Introduction
Colorectal cancer (CRC) is a major burden to
healthcare systems worldwide, accounting for approximately one
million novel cancer cases (1).
Although CRC mortality has decreased over the last 20 years, it
remains the third most common cause of cancer-associated mortality,
accounting for ~600,000 fatalities in 2008 worldwide (1). Therefore, identification of tumor
biomarkers for screening and early detection is imperative for
patients with colon cancer.
MicroRNAs (miRs) are endogenous small non-coding RNA
molecules that regulate gene expression in a sequence-specific
manner. This is primarily accomplished through binding to the 3′
untranslated region of target mRNAs, either targeting the
transcripts for degradation or inhibiting their translation
(2). miRs are crucial not only in
tissue development but also in the pathology of a number of
diseases. A reduction in miR-185 expression levels has been
demonstrated to alter dendritic and spinal development, and has
also been implicated in psychiatric disorders and cognitive
dysfunction (3). miR-185 regulates
numerous biological functions, including immune and inflammatory
responses, and glutathione metabolism in alcoholic liver disease
(4). miR-185 also represses
selective high-density lipoprotein cholesterol uptake through the
inhibition of scavenger receptor class B member 1 in human hepatic
cells, implicating an important role of miRs in modulating
cholesterol metabolism (5).
The deregulation of miR expression has been
identified in several types of cancer. miR-185 was observed to
exhibit significant differential expression in prostate cancer and
normal tissue, and has been shown to suppress the proliferation,
invasion, migration and tumorigenicity of prostate cancer cells
through targeting androgen receptors (6). Furthermore, miR-185 was found to
inhibit cell growth and cell cycle progression, and its expression
was reduced in non-small cell lung cancer (7). However, miR microarray analysis
revealed upregulation of miR-185 expression in gastric and bladder
cancer, which was validated by quantitative polymerase chain
reaction (qPCR), determining a novel stimulatory function of
miR-185 in cancer (8,9). In order to investigate the function
of miR-185 in colon cancer, the clinical significance of miR-185
expression in colon cancer was analyzed in the present study using
qPCR, and the effects of miR-185 overexpression on cell
proliferation and the invasive potential of SW620 colon cancer
cells were investigated, in order to evaluate miR-185 as a
potential therapeutic target in colon cancer.
Materials and methods
Materials
The SW620, SW480, LOVO, HCT8 and HT29 human colon
cancer cell lines used in the experiments was obtained from the
Institute of Biochemistry and Cell Biology (Shanghai, China). The
colon cancer tissue and the corresponding adjacent non-cancerous
tissues (ANCT) were collected from 30 patients at the Department of
General Surgery of Shanghai First People’s Hospital, Affiliated to
Shanghai Jiaotong University (Shanghai, China). This study was
approved by the Medical Ethics Committee of Shanghai Jiao Tong
University (Shanghai, China) and written informed consent was
obtained from the patients or their parents prior to sample
collection. Two pathologists reviewed each of the cases. miR-185
mimic and negative control vectors were provided by Shanghai
Genechem Co., Ltd. (Shanghai, China). The miR-185 and HIF-2α
primers were synthesized by Applied Biosystems (Foster City, CA,
USA). All antibodies were provided by Santa Cruz Biotechnology,
Inc. (Santa Cruz, CA, USA).
Drugs and reagents
Dulbecco’s modified Eagle’s medium (DMEM) and fetal
bovine serum (FBS) were purchased from Thermo Fisher Scientific Inc
(Waltham, MA, USA); TRIzol Reagent and Lipofectamine 2000 were from
Invitrogen Life Technologies (Carlsbad, CA, USA); M-MLV Reverse
Transcriptase was from Promega GmbH (Madison, WI, USA); SYBR Green
Master Mixture was from Takara Bio, Inc. (Shiga, Japan). The
Enhanced Chemiluminescence (ECL)-PLUS kit was from GE Healthcare
(Piscataway, NJ, USA).
Reverse-transcription (RT) and qPCR
RT-PCR was used to detect the expression of the
primary transcripts and mature products of miR-185, and qPCR was
used to determine the expression of miR-185 and HIF-2α. Briefly,
for the primary transcript, 11 μg total RNA was reversely
transcribed using oligo-dT primer (Takara Bio, Inc.), and 2 μl
reverse transcription reaction mix was amplified by PCR as follows:
Denaturation at 95°C for 2 min and 25 cycles at 95°C for 30 sec,
55°C for 30 sec and 72°C for 1 min. For the mature product, 1 μg
total RNA was reverse-transcribed using miR-185-specific stem-loop
RT primer, and 2 μl reverse transcription mix was amplified by PCR
as follows: Denaturation at 95°C for 2 min and 25 cycles
(semiquantitative RT-PCR) or 50 cycles (quantitative real-time PCR)
at 95°C for 10 sec and 60°C for 1 min. The average level of U6
small nuclear (sn)RNA served as an internal control. The SYBR green
(Takara Bio, Inc.) method and the IQ5 Real-time PCR detection
system (Bio-Rad, Hercules, CA) were used for qPCR. The primer
sequences for the qPCR were as follows: Forward:
5′-CAATGGAGAGAAAGGCAGTTCC-3′ and reverse:
5′-AATCCATGAGAGATCCCTACCG-3′ for miR-185; forward:
5′-ATTGGAACGATACAGAGAAGATT-3′ and reverse:
5′-GGAACGCTTCACGAATTTG-3′ for U6 snRNA; forward:
5′-GCGCTAGACTCCGAGAACAT-3′ and reverse:
5′-TGGCCACTTACTACCTGACCCTT-3′ for HIF-2α; and forward:
5′-CAACGAATTTGGCTACAGCA-3′ and reverse 5′-AGGGGTCTACATGGCAACTG-3′
for GAPDH.
Cell culture and infection
The SW620 cells were cultured in DMEM supplemented
with 10% heat-inactivated FBS, 100 U/ml penicillin and 100 μg/ml
streptomycin. This was placed in a humidified atmosphere containing
5% CO2 at 37°C. Either the miR-185 mimic or the negative
control was used to infect the SW620 cells. The cells were
subcultured at a 1:5 dilution in 300 μg/ml G418-containing medium.
Positive stable transfectants were selected and expanded for
further investigation. The clone transfected by the miR-185 mimic
was termed the miR-185 mimic group, the clone transfected by the
negative control was termed the NC group and SW620 cells without
transfection were termed the CON group.
Western blot analysis
Colon cancer cells were harvested and extracted
using lysis buffer (containing Tris-HCl, SDS, mercaptoethanol and
glycerol). The cell extracts were boiled for 5 min in loading
buffer and then equal quantities of cell extracts were separated on
15% SDS-PAGE gels. The separated protein bands were transferred to
polyvinylidene fluoride membranes and the membranes were then
blocked in 5% skimmed milk powder. The primary antibodies against
HIF-2α, proliferating cell nuclear antigen (PCNA) and matrix
metallopeptidase-2 (MMP-2) were diluted according to the
manufacturer’s instructions and incubated overnight at 4°C.
Horseradish peroxidase-linked goat anti-mouse and goat anti-rabbit
IgG secondary antibodies were then added at a dilution ratio of
1:1,000 and the membranes were incubated at room temperature for 2
h. The membranes were then washed with phosphate-buffered saline
(PBS) three times and the immunoreactive bands were visualized
using the ECL-PLUS kit according to the manufacturer’s
instructions. The relative protein levels of the different cell
lines were normalized to the GAPDH concentration. Three separate
experiments were performed for each clone.
Cell proliferation assay
Cell proliferation was analyzed with the MTT assay.
Briefly, cells infected with miR-185 mimic were incubated in
96-well-plates at a density of 1×105 cells per well in
DMEM supplemented with 10% FBS. The cells were treated with 20 μl
MTT dye for 0, 24, 48 and 72 h, and then incubated with 150 μl
dimethylsulfoxide for 5 min. The color reaction was measured at 570
nm with an Automated immunoassay analyzer (Bio-Rad Laboratories,
Hercules, CA, USA). The proliferation activity was calculated for
each clone.
Transwell invasion assay
Transwell filters were coated with Matrigel (BD
Biosciences, Franklin Lakes, NJ, USA; 3.9 μg/μl, 60–80 μl) on the
upper surface of a polycarbonic membrane (diameter, 6.5 mm; pore
size, 8 μm; Corning, Corning, NY, USA). Following incubation at
37°C for 30 min, the Matrigel solidified and served as the
extracellular matrix for analysis of tumor cell invasion. Harvested
cells (1×105) in 100 μl serum-free DMEM were added into
the upper compartment of the chamber. A total of 200 μl conditioned
medium derived from NIH3T3 cells served as a source of
chemoattractant and was placed in the bottom compartment of the
chamber. After 24 h incubation at 37°C with 5% CO2, the
medium was removed from the upper chamber. The non-invaded cells on
the upper side of the chamber were scraped off with a cotton swab.
The cells that had migrated from the Matrigel into the pores of the
inserted filter were fixed with 100% methanol, stained with
hematoxylin, mounted and dried at 80°C for 30 min. The number of
cells invading through the Matrigel was counted in three randomly
selected visual fields from the central and peripheral portions of
the filter using an inverted microscope (Olympus, Tokyo, Japan;
magnification, ×200). Each assay was repeated three times.
Subcutaneous tumor model and gene
therapy
Female immune-deficient nude mice (BALB/c-nu; age, 6
weeks) from the laboratory animal facility of the Hematology
Institute of Chinese Academy of Sciences (Shanghai, China) were
housed individually in microisolator ventilated cages with free
access to water and food. All experimental procedures were
performed according to the regulations and internal biosafety and
bioethics guidelines of Shanghai Tongji University and the Shanghai
Municipal Science and Technology Commission (Shanghai, China).
Four mice were injected subcutaneously with
1×108 SW620 cells in 50 μl PBS pre-mixed with an equal
volume of matrigel matrix (Becton-Dickinson, Franklin Lakes, NJ,
USA). The mice were monitored daily and three out of the four mice
developed a subcutaneous tumor. When the tumors reached ~5 mm in
length, they were surgically removed, cut into 1–2-mm3
sections and reseeded individually into six other mice. When the
reseeded tumors reached ~5 mm in length, the mice were randomly
assigned to either the NC group or the miR-185-treated group. In
the miR-185 group, 15 μl lentivirus was injected into the
subcutaneous tumors using a multi-site injection format. The mice
in the NC group received 15 μl PBS only. Injections were repeated
every other day following the initial treatment. The tumor volume
was measured every three days with a caliper, using the formula:
Volume = (length × width)2/2.
Statistical analysis
SPSS 21.0 software (IBM, Armonk, NY, USA) was used
for the statistical analysis. One-way analysis of variance (ANOVA)
was used to analyze the differences between groups. The least
significant difference method of multiple comparisons was used when
the probability for ANOVA was statistically significant and
P<0.05 was considered to indicate a statistically significant
difference.
Results
Correlation of miR-185 expression with
clinicopathologic characteristics of patients with colon
cancer
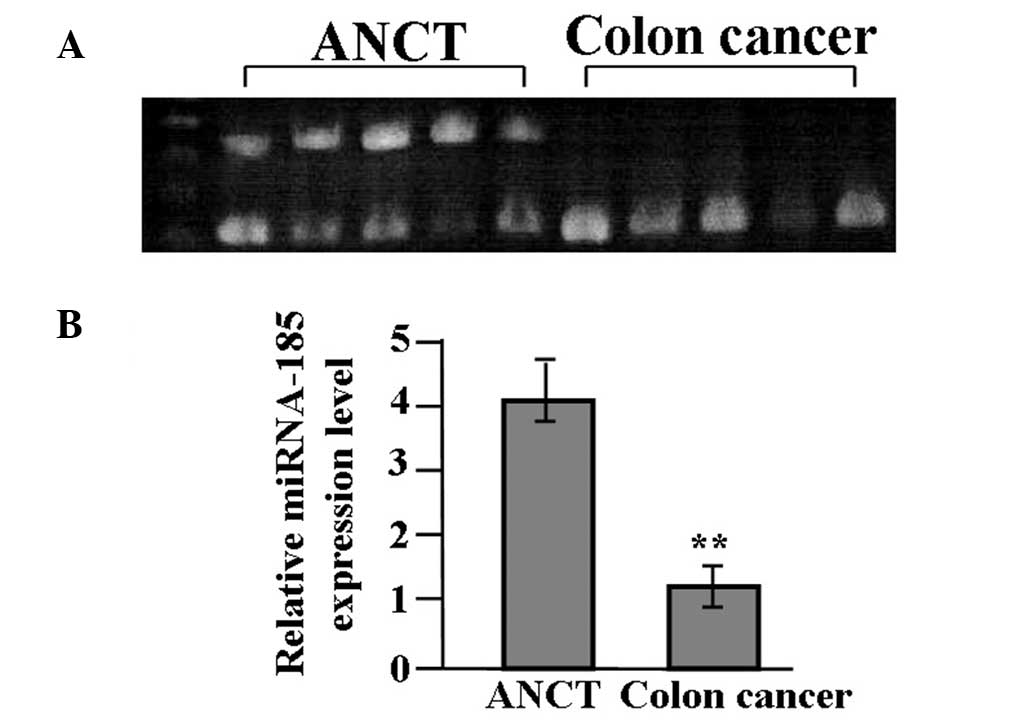
The expression of miR-185 in colon cancer tissues
and ANCT was evaluated by RT-PCR. As shown in Fig. 1, miR-185 expression was
significantly reduced in cancer tissues compared with the ANCT
(P<0.01).
The correlation of miR-185 expression with
clinicopathologic characteristics of patients with colon cancer was
further analyzed. As shown in Table
I, the reduced expression of miR-185 was closely associated
with lymph node metastasis of colon cancer (P<0.001), but was
not associated with the age or gender of the patients, tumor size,
degree of tumor differentiation or tumor-node-metastasis stage
(P>0.05).
 | Table ICorrelation of miR-185 expression with
clinicopathologic characteristics of patients with colon
cancer. |
Table I
Correlation of miR-185 expression with
clinicopathologic characteristics of patients with colon
cancer.
| Variable | Cases (n) | Relative expression
level of miR-185 (95% CI) | P-value |
|---|
| Age (years) | | | >0.05 |
| ≥60 | 16 | 0.36–22.35 | |
| <60 | 14 | 0.32–32.67 | |
| Sex | | | >0.05 |
| Male | 18 | 0.35–35.78 | |
| Female | 12 | 0.32–24.37 | |
| Tumor size (cm) | | | >0.05 |
| ≥5 | 20 | 0.32–35.47 | |
| <5 | 10 | 0.37–22.15 | |
| Degree of
differentiation | | | >0.05 |
| Well/Moderately | 22 | 0.36–22.35 | |
| Poorly | 8 | 0.36–22.35 | |
| TNM stage | | | >0.05 |
| I+II | 14 | 0.36–36.47 | |
| III+IV | 16 | 0.32–32.77 | |
| Lymph node
metastasis | | | <0.001 |
| Negative | 17 | 0.39–37.67 | |
| Positive | 13 | 0.31–9.12 | |
Modulation of miR-185 and HIF-2α
expression
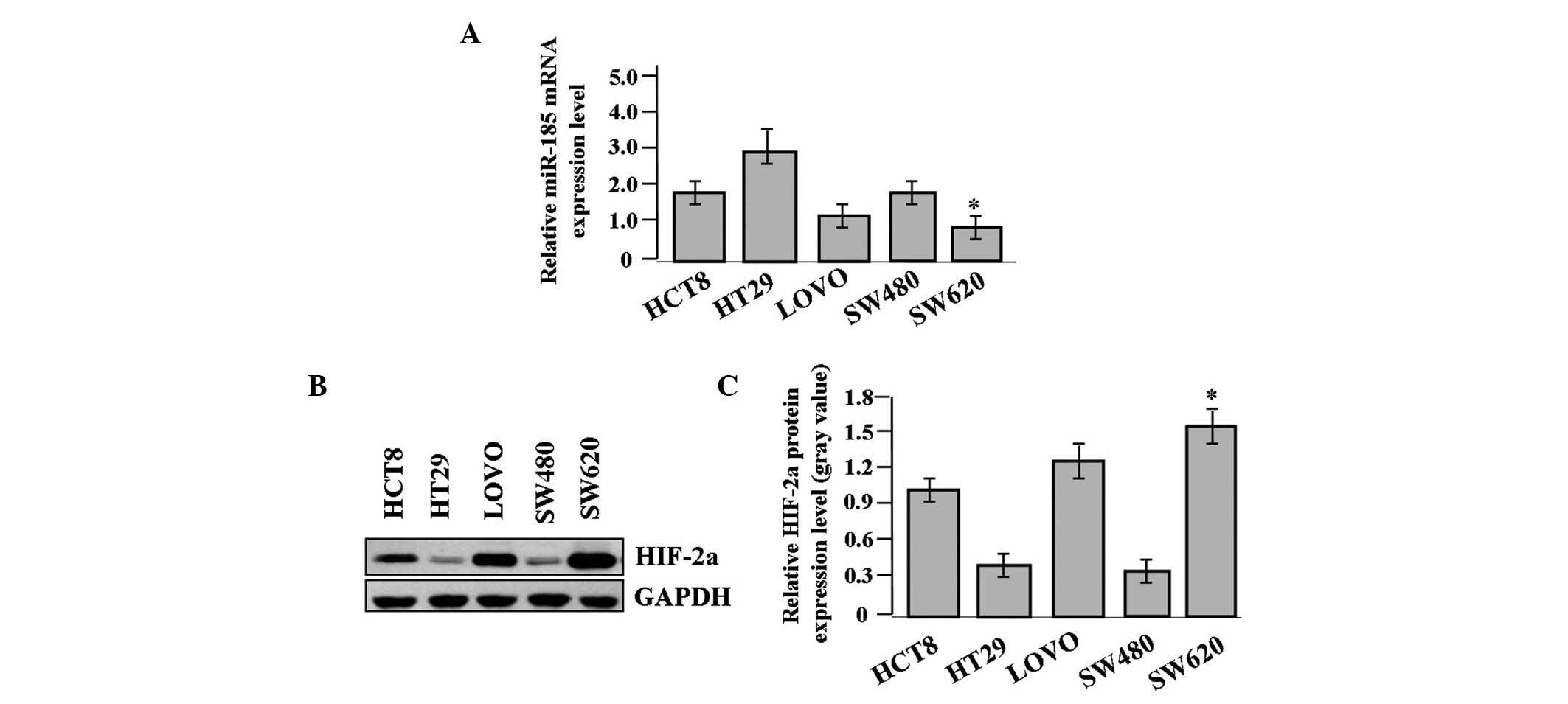
The expression of miR-185 and HIF-2α was examined in
different colon cancer cell lines. The expression level of miR-185,
indicated by real-time PCR, was significantly reduced (Fig. 2A), while that of HIF-2α was
increased in the SW620 colon cancer cells compared with the other
cell lines, as shown by western blot analysis (Fig. 2B and C).
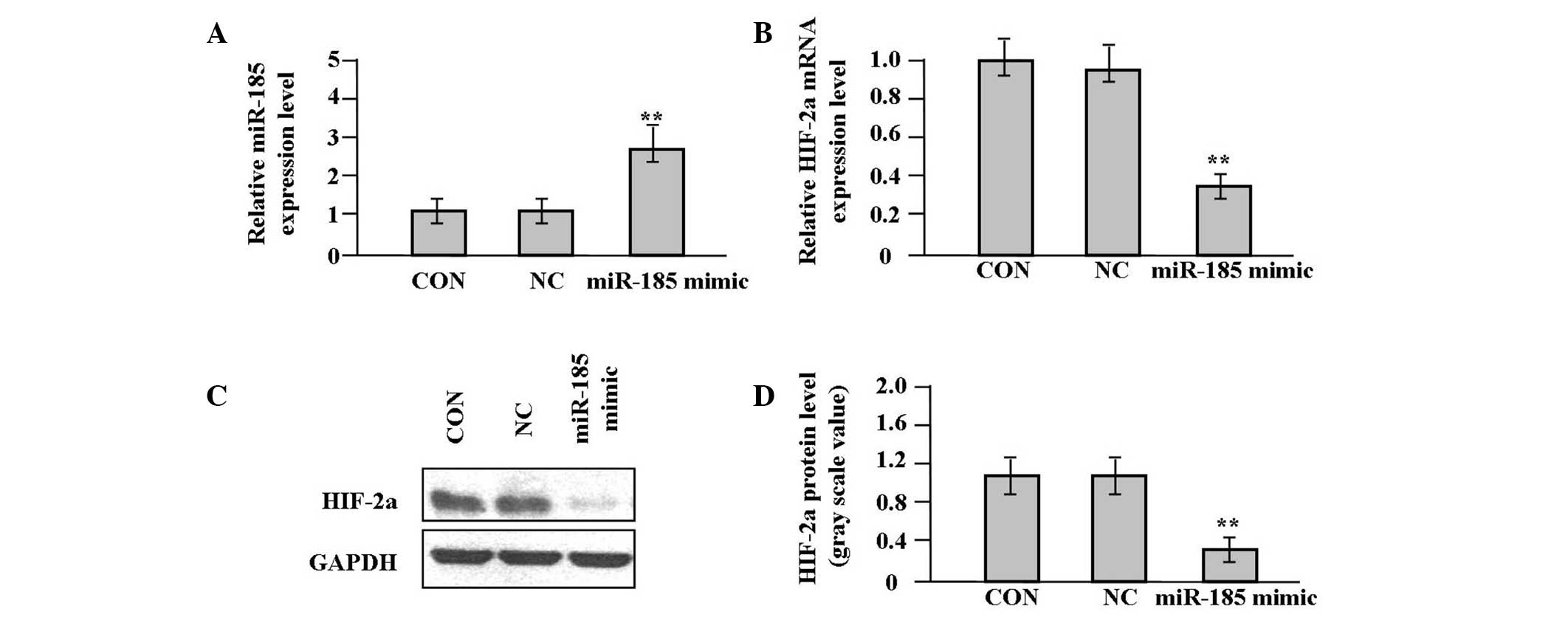
To monitor the effect of miR-185 overexpression on
HIF-2α expression in target cells expressing low levels of miR-185,
the miR-185 mimic and scrambled control were delivered into SW620
cells. The expression levels of miR-185 and HIF-2α following
miR-185 mimic infection were further examined for 48 h by qPCR. As
shown in Fig. 3A and B, a
significant increase in miR-185 expression and a significant
reduction in HIF-2α mRNA expression respectively were observed in
the miR-185 mimic group compared with the NC and CON groups (each
P<0.01). The expression level of HIF-2α protein, indicated by
western blot analysis, was significantly downregulated in the
miR-185 mimic group in comparison with the NC and CON groups
(P<0.01, Fig. 3C and D).
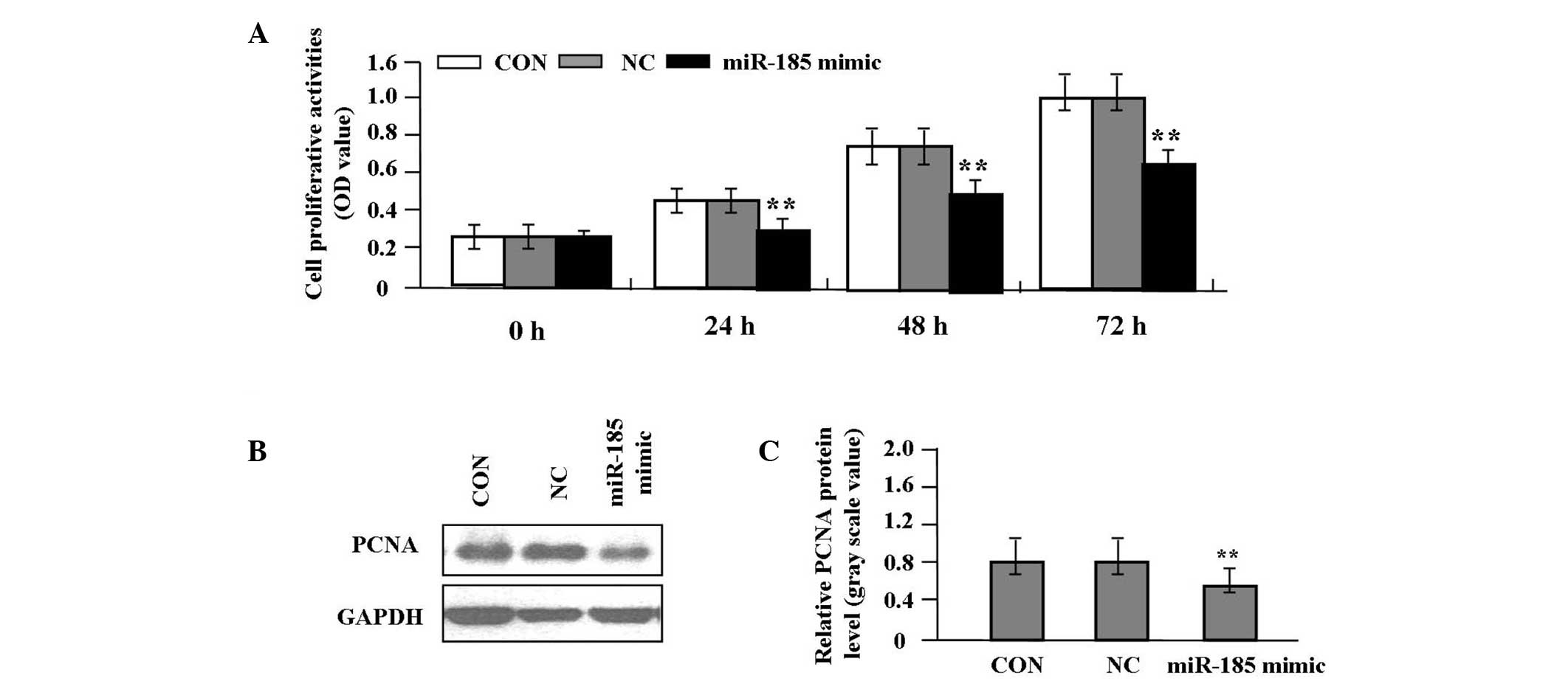
Effect of miR-185 overexpression on cell
proliferation
Deregulated cell proliferation is a hallmark of
cancer (10). In order to analyze
the effect of miR-185 overexpression on the growth of colon cancer
cells, the proliferative activities of SW620 cells were
investigated by an MTT assay. miR-185 overexpression significantly
diminished the proliferative activities of SW620 cells compared
with the NC and CON groups (P<0.01, Fig. 4A). In order to determine whether
miR-185 suppressed the endogenous expression of PCNA through
translational repression, the expression of PCNA protein was
examined by western blot analysis, which indicated that the
quantity of PCNA protein was significantly reduced in the miR-185
mimic group compared with the NC and CON groups (P<0.01,
Fig. 4B and C), suggesting that
miR-185 may inhibit the proliferation of colon cancer cells through
downregulation of PCNA expression.
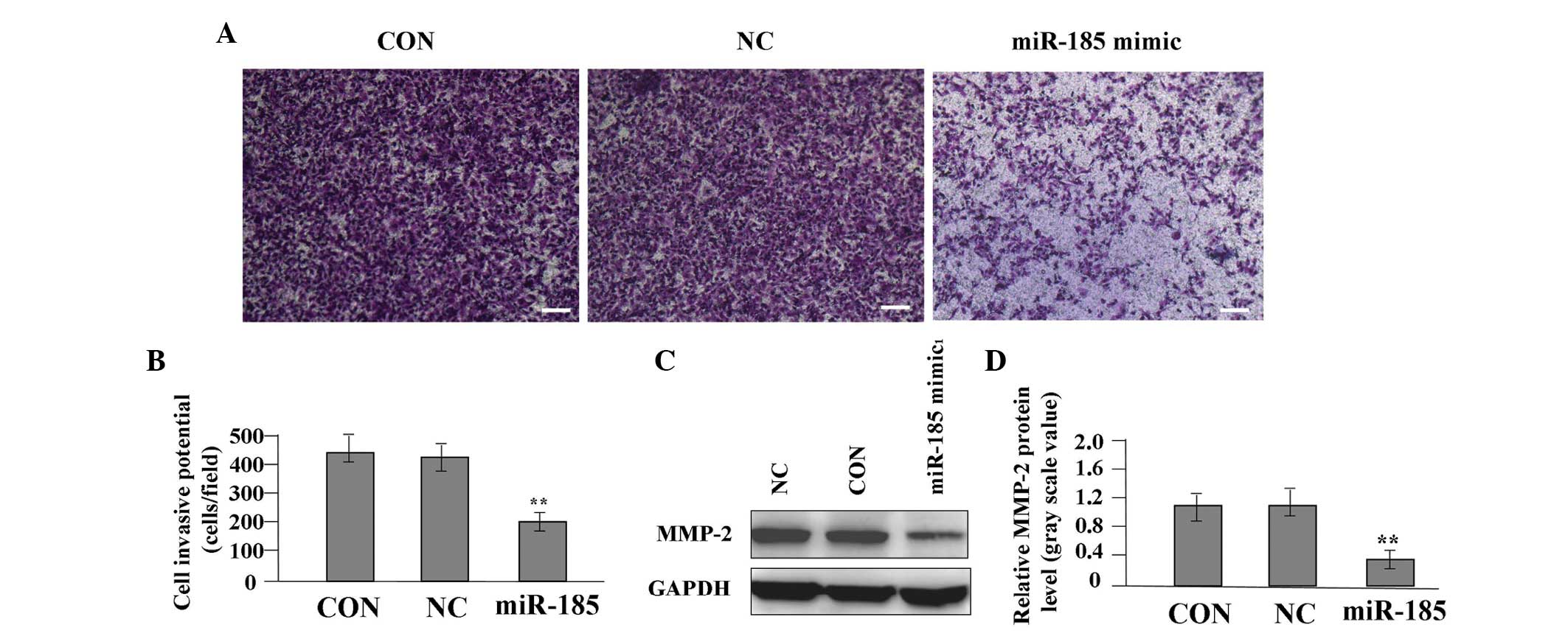
Effect of miR-185 overexpression on cell
invasion
To determine the effect of miR-185 overexpression on
the invasive potential of colon cancer cells, a Transwell assay was
performed. The invasive and metastatic potential was determined in
the Transwell assay on the basis of the ability of cells to invade
a matrix barrier containing laminin and type IV collagen, the
predominant components of the basement membrane. Representative
micrographs of Transwell filters are shown in Fig. 5A. The invasive potential of SW620
cells was significantly reduced in the miR-185 mimic group compared
with the NC and CON groups (P<0.01, Fig. 5B). A western blot analysis was
performed to investigate the effect of miR-185 overexpression on
the endogenous expression of the MMP-2 protein. The expression
level of MMP-2 was found to be significantly reduced in the miR-185
mimic group compared with the NC and CON groups (P<0.01,
Fig. 5C and D), indicating that
miR-185 may inhibit cell invasion of colon cancer through
downregulation of MMP-2 expression.
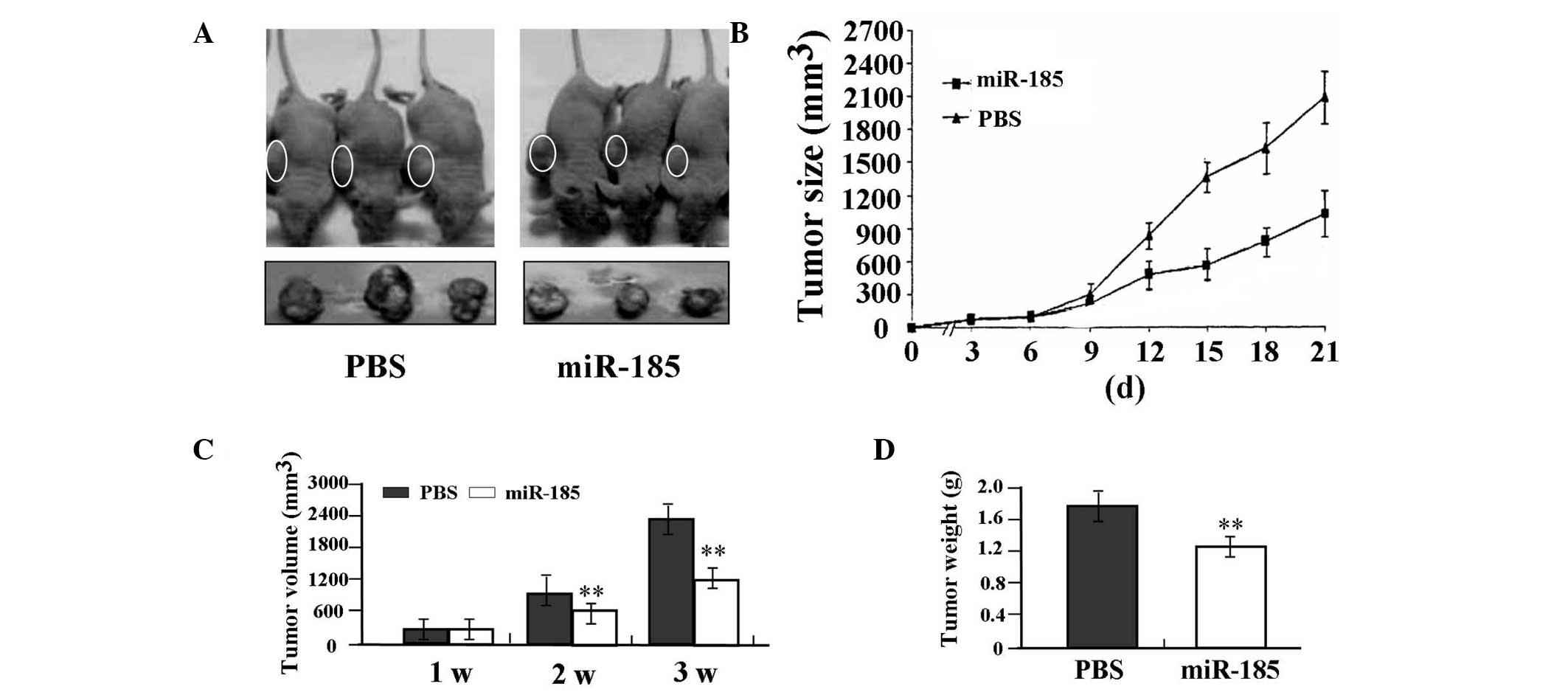
Antitumor effect of miR-185 in the SW620
xenograft model
The in vitro experiments demonstrated that
overexpression of miR-185 efficiently inhibited proliferation and
invasion in SW620 colon cancer cells. The antitumor effect of
miR-185 was further investigated in vivo using the SW620
xenograft model and lentivirus-mediated gene therapy. The mean
volume of tumors in all experimental mice prior to treatment was
51.65±8.33 mm3. Each mouse was administered either in
situ injection of PBS (n=3) or lentivirus-mediated miR-185
(n=3). During the first three weeks of recovery, the tumors in the
miR-185 group grew at a slower rate compared with those in the PBS
group (Fig. 6A and B). There was a
significant difference in tumor volumes and weight between the
miR-185 and PBS groups over the observation period (P<0.01,
Fig. 6C and D).
Discussion
miRs are small regulatory RNAs involved in various
physiological and cellular processes. Alterations of miRs are
crucial for tumorigenesis (11).
miRs exhibit potential as diagnostic and prognostic biomarkers for
colon cancer (12). Using miR
expression patterns and target prediction, previous studies have
observed that miR-185 expression is upregulated and results in loss
of function of tumor suppressors in clear cell renal cell carcinoma
(13) and lung squamous cell
carcinoma (14). However, in the
present study, miR-185 was found to have low expression in colon
cancer tissues compared with ANCT, suggesting that miR-185
expression may be different among types of tumor tissue. miR-185
expression was demonstrated to be negatively correlated with lymph
node metastasis of colon cancer. However, the clinical data
collected lacked information regarding the survival times of the
colon cancer patients, thus the association between miR-185
expression and survival time was not assessed. Nevertheless,
another study indicated the potential prognostic value of miR-185
expression levels for predicting clinical outcomes following
surgery (15).
Low expression of miR-185 is also associated with a
poor outcome in glioma patients. miR-185 was found to suppress
glioma cell invasion, indicating that it may be a potential
prognostic marker and therapeutic target (16). Previous studies have revealed that
miR-185 impedes anchorage-independent growth, cell migration and
invasion, in addition to suppressing tumor growth in vivo
and increasing cisplatin sensitivity by promoting apoptosis,
implicating it as a potent tumor suppressor (17–19).
In the present study, overexpression of miR-185 repressed the
growth and invasion of colon cancer cells in vitro and in
vivo, which was consistent with the results of previous
studies, indicating that miR-185 is a negative regulator of RhoA
and Cdc42 and their cellular activities, and can inhibit the
proliferation and invasion of CRC cells (20).
A number of studies have shown a link between the
microRNA pathway and hypoxia signaling. HIF-2α is regulated by the
Dicer-dependent miR-185, which is downregulated in hypoxia
(21). HIF-2α promotes hypoxic
tumor cell proliferation by enhancing c-myc transcriptional
activity (22) and results in CRC
progression by dysregulating iron homeostasis (23). Disruption of the HIF-2α gene
inhibits tumor angiogenesis and perfusion in human colon cancer
cells (24). In the present study,
overexpression of miR-185 suppressed proliferation and invasion,
and reduced expression levels of HIF-2α in colon cancer cells,
suggesting that miR-185 may be implicated in the development of
colon cancer cells through inhibition of the HIF-2α pathway.
However, whether HIF-2α is the target of miR-185 in colon cancer
cells requires further investigation.
The findings of the present study showed that
overexpression of miR-185 markedly reduced the expression of PCNA
and MMP-2 in colon cancer cells at the translational level,
indicating that miR-185 may suppress proliferation and invasion of
colon cancer cells via the downregulation of PCNA and MMP-2
expression. PCNA is a nuclear protein that is expressed in
proliferating cells and may be required for maintaining cell
proliferation, which is used as a marker to evaluate colon cancer
cell proliferation (25,26). MMP-2, a predictor of GM cell
invasion, is pivotal in the degradation of the extracellular
matrix, and thereby enhances the invasive, proliferative and
metastatic potential of colon cancer (27). Certain studies have confirmed that
HIF-1 signaling promotes tumor migration and invasion through
upregulation of MMP-2 and PCNA expression (28,29).
However, there was no direct evidence that HIF-2α promoted
proliferation and invasion of colon cancer cells through
upregulation of MMP-2 and PCNA expression from the present study.
Therefore, miR-185 may be involved in proliferation and invasion of
colon cancer cells through inhibition of HIF-2α-mediated PCNA and
MMP-2 expression.
In conclusion, these findings indicate that miR-185
as a tumor suppressor may be involved in the development of colon
cancer cells via inhibition of HIF-2α signaling, suggesting that
miR-185 may serve as a potential therapeutic target in the
treatment of cancer.
Acknowledgements
This study was supported by the project of Shanghai
First People’s Hospital (grant no. 12B01).
References
|
1
|
Antonic V, Stojadinovic A, Kester KE, et
al: Significance of infectious agents in colorectal cancer
development. J Cancer. 4:227–240. 2013. View Article : Google Scholar
|
|
2
|
He L and Hannon GJ: MicroRNAs: small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xu B, Hsu PK, Stark KL, et al:
Derepression of a neuronal inhibitor due to miRNA dysregulation in
a schizophrenia-related microdeletion. Cell. 152:262–275. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu Y, Chen SH, Jin X and Li YM: Analysis
of differentially expressed genes and microRNAs in alcoholic liver
disease. Int J Mol Med. 31:547–554. 2013.PubMed/NCBI
|
|
5
|
Wang L, Jia XJ, Jiang HJ, et al: MicroRNAs
185, 96, and 223 repress selective high density lipoprotein
cholesterol uptake through posttranscriptional inhibition. Mol Cell
Biol. 33:1956–1964. 2013. View Article : Google Scholar
|
|
6
|
Qu F, Cui X, Hong Y, et al: MicroRNA-185
suppresses proliferation, invasion, migration, and tumorigenicity
of human prostate cancer cells through targeting androgen receptor.
Mol Cell Biochem. 377:121–130. 2013. View Article : Google Scholar
|
|
7
|
Takahashi Y, Forrest AR, Maeno E, et al:
MiR-107 and MiR-185 can induce cell cycle arrest in human non small
cell lung cancer cell lines. PLoS One. 4:e66772009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yao Y, Suo AL, Li ZF, et al: MicroRNA
profiling of human gastric cancer. Mol Med Rep. 2:963–970.
2009.PubMed/NCBI
|
|
9
|
Gottardo F, Liu CG, Ferracin M, et al:
Micro-RNA profiling in kidney and bladder cancers. Urol Oncol.
25:387–392. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: the next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mannoor K, Liao J and Jiang F: Small
nucleolar RNAs in cancer. Biochim Biophys Acta. 1826:121–128.
2012.PubMed/NCBI
|
|
12
|
Yu G, Tang JQ, Tian ML, et al: Prognostic
values of the miR-17–92 cluster and its paralogs in colon cancer. J
Surg Oncol. 106:232–237. 2012.
|
|
13
|
Liu H, Brannon AR, Reddy AR, et al:
Identifying mRNA targets of microRNA dysregulated in cancer: with
application to clear cell Renal Cell Carcinoma. BMC Syst Biol.
4:512010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang Y, Li X, Yang Q, et al: The role of
microRNA in human lung squamous cell carcinoma. Cancer Genet
Cytogenet. 200:127–133. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Akçakaya P, Ekelund S, Kolosenko I, et al:
miR-185 and miR-133b deregulation is associated with overall
survival and metastasis in colorectal cancer. Int J Oncol.
39:311–318. 2011.PubMed/NCBI
|
|
16
|
Tang H, Wang Z, Liu X, et al: LRRC4
inhibits glioma cell growth and invasion through a
miR-185-dependent pathway. Curr Cancer Drug Targets. 12:1032–1042.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Imam JS, Buddavarapu K, Lee-Chang JS, et
al: MicroRNA-185 suppresses tumor growth and progression by
targeting the Six1 oncogene in human cancers. Oncogene.
29:4971–4979. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Greenberg E, Hershkovitz L, Itzhaki O, et
al: Regulation of cancer aggressive features in melanoma cells by
microRNAs. PLoS One. 6:e189362011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xiang Y, Ma N, Wang D, et al: MiR-152 and
miR-185 co-contribute to ovarian cancer cells cisplatin sensitivity
by targeting DNMT1 directly: a novel epigenetic therapy independent
of decitabine. Oncogene. 33:378–386. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu M, Lang N, Chen X, et al: miR-185
targets RhoA and Cdc42 expression and inhibits the proliferation
potential of human colorectal cells. Cancer Lett. 301:151–160.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ho JJ, Metcalf JL, Yan MS, et al:
Functional importance of Dicer protein in the adaptive cellular
response to hypoxia. J Biol Chem. 287:29003–29020. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gordan JD, Bertout JA, Hu CJ, et al:
HIF-2α promotes hypoxic cell proliferation by enhancing c-myc
transcriptional activity. Cancer Cell. 11:335–347. 2007.
|
|
23
|
Xue X, Taylor M, Anderson E, et al:
Hypoxia-inducible factor-2α activation promotes colorectal cancer
progression by dysregulating iron homeostasis. Cancer Res.
72:2285–2293. 2012.
|
|
24
|
Burkitt K, Chun SY, Dang DT, et al:
Targeting both HIF-1 and HIF-2 in human colon cancer cells improves
tumor response to sunitinib treatment. Mol Cancer Ther.
8:1148–1156. 2009. View Article : Google Scholar
|
|
25
|
Naryzhny SN: Proliferating cell nuclear
antigen: a proteomics view. Cell Mol Life Sci. 65:3789–3808. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wilson MS and Schofield PF: Markers to
study human colonic cell proliferation. Gut. 36:1521995. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Baker EA, Bergin FG and Leaper DJ: Matrix
metalloproteinases, their tissue inhibitors and colorectal cancer
staging. Br J Surg. 87:1215–1221. 2000. View Article : Google Scholar
|
|
28
|
Fujiwara S, Nakagawa K, Harada H, et al:
Silencing hypoxia-inducible factor-1α inhibits cell migration and
invasion under hypoxic environment in malignant gliomas. Int J
Oncol. 30:793–802. 2007.
|
|
29
|
Nakanishi K, Hiroi S, Tominaga S, et al:
Expression of hypoxia-inducible factor-1α protein predicts survival
in patients with transitional cell carcinoma of the upper urinary
tract. Clin Cancer Res. 11:2583–2590. 2005.
|