In conclusion, the present study
demonstrated that ABCB4 and ABCB11 may be promising candidate genes
for evaluating the risk of developing PIS. Mutations in these genes
have been shown to alter the expression and function of their
corresponding proteins and therefore may be associated with the
formation of PIS.
Introduction
Primary intrahepatic stones (PIS) are one of the
most prevalent hepatobiliary disorders in China and Japan; in
China, >20% of PIS patients were found to be located in the
South-West of the country (1,2). PIS
can significantly affect the health and quality of life of patients
and treatment of the disease is complex, requiring multiple
surgical interventions (2).
Long-term PIS may develop into biliary cirrhosis or cancer of the
liver bile duct. The pathogenesis of PIS was reported to be
associated with risk factors, including bile stasis, chronic
inflammation of the bile ducts, bacterial infection and
malnutrition (3,4) as well as environmental factors
(5–7). Studies have reported that the
variation of two genes, ABCB4 and ABCB11, may result in altered
bile composition and therefore contribute to the pathogenesis of
PIS (8,9).
ABCB4 encodes for a lipid translocator, multidrug
resistance protein 3 (MDR3), which transports phosphatidylcholine
from the inner to the outer leaflet of the canalicular membrane of
hepatocytes (10) and therefore
has a crucial role in maintaining normal phosphatidylcholine
concentrations in bile. Studies have shown that the hereditary
depletion of ABCB4 expression may result in progressive familial
intrahepatic cholestasis type 3 (PFIC3) (11) and intrahepatic cholestasis of
pregnancy (ICP) (12–19). Furthermore, variants of the ABCB4
gene have been identified and were reported to be associated with
cholecystolithiasis (20–21). However, the function of ABCB4 in
PIS remains to be elucidated.
ABCB11 encodes for the bile salt export pump (BSEP),
which mediates the canalicular secretion of bile salts from
hepatocytes into the bile, representing the primary driving force
for the generation of bile salts (8, 22–23).
ABCB11 gene mutations have been shown to decrease the secretion of
bile acid (24) and cause PFIC
type 2 (PFIC2) as well as benign recurrent intrahepatic cholestasi
type 2 (BRIC2) (25–27). Studies have also confirmed that
genetic variation in ABCB11 was associated with the pathogenesis of
ICP (27). However, an association
between ABCB11 and PIS has not yet been established.
Mutations of the ABCB4 and ABCB11 genes affect the
composition of bile and were associated with cholestasis and
cholelithiasis (17,28–29).
Therefore, the hypothesis of the present study was that mutations
of these genes may be responsible for the development of PIS. To
investigate whether variations in ABCB4 and ABCB11 are associated
with PIS, variations in these two genes were analyzed in PIS
patients and control subjects using exon sequencing and
subsequently evaluated to determine possible associations between
these variations and the clinical manifestations of PIS.
Materials and methods
Liver tissue and blood specimens
Samples were obtained from patients at the
Department of Hepatobiliary Surgery at Southwest Hospital of the
Third Military Medical University (Chongqing, China) between
February 2009 and December 2012. Paraffin-embedded liver tissue
blocks and blood specimens were obtained from all 176 patients.
Another 62 normal liver tissue blocks were obtained from patients
with hepatic hemangioma. A total of 178 healthy controls were
recruited at random from the Center for Health Inspection at the
same hospital. All patients (excluding patients with hepatic
hemangioma) were assessed every 3 months following being discharged
using ultrasonography and computed tomography. The occurrence of
new stones was considered indicative of a relapse. The median
follow-up period was 20 months (range, 3–57 months). During this
period, 38 patients experienced recurrences of PIS (21.6%). The
present study was approved by the local ethics committee of the
Third Military Medical University following the guidelines of the
Declaration of Helsinki.
Inclusion and exclusion criteria of study
patients and controls
The diagnostic criteria used to identify patients
with PIS were as follows: a) No history of chronic liver disease;
b) absence of current infection with hepatitis A, B or C virus and
cytomegalovirus; c) no previous liver resection for PIS; d) PIS
confirmed using abdominal B-mode ultrasound and computed tomography
or magnetic resonance cholangiopancreatography (Siemens., Berlin,
Germany); e) exclusion of cholecystolithiasis using abdominal
B-mode ultrasound; and f) only patients from the Han Chinese
population. Age, gender, and body mass index (BMI) were recorded
when patients were recruited to the study.
The 62 patients with hepatic hemangioma were all
identified by pathological diagnosis. All control subjects
underwent routine laboratory investigations and were not
genetically related to any of the patients in the study. Controls
were selected according to the following criteria a) PIS was not
detected during the health inspection at the time of recruitment;
b) ethnicities other than Han Chinese were excluded; and c) health
was inspected in the same time-frame as the recruitment of
patients.
Immunohistochemistry
BSEP and MDR3 proteins were determined
immunohistologically using an immunoperoxidase staining method.
Paraffin-embedded tissue blocks were cut into serial sections of
3–5 μm, which were then deparaffinized in xylene and rehydrated in
a graded ethanol series. Endogenous peroxidase activity was
quenched using a 3% hydrogen peroxide solution for 30 min. Antigens
for BSEP and MDR3 were retrieved by heating the tissue sections at
120°C for 10 min in citrate buffer (10 mmol/l; pH 6.0). Following
rinsing in phosphate-buffered saline (PBS) for five minutes,
sections were incubated at 37°C for 120 min with antibodies against
BSEP (HPA019035; Sigma, St. Louis, MO, USA) or MDR3 (ab111209;
Abcam, Cambridge, MA, USA). The sections were then incubated with a
secondary antibody for HPA019035 (K5007; Dako, Carpinteria, CA,
USA) and ab111209 (PV-9003; ZSGB-BIO, Beijing, China) at 37°C for
30 min. Negative controls were prepared by omitting the primary
antibodies. Immunoreactivity was detected using a Diaminobenzidine
Elite kit (K5007; Dako; Zhongshan Biotechnology Company, Beijing,
China) and positive staining was identified as brown staining of
immunopositive cells. The sections were dehydrated, cleared and
mounted.
The immunostained tissue slides were scored
according to stain intensity (0, no stain; 1, slight staining; 2,
moderate staining; 3, intense staining) multiplied by a
distribution score (1, staining of 0–33%; 2, staining of 33–66%; 3,
>66% staining). The final score was grouped as low expression
[negative, zero or low (1–2) scores], medium expression (moderate
score, 3–4), or high expression (high score, 6–9) for further
non-parametric tests (see Fig. 1).
Each immunostained slide was scored by two pathologists in a
double-blind manner using a microscope (Olympus, Tokyo, Japan).
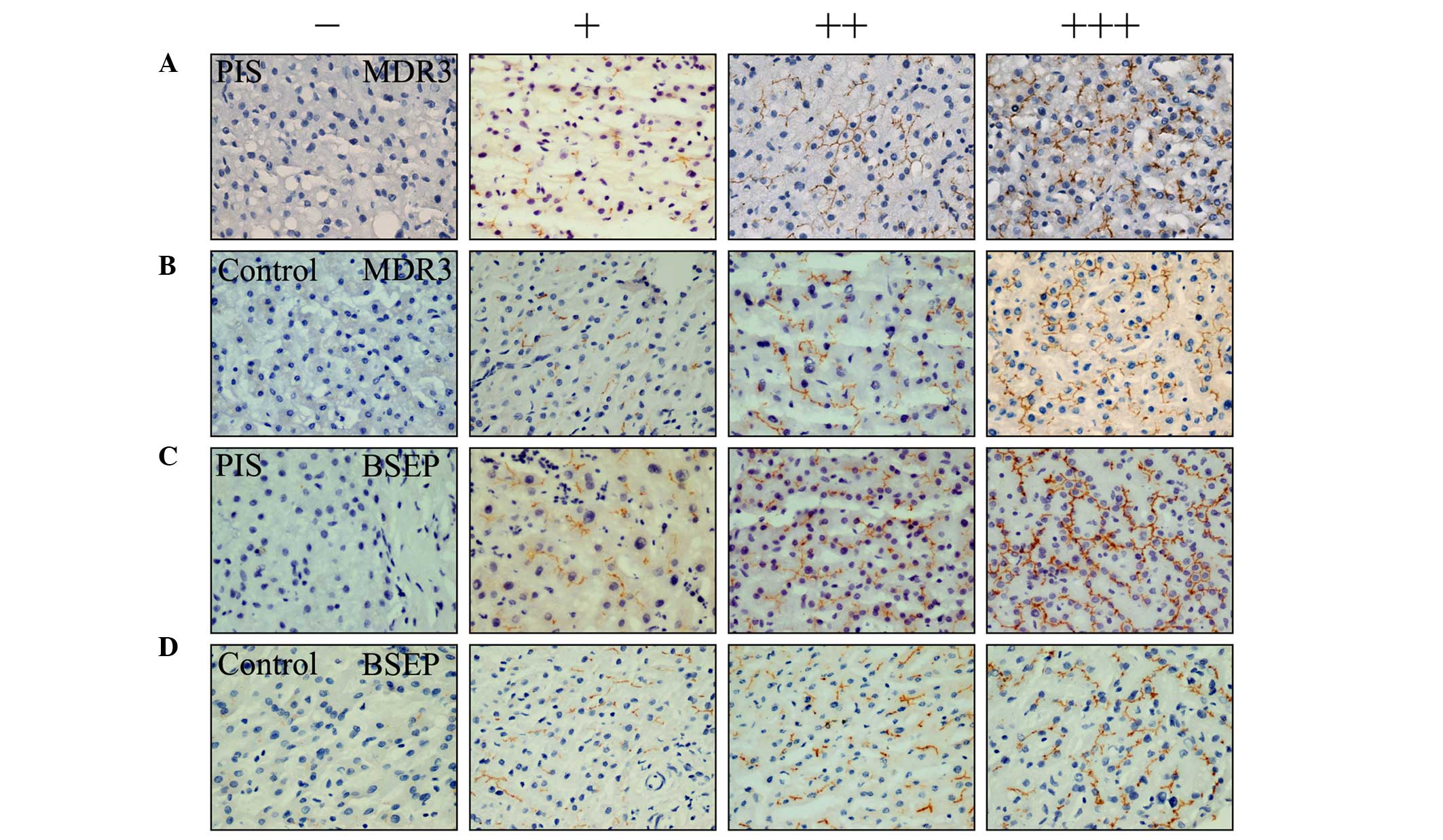 | Figure 1MDR3 and BSEP expression levels,
examined by immunohistochemistry in liver tissues of PIS patients
and healthy individuals. Immunohistochemical staining of liver
tissues of PIS: (A) Left two images indicate low expression of MDR3
and the right two indicate medium and high expression of MDR3,
respectively; (B) left two images indicate low expression of MDR3
and the right two indicate medium and high expression of MDR3,
respectively; and (C) left two images indicate low expression of
BSEP and the right two images indicate medium and high expression
of BSEP, respectively. (D) Immunohistochemical staining of liver
tissues of healthy controls: left two images indicate low
expression of BSEP, right two indicate medium and high expression
of BSEP, respectively. (magnification, ×400). −, negative or zero
score; +, score 1–2; ++ score 3–4; +++, score 6–9; MDR3, multidrug
resistance protein 3; BSEP, bile salt export pump; PIS, primary
intrahepatic stones. |
Genotyping
Genomic DNA was extracted from peripheral venous
blood leucocytes using a Blood Genome DNA Extraction kit (D9081;
Takara Bio, Inc., Shiga, Japan) according to the manufacturer’s
instructions. The concentration of extracted DNA was quantified
using spectrophotometry with a nucleic acid analyzer NV3000C
(Vastech, Inc., San Jose, CA, USA). 54 primer pairs were designed
in order to amplify all 54 exons in ABCB4 and ABCB11 (Table I) using standard PCR methods. DNA
polymorphism of the genes of interest was determined in all
subjects by direct sequencing of all 54 exons of ABCB4 and ABCB11
using an ABI 3730xl genetic analyzer (Applied Biosystems, Foster
City, CA, USA).
 | Table IPrimer pairs for amplification of
ABCB4 and ABCB11 coding sequences. |
Table I
Primer pairs for amplification of
ABCB4 and ABCB11 coding sequences.
| Exon | Forward primer | Reverse primer | Fragment length
(bp) |
|---|
| ABCB4 |
| 2 |
5′-GAGAGGGTGTACTTGGTTCTGA-3′ |
5′-AGACAAGGAGGAAGAGACATAACA-3′ | 600 |
| 3 |
5′-ACTTAGCGACACTGTTAGCATAC-3′ |
5′-ATCTCACTATGTTGTCCAGGCT-3′ | 537 |
| 4 |
5′-TAGCCGTTCTTTATTACCACAGG-3′ |
5′-GGAGTCAACCAGATATCCAAATCA-3′ | 590 |
| 5 |
5′-CTCCTCTGTGCCTCCTTAGAAG-3′ |
5′-GCCATGATGTGTGCATCTTACT-3′ | 535 |
| 6 |
5′-GTGGTGGCTCATGCTATAATCC-3′ |
5′-TAGGCTATAGATGCTGCTAGACAT-3′ | 691 |
| 7 |
5′-GGGAAGATGTTATTCAAGAGGCTA-3′ |
5′-GATACAGGAATGAGTCACCACAC-3′ | 537 |
| 8 |
5′-GTAGATGTGGAACTTGACAGTGT-3′ |
5′-GCCATCAGTAAAGGGTGCTTAT-3′ | 554 |
| 9 |
5′-GAATGTTGGTTGGCTTGGAATG-3′ |
5′-CTGGACAGTGGAAAGATTCACC-3′ | 559 |
| 10 |
5′-AAATAGACCCACTCAGGCAATA-3′ |
5′-GATTCTCAGTCTGGCTACAACA-3′ | 620 |
| 11 |
5′-ACACACAATCTTCATCTTGGGT-3′ |
5′-AGAACACACTTCACAACAAAGC-3′ | 455 |
| 12 |
5′-CCTAACTGAGGTTGATTGTTGA-3′ |
5′-GAAGGCATTATCCATCGGCATT-3′ | 462 |
| 13 |
5′-GGCAAATAATCCTTCCACGAAA-3′ |
5′-AGTTGGACAATCTTGCATCTCA-3′ | 538 |
| 14 |
5′-GCCTCTGATAAAGTAGTTGTCCTT-3′ |
5′-GCACTGGCAAGAATCTTCAATAG-3′ | 592 |
| 15 |
5′-CATTCATCCAAGTGCTTAACTGTG-3′ |
5′-GTCTCTGGCATTCCATATAACTCA-3′ | 615 |
| 16 |
5′-ATCCTTGATTGAGAAGCAGTTAGG-3′ |
5′-GCATCTCAGCGTAAAGACTACAT-3′ | 572 |
| 17 |
5′-GCTTGTCATTCTCTGCACCTAG-3′ |
5′-GTCTGACACCAGTTTCACCAAA-3′ | 545 |
| 18 |
5′-TGTAGATGGATACTGGTGAAGGTT-3′ |
5′-TTGGTAATGAATGTCTGCTGAGG-3′ | 570 |
| 19 |
5′-AGTTGAACATGATTGGTCTGGTG-3′ |
5′-GACATCCTGGAGTCTCAATGGA-3′ | 503 |
| 20 |
5′-CTACCACAGAGCAGTCAGGAAG-3′ |
5′-GGGATAAAGCCAAAGCCAATGA-3′ | 504 |
| 21 |
5′-CATTCTGACCAAGAGGCTAAGG-3′ |
5′-GGATGACATTTGTCCAAACAGTAG-3′ | 591 |
| 22 |
5′-ACCATACTTAGCACAGCCAGAA-3′ |
5′-GTCTTGGAGACCTATTCTTGTTGT-3′ | 528 |
| 23 |
5′-GGAGTTGAACATTGGAGTCATCA-3′ |
5′-CATCTTTGGACACAGGAGTCATTT-3′ | 542 |
| 24 |
5′-GGAAGTCACAGTAGTCCTAAGAA-3′ |
5′-ATGTCAGTCAAGTTGCCCAAAT-3′ | 589 |
| 25 |
5′-CTCTCACCTTCATTTCACACCAT-3′ |
5′-GAGCTAGACTGAATTCCAGACATT-3′ | 580 |
| 26 |
5′-TTGCCAGTTAGACCAAGATAGG-3′ |
5′-AAGTGCCTTGTCCAAGTTGTTA-3′ | 549 |
| 27 |
5′-GAACTGTCAACTGTTAAGCAAC-3′ |
5′-CCAACATAATGAAACCCTGTCTC-3′ | 532 |
| 28 |
5′-GCATGGGAACCCATTTGTGTTA-3′ |
5′-ACAGGTGTCACTTCTAACTCTCA-3′ | 607 |
| ABCB11 |
| 2 |
5′-GGCTCTTTCAGGGAGTTATTAACC-3′ |
5′-ACTTGACCAGCTTGTCCTACTT-3′ | 335 |
| 3 |
5′-AGAGACAATATGAGCAGGAAGA-3′ |
5′-CTGCTTTGTGCCTTTGATATGA-3′ | 266 |
| 4 |
5′-TCTGTGAATCGCTAGTGAACCT-3′ |
5′-ACACCCACTGCCATAAATCAAC-3′ | 313 |
| 5 |
5′-ATACGAACTCTGCCACTCAATT-3′ |
5′-GTTAGATACCACTCCAGCTCAG-3′ | 492 |
| 6 |
5′-AATGTAATCTCTGGTGGCTTGA-3′ |
5′-TGTAGTTCTTAGGGCTTCTGAT-3′ | 262 |
| 7 |
5′-CTTAGTTCCCAAGAAGAGGCATT-3′ |
5′-CACACCAAATTGCAGTACCTTG-3′ | 487 |
| 8 |
5′-GAGAGGCTGTTAATGCTATCCA-3′ |
5′-TGTTGCTAACTGTACTCAGGAA-3′ | 410 |
| 9 |
5′-TCTTCCTCCTGTCAATGATGTTAC-3′ |
5′-ATTACTCTGCTTAGCTCCCTCTT-3′ | 412 |
| 10 |
5′-TGCTCTGTGTTTGCGATGATTT-3′ |
5′-TGTTTCCACAGACAGACTCCATA-3′ | 435 |
| 11 |
5′-TCTCTGCGTTAACATGGAAGAC-3′ |
5′-CAAGAGCGAAACTCCATCTCAA-3′ | 418 |
| 12 |
5′-GCAGAGATACGCCAAAGATGTT-3′ |
5′-AAGACACCTCCATTCCCTATTACT-3′ | 337 |
| 13 |
5′-CACAGACACCGAGTATCAACAC-3′ |
5′-CCAGGACAGTCTCAATGTATGC-3′ | 332 |
| 14 |
5′-TTTCTGCCCATTGGTCAAGTAT-3′ |
5′-CTCTTAGTTTCTCCCAGGAATGTA-3′ | 331 |
| 15 |
5′-GATCACTGTCAGAAGCCATCAA-3′ |
5′-TATCAACTACTCCCATCCCTCC-3′ | 336 |
| 16 |
5′-TCTAATGTCTGCACAGCCTATT-3′ |
5′-GTTGGGAGAACAGTGAGTATTGA-3′ | 441 |
| 17 |
5′-TTGCTACTTCTGATGGACTTCTC-3′ |
5′-AGGATTAGGACTACAGAGGACTC-3′ | 437 |
| 18 |
5′-AACTTGGACACCAGTTGATCCT-3′ |
5′-TAGTCTGACTTGAAACACTGCTAG-3′ | 300 |
| 19 |
5′-CCATATCCCATAGACATTTGAGGT-3′ |
5′-ATGAGAAGAAGAAAGCTAGTCCAG-3′ | 335 |
| 20 |
5′-CCACCAGAATGATACATTTCCTAC-3′ |
5′-TGAAGAGGGAGATGTTAGAGAA-3′ | 405 |
| 21 |
5′-GCAATGGGCTGTGTATCTCTTT-3′ |
5′-GTCAGTGTTAGAAGCAGTGGAA-3′ | 444 |
| 22 |
5′-TCTGAGACGGGTTGATTGCTTT-3′ |
5′-GCTTCCTTCAGTCTCTTCGTACTA-3′ | 331 |
| 23 |
5′-CCACTGAAATGTCACGAAAGGA-3′ |
5′-TGGAGACAGAAGAATACACAGAAG-3′ | 524 |
| 24 |
5′-ATGCTTGTTCAGTCCTCTTCTT-3′ |
5′-CCTGTGTCCATGTGTTCTGTT-3′ | 560 |
| 25 |
5′-TGAAGGTATCTCAAGCAGGGATT-3′ |
5′-AAAGTGAGTCTGGCAAAGCAAA-3′ | 397 |
| 26 |
5′-TTAGCCTTGGGATTGTTAGTCTG-3′ |
5′-CACTCTGGTCATTCTACTTCTCC-3′ | 402 |
| 27 |
5′-GAGGAGACCTTGACATGAGTTC-3′ |
5′-GGTTCCACAAAGTATTGCCAAT-3′ | 362 |
| 28 |
5′-GGATTGTTATTCAGGTCGTGTT-3′ |
5′-TTAGCTTGGATTCCGATGTAGG-3′ | 462 |
Statistical analysis
Genotype frequencies were determined by direct
counting. The Chi-square test was used to assess the distribution
of gender, the Hardy-Weinberg equilibrium of all SNPs in the
control group, the difference of transporter protein expression
levels in different genotypic groups and the genotypic
distributions in different groups according to clinical
classification. Associations between polymorphisms and PIS were
estimated by odds ratios (ORs) with a 95% confidence interval (CI),
using an unconditional logistic regression model. The ORs were
adjusted for age, gender and BMI. All calculations were performed
using Statistical Analysis System software (version 9.0, SAS
Institute Inc., Cary, NC, USA). P<0.05 was considered to
indicate a statistically significant difference between values.
Results
ABCB4 and ABCB11 gene mutations detected
that may be associated with the pathogenesis of PIS
It has previously been shown that ABCB4 was involved
in the pathogenesis of intrahepatic cholestasis (11–19)
and cholecystolithiasis (20,21).
Exon sequencing of blood samples from 176 patients with PIS and 178
healthy controls was performed in order to analyze the presence of
mutations in the ABCB4 gene in patients with PIS (Table II). A missense mutation (no. 22677
G>T; Q151H) was detected in exon 6 of the ABCB4 gene in 20
heterozygous patients with PIS. In addition, a synonymous mutation
(no. 69233 G>A) was detected in exon 26 in 23 heterozygous
patients with PIS. These mutations were not detected in healthy
individuals or in the Single Nucleotide Polymorphism Database
(dbSNP; gene ID 5244). Two more synonymous mutations were
identified (rs1202283 in exon 6 and rs2109505 in exon 8); however,
there was no difference in their distributions between the PIS and
healthy groups.
 | Table IIDistributions of 11 SNPs and
association between SNPs and PIS. |
Table II
Distributions of 11 SNPs and
association between SNPs and PIS.
| Gene | dbSNP
Reference | Amino acid
change | Genotypea | Cases (%)
n=176 | Controls
(%)
n=178 | Adjusted OR (95%
CI)b | P-value | P for HWE in
control |
|---|
| ABCB4 |
| rs1202283 | Synonymous | CC | 67 (38.1) | 76 (42.7) | Ref. | Ref. | 0.58 |
| | | CT | 79 (44.9) | 78 (43.8) | 1.23
(0.72–2.12) | 0.45 | |
| | | TT | 30 (17.0) | 24 (13.5) | 1.31
(0.61–2.80) | 0.48 | |
| | | CT+TT | 109 (61.9) | 103 (57.3) | 1.25
(0.75–2.08) | 0.38 | |
| no. 22677c | Q151H | GG | 156 (88.6) | 178 (100.0) | Ref. | Ref. | NA |
| | | GT | 20 (11.4) | 0 (0.0) | NA | <0.001e | |
| | | TT | 0 (0.0) | 0 (0.0) | NA | NA | |
| | | GT+TT | 20 (11.4) | 0 (0.0) | NA | <0.001e | |
| rs2109505 | Synonymous | AA | 108 (61.4) | 108 (60.7) | Ref. | Ref. | 0.09 |
| | | AT | 55 (31.3) | 56 (31.5) | 1.07
(0.62–1.85) | 0.82 | |
| | | TT | 13 (7.4) | 14 (7.9) | 0.74
(0.29–1.86) | 0.52 | |
| | | AT+TT | 68 (38.6) | 70 (39.3) | 0.99
(0.59–1.64) | 0.96 | |
| no. 69233c | Synonymous | GG | 153 (86.9) | 178 (100.0) | Ref. | Ref. | NA |
| | | GA | 23 (13.1) | 0 (0.0) | NA | <0.001e | |
| | | AA | 0 (0.0) | 0 (0.0) | NA | NA | |
| | | GA+AA | 23 (13.1) | 0 (0.0) | NA | <0.001e | |
| ABCB11 |
| rs3815675 | Synonymous | TT | 90 (51.1) | 89 (50.0) | Ref. | Ref. | 0.10 |
| | | TC | 73 (41.5) | 67 (37.6) | 1.19
(0.70–2.02) | 0.52 | |
| | | CC | 13 (7.4) | 22 (12.4) | 0.47
(0.19–1.16) | 0.10 | |
| | | CC | 13 (7.4) | 22 (12.4) | Ref. | Ref. | |
| | | TC+TT | 163 (92.6) | 156 (87.6) | 2.28
(0.96–5.43) | 0.06 | |
| rs4148777 | Synonymous | TT | 159 (90.3) | 161 (90.4) | Ref. | Ref. | 0.50 |
| | | TC | 16 (9.1) | 17 (9.6) | 0.74
(0.31–1.79) | 0.50 | |
| | | CC | 1 (0.6) | 0 (0.0) | NA | 0.99 | |
| | | TC+CC | 17 (9.7) | 17 (9.6) | 0.76
(0.32–1.83) | 0.55 | |
| rs2287616 | Synonymous | TT | 83 (47.2) | 94 (52.8) | Ref. | Ref. | 0.08 |
| | | TC | 79 (44.9) | 64 (36.0) | 1.58
(0.92–2.70) | 0.07 | |
| | | CC | 14 (8.0) | 20 (11.2) | 0.40
(0.16–0.96) | 0.04d | |
| | | CC | 14 (8.0) | 24 (11.2) | Ref. | Ref. | |
| | | TC+TT | 162 (92.0) | 154 (88.8) | 3.06
(1.31–7.18) | 0.01d | |
| rs2287617 | R299K | GG | 173 (98.3) | 176 (98.9) | Ref. | Ref. | 0.94 |
| | | GA | 3 (1.7) | 2 (1.1) | 0.58
(0.06–5.20) | 0.62 | |
| | | AA | 0 (0.0) | 0 (0.0) | NA | NA | |
| | | GA+AA | 3 (1.7) | 2 (1.1) | 0.58
(0.06–5.20) | 0.62 | |
| rs2287622 | V444A | TT | 16 (9.1) | 16 (9.0) | Ref. | Ref. | 0.47 |
| | | TC | 78 (44.3) | 68 (38.2) | 1.18
(0.48–2.89) | 0.73 | |
| | | CC | 82 (46.6) | 94 (52.8) | 0.83
(0.34–2.02) | 0.68 | |
| | | CC | 82 (46.6) | 94 (52.8) | Ref. | Ref. | |
| | | TC+TT | 94 (53.4) | 84 (47.2) | 1.37
(0.83–2.27) | 0.22 | |
| rs118109635 | A865V | CC | 166 (94.3) | 177 (99.4) | Ref. | Ref. | 0.97 |
| | | CT | 10 (5.7) | 1 (0.6) | 9.21
(1.09–77.72) | 0.04d | |
| | | TT | 0 (0.0) | 0 (0.0) | NA | NA | |
| | | CT+TT | 10 (5.7) | 1 (0.6) | 9.21
(1.09–77.72) | 0.04d | |
| rs497692 | Synonymous | AA | 21 (11.9) | 40 (22.5) | Ref. | Ref. | 0.22 |
| | | AG | 75 (42.6) | 80 (44.9) | 1.78
(0.87–3.64) | 0.12 | |
| | | GG | 80 (45.5) | 58 (32.6) | 2.43
(1.17–5.06) | 0.02d | |
| | | AG+GG | 155 (88.1) | 138 (77.5) | 2.06
(1.05–4.01) | 0.04d | |
ABCB11 has also been associated with the formation
of intrahepatic cholestasis (25,27),
therefore indicating that it may be involved in the pathogenesis of
PIS. The present study analyzed the presence of mutations in the
ABCB11 gene in patients with PIS (Table II). As shown in Table II, three nonsynonymous and four
synonymous mutations were detected in the ABCB11 gene of PIS
patients. In this gene, two missense (rs2287617 and rs2287622) and
two synonymous (rs3815675 and rs4148777) mutations were detected in
exons 9, 13, 4 and 5, respectively. However, the distributions of
allele frequencies for these four mutations were not significantly
different between the patients and controls (P>0.05).
Conversely, one missense mutation (rs118109635) and two synonymous
mutations (rs2287616 and rs497692) were also detected,
respectively, in exons 21, 9, and 24. The distributions of these
allele frequencies were significantly different in PIS patients
compared to those in the healthy group (P=0.04, P=0.01 and P=0.02,
respectively).
Overall, two novel mutations (no. 22677 G>T and
no. 69233 G>A) were identified in the ABCB4 gene of PIS
patients. In addition, three mutations in the ABCB11 gene
(rs118109635, rs2287616 and rs497692) were found to be associated
with the pathogenesis of PIS.
Altered protein expression in PIS
patients with ABCB4 and ABCB11 gene mutations
Immunohistochemical staining was used to evaluate
whether ABCB4 and ABCB11 gene mutations in PIS patients affected
the expression of their corresponding proteins (Table III). The expression levels of
ABCB4 as well as ABCB11 proteins were significantly reduced in the
liver tissue of patients with PIS compared to those in normal liver
tissue. In 20 PIS patients with the GT genotype of the ABCB4
mutation at exon 6 (no. 22677 G>T), the protein expression
levels of ABCB4 were significantly higher in individuals with the
GT genotype compared with those with the GG genotype (P=0.019). By
contrast, the ABCB4 mutations at rs1202283, rs2109505 and no. 69233
G>A did not significantly affect protein expression levels in
patients with PIS.
 | Table IIIAssociation between SNPs and protein
expression. |
Table III
Association between SNPs and protein
expression.
| SNP | Cases, n (%) | Low expression, n
(%) | Medium expression,
n (%) | High expression, n
(%) | P |
|---|
| 1) ABCB4 (MDR3
Protein) | n=176 | n=120 | n=39 | n=17 | |
| Alleles (2n) | 2n=352 (100) | 2n=240 (100%) | 2n=78 (100%) | 2n=34 (100%) | |
| a) rs1202283
(Synonymous) | | | | | 0.618 |
| Genotypes: |
| CC | 67 (100) | 48 (72) | 13 (19) | 6 (9) | |
| CT | 79 (100) | 55 (70) | 16 (20) | 8 (10) | |
| TT | 30 (100) | 17 (57) | 10 (33) | 3 (10) | |
| b) no. 22677
(Q151H) | | | | | 0.019a |
| Genotypes: |
| GG | 156 (100) | 111 (71) | 33 (21) | 12 (8) | |
| GT | 20 (100) | 9 (45) | 6 (30) | 5 (25) | |
| TT | 0 | 0 | 0 | 0 | |
| c) rs2109505
(Synonymous) | | | | | 0.839 |
| Genotypes: |
| AA | 108 (100) | 72 (67) | 24 (22) | 12 (11) | |
| AT | 55 (100) | 40 (73) | 11 (20) | 4 (7) | |
| TT | 13 (100) | 8 (62) | 4 (31) | 1 (7) | |
| d) no. 69233
(Synonymous) | | | | | 0.063 |
| Genotypes: |
| GG | 153 (100) | 104 (68) | 32 (21) | 17 (11) | |
| GA | 23 (100) | 16 (70) | 7 (30) | 0 | |
| AA | 0 | 0 | 0 | 0 | |
| 2) ABCB11 (BSEP
Protein) | n=176 | n=112 | n=36 | n=28 | |
| Alleles (2n) | | 2n=224 (100) | 2n=72 (100) | 2n=56 (100) | |
| a) rs3815675
(Synonymous) | | | | | 0.089 |
| Genotypes: |
| TT | 90 (100) | 57 (63) | 18 (20) | 15 (17) | |
| TC | 73 (100) | 46 (63) | 18 (25) | 9 (12) | |
| CC | 13 (100) | 9 (69) | 0 | 4 (31) | |
| b) rs4148777
(Synonymous) | | | | | 0.510 |
| Genotypes: |
| TT | 159 (100) | 102 (64) | 32 (20) | 25 (16) | |
| TC | 16 (100) | 10 (62) | 3 (19) | 3 (19) | |
| CC | 1 (100) | 0 | 1 (100) | 0 | |
| c) rs2287616
(Synonymous) | | | | | 0.088 |
| Genotypes: |
| TT | 83 (100) | 50 (60) | 19 (23) | 14 (17) | |
| TC | 79 (100) | 52 (66) | 17 (22) | 10 (13) | |
| CC | 14 (100) | 10 (71) | 0 | 4 (29) | |
| d) rs2287617
(R299K) | | | | | 0.173 |
| Genotypes: |
| GG | 173 (100) | 111 (64) | 34 (20) | 28 (16) | |
| GA | 3 (100) | 1 (33) | 2 (67) | 0 | |
| AA | 0 | 0 | 0 | 0 | |
| e) rs2287622
(V444A) | | | | | 0.003b |
| Genotypes: |
| TT | 16 (100) | 3 (19) | 8 (50) | 5 (31) | |
| TC | 78 (100) | 53 (68) | 14 (18) | 11 (14) | |
| CC | 82 (100) | 56 (68) | 14 (17) | 12 (15) | |
| f) rs118109635
(A865V) | | | | | 0.032a |
| Genotypes: |
| CC | 166 (100) | 107 (64) | 31 (19) | 28 (17) | |
| CT | 10 (100) | 5 (50) | 5 (50) | 0 | |
| TT | 0 | 0 | 0 | 0 | |
| g) rs497692
(Synonymous) | | | | | 0.001b |
| Genotypes: |
| AA | 21 (100) | 6 (28) | 5 (24) | 10 (48) | |
| AG | 75 (100) | 52 (69) | 14 (19) | 9 (12) | |
| GG | 80 (100) | 54 (67) | 17 (21) | 9 (11) | |
In 10 PIS patients with the ABCB11 CT genotype at
exon 21 (rs118109635 C>T), the protein expression levels of
ABCB11 were increased compared to those with the CC genotype
(P=0.032). By contrast, the protein expression levels of ABCB11
were decreased in individuals with the rs2287622 TC or CC genotype,
as well as those with the rs497692 AG or GG genotype compared to
patients with wild type ABCB11 gene expression (P=0.003 and
P=0.001, respectively). However, the ABCB11 mutations, rs3815675,
rs4148777, rs2287616 and rs2287617, did not alter protein
expression levels.
These results therefore indicated that the GT
genotype at no.22677 G>T of the ABCB4 gene may lead to increased
protein expression and the ABCB11 gene mutations rs118109635,
rs2287622 and rs497692 may also influence protein expression.
Mutations in ABCB4 and ABCB11 correlate
with recurrence of PIS, cholangitis and preoperative jaundice
The associations between the clinical manifestations
of PIS and mutations of ABCB4 and ABCB11 were analyzed. As shown in
Table V, for mutations of ABCB4,
rs2109505 was associated with types of gallstone (P=0.03), and no.
69233 G>A was associated with the recurrence of PIS (P=0.01) and
recurrent episodes of cholangitis (P=0.04). Similarly, ABCB11
mutations rs4148777 and rs2287617 were associated with the
recurrence of PIS (P=0.02, P<0.001, respectively); furthermore,
rs118109635 and rs497692 mutations of ABCB11 were associated with
preoperative jaundice (P<0.001 and P=0.03, respectively). These
results suggested that mutations in ABCB4 and ABCB11 may be
associated with the recurrence of PIS, cholangitis and preoperative
jaundice.
 | Table VGenotype distributions in groups of
jaundice, recurrence of cholangitis, types of gallstone and
recurrence of PIS. |
Table V
Genotype distributions in groups of
jaundice, recurrence of cholangitis, types of gallstone and
recurrence of PIS.
| Jaundice | Recurrence of
cholangitis | Types of
gallstone | Recurrence of
PIS |
|---|
|
|
|
|
|
|---|
| SNP (n) | No
85 (%) | Yes
91 (%) | P | No
87 (%) | Yes
99 (%) | P | CBS
169 (%) | CS
7 (%) | P | No
138 (%) | Yes
38 (%) | P |
|---|
| 1) ABCB4
(MDR3) |
| a) rs1202283
C>T |
| CC (n=67) | 32 (37.6) | 35 (38.5) | 0.14 | 37 (42.5) | 30 (30.3) | 0.48 | 63 (37.3) | 4 (57.1) | 0.22 | 52 (37.7) | 15 (39.5) | 0.92 |
| CT (n=79) | 43 (50.6) | 36 (39.6) | | 36 (41.4) | 43 (43.4) | | 76 (45.0) | 3 (42.9) | | 63 (45.7) | 16 (42.1) | |
| TT (n=30) | 10 (11.8) | 20 (22.0) | | 14 (16.1) | 16 (16.2) | | 30 (17.8) | 0 (0.0) | | 23 (16.7) | 7 (18.4) | |
| b) no. 22677
G>T |
| GG (n=156) | 78 (91.8) | 78 (85.7) | 0.21 | 79 (90.8) | 77 (77.8) | 0.37 | 151 (89.3) | 5 (71.4) | 0.20 | 123 (89.1) | 33 (86.8) | 0.70 |
| GT (n=20) | 7 (8.2) | 13 (14.3) | | 8 (9.2) | 12 (12.1) | | 18 (10.7) | 2 (28.6) | | 15 (10.9) | 5 (13.2) | |
| TT (n=0) | 0 (0.0) | 0 (0.0) | | 0 (0.0) | 0 (0.0) | | 0 (0.0) | 0 (0.0) | | 0 (0.0) | 0 (0.0) | |
| c) rs2109505
A>T |
| AA( n=108) | 51 (60.0) | 57 (62.6) | 0.61 | 49 (56.3) | 59 (59.6) | 0.36 | 103 (60.9) | 5 (71.4) | 0.03a | 85 (61.6) | 23 (60.5) | 0.71 |
| AT( n=55) | 26 (30.6) | 29 (31.9) | | 30 (34.5) | 25 (25.3) | | 55 (32.5) | 0 (0.0) | | 44 (31.9) | 11 (28.9) | |
| TT( n=13) | 8 (9.4) | 5 (5.5) | | 8 (9.2) | 5 (5.1) | | 11 (6.5) | 2 (28.6) | | 9 (6.5) | 4 (10.5) | |
| d) no. 69233
G>A |
| GG( n=153) | 74 (87.1) | 79 (86.8) | 0.96 | 71 (81.6) | 82 (82.8) | 0.04a | 147 (87.0) | 6 (85.7) | 0.92 | 111 (80.4) | 37 (97.4) | 0.01a |
| GA (n=23) | 11 (12.9) | 12 (13.2) | | 16 (18.4) | 7 (7.1) | | 22 (13.0) | 1 (14.3) | | 22 (15.9) | 1 (2.6) | |
| AA (n=0) | 0 (0.0) | 0 (0.0) | | 0 (0.0) | 0 (0.0) | | 0 (0.0) | 0 (0.0) | | 0 (0.0) | 0 (0.0) | |
| 2) ABCB11
(BSEP) |
| a) rs3815675
T>C |
| TT (n=90) | 42 (49.4) | 48 (52.7) | 0.29 | 46 (52.9) | 44 (44.4) | 0.50 | 86 (50.9) | 4 (57.1) | 0.68 | 71 (51.4) | 19 (50.0) | 0.98 |
| TC (n=73) | 39 (45.9) | 34 (37.4) | | 33 (37.9) | 40 (40.4) | | 71 (42.0) | 2 (28.6) | | 57 (41.3) | 16 (42.1) | |
| CC (n=13) | 4 (4.7) | 9 (9.9) | | 8 (9.2) | 5 (5.1) | | 12 (7.1) | 1 (14.3) | | 10 (7.2) | 3 (7.9) | |
| b) rs4148777
T>C |
| TT (n=159) | 78 (91.8) | 81 (89) | 0.32 | 76 (87.4) | 83 (83.8) | 0.26 | 152 (89.9) | 7 (100) | 0.48 | 129 (93.5) | 30 (78.9) | 0.02a |
| TC (n=16) | 6 (7.1) | 10 (11.0) | | 10 (11.5) | 6 (6.1) | | 16 (9.5) | 0 (0.0) | | 9 (6.5) | 7 (18.4) | |
| CC( n=1) | 1 (1.2) | 0 (0.0) | | 1 (1.1) | 0 (0.0) | | 1 (0.6) | 0 (0.0) | | 0 (0.0) | 1 (2.6) | |
| c) 2287616
T>C |
| TT (n=83) | 38 (44.7) | 45 (49.5) | 0.67 | 43 (49.4) | 40 (40.4) | 0.32 | 79 (46.7) | 4 (57.1) | 0.62 | 63 (45.7) | 20 (52.6) | 0.73 |
| TC (n=79) | 41 (48.2) | 38 (41.8) | | 35 (40.2) | 44 (44.4) | | 77 (45.6) | 2 (28.6) | | 64 (46.4) | 15 (39.5) | |
| CC( n=14) | 6 (7.1) | 8 (8.8) | | 9 (10.3) | 5 (5.1) | | 13 (7.7) | 1 (14.3) | | 11 (8.0) | 3 (7.9) | |
| d) 2287617
G>A |
| GG (n=173) | 84 (98.8) | 89 (97.8) | 0.59 | 85 (97.7) | 88 (88.9) | 0.54 | 166 (98.2) | 7 (100.0) | 0.62 | 138 (100.0) | 35 (92.1) | 0.00b |
| GA (n=3) | 1 (1.2) | 2 (2.2) | | 2 (2.3) | 1 (1.0) | | 3 (1.8) | 0 (0.0) | | 0 (0.0) | 3 (7.9) | |
| AA( n=0) | 0 (0.0) | 0 (0.0) | | 0 (0.0) | 0 (0.0) | | 0 (0.0) | 0 (0.0) | | 0 (0.0) | 0 (0.0) | |
| e) rs2287622
T>C |
| TT (n=16) | 6 (7.1) | 10 (11.0) | 0.24 | 10 (11.5) | 6 (6.1) | 0.29 | 16 (9.5) | 0 (0.0) | 0.23 | 11 (8.0) | 5 (13.2) | 0.19 |
| TC( n=78) | 43 (50.6) | 35 (38.5) | | 34 (39.1) | 44 (44.4) | | 73 (43.2) | 5 (71.4) | | 58 (42.0) | 20 (52.6) | |
| CC (n=82) | 36 (42.4) | 46 (50.5) | | 43 (49.4) | 39 (39.4) | | 80 (47.3) | 2 (28.6) | | 59 (42.8) | 13 (34.2) | |
| f) rs118109635
C>T |
| CC( n=166) | 84 (98.8) | 82 (90.1) | <0.001b | 82 (94.3) | 84 (84.8) | 0.97 | 159 (94.1) | 7 (100.0) | 0.36 | 131 (94.9) | 35 (92.1) | 0.52 |
| CT (n=10) | 1 (1.2) | 9 (9.9) | | 5 (5.7) | 5 (5.1) | | 10 (5.9) | 0 (0.0) | | 7 (5.1) | 3 (7.9) | |
| TT (n=0) | 0 (0.0) | 0 (0.0) | | 0 (0.0) | 0 (0.0) | | 0 (0.0) | 0 (0.0) | | 0 (0.0) | 0 (0.0) | |
| g) rs497692
A>G |
| AA (n=21) | 14 (16.5) | 7 (7.7) | 0.03a | 8 (9.2) | 13 (13.1) | 0.50 | 20 (11.8) | 1 (14.3) | 0.73 | 14 (10.1) | 7 (18.4) | 0.38 |
| AG( n=75) | 40 (47.1) | 35 (38.5) | | 37 (42.5) | 38 (38.4) | | 73 (43.2) | 2 (28.6) | | 59 (42.8) | 16 (42.1) | |
| GG( n=80) | 31 (36.5) | 49 (53.8) | | 42 (48.3) | 38 (38.4) | | 76 (45.0) | 4 (57.1) | | 65 (47.1) | 15 (39.5) | |
Decreased expression of ABCB11 protein is
associated with cholangitis and jaundice
Further analysis of the associations between protein
expression levels of ABCB11 and the clinical manifestations of PIS
revealed an increased recurrence of cholangitis (P=0.006) and
preoperative jaundice (P=0.015) when ABCB11 protein expression was
decreased (Table VI). In
addition, it was observed that decreased ABCB11 protein expression
was associated with gender, as significantly more females had
decreased BSEP expression (P=0.016). By contrast, the expression of
ABCB4 protein was not associated with the clinical characteristics
of PIS. This therefore indicated that altered expression of the
ABCB11 protein may be indicative of an increased risk of
cholangitis and preoperative jaundice.
 | Table VIBSEP expression levels in groups of
gender, preoperative jaundice, recurrence of cholangitis. |
Table VI
BSEP expression levels in groups of
gender, preoperative jaundice, recurrence of cholangitis.
| BSEP
Expression | |
|---|
|
| |
|---|
| Low n=112 (%) | Medium n=36
(%) | High n=28 (%) | P-value |
|---|
| Gender | | | | 0.016a |
| Male (n=50) | 25 (50) | 17 (34) | 8 (16) | |
| Female
(n=126) | 87 (69) | 19 (15) | 20 (16) | |
| Preoperative
jaundice | | | | 0.015a |
| No (n=85) | 58 (68) | 10 (12) | 17 (20) | |
| Yes (n=91) | 54 (59) | 26 (29) | 11 (12) | |
| Recurrence of
cholangitis | | | | 0.006b |
| No (n=87) | 53 (61) | 13 (15) | 21 (24) | |
| Yes (n=89) | 59 (66) | 23 (26) | 7 (8) | |
Discussion
The present study identified one novel mutation in
ABCB4 (no. 69233 G>A) and two novel mutations in ABCB11
(rs118109635 and rs497692) associated with the risk of PIS
pathogenesis. In addition, these mutations in ABCB4 and ABCB11 were
associated with the recurrence of PIS, cholangitis and preoperative
jaundice. Altered expression of the ABCB11 protein was found to be
associated with the risk of cholangitis and preoperative jaundice,
whereas the expression of ABCB4 was not associated with the
clinical characteristics of PIS. By contrast, BSEP and MDR3 levels
were decreased in the liver tissue of PIS patients compared to
those in normal liver tissue.
Studies have indicated that the formation of
intrahepatic stones was attributable to the downregulation of bile
acid-synthesis or defective secretion of phospholipids in the liver
(30,31). This therefore implied that ABCB4
and ABCB11 may represent promising candidate genes for evaluating
the risks of PIS pathogenesis. The present study identified two
mutations in ABCB4, not previously reported in the dbSNP, in
patients with PIS. The mutation at no. 22677 G>T was a missense
mutation that altered the amino acid sequence. A synonymous
mutation (no. 69233 G>A) was found to be located at the boundary
of the coding region and an intron, suggesting that this mutation
may affect the splicing process, resulting in an abnormal messenger
RNA (mRNA) transcript. Three mutations were detected in the ABCB11
gene of PIS patients. One was a missense mutation (rs118109635
C>T), and the other two were synonymous mutations (rs497692 and
rs497616). The rs118109635 mutation has previously been reported to
be associated with transient neonatal cholestasis (32,33);
and the rs497692 mutation was reported to result in defects at
either the protein level, mRNA level or a combination of the two,
which significantly contributed to BSEP deficiency (34). In addition, the rs497692 mutation
was observed in patients with primary biliary cirrhosis and ICP
(35,36). To the best of our knowledge, the
rs497616 mutation has not been reported to be associated with any
disease. The present study performed computational functional
analysis of rs497616, and the results indicated that this mutation
altered the mRNA splicing process (Table VII). However, further studies are
required in order to confirm the association of this functional
change with the risk of developing PIS.
 | Table VIIFunctional analysis of exonic
variants in ABCB4 and ABCB11. |
Table VII
Functional analysis of exonic
variants in ABCB4 and ABCB11.
| Effects Identified
by Software | |
|---|
|
| |
|---|
| SNP | Amino acid
change | PolyPhen2 | BLOSUM62 | EC/EU | FAS-ESS | ESE finder | RESCUE-ESE | Effects
reported |
|---|
| ABCB4 |
| rs1202283 | N168N | 0 | NA | NA | NO | Increase of the
score of the branch site (2.11 VS 3.53); Decrease of the score of
the; branch site (3.32 VS 1.87) | Loss of one ESE,
Introduction of two ESEs | |
| no. 22677 | Q151H | 0.708 | 0 | EC | A New ESS | Loss of a binding
site of SF2/ASF (score 3.47); Lose of two branch sites | Loss of four
ESEs | |
| rs2109505 | I237I | 0 | NA | NA | NO | Loss of two branch
sites; Enhancement of the SRp40 (score 3.68 VS 4.73) | NO | |
| no. 69233 | Synony. | 0 | NA | NA | NO | NO | Loss of five ESEs,
Introduction of one ESEs | |
| ABCB11 |
| rs3815675 | D36D | 0 | NA | NA | NO | A new binding site
of SF2/ASF (score 3.84); Decrease of the score SRP40(3.01 vs
2.85) | Loss of six ESEs,
Introduction of one ESEs | |
| rs4148777 | F90F | 0 | NA | NA | Loss of one
ESS | Increase of the
score of the branch site (0.75 VS 2.49) | NO | No impact on mRNA
splicing or BSEP expression32 |
| rs2287616 | Y269Y | 0 | NA | NA | NO | Introduction of a
new binding site of SC35 (score 2.62); decrease of the score of
SC35 (score 3.40 vs 2.76). | NO | |
| rs2287617 | R299K | 0 | 2 | EU | NO | Loss of a SRp40
binding site (score 2.73) | Loss of one ESEs,
Introduction of two ESEs | |
| rs2287622 | V444A | 0.001 | 0 | EC | NO | Enhancement of the
SC35 (score 3.23 vs 3.80) | Loss of one
ESEs | Decreasing BSEP
Expression35 |
| rs118109635 | A865V | 0.88 | 0 | EC | Two new ESS | A new binding site
of SF2/ASF; (score 3.47) Enhancement of the SC35 (score 2.52 vs
3.90) Lose of a SRp55 binding site (score 3.83) | Loss of one ESEs,
introduction of one ESEs | |
| rs497692 | A1028A | NA | NA | NA | NO | Increase of the
score of the branch site (1.93 VS 2.34); Enhancement of the SRp40
(score 2.86 VS 4.30). | Loss of one ESEs of
mRNA splicing32 | Severe
disturbance |
ABCB4 and ABCB11 are responsible for the biliary
secretion of phosphatidylcholine and bile acids, respectively.
Mutations of these genes may affect the composition of bile and are
reported to be associated with cholestasis and cholelithiasis
(17,28,29).
Therefore, mutations of the ABCB4 or ABCB11 genes may be
responsible for the pathogenesis of PIS. The present study reported
that PIS patients with ABCB4 and ABCB11 mutations have different
clinical prognoses. The mutation of the ABCB4 gene at no. 69233
G>A was a synonymous mutation, which did not alter the levels or
function of the corresponding protein; however, this mutation was
associated with the decreased recurrence of PIS. Patients with
ABCB11 rs118109635 and rs497692 mutations had a significantly
increased risk of preoperative jaundice. These results suggested
that these mutations may cause BSEP deficiency and increased
intrahepatic cholestasis, therefore promoting the formation of PIS.
Previous studies on neonatal cholestasis reported comparable
results (32,33). Although rs497692 is a synonymous
mutation, it resulted in decreased protein expression levels of
ABCB11. This result is consistent with observations of Byrne et
al (35), which showed that
rs497692 was located at the boundary of a coding sequence and an
intron, resulting in the loss of one exonic splicing enhancer and
the severe disturbance of mRNA splicing. The present study also
reported that rs118109635, a missense mutation, was associated with
increased levels of ABCB11 protein. Computational functional
analysis revealed that this type of mutation may affect the
function of BSEP (Table VII). In
addition, it has been reported that the most common ABCB11 mutation
rs2287622 resulted in decreased ABCB11 protein expression (37); this mutation has been associated
with intrahepatic cholestasis (20,38–40),
ICP (35,41) and benign recurrent intrahepatic
cholestasis (BRIC) (42). This
mutation was also detected at high rates in the cohort of PIS
patients from the present study (90.9%) and was associated with
decreased levels of ABCB11 protein. However, statistical analysis
did not show any differences in the distribution between PIS
patients and healthy individuals, nor the association of this
mutation with clinical characteristics of patients with PIS. Of
note, the ABCB4 missense mutation at no. 22677 G>T was found to
be associated with increased levels of the corresponding protein;
however, the functional significance of this remains to be
elucidated. Computational functional analysis of no. 22677 G>T
(Table VII) revealed that this
type of mutation may affect the function of MDR3. However, the
results of the present study showed no association between this
mutation and the clinical manifestations of PIS. Therefore, the
functional roles of this novel variant in PIS patients requires
further characterization. Immunohistochemical analysis demonstrated
that decreased expression of ABCB11 protein was associated with
cholangitis and preoperative jaundice, whereas ABCB4 expression was
not associated with the clinical information of PIS patients.
Furthermore, the levels of BSEP and MDR3 were decreased in the
liver tissue of PIS patients compared to those in normal liver
tissue. The results of the present study, in accordance with
previous studies, suggested that the reduction of ABCB4 and ABCB11
proteins may alter the composition of bile, including decreased
levels of phospholipids (10) and
bile salts (9,43). This therefore indicated that the
expression of ABCB4 and ABCB11 proteins may be responsible for the
pathogenesis of PIS.
In conclusion, the results of the present study
provided evidence that ABCB4 and ABCB11 are promising candidate
genes, whose mutation may alter the expression and functions of
their corresponding proteins and therefore may be indicative of the
risk of PIS. This therefore expands the current understanding of
the pathogenesis of the PIS and provides the basis for further
clinical studies.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation of China (no. 81270535), the National
Science and Technology Major Project of China (no. 2008ZX10002-026)
and the Third Military Medical University Science Foundation (no.
2009xlc05).
References
|
1
|
Neuhaus H: Intrahepatic stones: the
percutaneous approach. Can J Gastroenterol. 13:467–472.
1999.PubMed/NCBI
|
|
2
|
Mori T, Sugiyama M and Atomi Y: Gallstone
disease: Management of intrahepatic stones. Best Prac Res Clin
Gastroenterol. 20:1117–1137. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kaufman HS, Magnuson TH, Lillemoe KD,
Frasca P and Pitt HA: The role of bacteria in gallbladder and
common duct stone formation. Ann Surg. 209:584–592. 1989.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cetta FM: Bile infection documented as
initial event in the pathogenesis of brown pigment biliary stones.
Hepatology. 6:482–489. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nakayama F and Koga A: Hepatolithiasis:
present status. World J Surg. 8:9–14. 1984. View Article : Google Scholar
|
|
6
|
Chang TM and Passaro E Jr: Intrahepatic
stones: the Taiwan experience. Am J Surg. 146:241–244. 1983.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nakayama F, Soloway RD, Nakama T, et al:
Hepatolithiasis in East Asia. Retrospective study. Dig Dis Sci.
31:21–26. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Oude Elferink RP and Paulusma CC: Function
and pathophysiological importance of ABCB4 (MDR3 P-glycoprotein).
Pflugers Arch. 453:601–610. 2007.PubMed/NCBI
|
|
9
|
Arrese M and Ananthanarayanan M: The bile
salt export pump: molecular properties, function and regulation.
Pflugers Arch. 449:123–131. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Meier Y, Pauli-Magnus C, Zanger UM, et al:
Interindividual variability of canalicular ATP-binding-cassette
(ABC)-transporter expression in human liver. Hepatology. 44:62–74.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jacquemin E: Role of multidrug resistance
3 deficiency in pediatric and adult liver disease: one gene for
three diseases. Semin Liver Dis. 21:551–562. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jacquemin E, Cresteil D, Manouvrier S,
Boute O and Hadchouel M: Heterozygous non-sense mutation of the
MDR3 gene in familial intrahepatic cholestasis of pregnancy.
Lancet. 353:210–211. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dixon PH, Weerasekera N, Linton KJ, et al:
Heterozygous MDR3 missense mutation associated with intrahepatic
cholestasis of pregnancy: evidence for a defect in protein
trafficking. Hum Mol Genet. 9:1209–1217. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lucena JF, Herrero JI, Quiroga J, et al: A
multidrug resistance 3 gene mutation causing cholelithiasis,
cholestasis of pregnancy, and adulthood biliary cirrhosis.
Gastroenterology. 124:1037–1042. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gendrot C, Bacq Y, Brechot MC, Lansac J
and Andres C: A second heterozygous MDR3 nonsense mutation
associated with intrahepatic cholestasis of pregnancy. J Med Genet.
40:e322003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Müllenbach R, Linton KJ, Wiltshire S, et
al: ABCB4 gene sequence variation in women with intrahepatic
cholestasis of pregnancy. J Med Genet. 40:e702003.PubMed/NCBI
|
|
17
|
Pauli-Magnus C, Lang T, Meier Y, et al:
Sequence analysis of bile salt export pump (ABCB11) and multidrug
resistance P-glycoprotein 3 (ABCB4, MDR3) in patients with
intrahepatic cholestasis of pregnancy. Pharmacogenetics. 14:91–102.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Floreani A, Carderi I, Paternoster D, et
al: Hepatobiliary phospholipid transporter ABCB4, MDR3 gene
variants in a large cohort of Italian women with intrahepatic
cholestasis of pregnancy. Dig Liver Dis. 40:366–370. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schneider G, Paus TC, Kullak-Ublick GA, et
al: Linkage between a new splicing site mutation in the MDR3 alias
ABCB4 gene and intrahepatic cholestasis of pregnancy. Hepatology.
45:150–158. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Poupon R, Barbu V, Chamouard P, Wendum D,
Rosmorduc O and Housset C: Combined features of low
phospholipid-associated cholelithiasis and progressive familial
intrahepatic cholestasis 3. Liver Int. 30:327–331. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nakken KE, Labori KJ, Rødningen OK, et al:
ABCB4 sequence variations in young adults with cholesterol
gallstone disease. Liver Int. 29:743–747. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Noé J, Stieger B and Meier PJ: Functional
expression of the canalicular bile salt export pump of human liver.
Gastroenterology. 123:1659–1666. 2002.PubMed/NCBI
|
|
23
|
Byrne JA, Strautnieks SS, Mieli-Vergani G,
Higgins CF, Linton KJ and Thompson RJ: The human bile salt export
pump: characterization of substrate specificity and identification
of inhibitors. Gastroenterology. 123:1649–1658. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lam P, Pearson CL, Soroka CJ, Xu S,
Mennone A and Boyer JL: Levels of plasma membrane expression in
progressive and benign mutations of the bile salt export pump
(Bsep/Abcb11) correlate with severity of cholestatic diseases. Am J
Physiol Cell Physiol. 293:C1709–C1716. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
van Mil SW, van der Woerd WL, van der
Brugge G, et al: Benign recurrent intrahepatic cholestasis type 2
is caused by mutations in ABCB11. Gastroenterology. 127:379–384.
2004.PubMed/NCBI
|
|
26
|
Strautnieks SS, Bull LN, Knisely AS, et
al: A gene encoding a liver-specific ABC transporter is mutated in
progressive familial intrahepatic cholestasis. Nat Genet.
20:233–238. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dixon PH, van Mil SW, Chambers J, et al:
Contribution of variant alleles of ABCB11 to susceptibility to
intrahepatic cholestasis of pregnancy. Gut. 58:537–544. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Acalovschi M, Tirziu S, Chiorean E,
Krawczyk M, Grünhage F and Lammert F: Common variants of ABCB4 and
ABCB11 and plasma lipid levels: a study in sib pairs with
gallstones, and controls. Lipids. 44:521–526. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Marschall HU, Katsika D, Rudling M and
Einarsson C: The genetic background of gallstone formation: an
update. Biochem Biophys Res Commun. 396:58–62. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shoda J, Inada Y and Osuga T: Molecular
pathogenesis of hepatolithiasis - a type of low
phospholipid-associated cholelithiasis. Front Biosci. 11:669–675.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tazuma S: Gallstone disease: Epidemiology,
pathogenesis, and classification of biliary stones (common bile
duct and intrahepatic). Best Pract Res Clin Gastroenterol.
20:1075–1083. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen HL, Liu YJ, Su YN, et al: Diagnosis
of BSEP/ABCB11 mutations in Asian patients with cholestasis using
denaturing high performance liquid chromatography. J Pediatr.
153:825–832. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu LY, Wang XH, Lu Y, Zhu QR and Wang JS:
Association of variants of ABCB11 with transient neonatal
cholestasis. Pediatr Int. 55:138–144. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Byrne JA, Strautnieks SS, Ihrke G, et al:
Missense mutations and single nucleotide polymorphisms in ABCB11
impair bile salt export pump processing and function or disrupt
pre-messenger RNA splicing. Hepatology. 49:553–567. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Keitel V, Vogt C, Häussinger D and Kubitz
R: Combined mutations of canalicular transporter proteins cause
severe intrahepatic cholestasis of pregnancy. Gastroenterology.
131:624–629. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pauli-Magnus C, Kerb R, Fattinger K, et
al: BSEP and MDR3 haplotype structure in healthy Caucasians,
primary biliary cirrhosis and primary sclerosing cholangitis.
Hepatology. 39:779–791. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ho RH, Leake BF, Kilkenny DM, et al:
Polymorphic variants in the human bile salt export pump (BSEP;
ABCB11): functional characterization and interindividual
variability. Pharmacogenet Genomics. 20:45–57. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fattinger K, Funk C, Pantze M, et al: The
endothelin antagonist bosentan inhibits the canalicular bile salt
export pump: a potential mechanism for hepatic adverse reactions.
Clin Pharmacol Ther. 69:223–231. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Eloranta ML, Häkli T, Hiltunen M,
Helisalmi S, Punnonen K and Heinonen S: Association of single
nucleotide polymorphisms of the bile salt export pump gene with
intrahepatic cholestasis of pregnancy. Scand J Gastroenterol.
38:648–652. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lang C, Meier Y, Stieger B, et al:
Mutations and polymorphisms in the bile salt export pump and the
multidrug resistance protein 3 associated with drug-induced liver
injury. Pharmacogenet Genomics. 17:47–60. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Muehlenberg K, Wiedmann K, Keppeler H,
Sauerbruch T and Lammert F: Recurrent intrahepatic cholestasis of
pregnancy and chain-like choledocholithiasis in a female patient
with stop codon in the ABDC4-gene of the hepatobiliary phospholipid
transporter. Z Gastroenterol. 46:48–53. 2008.(In German).
|
|
42
|
Kubitz R, Keitel V, Scheuring S, Köhrer K
and Häussinger D: Benign recurrent intrahepatic cholestasis
associated with mutations of the bile salt export pump. J Clin
Gastroenterol. 40:171–175. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Higuchi H and Gores GJ: Bile acid
regulation of hepatic physiology: IV. Bile acids and death
receptors. Am J Physiol Gastrointest Liver Physiol. 284:G734–G738.
2003.PubMed/NCBI
|















