Introduction
MicroRNAs (miRNA) are a type of short, non-coding
RNAs that suppress the expression of protein-coding genes by
partial complementary binding, especially to the 3′ untranslated
regions (UTRs), of messenger RNAs (mRNAs). Alterations in miRNA
expression are involved in the initiation, progression and
metastasis of human cancer, and it is believed that miRNAs function
as tumor suppressors and oncogenes in cancer development (1,2).
A number of studies have shown that miR-125a is an
important tumor suppressor gene, and reduced miR-125a expression
has been detected in many types of human cancer, including breast
(3,4), lung (5,6) and
ovarian cancer (7), as well as
glioblastoma (8). Nishida et
al (9) reported that a reduced
expression of miR-125a-5p is associated with enhanced potential to
develop gastric cancer. Furthermore, Hashiguchi et al
(10) reported that miR-125a-3p,
commonly ignored by investigators, has almost the same function in
gastric cancer as miR-125a-5p. There are also reports that germline
mutations in the miR-125a coding region can reduce miR-125a
expression and are associated with human breast cancer (3,11).
These findings strongly suggest that miR-125a-5p and -3p variants
can act as tumor suppressors and reduce miR-125a expression,
thereby serving as genetic markers for gastric cancer diagnosis and
treatment.
In this study, we first examined the expression
level of miR-125a-5p and -3p in gastric cancer tissue and adjacent
healthy gastric tissue. We then genotyped the miR-125a coding
region in gastric cancer patients and healthy controls. We found a
germline mutation in the pri-miR-125a coding region that was
associated with gastric cancer and the reduction of miR-125a
expression, suggesting that miR-125a is likely to function as a
tumor suppressor gene in human gastric cancer.
Materials and methods
Study population, tissue samples and cell
lines
A total of 75 pairs of histopathologically confirmed
gastric cancer and adjacent non-cancer tissue samples were obtained
from patients in the Department of Gastroenterology, Qilu Hospital
of Shangdong University, China. Informed consent was obtained from
all patients and the procedure was approved by the Medical Ethics
Committee of the hospital. Control samples from a total of 287
healthy Han-Chinese individuals were also collected at the Central
Hospital of Zibo, Shandong, China. Human gastric adenoma cell lines
(MGC-803 and BGC-823) were purchased from the Cell Bank of Shanghai
Institute of Cell Biology, Chinese Academy of Sciences (Shanghai,
China). Cells were routinely cultured in RPMI-1640 medium
supplemented with 10% fetal bovine serum at 37°C in a humidified
atmosphere with 5% CO2.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
The expression level of miR-125a-5p and -3p was
assessed by TaqMan miRNA real-time (quantitative) RT-PCR. Total RNA
was extracted from tissues and cells using TRIzol (Invitrogen Life
Technologies, Carlsbad, CA, USA) according to the manufacturer’s
instructions. Single-stranded cDNA was synthesized using the TaqMan
MicroRNA Reverse Transcription kit (Applied Biosystems, Foster
City, CA, USA) and then amplified using the TaqMan Universal PCR
Master mix together with miRNA-specific TaqMan minor groove binder
(MGB) probes for miR-125a-5p and -3p (all from Applied Biosystems).
The small nuclear (sn) RNA for U6 was used for normalization. Each
sample of a group was measured in triplicate and the experiment was
repeated at least three times.
DNA collection and genotyping
DNA from tumor and adjacent healthy tissues of the
gastic cancer cohort was isolated using the TIANamp Genomic DNA kit
(Tiangen Biotech, Beijing, China). DNA from blood samples was
extracted using the TIANamp Blood DNA kit (Tiangen Biotech). DNA
samples were amplified using standard PCR protocols. The PCR
products were sequenced in the forward direction on an ABI 3730xl
sequencing platform. The sequencing results were analyzed using the
open-source software DNAMAN and Chromas Lite. The PCR primers used
for miR-125a sequencing were: 5′-TGT GTC TCT TTC ACA GTG GAT C-3′
and 5′-CCA TCG TGT GGG TCT CAA G-3′.
Secondary structure prediction
The secondary structure of the 217-bp pri-miR-125a
sequence, which includes the mutation site, was predicted using the
RNAfold web server (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi).
miR-125a expression vectors
To construct miR-125a expression vectors, 1,016-nt
fragments corresponding to pri-miR-125a and its flanking regions
(previously confirmed to bear the two alleles) were amplified from
cDNA and cloned into the pcDNA3.1 vector (Invitrogen Life
Technologies). The sequences of the two resulting vectors were
confirmed by direct sequencing, with the only difference being in
the mutation site. The primers used were: miR125a-F/XhoI,
5′-CCG CTC GAG GGT AGG AGG TTG TAT AGT TGA GGA GG-3′ and
miR-125a-R/XbaI, 5′-GCT CTA GAC CTC TGG GCC TCT CCT
GC-3′.
Dual luciferase assay
The full-length region (618 bp) of the 3′ UTR of the
erythroblastic leukemia viral oncogene homolog 2 (ERBB2)
gene was cloned downstream of the coding region of the firefly
luciferase gene within the pmirGLO Dual-Luciferase miRNA Target
vector (Promega Corp., Madison, WI, USA) to generate the luciferase
reporter vector. For luciferase reporter assays, MGC-803 and
BGC-823 cells were seeded into 48-well plates. The miR-125a
expression and the luciferase reporter vectors were co-transfected
using Lipofectamine 2000 (Invitrogen Life Technologies). Two days
later, the cells were harvested and assayed with the
Dual-Luciferase Assay system (Promega Corp.). Each assay was
performed in triplicate, in three independent experiments. The
results were expressed as relative luciferase (LUC) activity
(firefly LUC/Renilla LUC).
Cell proliferation assay
MGC-803 and BGC-823 cells were seeded into 96-well
plates at low density (5×103) in Dulbecco’s modified
Eagle’s medium (DMEM) and were allowed to attach overnight. The
cells were then transfected with different haplotype miR-125a
expression vectors, with the empty pcDNA3.1 vector used as the
control. Twenty microliters MTT (5 mg/ml) (Sigma-Aldrich, St.
Louis, MO, USA) were added into each well 48 h following
transfection, and the cells were incubated for an additional 4 h.
Following the addition of dimethylsulfoxide (DMSO) to the samples,
their absorbance was recorded at 570 nm on a 96-well plate
reader.
Statistical analysis
Data were analyzed using the SPSS statistical
package version 16 (SPSS, Inc. Chicago, IL, USA). Comparisons
between two independent groups were performed with the Student’s
t-test. Expression data for miR-125a were compared using the
Mann-Whitney U test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Increased miR-125a-5p and -3p in gastric
cancer tissues
In order to explore the role of miR-125a in gastric
carcinogenesis, the expression patterns of miR-125a were analyzed
in 75 pairs of human gastric cancer and adjacent healthy gastric
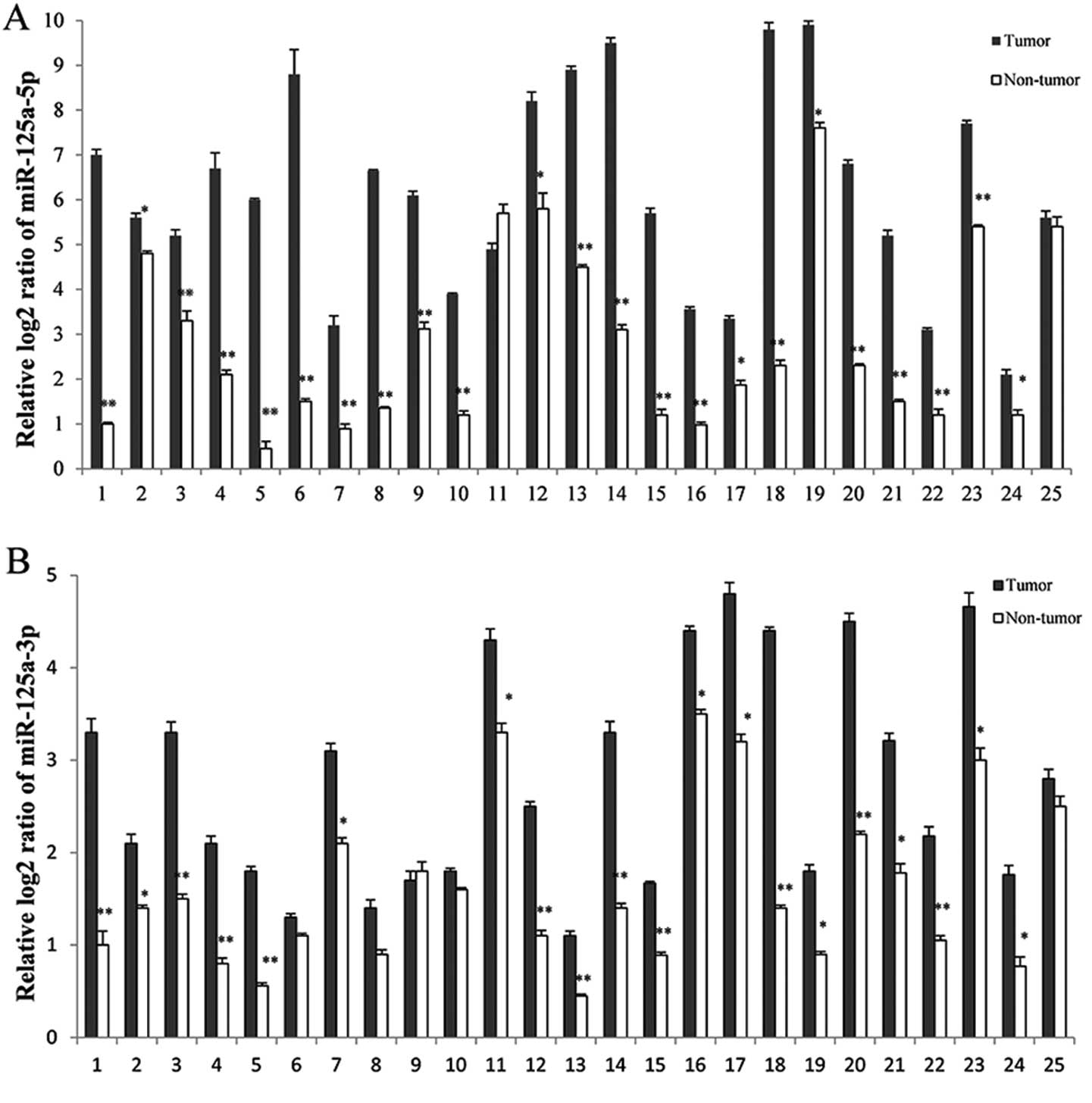
tissues using RT-qPCR (Fig. 1).
Each sample consisted of pooled RNAs from cancer tissues of three
patients (25 samples for 75 pairs). The levels of miR-125a-5p and
-3p were significantly increased in 92 (23/25) and 80% (20/25
samples) of gastric cancer tissues, respectively (Fig. 1).
A germline mutation was detected in the
pri-miR-125a coding region
Since nucleotide variants can alter miRNA expression
and are associated with many types of human diseases, we genotyped
the coding region of pri-miR-125a by sequencing the DNA extracted
from gastric cancer tissues. We found five patients carrying the
minor G allele (Table I), which
existed in the +43 relative to the mature miR-125a-5p and +29
relative to pre-miR-125a. Furthermore, we sequenced genomic DNAs
isolated from adjacent healthy gastric tissues and found the same
genotypes as the ones from gastric cancer tissues (data not
shown).
 | Table IThe minor G allele of pri-miR-125a is
detected in gastric cancer patients. |
Table I
The minor G allele of pri-miR-125a is
detected in gastric cancer patients.
| Genotype
distribution |
|---|
|
|
|---|
| Samples | AA | AG | GG |
|---|
| Gastric cancer (Qilu
Hospital) | 70 (93.3%) | 5 (6.7%) | 0 (0%) |
| Healthy (Zibo Central
Hospital) | 287 (100%) | 0 (0%) | 0 (0%) |
The frequency of the minor G allele was examined in
the population of 287 healthy individuals (control group) collected
in the same region. We found no individual carrying the G allele in
this population, while this allele was also not found in the 1000
Genomes database (http://www.1000genomes.org/), suggesting that the T
allele is a mutation. Furthermore, since no individual in the
control group from the same area carried the G allele, the presence
of the G allele among gastric cancer patients is unlikely to be due
to a founder effect. Together, these results suggest that a
germline mutation in pri-miR-125a is associated with gastric cancer
tumorigenesis.
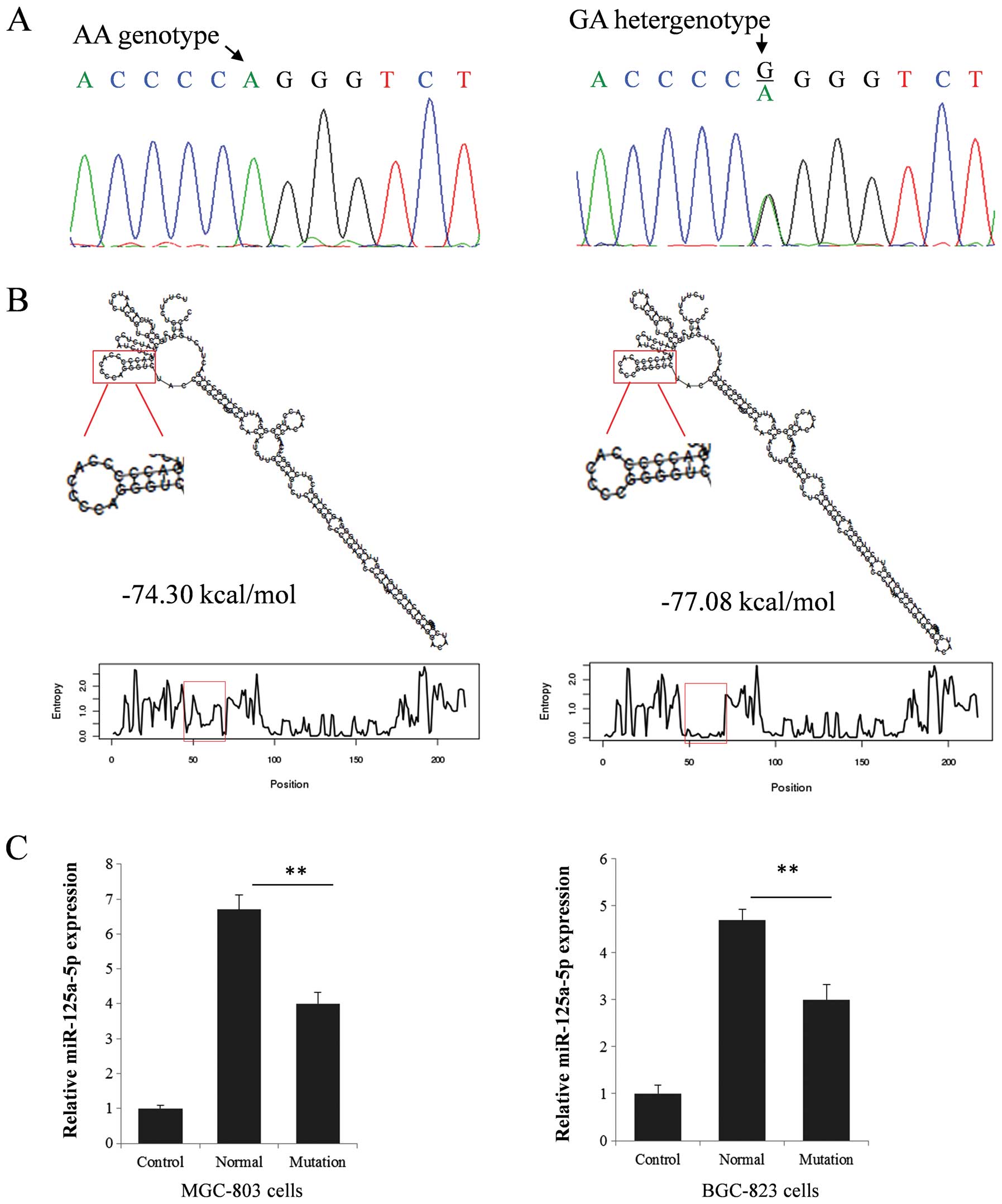
The G mutation can enhance the predicted
stability of pri-miR-125a and reduce miR-125a expression
To explore the function of the mutation (Fig. 2A), we first compared the predicted
secondary structure of pri-miR-125a molecules bearing or not the
minor allele. As shown in Fig. 2B,
the minor allele G causes an apparent change in loop size (from 8
nucleotides loop to 6 nucleotides loop and from 10 paired
nucleotides stem to 12 paired nucleotides stem) and a reduction of
the predicted ΔG from −74.30 to −77.08 kcal/mol. Using RT-qPCR and
two different expression vectors carrying the alternative
miR-125a-5p alleles, we quantified the expression level of the
mature miR-125a-5p in the two gastric cancer cell lines MGC-803 and
BGC-823. The presence of the mutation was associated with an ~40%
reduction in mature miR-125a-5p expression (Fig. 2C), in agreement with RT-qPCR
analyses on gastric cancer tissue samples (Fig. 3B).
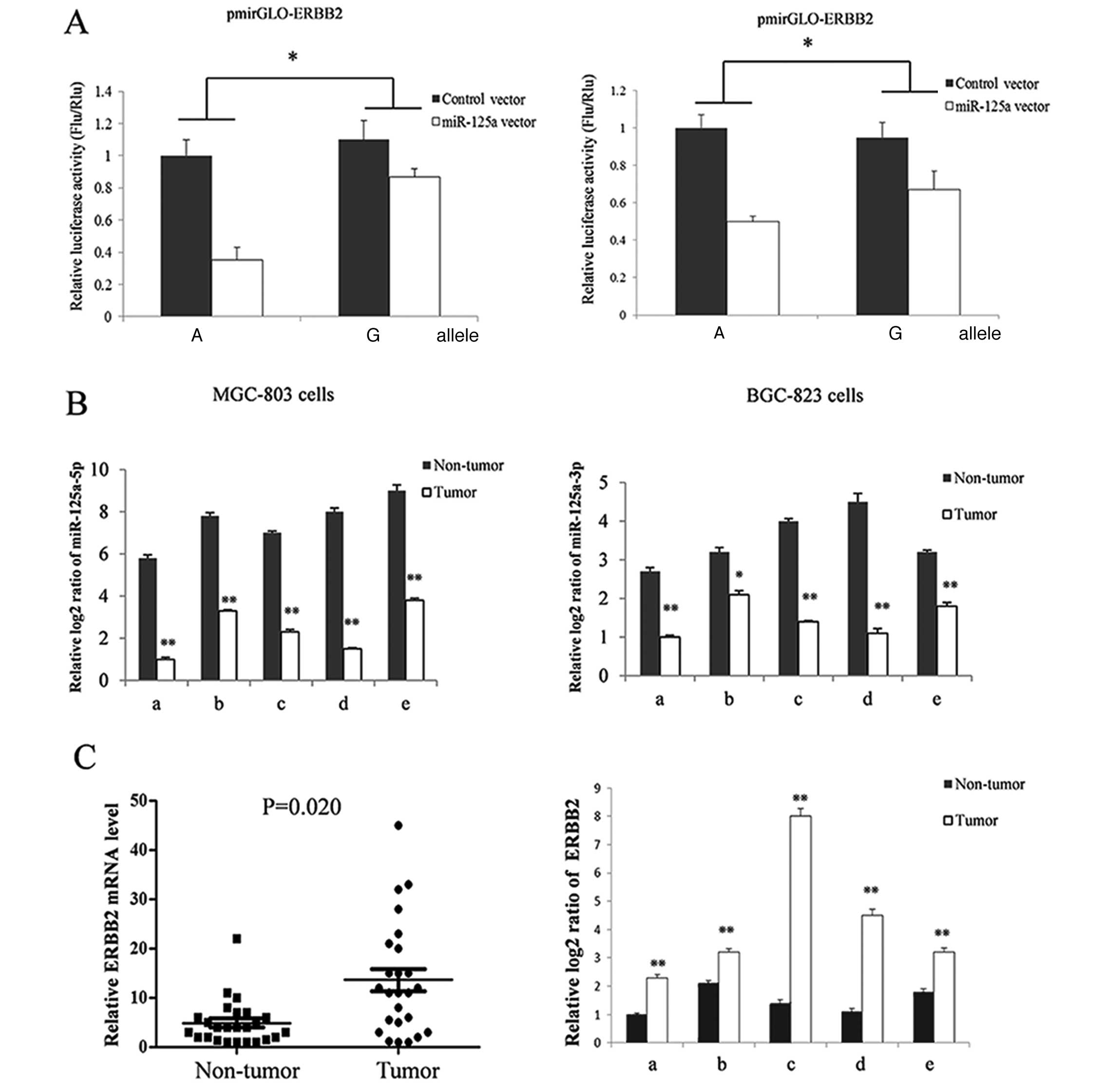
Altered miR-125a expression attenuates
the inhibitory effect of miR-125a on ERBB2 expression and gastric
cancer cell proliferation
The ERBB2 gene, a confirmed target of
miR-125a-5p (12,13), is a member of the epidermal growth
factor receptor (EGFR/ERBB) family. The protein is commonly
overexpressed in numerous types of cancer cells and has been
suggested to promote cell proliferation (14,15).
We used the ERBB2 3′ UTR reporter system to study the effect
of the identified mutation on ERBB2 gene expression. As
shown in Fig. 3A, the reduction in
ERBB2 expression caused by transfection with the miR-125a
vector was significantly attenuated in cells bearing the G compared
to those bearing the A allele. To explore the in vivo
expression pattern of ERBB2, we measured the ERBB2
mRNA level by using RT-qPCR in gastric cancer and adjacent healthy
gastric tissues. The ERBB2 expression level was
significantly higher (Fig. 3C) in
cancer tissues compared to healthy controls, which had reduced
miR-125a expression (Fig. 1).
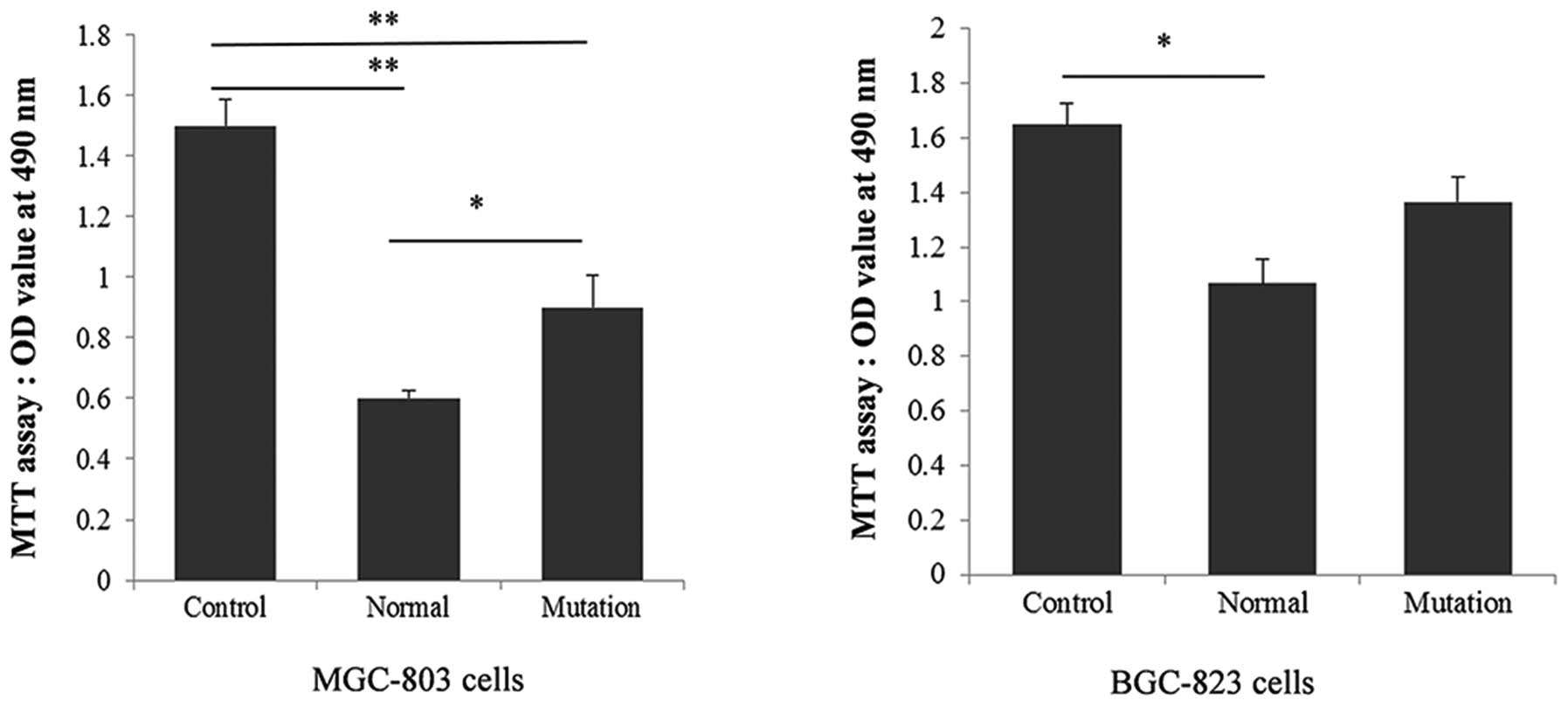
Based on the predicted function of ERBB2, the
reduction of miR-125a expression is expected to promote cell
proliferation. Therefore, a proliferation assay was carried out to
explore potential differences in antitumor activity caused by
different miR-125a genotypes in gastric cancer cells. MGC-803 and
BGC-823 cells were transfected with different pri-miR-125a
expression vectors, and, as expected, the proliferation of MGC-803
cells was significantly reduced (P=0.0008 and 0.0067 compared to
the control) in pri-miR-125a genotypic backgrounds (bearing the A
and the mutant G allele), while the A to G mutation attenuated this
effect by almost 20% (P=0.023 compared to the A allelic
background). In BGC-823 cells, proliferation was also reduced in
the pri-miR-125a genotype bearing the A allele (P=0.0075), but this
effect was not significantly attenuated by the G mutation (Fig. 4).
Discussion
Growing evidence indicates that altered patterns of
miRNA expression correlate with various human diseases, and
especially with numerous cancer types (16,17).
The effects of miRNAs are complex, owing to the fact that these
molecules regulate hundreds of targets, resulting in the
downregulation of numerous genes, including oncogenes and tumor
suppressors. Therefore, exploring the clinical potential of miRNAs
is of particular value.
In this study, we first detected a link between high
miR-125a expression and gastric carcinogenesis. Nucleotide variants
in coding regions can disturb the expression of miRNAs and relate
to many types of human disease, especially cancer (3,11,18–20).
We therefore sequenced the coding region of miR-125a in 75 gastric
cancer patients. To exclude the possibility of false negatives
caused by a founder effect, we sequenced the same region in a
population of 287 healthy individuals collected in the same region.
The 1000 Genomes database was used to exclude known variants and
false negatives in the two different populations. We identified a
nucleotide mutation (+29A>G, position relative to the
pri-miR-125a start codon) that was only detected in gastric cancer
patients. The G mutation at this site attenuated the inhibitory
effect of miR-125a on ERBB2 gene expression by reducing the
expression of the mature miR-125a both in vitro and in
vivo. Reduced miR-125a expression further attenuated the
suppressive effect of miR-125a on gastric cancer cell
proliferation, especially in MGC-803 cells. This result indicated
that miR-125a acts as a tumor suppressor and that the identified
germline mutation in pri-miR-125a is associated with gastric cancer
tumorigenesis.
Our data indicate that miR-125a is likely to
function as a tumor suppressor gene in human gastric cancer. The
reduction in the miR-125a expression level can lead to upregulation
of miR-125a target genes, such as ERBB2, and predispose
individuals carrying the G allele to gastric cancer. To the best of
our knowledge, this is the first mutation in the miR-125a coding
region reported to associate with human gastric cancer. Systematic
sequencing of known miRNAs in a high number of distinct populations
is expected to provide valuable insights in the regulation of
various diseases.
References
|
1
|
Wu WK, Lee CW, Cho CH, et al: MicroRNA
dysregulation in gastric cancer: a new player enters the game.
Oncogene. 29:5761–5771. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nicoloso MS, Spizzo R, Shimizu M, Rossi S
and Calin GA: MicroRNAs - the micro steering wheel of tumour
metastases. Nat Rev Cancer. 9:293–302. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li W, Duan R, Kooy F, Sherman SL, Zhou W
and Jin P: Germline mutation of microRNA-125a is associated with
breast cancer. J Med Genet. 46:358–360. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Guo X, Wu Y and Hartley RS: MicroRNA-125a
represses cell growth by targeting HuR in breast cancer. RNA Biol.
6:575–583. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang G, Mao W, Zheng S and Ye J: Epidermal
growth factor receptor-regulated miR-125a-5p - a metastatic
inhibitor of lung cancer. FEBS J. 276:5571–5578. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jiang L, Huang Q, Zhang S, et al:
Hsa-miR-125a-3p and hsa-miR-125a-5p are downregulated in non-small
cell lung cancer and have inverse effects on invasion and migration
of lung cancer cells. BMC Cancer. 10:3182010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cowden Dahl KD, Dahl R, Kruichak JN and
Hudson LG: The epidermal growth factor receptor responsive miR-125a
represses mesenchymal morphology in ovarian cancer cells.
Neoplasia. 11:1208–1215. 2009.PubMed/NCBI
|
|
8
|
Cortez MA, Nicoloso MS, Shimizu M, et al:
miR-29b and miR-125a regulate podoplanin and suppress invasion in
glioblastoma. Genes Chromosomes Cancer. 49:981–990. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nishida N, Mimori K, Fabbri M, et al:
MicroRNA-125a-5p is an independent prognostic factor in gastric
cancer and inhibits the proliferation of human gastric cancer cells
in combination with trastuzumab. Clin Cancer Res. 17:2725–2733.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hashiguchi Y, Nishida N, Mimori K, et al:
Down-regulation of miR-125a-3p in human gastric cancer and its
clinicopathological significance. Int J Oncol. 40:1477–1482.
2012.PubMed/NCBI
|
|
11
|
Duan R, Pak C and Jin P: Single nucleotide
polymorphism associated with mature miR-125a alters the processing
of pri-miRNA. Hum Mol Genet. 16:1124–1131. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Scott GK, Goga A, Bhaumik D, Berger CE,
Sullivan CS and Benz CC: Coordinate suppression of ERBB2 and ERBB3
by enforced expression of micro-RNA miR-125a or miR-125b. J Biol
Chem. 282:1479–1486. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang S, Huang J, Lyu H, et al: Functional
cooperation of miR-125a, miR-125b, and miR-205 in
entinostat-induced downregulation of erbB2/erbB3 and apoptosis in
breast cancer cells. Cell Death Dis. 4:e5562013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schade B, Lesurf R, Sanguin-Gendreau V, et
al: β-Catenin signaling is a critical event in ErbB2-mediated
mammary tumor progression. Cancer Res. 73:4474–4487. 2013.
|
|
15
|
Conesa-Zamora P, Torres-Moreno D, Isaac MA
and Pérez-Guillermo M: Gene amplification and immunohistochemical
expression of ERBB2 and EGFR in cervical carcinogenesis.
Correlation with cell-cycle markers and HPV presence. Exp Mol
Pathol. 95:151–155. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Di Leva G, Garofalo M and Croce CM:
MicroRNAs in cancer. Annu Rev Pathol. 9:287–314. 2014.
|
|
17
|
Gargalionis AN and Basdra EK: Insights in
microRNAs biology. Curr Top Med Chem. 13:1493–1502. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hu Y, Liu CM, Qi L, et al: Two common SNPs
in pri-miR-125a alter the mature miRNA expression and associate
with recurrent pregnancy loss in a Han-Chinese population. RNA
Biol. 8:861–872. 2011. View Article : Google Scholar
|
|
19
|
Jazdzewski K, Murray EL, Franssila K,
Jarzab B, Schoenberg DR and de la Chapelle A: Common SNP in
pre-miR-146a decreases mature miR expression and predisposes to
papillary thyroid carcinoma. Proc Natl Acad Sci USA. 105:7269–7274.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Saunders MA, Liang H and Li WH: Human
polymorphism at microRNAs and microRNA target sites. Proc Natl Acad
Sci USA. 104:3300–3305. 2007. View Article : Google Scholar : PubMed/NCBI
|