Introduction
The circadian clock is an inner rhythm, which
regulates daily rhythmic fluctuations in several physiological
processes in organisms (1,2). In humans, the circadian clock is
regulated by a transcription-translation feedback loop, which
consists of multiple biological clock genes, including circadian
locomotor output cycles kaput (Clock), brain and muscle
Arnt-like-1 (Bmal1), period (Per)1,
Per2, Per3, cryptochrome (Cry)1,
Cry2 and casein kinase Iɛ (CKIɛ) (3). Clock and Bmal1 form
heterodimers and bind to E-boxes, which are a CACGTG nucleotide
sequence in the promoter, driving the rhythmic transcription of the
Per and Cry genes. The Per and Cry proteins are
translated in the cytoplasm and form Per-Cry complexes, which
translocate into the nucleus to suppress the further transcription
of Per and Cry, which is mediated by Bmal1 and
Clock (4). Another
transcriptional loop is to modulate the protein stability of Per
and Bmal1 by CKIɛ-induced phosphorylation (5).
The potential association between the disruption of
circadian rhythm and tumor development has prompted widespread
concern (6,7). An increasing number of studies have
demonstrated that disruption of the circadian rhythm is associated
with the development and progression of several types of tumor,
including colorectal cancer (8),
breast cancer (9) and pancreatic
cancer (10). Altered expression
of the circadian genes has also been observed in hepatocellular
carcinoma (HCC) (11), however,
the predominant factors that disturb the circadian clock in HCC
remain to be elucidated. Liver cancer and other types of solid
tumor are generally in a hypoxic state (12–14),
and an association between hypoxia and the disturbance of the
circadian clock has been reported (15). The present study aimed to
investigate the causal association between hypoxia and the abnormal
expression of circadian genes in HCC cells.
Materials and methods
Cell culture and transfection
The normal human HCC cell line, PLC/PRF/5, was
purchased from the Institute of Biochemistry and Cell Biology
(SIBS) of the Chinese Academy of Sciences (Shanghai, China).
Expression plasmids containing hypoxia-inducible factor (HIF)-1α
and HIF-2α, and a control plasmid, pcDNA3.1, were purchased from
Shanghai GeneChem Co., Ltd. (Shanghai, China). The cells were grown
in Dulbecco’s modified Eagle’s medium (DMEM; HyClone, Logan, UT,
USA) containing 10% fetal bovine serum (FBS; Invitrogen Life
Technologies, Carlsbad, CA, USA), 2 mmol/l L-glutamine (HyClone),
50 U/ml penicillin and 50 g/ml streptomycin (HyClone) at 37°C in an
atmosphere of 5% CO2 in air. PLC/PRF/5 cells at between
70 and 80% confluence were then transfected with the different
plasmids using Lipofectamine 2000 (Invitrogen Life Technologies)
according to the manufacturer’s instructions. In brief, 4 μg
plasmid pcDNA3.1, pcDNA3.1-HIF-1α and pcDNA3.1-HIF-2α were diluted
in 250 μl Opti-MEM medium (Invitrogen Life Technologies) without
serum, and mixed. In addition, 10 μl Lipofectamine 2000 was diluted
in 250 μl serum-free Opti-MEM medium, mixed gently and incubated
for 5 min at room temperature. Following incubation, the diluted
plasmids were combined with the diluted Lipofectamine 2000 (total
volume, 500 μl), mixed gently and incubated for 20 min at room
temperature. Subsequently, 500 μl dilution mixture was added to
each well of a 6-well plate. The transfected cells were incubated
at 37°C for 6 h prior to the medium being replaced with fresh DMEM
containing 10% FBS and the cells were cultured for a further 18 h
at 37°C with 5% CO2. The protein and mRNA expression
levels of the target genes in the transfected cells were analyzed
by either reverse transcription quantitative polymerase chain
reaction (RT-qPCR) or westernblotting.
Protein preparation and western blot
analysis
The PLC/PRF/5 cells were treated with either a
vehicle (phosphate-buffered saline; Boster Biological Technology,
Ltd, Wuhan, China) or CoCl2 at 50, 100, or 200 μM for 24
h in a 6-well plate at a cell density of 7×105
cells/well at 37°C with 5% CO2. Cells were cultured with
DMEM containing 10% FBS. The cells were collected and homogenized
in lysis buffer (Boster Biological Technology, Ltd) containing 50
mmol/l Tris-HCl (pH 8.5), 150 mol/l NaCl, 0.2 g/l NaN3,
0.1 g/l sodium dodecyl sulphate (SDS), 100 μg/ml
phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, 10 ml/l NP-40 and
5 g/l sodium deoxycholate. The cells were then centrifuged at
14,000 × g for 15 min to remove the cellular debris and the protein
concentrations were determined using the Bradford method (16). Protein expression was quantified
using a Pierce BCA Protein Assay kit (Thermo Fisher Scientific
Inc., Rockford, IL, USA) according to the manufacturer’s
instructions. Western blotting was performed, as described
previously (17). The proteins
(30–50 μg) were separated by SDS-polyacrylamide gel electrophoresis
using 10% SDS polyacrylamide gels (Boster Biological Technology,
Ltd), they were then transferred onto polyvinylidene fluoride
membranes (Invitrogen Life Technologies) and subsequently blocked
in 5% nonfat milk (Boster Biological Technology, Ltd) in
Tris-buffered saline containing 0.1% Tween-20 (Boster Biological
Technology, Ltd) for 2 h. The membranes were then incubated with
primary mouse anti-human monoclonal antibodies against HIF-1α
(1:500; sc-53546; Santa Cruz Biotechnology, Inc., Santa Cruz, CA,
USA) and HIF-2α (1:500, sc-13596; Santa Cruz Biotechnology, Inc.)
or primary mouse anti-gizzard monoclonal antibodies against β-actin
(1:2,000; sc-47778; Santa Cruz Biotechnology, Inc.) for 1 h at 37°C
and, followed by incubation overnight at 4°C. The membranes were
subsequently incubated with the appropriate horseradish
peroxidase-conjugated monoclonal goat anti-mouse secondary
immunoglobulin G antibodies for 1 h at room temperature and the
bands were then visualized using an enhanced chemiluminescence
detection system (SuperSignal West Pico substrate, cat. no. 34080;
Thermo Fisher Scientific Inc.).
RNA extraction and RT first-strand cDNA
synthesis
The transiently transfected cells (6-well plate at a
cell density of 1×106 cells/well) were used to isolate
the total RNA using TRIzol reagent (Invitrogen Life Technologies).
The concentration and quality of the RNA were determined using a
Nano Drop Spectrophotometer (Nano Drop Technologies, Inc.,
Wilmington, DE, USA) to measure absorbance at 200–350 nm. cDNA
synthesis was performed at 42°C for 60 min in a reaction mixture
(25 μl; Promega Corp., Madison, WI, USA) containing 2 μg RNA, 1.6
μM Oligo (dT)18, 0.6 μM dNTP, 200 U/μl M-MLV reverse transcriptase
and the reaction buffer supplied.
qPCR
Following RT, the cDNA samples were diluted 1:5 with
RNase-free water. The primers were designed, according to the cDNA
sequences in the GeneBank database (http://www.ncbi.nlm.nih.gov/genbank/) using Primers
Express 3.0 software (PE Applied Biosystems, Foster City,. CA, USA)
(18) and are listed in Table I. Each reaction contained 10 μl 2X
SYBR Green mix (Invitrogen Life Technologies), 2 μl cDNA template,
0.6 μl forward primer (10 μM), 0.6 μl reverse primer (10 μM) and
double distilled H2O in a total volume of 20 μl. qPCR
was performed on a Real-Time PCR system 7500 (Applied Biosystems,
Foster City, CA, USA) with the following cycling program: One cycle
at 94°C for 1 min for denaturation and 40 cycles at 94°C for 60
sec, 55°C for 60 sec and 72°C for 60 sec (19). The average fluorescence was
automatically recorded and the baseline and threshold were adjusted
using the ABI 7500 software system (excitation, 497 nm; and
emision, 520 nm). The cycle threshold (Ct) values were determined
and the data were analyzed using the 2−ΔΔCT method and
were normalized against the expression of β-actin in each sample
(20).
 | Table IPolymerase chain reaction primers and
conditions. |
Table I
Polymerase chain reaction primers and
conditions.
| Gene | Primer (5′-3′) | Temperature
(°C) | Product size
(bp) |
|---|
| Per1 |
| Forward |
CCATTGTCCGCATCCTTCC | 60.4 | 142 |
| Reverse |
TGTTCCCTCCCAACCTTCG | | |
| Per2 |
| Forward |
CTATTCTCCCATTCGGTTTCG | 60.0 | 128 |
| Reverse |
CCACCCTGACTTTGTGCCTC | | |
| Per3 |
| Forward |
GTGGAGGTGAAGACAGAAAGCA | 59.7 | 117 |
| Reverse |
TGAGACAGCAAGGTTCCGATT | | |
| Cry1 |
| Forward |
CAACCTCCATTCATCTTTCC | 58.9 | 151 |
| Reverse |
CTCATAGCCGACACCTTC | | |
| Cry2 |
| Forward |
AACCACGACGAGACCTACGG | 61.0 | 178 |
| Reverse |
GGGAGTTGGCGTTCATTCG | | |
| Clock |
| Forward |
GCAGCAGCAGCAGCAGAG | 61.9 | 149 |
| Reverse |
CAGCAGAGAGAATGAGTTGAGTTG | | |
| Bmal1 |
| Forward |
TGCCACCAATCCATACACAGAAG | 60.9 | 123 |
| Reverse |
TTCCCTCGGTCACATCCTACG | | |
|
CKIɛ |
| Forward |
TCAGCGAGAAGAAGATGTC | 58.9 | 149 |
| Reverse |
GAAGAGGTTGCGGAAGAG | | |
| β-actin |
| Forward |
AGTTGCGTTACACCCTTTCTTGAC | 63.9 | 171 |
| Reverse |
GCTCGCTCCAACCGACTGC | | |
|
HIF-1α |
| Forward |
CATCTCCATCTCCTACCCACA | 58.3 | 105 |
| Reverse |
CTTTTCCTGCTCTGTTTGGTG | | |
|
HIF-2α |
| Forward |
TCATGCGACTGGCAATCAGC | 61.3 | 141 |
| Reverse |
GTCACCACGGCAATGAAACC | | |
Statistical analysis
The data were analyzed using SPSS version 13.0
software (SPSS, Inc., Chicago, IL, USA). Student’s t-test was used
to compare the differences between two groups and one-way analysis
of variance was used to compare the differences among multiple
groups. The data are expressed as the mean ± standard deviation and
P<0.05 was considered to indicate a statistically significant
difference.
Results
A hypoxic environment disrupts the
expression levels of circadian genes in HCC cells
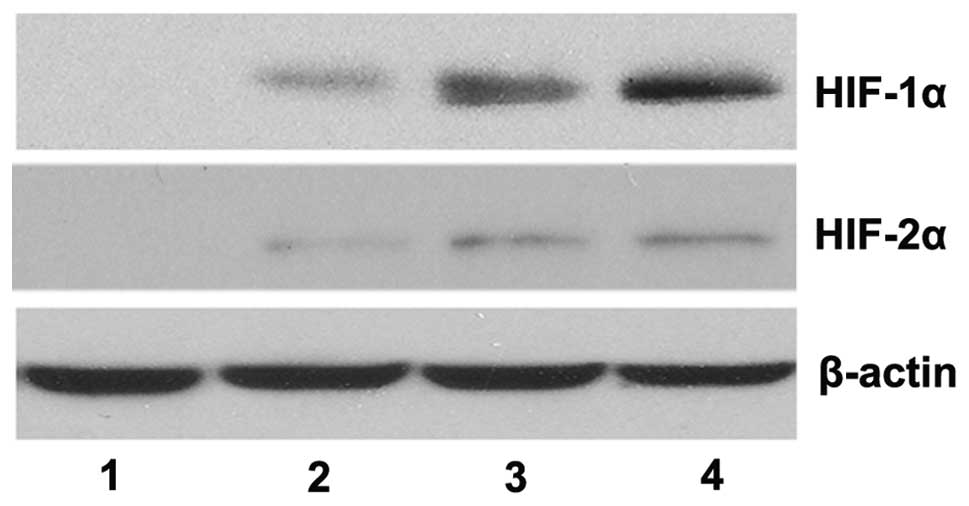
The western blotting results revealed that, in the
absence of CoCl2, the protein expression levels of
HIF-1α and HIF-2α were not detectable, however, their expression
levels were significantly upregulated in the presence of
CoCl2 in a dose-dependent manner (Fig. 1). The qPCR results demonstrated
that treatment with CoCl2 increased the mRNA expression
levels of Clock, Bmal1 and Cry2 and decreased
the mRNA expression levels of Per1, Per2,
Per3, Cry1 and CKIɛ (Fig. 2). Notably, the mRNA expression
levels of Clock, Bmal1, Cry1 and CKIɛ
were dysregulated by treatment with CoCl2 in a
concentration-dependent manner (P<0.05; Fig. 2).
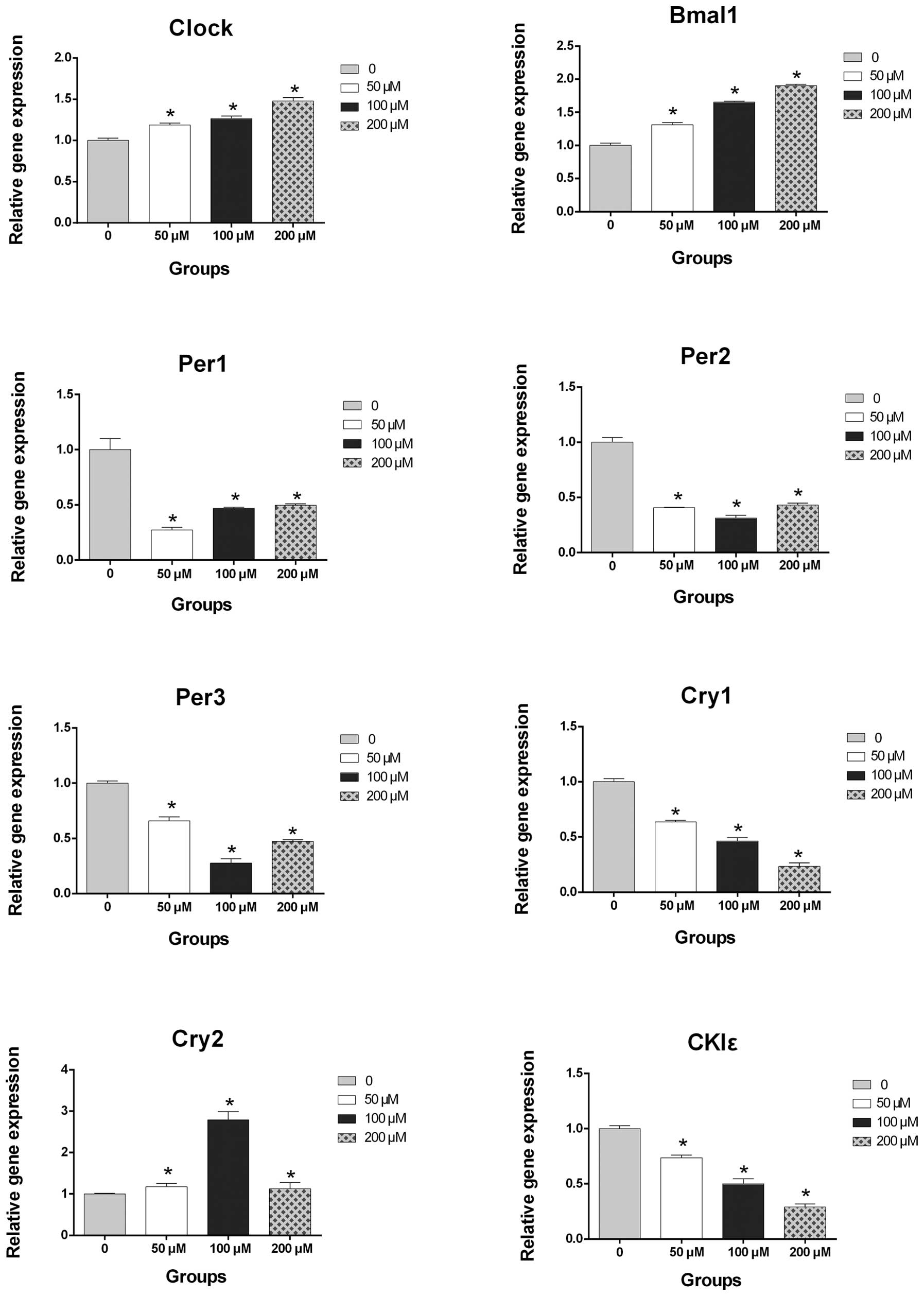 | Figure 2Reverse transcription quantitative
polymerase chain reaction detection of the mRNA expression levels
of Clock, Bmal1, Per1, Per2, Per3, Cry1, Cry2 and
CKIɛ in the PLC/PRF/5 cells exposed to a vehicle
(phosphate-buffered saline) or various concentrations of
CoCl2. mRNA expression levels of Clock, Bmal1 and
Cry2 were increased and the expression levels of Per1,
Per2, Per3, Cry1 and CKIɛ were reduced, in a
CoCl2 concentration-dependent manner. Values are
presented as the mean ± standard deviation (*P<0.05,
vs. vehicle control). Clock, circadian locomotor output
cycles kaput; Bmal, brain and muscle Arnt-like-1;
Per, period; Cry, cryptochrome; CKIɛ, casein
kinase Iɛ. |
HIF-1α and HIF-2α disrupt the expression
levels of circadian genes in HCC cells
HIF-1α and HIF-2α are the predominant transcription
factors in a hypoxic microenvironment. In order to examine the
effects of HIF-1α and HIF-2α on the expression levels of circadian
genes, PLC/PRF/5 cells were transfected with HIF-1α or HIF-2α
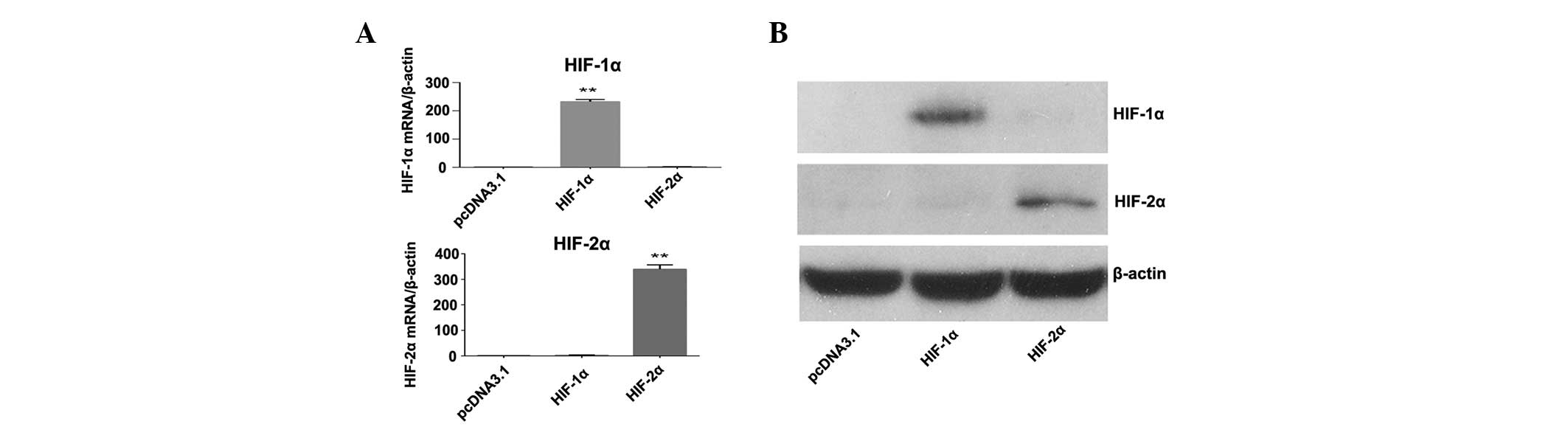
expression plasmids. The mRNA and protein expression levels of
HIF-1α and HIF-2α in the transfected cells were confirmed by
RT-qPCR (Fig. 3A) and western
blotting (Fig. 3B). Subsequently,
the expression levels of the circadian genes were determined in the
transfected cells. The mRNA expression levels of Clock,
Bmal1 and Cry2 were increased and the expression
levels of Per1, Per2, Per3, Cry1 and
CKIɛ were decreased in the PLC/PRF/5 cells transfected with
the HIF-1α plasmid compared with the control cells (Fig. 4). Transfection with the HIF-2α
plasmid increased the mRNA expression levels of Clock,
Bmal1, Per1, Cry1, Cry2 and
CKIɛ, and decreased the expression levels of Per2 and
Per3 (P<0.05; Fig.
4).
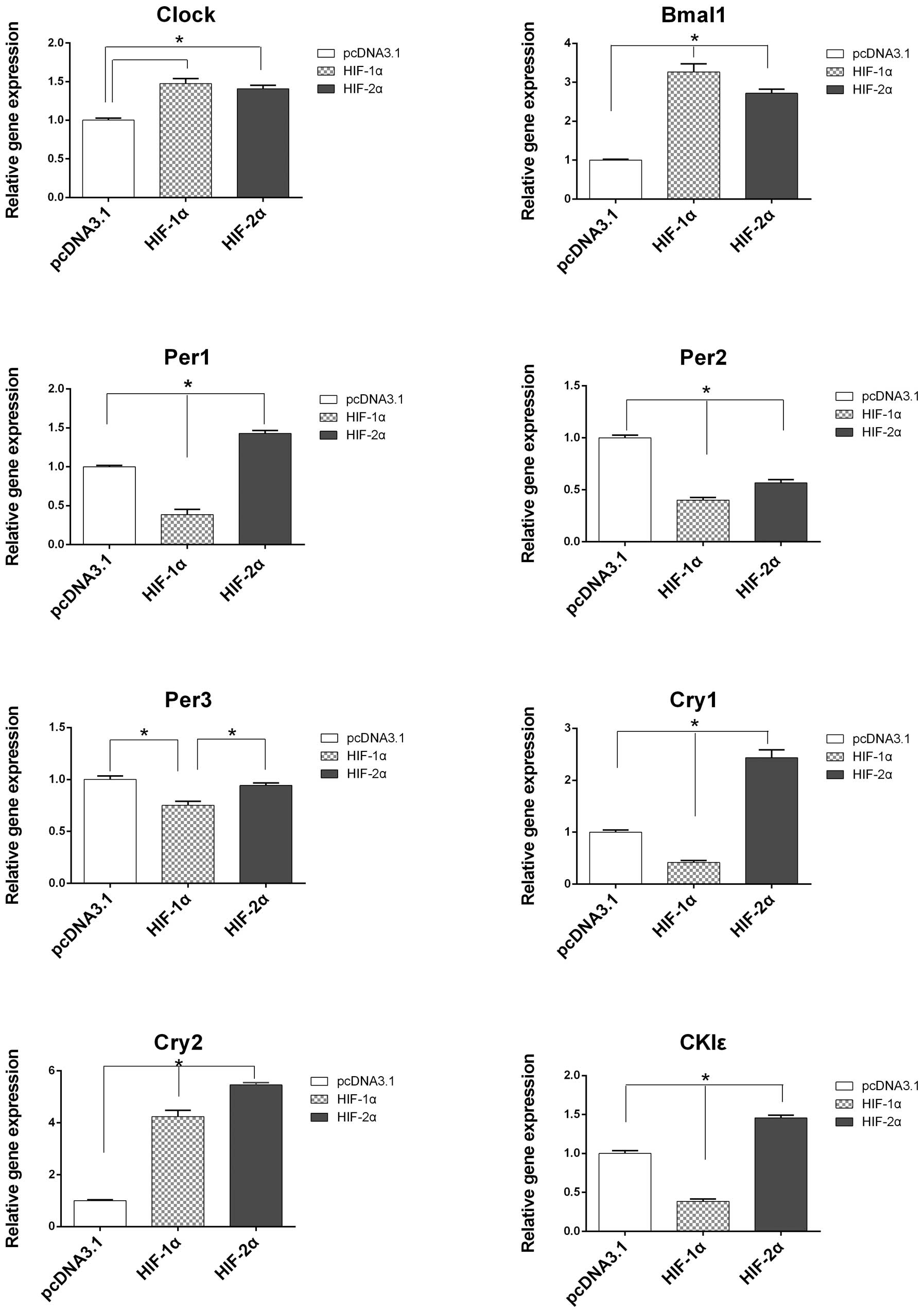 | Figure 4Comparisons of the mRNA expression
levels of circadian genes in HCC cells following transfection with
either the pcDNA3.1 (control), HIF-1α or HIF-2α plasmids. The mRNA
expression levels of Clock, Bmal1 and Cry2 were increased and the
expression levels of Per1, Per2, Per3,
Cry1 and CKIɛ were decreased in the PLC/PRF/5 cells
following transfection with the HIF-1α plasmid, compared with the
control. Following transfection with the HIF-2α plasmid, the mRNA
expression levels of Clock, Bmal1, Per1,
Cry1, Cry2 and CKIɛ were upregulated and the
mRNA expression levels of Per2, Per3 were
downregulated. The mRNA expression levels were normalized against
β-actin and are presented as the relative mRNA expression levels
compared with the control. Values are presented as the mean ±
standard deviation. The results are an average of three independent
experiments (*P<0.05, vs. control). Clock,
circadian locomotor output cycles kaput; Bmal, brain and
muscle Arnt-like-1; Per, period; Cry, cryptochrome;
CKIɛ, casein kinase Iɛ; HIF, hypoxia-inducible factor; PC,
plasmid control. |
Discussion
Investigations into the association between
circadian rhythm and cancer originated from several large
epidemiological studies, which revealed that night-shift workers
have higher incidences of breast, colon and prostate cancer
(8,21,22).
Further studies have demonstrated that disturbances in the
expression of circadian genes is common in several types of cancer
(7,10,23–28).
The expression levels of between 5 and 15% of genes, including key
cell cycle regulators, tumor suppressor genes and oncogenes, are
regulated by circadian rhythm and are driven by clock genes
(29,30). Therefore, circadian genes regulate
the timing of DNA repair, apoptosis and cell proliferation
(29,30). Disruption of the circadian rhythm
may affect cellular proliferation and promote tumor formation. It
has been demonstrated that disruptions to the circadian clock
accelerate carcinogenesis in murine cell models (31,32).
Although disturbances in the expression of circadian
rhythm genes have been found to be closely associated with the
occurrence and development of HCC and other types of tumor
(33,34), the mechanisms underlying how
circadian rhythm affects tumor growth remain to be elucidated. It
was reported that there is a bidirectional interaction between the
hypoxic signaling pathway and the circadian clock (15). Although HCC is one of the most
hypervascularized types of tumor, with rich blood perfusion, it
contains hypoxic regions due to rapid cell proliferation and the
formation of aberrant blood vessels, particularly in patients with
liver cirrhosis (12). The present
study demonstrated, in a CoCl2-induced hypoxic
environment that the mRNA expression levels of all the circadian
clock genes were altered, with upregulation of Clock,
Bmal1 and Cry2, and downregulation of Per1,
Per2, Per3, Cry1 and CKIɛ. Lin et
al (11) demonstrated that the
expression levels of Per1, Per2, Per3 and
Cry2 in HCC cancerous tissues were significantly reduced
compared with their expression levels in paired peritumoral
tissues, whereas no significant differences were observed in the
expression levels of Clock, Bmal1, Cry1 and
CK1ɛ. Comparing the two studies revealed that the expression
pattern of circadian genes in HCC cells in a hypoxic environment
was similar to their expression pattern in HCC tissues. Therefore,
hypoxia is one of the causes of abnormal expression of the clock
genes in HCC.
Two hypoxia-specific transcription factors, HIF-1α
and HIF-2α, are important in the response to hypoxia. These
proteins form heterodimers, which consist of a constitutively
expressed HIF-1β subunit and an O2-regulated HIF-1α or
HIF-2α subunit. HIF-1α and HIF-2α function as transcription factors
only under hypoxic conditions and, in well-oxygenated cells,
hydroxylation of the proline residues by prolyl hydroxylase domain
protein 2 promotes the interaction of HIF-1α and HIF-2α with the
von-Hippel-Lindau tumor suppressor protein, which recruits E3
ubiquitin-protein ligase, targetingHIF-1α and HIF-2α for
degradation by the ubiquitin-proteasome system (35). Although HIF-1α and HIF-2α have
similar structures and common hypoxia-response elements, their
target genes are not identical (36). In addition, the transcriptional
activities of HIF-1α and HIF-2α are different, even when targeting
an identical set of genes (36,37).
The present study revealed that HIF-1α and HIF-2α altered the mRNA
expression of the circadian clock genes in HCC cells. HIF-1α
upregulated the expression levels of Clock, Bmal1 and
Cry2, and downregulated the expression levels of
Per1, Per2, Per3, Cry1 and CKIɛ.
HIF-2α increased the expression levels of Clock,
Bmal1, Per1, Cry1, Cry2 and
CKIɛ, and decreased the expression levels of Per2 and
Per3. Therefore, it was observed that HIF-1α and HIF-2α have
the opposite regulatory effects on the mRNA expression levels of
Per1, Cry1 and CKIɛ. Among these circadian
genes, Per1 is involved in the DNA damage response pathways,
as a cofactor of checkpoint kinase 2 for the activation of ataxia
telangiectasia mutated and is considered to be a potential tumor
suppressor gene (23,34,38).
The results of the present study suggested that HIF-1α and HIF-2α
were involved in modulating the circadian clock by exhibiting
similar, but not identical, effects and further supports our
previous findings that HIF-1α and HIF-2α may have different effects
in the occurrence and development of HCC (39).
In conclusion, the expression levels of circadian
genes were disrupted in the hypoxic environment and the
overexpression of HIF-1α and HIF-2α altered the expression pattern
of circadian genes. Further investigations are required to confirm
the effect of hypoxia on the circadian clock and the association
between hypoxia, circadian rhythm and HCC carcinogenesis. The
present study suggested that abnormal circadian rhythm has a
detrimental role in the occurrence and development of liver cancer.
Therefore, maintaining a normal circadian rhythm may be a novel
therapeutic strategy for the treatment of liver cancer.
Acknowledgements
This study was supported by a grant from The
National Natural Science Foundation of China (no. 81160311).
References
|
1
|
Barclay JL, Tsang AH and Oster H:
Interaction of central and peripheral Clocks in physiological
regulation. Prog Brain Res. 199:163–181. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Eckel-Mahan K and Sassone-Corsi P:
Metabolism and the circadian Clock converge. Physiol Rev.
93:107–135. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mazzoccoli G, Pazienza V and Vinciguerra
M: Clock genes and Clock-controlled genes in the regulation of
metabolic rhythms. Chronobiol Int. 29:227–251. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Isojima Y, Okumura N and Nagai K:
Molecular mechanism of mammalian circadian Clock. J Biochem.
134:777–784. 2003. View Article : Google Scholar
|
|
5
|
Eide EJ and Virshup DM: Casein kinase I:
another cog in the circadian Clockworks. Chronobiol Int.
18:389–398. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Greene MW: Circadian rhythms and tumor
growth. Cancer Lett. 318:115–123. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Savvidis C and Koutsilieris M: Circadian
rhythm disruption in cancer biology. Mol Med. 18:1249–1260. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Brudnowska J and Peplonska B: Night shift
work and cancer risk: a literature review. Med Pr. 62:323–338.
2011.(In Polish).
|
|
9
|
Leonardi GC, Rapisarda V, Marconi A, et
al: Correlation of the risk of breast cancer and disruption of the
circadian rhythm (Review). Oncol Rep. 28:418–428. 2012.PubMed/NCBI
|
|
10
|
Relles D, Sendecki J, Chipitsyna G, Hyslop
T, Yeo CJ and Arafat HA: Circadian gene expression and
clinicopathologic correlates in pancreatic cancer. J Gastrointest
Surg. 17:443–450. 2013. View Article : Google Scholar
|
|
11
|
Lin YM, Chang JH, Yeh KT, et al:
Disturbance of circadian gene expression in hepatocellular
carcinoma. Mol Carcinog. 47:925–933. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Aravalli RN, Cressman EN and Steer CJ:
Cellular and molecular mechanisms of hepatocellular carcinoma: an
update. Arch Toxicol. 87:227–247. 2013. View Article : Google Scholar
|
|
13
|
Yang Y, Sun M, Wang L and Jiao B: HIFs,
angiogenesis, and cancer. J Cell Biochem. 114:967–974. 2013.
View Article : Google Scholar
|
|
14
|
Tang CM and Yu J: Hypoxia-inducible
factor-1 as a therapeutic target in cancer. J Gastroenterol
Hepatol. 28:401–405. 2013. View Article : Google Scholar
|
|
15
|
Egg M, Köblitz L, Hirayama J, et al:
Linking oxygen to time: the bidirectional interaction between the
hypoxic signaling pathway and the circadian Clock. Chronobiol Int.
30:510–529. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bradford MM: A rapid and sensitive method
for the quantitation of microgram quantities of protein utilizing
the principle of protein-dye binding. Analytical biochemistry.
72:248–254. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
He YW, Wang HS, Zeng J, et al: Sodium
butyrate inhibits interferon-gamma induced indoleamine
2,3-dioxygenase expression via STAT1 in nasopharyngeal carcinoma
cells. Life Sci. 93:509–515. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fang X, Dong W, Thornton C, Scheffler B
and Willett KL: Benzo(a)pyrene induced glycine N-methyltransferase
messenger RNA expression in Fundulus heteroclitus embryos. Mar
Environ Res. 69(Suppl): S74–S76. 2010. View Article : Google Scholar
|
|
19
|
Fang X, Thornton C, Scheffler BE and
Willett KL: Benzo[a]pyrene decreases global and gene specific DNA
methylation during zebrafish development. Environ Toxicol
Pharmacol. 36:40–50. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Buscariollo DL, Fang X, Greenwood V, Xue
H, Rivkees SA and Wendler CC: Embryonic caffeine exposure acts via
A1 adenosine receptors to alter adult cardiac function and DNA
methylation in mice. PLoS One. 9:e875472014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Menegaux F, Truong T, Anger A, et al:
Night work and breast cancer: a population-based case-control study
in France (the CECILE study). Int J Cancer. 132:924–931. 2013.
View Article : Google Scholar
|
|
22
|
Lahti T, Merikanto I and Partonen T:
Circadian Clock disruptions and the risk of cancer. Ann Med.
44:847–853. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen R, Yang K, Zhao NB, et al: Abnormal
expression of PER1 circadian-Clock gene in oral squamous cell
carcinoma. Onco Targets Ther. 5:403–407. 2012.PubMed/NCBI
|
|
24
|
Mazzoccoli G, Panza A, Valvano MR, et al:
Clock gene expression levels and relationship with clinical and
pathological features in colorectal cancer patients. Chronobiol
Int. 28:841–851. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Krugluger W, Brandstaetter A, Kállay E, et
al: Regulation of genes of the circadian Clock in human colon
cancer: reduced period-1 and dihydropyrimidine dehydrogenase
transcription correlates in high-grade tumors. Cancer Res.
67:7917–7922. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang X, Yan D, Teng M, et al: Reduced
expression of PER3 is associated with incidence and development of
colon cancer. Ann Surg Oncol. 19:3081–3088. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Oshima T, Takenoshita S, Akaike M, et al:
Expression of circadian genes correlates with liver metastasis and
outcomes in colorectal cancer. Oncol Rep. 25:1439–1446. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang Y, Hua L, Lu C and Chen Z: Expression
of circadian Clock gene human Period2 (hPer2) in human colorectal
carcinoma. World J Surg Oncol. 9:1662011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Canaple L, Kakizawa T and Laudet V: The
days and nights of cancer cells. Cancer Res. 63:7545–7552.
2003.PubMed/NCBI
|
|
30
|
Schibler U: The daily timing of gene
expression and physiology in mammals. Dialogues Clin Neurosci.
9:257–272. 2007.PubMed/NCBI
|
|
31
|
Filipski E, Subramanian P, Carrière J,
Guettier C, Barbason H and Lévi F: Circadian disruption accelerates
liver carcinogenesis in mice. Mutat Res. 680:95–105. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Logan RW, Zhang C, Murugan S, et al:
Chronic shift-lag alters the circadian Clock of NK cells and
promotes lung cancer growth in rats. J Immunol. 188:2583–2591.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang SL, Yu C, Jiang JX, Liu LP, Fang X
and Wu C: Hepatitis B virus X protein disrupts the balance of the
expression of circadian rhythm genes in hepatocellular carcinoma.
Oncology letters. 8:2715–2720. 2014.PubMed/NCBI
|
|
34
|
Kelleher FC, Rao A and Maguire A:
Circadian molecular Clocks and cancer. Cancer Lett. 342:9–18. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Haase VH: The VHL tumor suppressor: master
regulator of HIF. Curr Pharm Des. 15:3895–3903. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Keith B, Johnson RS and Simon MC: HIF1α
and HIF2α: sibling rivalry in hypoxic tumour growth and
progression. Nat Rev Cancer. 12:9–22. 2012.
|
|
37
|
Loboda A, Jozkowicz A and Dulak J: HIF-1
versus HIF-2 - is one more important than the other? Vascul
Pharmacol. 56:245–251. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hsu CM, Lin PM, Lai CC, Lin HC, Lin SF and
Yang MY: PER1 and Clock are potential circulating biomarkers for
head and neck squamous cell carcinoma. Head Neck. 36:1018–1026.
2013. View Article : Google Scholar
|
|
39
|
Yang SL, Liu LP, Jiang JX, Xiong ZF, He QJ
and Wu C: The correlation of expression levels of HIF-1α and HIF-2α
in hepatocellular carcinoma with capsular invasion, portal vein
tumor thrombi and patients’ clinical outcome. Jpn J Clin Oncol.
44:159–167. 2014. View Article : Google Scholar : PubMed/NCBI
|