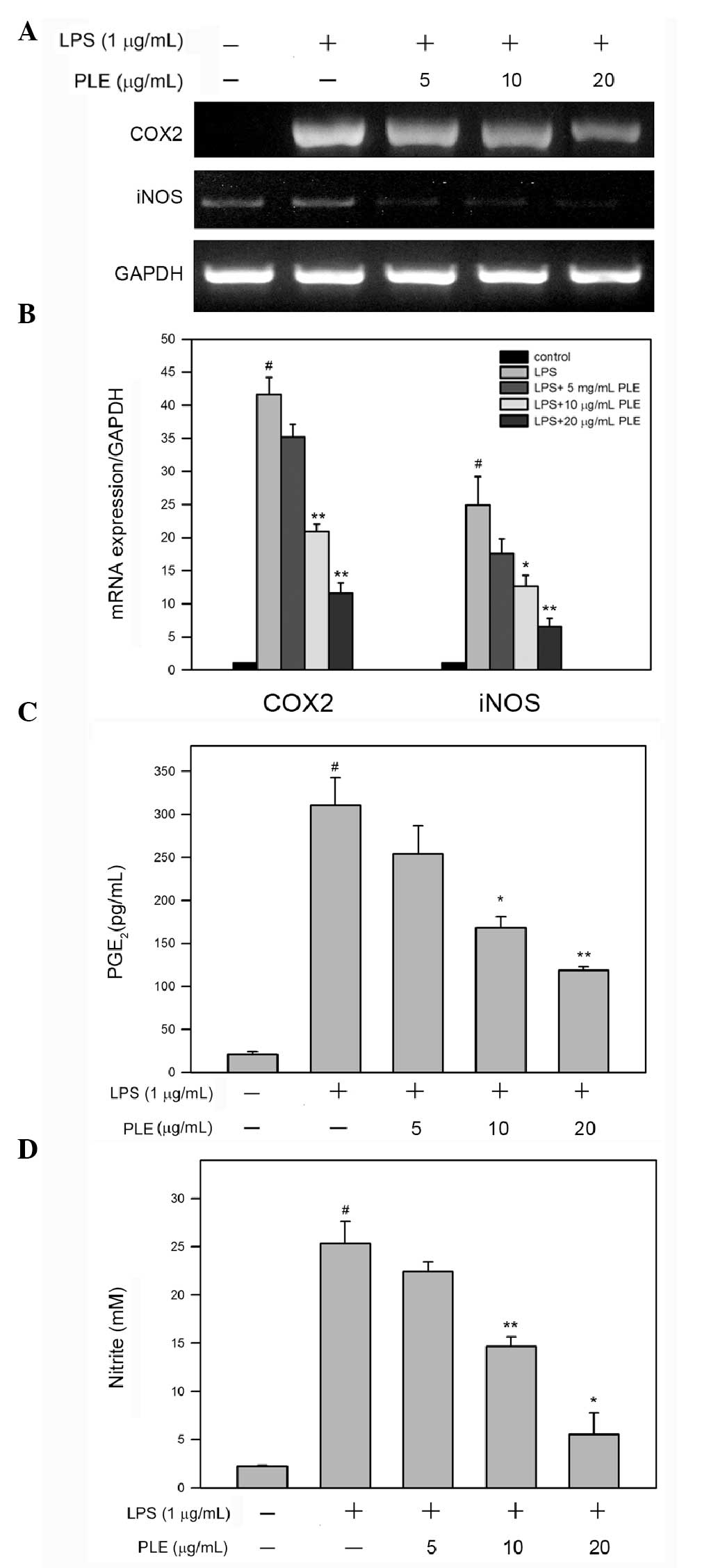
PLE inhibits LPS-induced mRNA expression
of COX-2 and iNOS and the production of PGE2 and NO in
RAW264.7 cells
COX-2 and iNOS are both critical enzymes associated
with macrophage-mediated development and progression of
inflammation (19–21); therefore, effects of PLE on the
mRNA level of COX-2 and iNOS were analyzed and
quantified in LPS-stimulated RAW264.7 cells. RT-PCR analysis showed
that LPS treatment alone enhances the expression of COX-2
and iNOS, and this increase was reverted by pretreatment
with PLE in a dose-dependent manner (Fig. 4A). Further quantitative analysis
using qRT-PCR revealed that LPS alone significantly increases the
mRNA levels of COX-2 and iNOS, 41.6±2.6- and
24.9±4.3-fold, respectively, as compared to the control
(P<0.05). In RAW264.7 cells pretreated with 20 μg/ml PLE and
then treated with 1 μg/ml LPS, the mRNA levels of COX-2 and
iNOS were dose-dependently reduced (Fig. 4B) 11.6±1.5- and 6.6±1.2-fold as
compared to the control, respectively (P<0.05 as compared to LPS
alone).
Since mRNA expression of COX-2 and
iNOS in RAW264.7 cells treated with LPS was inhibited by PLE
pretreatment, the production of PGE2 and NO was further
assessed. As shown in Fig. 4C and
D, treatment with LPS alone significantly increased the
concentrations of PGE2 and NO up to 310.9±22.5 pg/ml and
25.3±1.6 μM, respectively. Upon pretreatment with 10 or 20 μg/ml
PLE, the concentration of PGE2 was reduced down to
168.1±9.2 pg/ml and 118.7±3.1 pg/ml, respectively, and the
concentration of NO was decreased to 14.7±0.7 and 5.5±1.3 μM,
respectively (P<0.05 as compared to LPS alone).
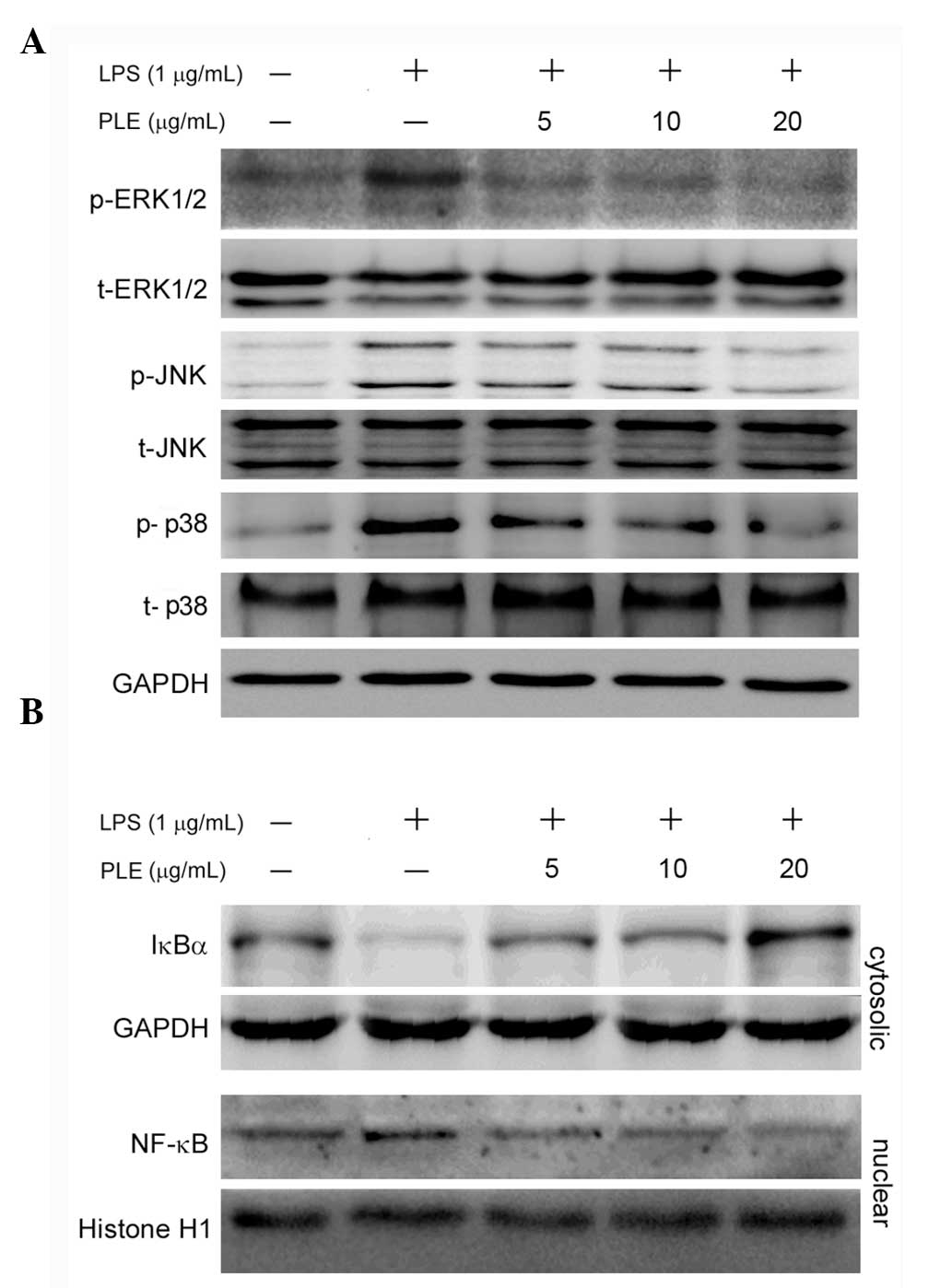
PLE inhibits phosphorylation of MAPKs,
the degradation of IκBα and the nuclear translocation of NF-κB in
LPS-stimulated RAW264.7 cells
Activation of MAPKs is known to be involved in
LPS-induced production of pro-inflammatory cytokines (22–24).
To investigate whether PLE regulates activation of MAPKs in
response to LPS, the levels of phosphorylated ERK1/2, JNK and p38
were determined by immunoblotting in LPS-stimulated RAW264.7 cells.
As shown in Fig. 5A, LPS alone
considerably increased the phosphorylation of ERK1/2, JNK and p38
in RAW264.7 cells, and pretreatment with 5, 10 or 20 μg/ml PLE
inhibited the LPS-induced phosphorylation of ERK1/2, JNK and p38 in
a dose-dependent manner.
The transcription factor NF-κB plays a pivotal role
in regulation of pro-inflammatory factors, and its nuclear
translocation is associated with the expression of these factors
(25). Thus, the effects of PLE on
the cytosolic level of IκBα (that inhibits the nuclear
translocation of NF-κB) and the nuclear level of NF-κB were
subsequently investigated in RAW264.7 cells exposed to LPS. As
shown in Fig. 5B, exposure of
RAW264.7 cells to LPS led to an important decrease in the cytosolic
IκBα level, accompanied by the expected increase in the nuclear
NF-κB level. However, PLE pretreatment restored the LPS-decreased
cytosolic IκBα level and reduced the nuclear NF-κB level that had
been increased by LPS treatment, in a dose-dependent manner. These
findings revealed that PLE inhibits not only the degradation of
IκBα, but also the nuclear translocation of NF-κB.
Discussion
Macrophages were reported to be directly involved in
the inflammatory response (26).
One of the most important roles of macrophages is the production of
various cytokines, reactive oxygen and nitrogen species, growth
factors and chemokines as a response to signals such as bacterial
LPS (27). Although the bioactive
molecules produced by macrophages play important roles in
inflammation, these molecules were also shown to exert undesirable
effects to the cells (28).
Therefore, modulation of these products may provide potential
targets for the control of inflammatory diseases. IL-6 has various
biological effects in a number of chronic endothelial dysfunctions
such as modulation of hematopoiesis, proliferation and
differentiation of lymphocytes, and induction of acute-phase
reactions (29). IL-8 is generally
recognized as a key intermediate regulator in acute inflammatory
responses (30). TNF-α is an
important mediator produced by activated macrophages and was shown
to affect various biological processes, including the regulation
and the production of other cytokines (31,32).
Our findings revealed that PLE inhibits the dendritic
transformation of RAW264.7 cells and the expression of IL-6, IL-8
and TNF-α in LPS-stimulated RAW264.7 cells, suggesting that PLE may
attenuate the LPS-induced activation of macrophages and the
consequent production of inflammatory cytokines.
NO is an important mediator and regulator involved
in inflammatory responses. In activated inflammatory cells, NO is
produced at high levels by iNOS. COX-2 is regarded as a central
mediator of inflammation, and regulation of COX-2 was suggested to
be useful in the development of a therapeutic target (33). Moreover, the enzymatic activity of
COX-2 is directly affected by NO and iNOS (34,35).
Our results showed that PLE inhibits both COX-2 and
iNOS expression, as well as NO production, which further
supports that PLE can attenuate the inflammatory response.
LPS is known to activate several signaling kinases,
including ERK1/2, MEK (36), JNK,
AP-1 (22), p38 and other MAP
kinase family members (37). Our
findings show that PLE inhibits iNOS expression and NO
production in LPS-stimulated RAW264.7 cells, which may relate to
the downregulation of NF-κB signaling components, including ERK1/2,
JNK and p38. The inhibitory effect of PLE on NF-κB, ERK1/2 and JNK
expression is crucial for the expression of iNOS.
In conclusion, the results from the present study
indicate that PLE displays important anti-inflammatory activity in
LPS-stimulated macrophage RAW264.7 cells, through inhibition of the
expression of pro-inflammatory cytokines, inhibition of MAPK
activation, and of NF-κB nuclear translocation in response to
LPS.
Acknowledgements
This study was partly supported by grants from the
National Science Council, Taiwan (nos. NSC99-2320-B-040-003-MY3 and
NSC99-2632-B-040-001-MY3).
References
|
1
|
Ueda H and Yamazaki M: Inhibition of tumor
necrosis factor-alpha production by orally administering a
Perilla leaf extract. Biosci Biotechnol Biochem.
61:1292–1295. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Makino T, Ono T, Muso E, Honda G and
Sasayama S: Suppressive effects of Perilla frutescens on
spontaneous IgA nephropathy in ddY mice. Nephron. 83:40–46.
1999.
|
|
3
|
Ueda H and Yamazaki M: Anti-inflammatory
and anti-allergic actions by oral administration of a
Perilla leaf extract in mice. Biosci Biotechnol Biochem.
65:1673–1675. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shin TY, Kim SH, Kim SH, et al: Inhibitory
effect of mast cell-mediated immediate-type allergic reactions in
rats by Perilla frutescens. Immunopharmacol Immunotoxicol.
22:489–500. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hortelano S, Zeini M and Bosca L: Nitric
oxide and resolution of inflammation. Methods Enzymol. 359:459–465.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Paige JS and Jaffrey SR: Pharmacologic
manipulation of nitric oxide signaling: targeting NOS dimerization
and protein-protein interactions. Curr Top Med Chem. 7:97–114.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gao YT, Panda SP, Roman LJ, Martasek P,
Ishimura Y and Masters BS: Oxygen metabolism by neuronal
nitric-oxide synthase. J Biol Chem. 282:7921–7929. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Breitbach K, Klocke S, Tschernig T, van
Rooijen N, Baumann U and Steinmetz I: Role of inducible nitric
oxide synthase and NADPH oxidase in early control of
Burkholderia pseudomallei infection in mice. Infect Immun.
74:6300–6309. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cunha IW, Lopes A, Falzoni R and Soares
FA: Sarcomas often express constitutive nitric oxide synthases
(NOS) but infrequently inducible NOS. Appl Immunohistochem Mol
Morphol. 14:404–410. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Farley KS, Wang LF, Razavi HM, et al:
Effects of macrophage inducible nitric oxide synthase in murine
septic lung injury. Am J Physiol Lung Cell Mol Physiol.
290:L1164–L1172. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sacco RE, Waters WR, Rudolph KM and Drew
ML: Comparative nitric oxide production by LPS-stimulated
monocyte-derived macrophages from Ovis canadensis and Ovis aries.
Comp Immunol Microbiol Infect Dis. 29:1–11. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rahman A, Yatsuzuka R, Jiang S, Ueda Y and
Kamei C: Involvement of cyclooxygenase-2 in allergic nasal
inflammation in rats. Int Immunopharmacol. 6:1736–1742. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Geng Y, Zhang B and Lotz M: Protein
tyrosine kinase activation is required for lipopolysaccharide
induction of cytokines in human blood monocytes. J Immunol.
151:6692–6700. 1993.PubMed/NCBI
|
|
14
|
Kim HS, Ye SK, Cho IH, et al:
8-hydroxydeoxyguanosine suppresses NO production and COX-2 activity
via Rac1/STATs signaling in LPS-induced brain microglia. Free Radic
Biol Med. 41:1392–1403. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Park HJ, Kim IT, Won JH, et al:
Anti-inflammatory activities of
ent-16alphaH,17-hydroxy-kauran-19-oic acid isolated from the roots
of Siegesbeckia pubescens are due to the inhibition of iNOS
and COX-2 expression in RAW 264.7 macrophages via NF-kappaB
inactivation. Eur J Pharmacol. 558:185–193. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nakano H, Shindo M, Sakon S, et al:
Differential regulation of IkappaB kinase alpha and beta by two
upstream kinases, NF-kappaB-inducing kinase and mitogen-activated
protein kinase/ERK kinase kinase-1. Proc Natl Acad Sci USA.
95:3537–3542. 1998. View Article : Google Scholar
|
|
17
|
Green LC, Wagner DA, Glogowski J, Skipper
PL, Wishnok JS and Tannenbaum SR: Analysis of nitrate, nitrite, and
[15N]nitrate in biological fluids. Anal Biochem. 126:131–138.
1982.
|
|
18
|
Saxena RK, Vallyathan V and Lewis DM:
Evidence for lipopolysaccharide-induced differentiation of RAW264.7
murine macrophage cell line into dendritic like cells. J Biosci.
28:129–134. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Onuchic AC, Machado CM, Saito RF, Rios FJ,
Jancar S and Chammas R: Expression of PAFR as part of a prosurvival
response to chemotherapy: a novel target for combination therapy in
melanoma. Mediators Inflamm. 2012:1754082012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen H, Ma F, Hu X, Jin T, Xiong C and
Teng X: Elevated COX2 expression and PGE2 production by
downregulation of RXRα in senescent macrophages. Biochem Biophys
Res Commun. 440:157–162. 2013.PubMed/NCBI
|
|
21
|
Adesso S, Popolo A, Bianco G, et al: The
uremic toxin indoxyl sulphate enhances macrophage response to LPS.
PloS One. 8:e767782013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hambleton J, Weinstein SL, Lem L and
DeFranco AL: Activation of c-Jun N-terminal kinase in bacterial
lipopolysaccharide-stimulated macrophages. Proc Natl Acad Sci USA.
93:2774–2778. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
An H, Yu Y, Zhang M, et al: Involvement of
ERK, p38 and NF-kappaB signal transduction in regulation of TLR2,
TLR4 and TLR9 gene expression induced by lipopolysaccharide in
mouse dendritic cells. Immunology. 106:38–45. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang W, Deng M, Liu X, Ai W, Tang Q and Hu
J: TLR4 activation induces nontolerant inflammatory response in
endothelial cells. Inflammation. 34:509–518. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tak PP and Firestein GS: NF-kappaB: a key
role in inflammatory diseases. J Clin Invest. 107:7–11. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Murakami A, Nishizawa T, Egawa K, et al:
New class of linoleic acid metabolites biosynthesized by corn and
rice lipoxygenases: suppression of proinflammatory mediator
expression via attenuation of MAPK- and Akt-, but not PPARgamma-,
dependent pathways in stimulated macrophages. Biochem Pharmacol.
70:1330–1342. 2005. View Article : Google Scholar
|
|
27
|
Fujiwara N and Kobayashi K: Macrophages in
inflammation. Curr Drug Targets Inflamm Allergy. 4:281–286. 2005.
View Article : Google Scholar
|
|
28
|
Heumann D and Roger T: Initial responses
to endotoxins and Gram-negative bacteria. Clin Chim Acta.
323:59–72. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Desai TR, Leeper NJ, Hynes KL and Gewertz
BL: Interleukin-6 causes endothelial barrier dysfunction via the
protein kinase C pathway. J Surg Res. 104:118–123. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Noda A, Kinoshita K, Sakurai A, Matsumoto
T, Mugishima H and Tanjoh K: Hyperglycemia and lipopolysaccharide
decrease depression effect of interleukin 8 production by
hypothermia: an experimental study with endothelial cells.
Intensive Care Med. 34:109–115. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hume DA, Underhill DM, Sweet MJ, Ozinsky
AO, Liew FY and Aderem A: Macrophages exposed continuously to
lipopolysaccharide and other agonists that act via toll-like
receptors exhibit a sustained and additive activation state. BMC
Immunol. 2:112001. View Article : Google Scholar
|
|
32
|
Aggarwal BB, Shishodia S, Ashikawa K and
Bharti AC: The role of TNF and its family members in inflammation
and cancer: lessons from gene deletion. Curr Drug Targets Inflamm
Allergy. 1:327–341. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tsatsanis C, Androulidaki A, Venihaki M
and Margioris AN: Signalling networks regulating cyclooxygenase-2.
Int J Biochem Cell Biol. 38:1654–1661. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Aggarwal BB, Shishodia S, Sandur SK,
Pandey MK and Sethi G: Inflammation and cancer: how hot is the
link? Biochem Pharmacol. 72:1605–1621. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Korhonen R, Lahti A, Kankaanranta H and
Moilanen E: Nitric oxide production and signaling in inflammation.
Curr Drug Targets Inflamm Allergy. 4:471–479. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Geppert TD, Whitehurst CE, Thompson P and
Beutler B: Lipopolysaccharide signals activation of tumor necrosis
factor biosynthesis through the ras/raf-1/MEK/MAPK pathway. Mol
Med. 1:93–103. 1994.PubMed/NCBI
|
|
37
|
Han J, Lee JD, Bibbs L and Ulevitch RJ: A
MAP kinase targeted by endotoxin and hyperosmolarity in mammalian
cells. Science. 265:808–811. 1994. View Article : Google Scholar : PubMed/NCBI
|