Introduction
Bladder cancer (BC) is one of the most frequently
occurring types of malignancy worldwide (1). In 2012, 73,510 new cases of BC were
diagnosed in the United States, resulting in mortality of 14,880
individuals (2). In patients with
bladder cancer, ~70% are diagnosed with non-muscle-invasive bladder
cancer (NMIBC), which is a type of BC with a good survival rate,
but high rate of recurrence. The remaining 30% of BC patients have
muscle-invasive bladder cancer (MIBC), which often metastasizes and
leads to mortality (3).
Furthermore, BC is one of the most expensive types of cancer to
treat in the United States, accounting for ~3.7 billion US dollars
(2001 values) in direct costs (4).
However, the current treatment strategies do not always provide
satisfactory results, particularly for disease extending outside
the bladder (5). Since the
detection of BC in its early stages improves the chance of
successful treatment, early detection and diagnosis of BC will
significantly improve survival rates. At present, urine cytology
screening has been widely adopted for the detection of BC. However,
it can produce false-positive results due to the subjective
morphological criteria (6).
Although alternative diagnostic methods including cystoscopic and
histological evaluation of multiple bladder biopsies have been used
for diagnosis, these methods are impractical for population
screening due to their invasiveness and inconvenience (7). Therefore, the development of
non-invasive screening assessment, with high sensitivity and
specificity for the early detection of BC, has become a major
challenge.
MicroRNAs (miRNAs) are endogenous single-stranded
small RNA molecules of 19–24 nucleotides in length, which are
transcribed from DNA, but are not translated into proteins
(8,9). By binding to the 3′-untranslated
region of target mRNAs, miRNAs are able to negatively regulate gene
expression at the post-transcriptional level (10). Previous studies have demonstrated
that miRNAs are frequently dysregulated in human cancer and exert
significant effects on cancer development (11). Previous studies have suggested that
serum/plasma miRNAs exist in a cell-free form (12,13)
and are resistant to the degradation of RNases (12,14).
Reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) is an effective technique to examine the gene expression
levels of circulating miRNAs, and the interpretation of RT-qPCR
results depends largely on normalization of the data. At present,
the use of reference genes is the most widely used method used for
normalization, however, normalization to inappropriate reference
genes may lead to misinterpretation, confounding or significant
deviation of the data (15,16),
thus highlighting the importance of selecting suitable reference
genes. In previous studies of circulating miRNAs, reference genes
have been selected predominantly on the basis of previous
literature or their own experience, and a method of selection
remains to be fully established. Therefore, it is necessary to
systemically evaluate the suitable reference genes for the
investigation of circulating miRNAs in different types of human
cancer. In a previous study, the combination of miR-191-5p and U6
was identified as suitable for use as reference genes in the
investigation of serum microRNAs in patients with colorectal
adenocarcinoma and adenoma (17).
However, few studies have been performed concerning the
identification of reference genes for RT-qPCR analysis of serum
miRNAs in BC.
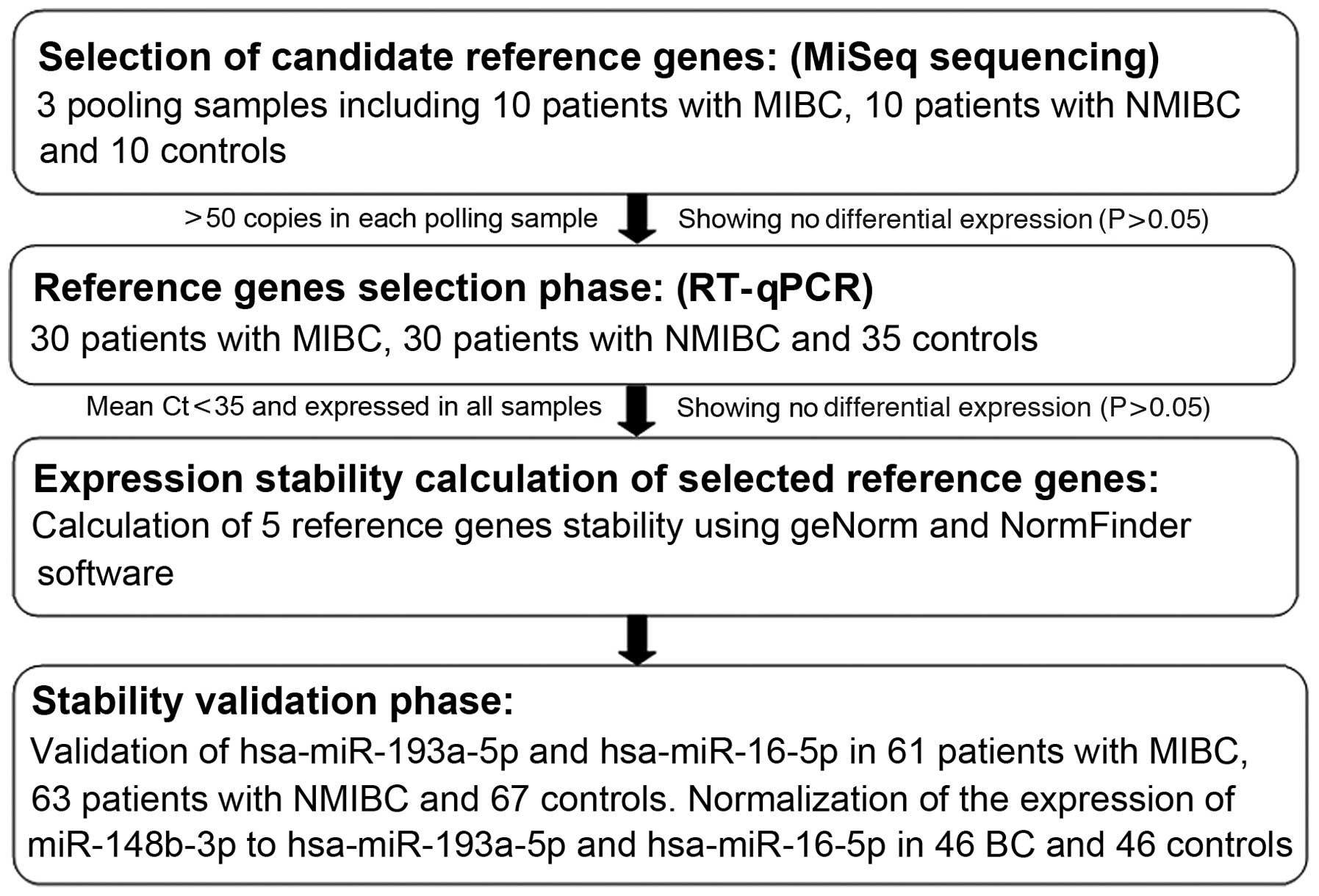
In the present multiphase, case-control study,
candidate reference genes were screened using MiSeq sequencing in
the first screening phase. In the second selection phase, RT-qPCR
and two different algorithms were used to evaluate their expression
stability. In the third validation phase, the selected reference
genes were confirmed in an independent cohort. In addition,
miR-148b-3p was selected as a target miRNA for further
validation.
Materials and methods
Study population and design
The present study enrolled 250 patients with newly
diagnosed BC in the Department of Urologic Surgery, Qilu Hospital,
Shandong University (Jinan, China) between 2007 and 2013. A total
of 176 patients with BC underwent transurethral resection of
bladder tumor, 13 patients with BC underwent partial excision of BC
and 61 patients with BC underwent total removal of BC. The tumors
of 64 patients were on the left bladder wall, 56 were on the right
bladder wall, 25 were on the anterior wall, 29 were on the
posterior wall, 19 were in the neck of the bladder, 8 were in the
apex of bladder and 49 patients had multiple bladder tumors. The
size of tumor specimens ranges from 0.1–4 cm2. During
the operation, the patients were administered the following
anaesthesia: Intravenous drip of midazolam, 0.1 mg/kg; propofol
injection, 1.5 mg/kg; cis-benzenesulfonic acid with
atracurium, 0.15 mg/kg; sufentanil citrate injection, 2
μg/kg. Prior to surgery, fentanyl (2 μg/kg) was
administered to the patients and during the surgery, patients
received oral sevoflurane. Histological specimens from all the
patients were reviewed to confirm the diagnosis of BC, and the
tumors were staged based on the following criteria of the Union for
International Cancer Control (Geneva, Switzerland): pTa,
noninvasive papillary urothelial carcinoma; pT1, tumor invading
into the lamina propria; pT2, tumor invading into muscularis
propria; pT3, tumor invading into perivesical soft tissue; pT4,
tumor invading into an adjacent organ, including the uterus,
vagina, prostate, pelvic wall or abdominal wall (18). A group of 158 healthy participants
were selected as controls from a large pool of individuals for
general health checks at the Healthy Physical Examination Centre of
Qilu Hospital (Jinan, China). All the controls were matched to the
patients by age and sex (Tables I
and II). The present study was
performed with the approval of the ethics committee of Qilu
Hospital, Shandong University (Jinan, China) and written informed
consent was obtained from each participant.
 | Table ICharacteristics of patients and
controls. |
Table I
Characteristics of patients and
controls.
| Characteristic | Screening
phase | Selection
phase | Validation
phase |
|---|
| Controls (n) | 10 | 35 | 67 |
| Age (mean ±
SD) | 65.38±14.31 | 62.15±12.18 | 63.30±12.54 |
| Gender (n) |
| Male | 6 (60.0%) | 19 (54.3%) | 41 (61.2%) |
| Female | 4 (40.0%) | 16 (45.7%) | 26 (38.8%) |
| NMIBC (n) | 10 | 30 | 63 |
| Age (mean ±
SD) | 66.72±13.28 | 65.31±14.23 | 63.46±12.66 |
| Gender (n) |
| Male | 6 (60.0%) | 17 (56.7%) | 39 (61.9%) |
| Female | 4 (40.0%) | 13 (43.3%) | 24 (38.1%) |
| Pathological stage
(n) |
| pTa | 7 (70.0%) | 22 (73.3%) | 38 (60.3%) |
| pT1 | 3 (30.0%) | 8 (26.7%) | 25 (39.7%) |
| MIBC (n) | 10 | 30 | 61 |
| Age (mean ±
SD) | 63.27±13.29 | 64.53±11.36 | 64.71±14.65 |
| Gender (n) |
| Male | 7 (70.0%) | 19 (63.3%) | 34 (55.7%) |
| Female | 3 (30.0%) | 11 (36.7%) | 27 (44.3%) |
| Pathological stage
(n) |
| pT2 | 2 (20.0%) | 5 (16.7%) | 9 (14.8%) |
| pT3 | 5 (50.0%) | 14 (46.7%) | 31 (50.8%) |
| pT4 | 3 (30.0%) | 11 (36.7%) | 21 (34.1%) |
 | Table IICharacteristics of patients and
controls in the target miRNA validation phase. |
Table II
Characteristics of patients and
controls in the target miRNA validation phase.
| Characteristic | MIBC | NMIBC | Control |
|---|
| Number | 20 | 26 | 46 |
| Age (mean ±
SD) | 67±13.59 | 65±15.21 | 61±11.33 |
| Gender (n) |
| Male | 14 (70.0%) | 17 (65.4%) | 27 (58.7%) |
| Female | 6 (30.0%) | 9 (34.6%) | 19 (41.3%) |
| Pathological stage
(n) |
| pTa | N/A | 21 (80.8%) | N/A |
| pT1 | N/A | 5 (19.2%) | N/A |
| pT2 | 4 (20.0%) | N/A | N/A |
| pT3 | 11 (55.0%) | N/A | N/A |
| pT4 | 5 (25.0%) | N/A | N/A |
A multiphase, case-control study was designed to
identify suitable candidate reference genes for normalizing the
RT-qPCR data of the serum miRNAs in BC (Fig. 1). In the first phase, MiSeq
sequencing was performed to screen candidate reference miRNAs in
three pooled serum samples from 10 patients with MIBC (stages
pT2–pT4), 10 patients with NMIBC (stages pTa and pT1) and 10
controls. Only miRNAs with a minimum of 50 copies in all three
pooled samples, and exhibiting no differential expression among the
three pooled samples (P>0.05) were selected as candidate
reference genes. In the second phase, RT-qPCR was performed in
additional participants (30 patients with MIBC, 30 patients with
NMIBC and 35 controls) in order to examine the expression levels of
the selected genes, to further validate the most suitable genes. In
the third phase, the selected genes were validated in an
independent cohort consisting of 61 patients with MIBC, 63 patients
with NMIBC and 67 controls. miR-148b-3p was selected as a target
miRNA for further validation in 46 patients with BC and 46
controls.
Sample preparation
Venous blood samples (5 ml) were collected from 158
controls and 250 patients with BC who had not undergone surgery,
chemotherapy or radiotherapy. Venous blood samples were collected
using a single use Vacutainer® blood collection tube (BD
Biosciences, Franklin Lakes, NJ, USA). The serum was separated
within 2 h of blood collection by centrifugation at 1,500 × g for
10 min, followed by 15 min high-speed centrifugation (13,800 × g;
Thermo Electro LED GmbH, Osterode, Germany) to completely remove
the cell debris. The serum samples were then stored at −80°C until
further analysis.
MiSeq sequencing
Equal 1 ml volumes of the serum from 10 patients
with MIBC, 10 patients with NIMBC and 10 sex- and age-matched
controls were pooled, respectively. An miRNeasy mini kit (Qiagen,
Valencia, CA, USA) was used to extract and purify total RNA,
including miRNA, according to the manufacturer’s instructions.
Briefly, the 3′ adaptor was ligated sequentially to the miRNA and
then selected using PAGE electrophoresis (adaptors and PAGE from
Berry Genomics Co., Ltd., Shanghai, China). Subsequently, the 5′
adaptor was ligated sequentially to the miRNA. Thus, a pair of
adaptors was ligated to the 3′ and 5′ ends of the miRNA and the
ligated miRNA molecules were used as templates for cDNA synthesis.
The cDNAs were then amplified to establish a cDNA library. A KAPA
SYBR FAST Universal qPCR kit (Kapa Biosystems, Inc., Wilmington,
MA, USA) was used to determine the library quality based on the
following criteria: i) cDNA concentration >1 nM, ii) no dimer
contamination. The sequencing analysis was performed using the
purified cDNA with a MiSeq Sequencing system (Illumina, Inc., San
Diego, CA, USA), according to the manufacturer’s instructions. For
the MiSeq sequencing, the final reads of each miRNA were determined
by normalization with the total reads of all the miRNAs in the
sample. Bioinformatics analysis was performed by searching against
the miRBase, version 17.0 (http://www.mirbase.org/) to identify known mature
miRNAs.
cDNA synthesis and RT-qPCR analysis
The cDNA of the miRNAs was synthesized using a One
Step PrimeScript miRNA cDNA Synthesis kit (Takara Bio Inc., Otsu,
Japan). The reaction mixture (20 μl) contained 10 μl
2X miRNA Reaction Buffer mix, 2 μl miRNA Primescript RT
Enzyme mix and 2 μl 0.1% bovine serum albumin, which were
provided in the kit, as well as 3 μl serum mixed with 3
μl serum buffer (2.5% Tween 20, 50 mmol/l Tris and 1 mmol/l
EDTA; Solarbio Science and Technology Co., Ltd, Beijing, China)
(19). The mixture was
subsequently incubated at 37°C for 60 min, at 85°C for 5 sec and
then at 4°C for 60 min. Following centrifugation at 9,600 × g for
10 min at 4°C, the cDNA was stored at −20°C until use.
The RT-qPCR reaction for the detection of miRNAs was
performed in a final volume of 25 μl using an ABI PRISM 7500
Sequence Detection system (Applied Biosystems Life Technologies,
Foster City, CA, USA) using a SYBR PrimeScript miRNA qPCR kit
(Takara Bio Inc.). The reaction mixture contained 2.0 μl
template cDNA, 12.5 μl SYBR Premix Ex Taq II, 0.5 μl
DyeII, 2 μl 5 μM forward primer (Ribobio Co., Ltd,
Guangzhou, China), 1 μl 10 μM Uni-miR qPCR primer and
7 μl ddH2O. The thermal cycling protocol was as
follows: An initial 30 sec denaturation step at 95°C, followed by
45 cycles of 95°C for 5 sec and 57°C for 34 sec. Melting curve
analysis was performed at the end of the RT-qPCR run between 65 and
95°C to verify the specific amplification. Each measurement was
performed in triplicate and the threshold cycle (Ct) value was
obtained by the ABI PRISM 7500 software. The candidate reference
genes were identified based on the criteria that: i) miRNA was
expressed in all samples; ii) the mean Ct value was <35 and iii)
the expression of miRNA was not significantly different among the
three groups.
Statistical analysis
The average Ct values were converted into relative
quantities for analysis of the overall stability using geNorm and
intergroup stability using NormFinder software. One-way analysis of
variance was used to calculate the differences in candidate
reference genes between the groups. Comparison of the expression
levels of miR-148b-3p between different groups was estimated using
the Mann-Whitney U test. Statistical analyses and graph
construction were performed using SPSS 17.0 (SPSS, Inc., Chicago,
IL, USA) and Minitab 15 software (Minitab, Inc., University Park,
PA, USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
Selection of candidate reference
genes
In the MiSeq sequencing results, 206, 259 and 180
miRNAs were detected (>10 copies) in the serum from the MIBC
patients, NMIBC patients and controls, respectively. Following
normalization of the final reads for each miRNA in the pooled
sample of total miRNAs, 23, 16 and 27 miRNAs with >50 copies
were identified, with no differential expression identified between
the MIBC and NMIBC, the MIBC and control or the NMIBC and control
groups. According to the selection criteria, described above in
‘study population and design’, 10 miRNAs were selected as candidate
reference genes (Table III).
 | Table IIICharacteristics of the candidate
reference miRNAs. |
Table III
Characteristics of the candidate
reference miRNAs.
| miRNA | Mature miRNA
sequence (5′-3′) | Copies in MIBC
(n) | Copies in NMIBC
(n) | Copies in control
(n) |
|---|
|
hsa-miR-193a-5p |
UGGGUCUUUGCGGGCGAGAUGA | 5911 | 7507 | 5759 |
| hsa-miR-191-5p |
CAACGGAAUCCCAAAAGCAGCUG | 2714 | 3320 | 2937 |
| hsa-miR-16-5p |
UAGCAGCACGUAAAUAUUGGCG | 1110 | 1422 | 1226 |
| hsa-miR-10a-5p |
UACCCUGUAGAUCCGAAUUUGUG | 462 | 559 | 431 |
| hsa-miR-345-5p |
GCUGACUCCUAGUCCAGGGCUC | 435 | 412 | 314 |
| hsa-miR-143-3p |
UGAGAUGAAGCACUGUAGCUC | 312 | 394 | 272 |
| hsa-miR-140-3p |
UACCACAGGGUAGAACCACGG | 192 | 245 | 263 |
| hsa-miR-502-3p |
AAUGCACCUGGGCAAGGAUUCA | 121 | 156 | 184 |
| hsa-let-7d-3p |
CUAUACGACCUGCUGCCUUUCU | 67 | 72 | 67 |
| hsa-miR-141-3p |
UAACACUGUCUGGUAAAGAUGG | 69 | 58 | 55 |
RT-qPCR analysis of candidate reference
genes
In this second phase of the present study, RT-qPCR
was performed in an independent cohort to examine the expression
levels of 11 genes, including the U6 and 10 selected candidate
reference genes. The hsa-miR-10a-5p and hsa-miR-345-5p miRNAs were
excluded from further examination as they were not detected in all
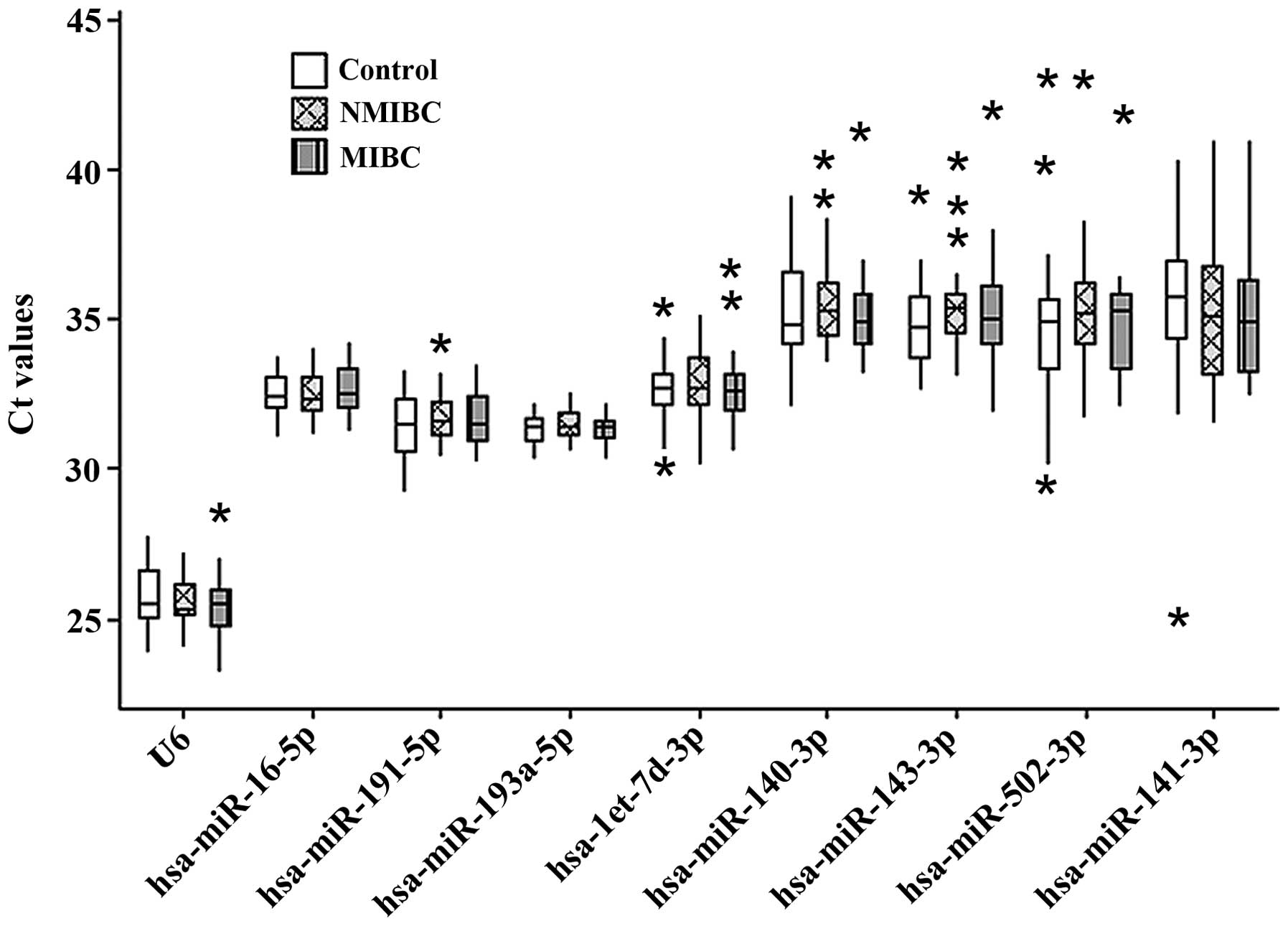
samples. Based on the Ct values of the remaining nine genes, no
significant differences were observed in the gene expression levels
among the three groups (P>0.05; Table IV and Fig. 2). Of the nine candidate reference
genes, the Ct values varied markedly, ranging between 23.19
and42.97. The expression levels of four miRNAs (hsa-miR-143-3p,
hsa-miR-140-3p, hsa-miR-502-3p and hsa-miR-141-3p) were deemed too
low, with median Ct values>35. Therefore, five genes
(hsa-miR-193a-5p, hsa-miR-16-5p, U6, hsa-miR-191-5p and
hsa-let-7d-3p) were included for subsequent stability analysis.
 | Table IVCt values of the candidate reference
miRNAs. |
Table IV
Ct values of the candidate reference
miRNAs.
| Name | Ct Range | Ct min | Ct max | Mean Ct ± SD
| P-value |
|---|
| MIBC | NMIBC | Control |
|---|
| U6 | 5.42 | 23.19 | 28.61 | 25.50±1.03 | 25.56±0.79 | 25.69±0.94 | 0.701 |
| hsa-miR-16-5p | 3.25 | 31.03 | 34.28 | 32.65±0.78 | 32.43±0.78 | 32.50±0.74 | 0.544 |
| hsa-miR-191-5p | 5.05 | 29.22 | 34.27 | 31.66±0.91 | 31.78±0.95 | 31.45±1.05 | 0.391 |
|
hsa-miR-193a-5p | 2.30 | 30.31 | 32.61 | 31.37±0.51 | 31.55±0.50 | 31.33±0.55 | 0.238 |
| hsa-let-7d-3p | 5.64 | 30.17 | 35.81 | 32.64±1.18 | 32.92±1.14 | 32.69±1.12 | 0.599 |
| hsa-miR-140-3p | 9.25 | 32.07 | 41.32 | 35.21±1.50 | 35.69±1.65 | 35.39±1.67 | 0.508 |
| hsa-miR-143-3p | 10.01 | 31.94 | 41.95 | 35.31±1.89 | 35.48±1.53 | 34.89±1.46 | 0.320 |
| hsa-miR-502-3p | 13.34 | 29.63 | 42.97 | 35.02±1.84 | 35.35±2.17 | 34.72±2.53 | 0.527 |
| hsa-miR-141-3p | 16.04 | 24.99 | 41.03 | 35.14±2.07 | 35.36±2.46 | 35.37±2.57 | 0.916 |
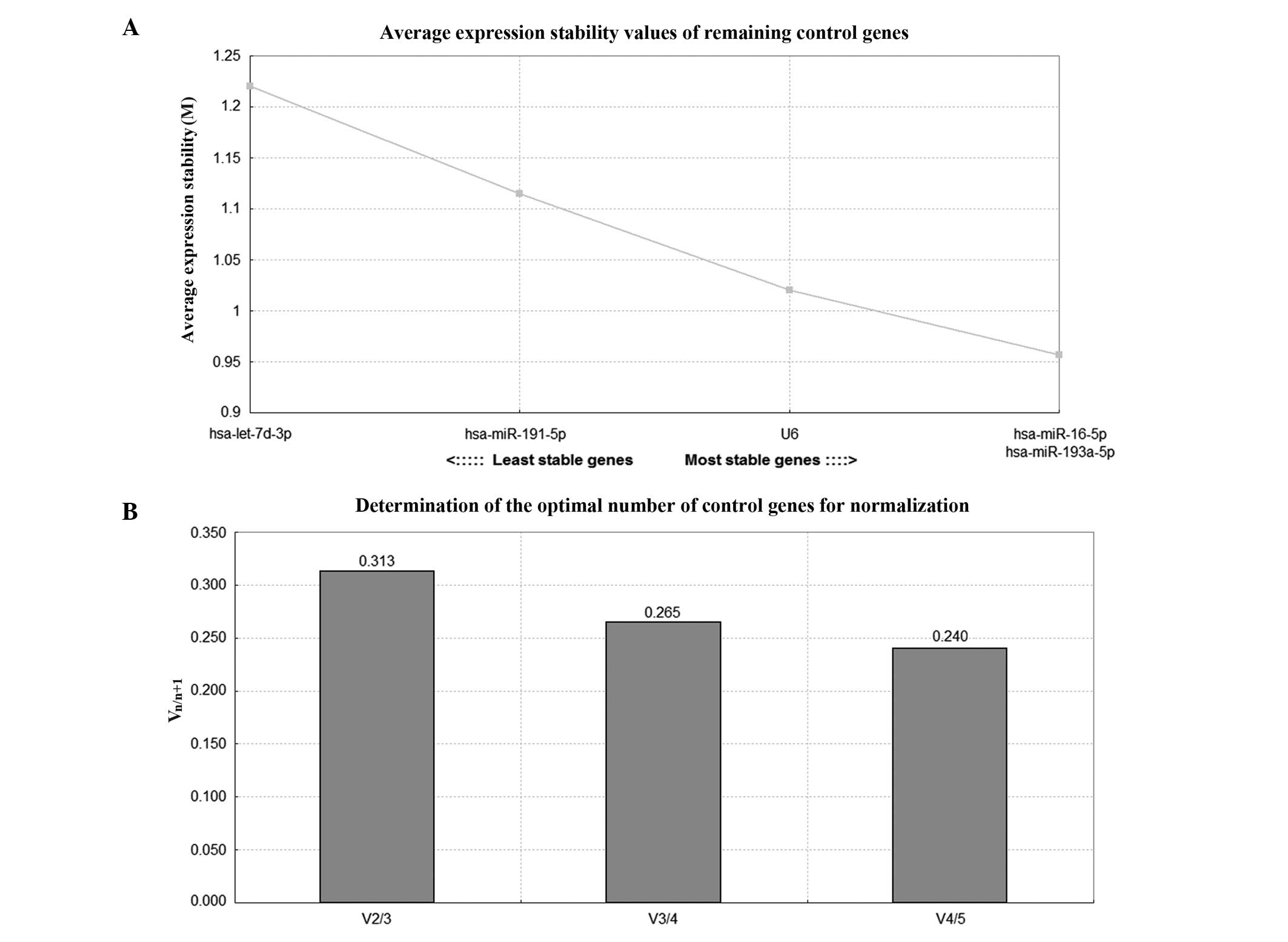
geNorm and NormFinder analysis
Table V lists the
stability of the five genes analyzed by geNorm and NormFinder
(20). geNorm calculates the
stability measure (M) of each gene, and lower values indicate the
genes with higher stability. Subsequently, geNorm excludes the
least stable miRNA following each calculation and recalculates the
M values of the remaining candidate reference genes, with the two
most stable miRNAs remaining (Fig.
3). It is also able to calculate the optimal number of
reference genes needed for normalization by generating a
normalization factor (NF), which is first calculated with the two
most stable genes, and then subsequently recalculated in a
stepwise-manner by including the most stable remaining gene. The
pair-wise variation (Vn/n+1) of two sequential NFs
(NFn and NFn+1) is then calculated, with the
lowest Vn/n+1 indicating the optimal number of reference
genes. The cut-off value for the lowest Vn/n+1 is 0.15,
above which the additional gene is necessary for normalization.
However, 0.15 is not an absolute cut-off value, and the number of
reference genes can be determined according to the requirement of
the investigation (21). In the
present study, geNorm analysis suggested that all of the five genes
(hsa-miR-193a-5p, hsa-miR-16-5p, U6, hsa-miR-191-5p and
hsa-let-7d-3p) were suitable as candidate reference genes.
NormFinder and geNorm identified hsa-miR-193a-5p as the most stably
expressed reference gene and selected hsa-miR-193a-5p and
hsa-miR-16-5p as the pair of reference genes with the highest
stability.
 | Table VStability ranking of the candidate
reference genes and the most stable combination. |
Table V
Stability ranking of the candidate
reference genes and the most stable combination.
| Rank | NormFinder
| geNorm
|
|---|
| Gene | Stability | Gene | Stability (M) |
|---|
| 1 (highest) |
hsa-miR-193a-5p | 0.088 |
hsa-miR-193a-5p | 1.065 |
| 2 | hsa-miR-16-5p | 0.131 | hsa-miR-16-5p | 1.164 |
| 3 | U6 | 0.158 | U6 | 1.223 |
| 4 | hsa-miR-191-5p | 0.169 | hsa-miR-191-5p | 1.271 |
| 5 (lowest) | hsa-let-7d-3p | 0.204 | hsa-let-7d-3p | 1.378 |
| Most stable
combination |
hsa-miR-193a-5p/16-5p | 0.080 |
hsa-miR-193a-5p/16-5p | 0.957 |
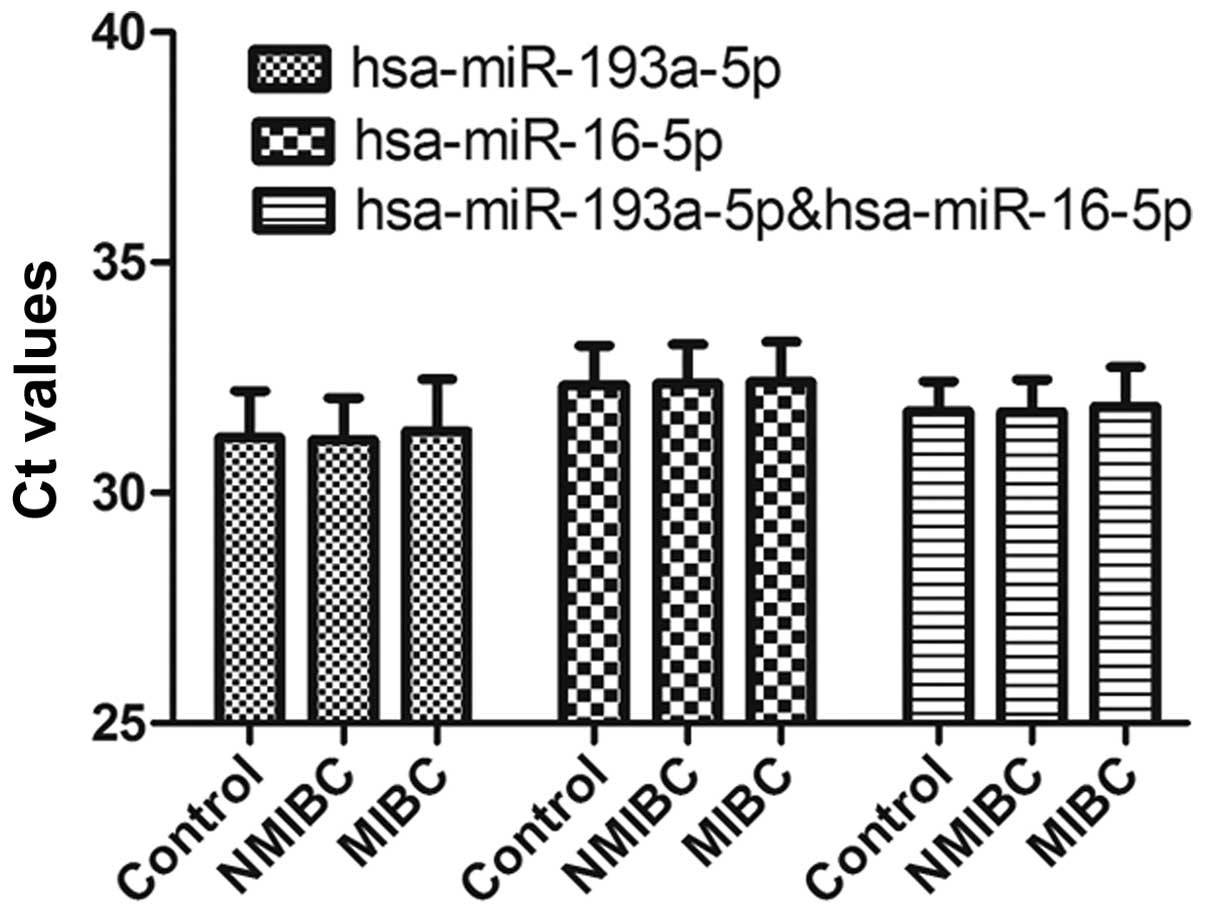
Gene validation
The stability of hsa-miR-193a-5p, hsa-miR-16-5p and
the combination of hsa-miR-193a-5p and hsa-miR-16-5p were further
confirmed with RT-qPCR in an independent cohort (Fig. 4). No significant differences in
expression levels were observed among the three groups (P>0.05).
In addition, to determine the effect of the two reference genes on
the quantification of miRNAs, circulating miR-148b-3p was selected
as the target miRNA, according to the sequencing results, and it
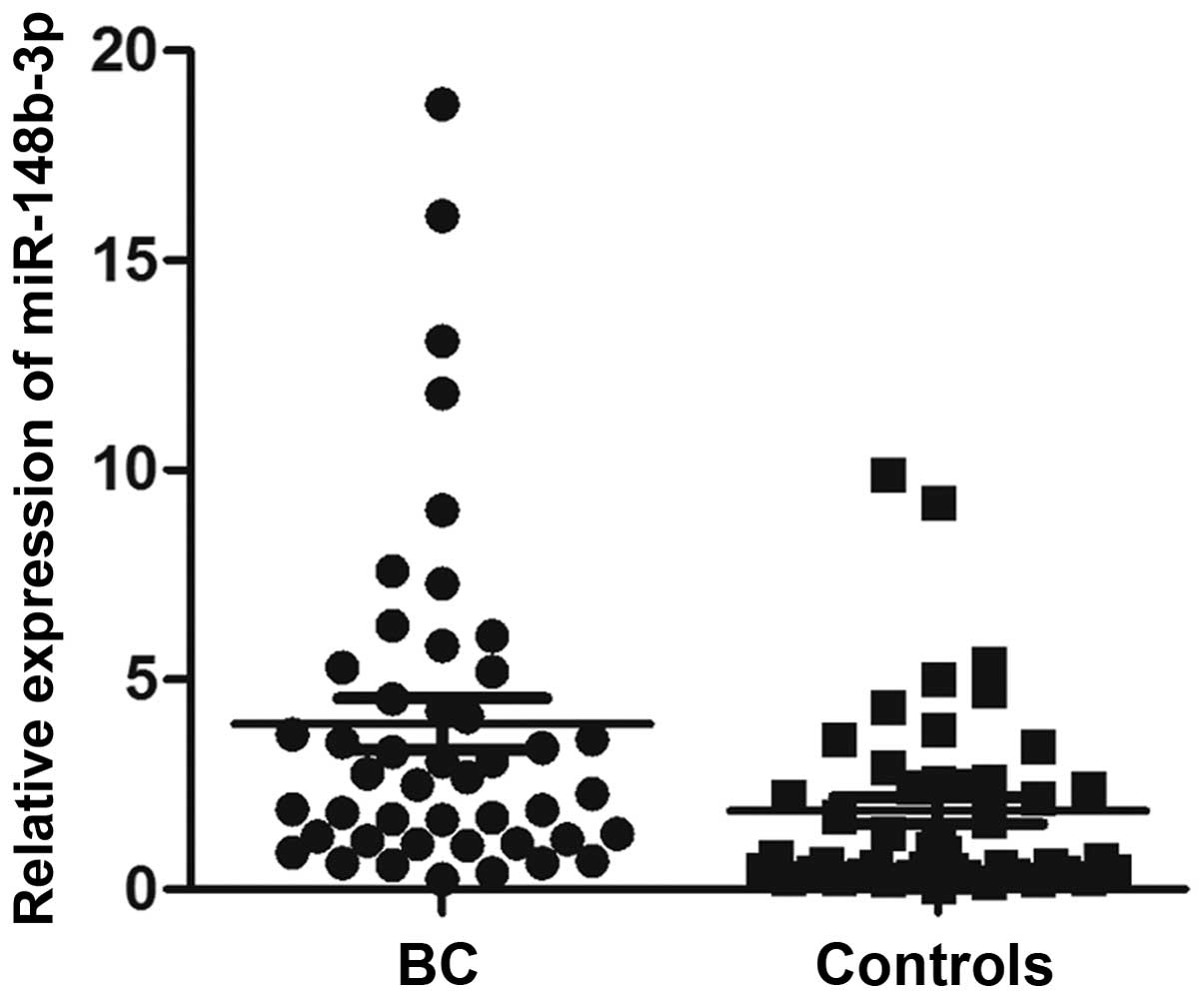
has been demonstrated to have potential in evaluating BC (22). The expression of miR-148b-3p was
significantly higher in patients with BC compared with the
controls, using hsa-miR-193a-5p or hsa-miR-16-5p as reference genes
(P<0.05; data not shown). This difference remained significant
following normalization of the data with the combination of
hsa-miR-193a-5p and hsa-miR-16-5p (P<0.05; Fig. 5).
Discussion
Cell free miRNAs have been reported to be released
and circulate in the blood of patients with cancer. Previous
studies have also demonstrated that there are distinct miRNA
expression patterns associated with specific types of cancer
(23,24). The dysregulation of miRNA
expression may be an early event during tumorigenesis, thus the
detection of differentially expressed circulating miRNAs in easily
accessible surrogate tissues, including serum or plasma, may be of
value for the early detection of cancer and real-time monitoring of
disease progression, including BC (25).
There are currently three methods, including next
generation sequencing, chip-based microarrays and RT-qPCR, which
are widely used for the identification of miRNA expression patterns
(26,27). Although next generation sequencing
and chip-based microarrays can be used to simultaneously filter or
screen hundreds of miRNAs in a single sample, the results from
these high-throughput methods require further validation by RT-qPCR
(28,29). In order to obtain reliable RT-qPCR
results, a valid standard for normalization is required to minimize
technical-associated effects, and remove systematic bias and
experimental variation. In previous studies, input volume and
spike-in synthetic RNA have been used for normalization of RT-qPCR
results (30,31). However, adding the same volume of
samples does not ensure an equal quantity of total RNA, while
adding spiked-in RNAs only corrects technical variations in miRNA
isolation (32). Therefore,
selecting appropriate reference genes as endogenous controls has
become the most common normalization method for accurate RT-qPCR
analysis (33).
It has been demonstrated that inappropriate
reference genes can markedly affect overall RT-qPCR results
(34). In a previous study of
colorectal cancer, the use of inappropriate reference genes
resulted in alterations in gene expression, which are biological
meaningful (16). In addition,
sample type and study design require consideration when selecting a
reference gene, as a previous study did not identify any reference
genes with constant expression levels in all sample types when
exposed to different experimental conditions (21). As a consequence, the expression
stability of reference genes requires verification in experiments
with different designs (32,35).
In previous studies, several miRNAs have been identified as
reference genes for the RT-qPCR analysis of serum miRNAs, including
miR-484 and miR-191 in breast cancer (36), and miR-16 and miR-93 in gastric
cancer (34). To the best of our
knowledge, no widely accepted reference genes have been reported
for normalizing serum miRNA RT-qPCR data in BC.
U6 is a gene, which is used as a stable reference
gene in the investigation of miRNA in different tissues. However,
several studies have observed that U6 may be not reliable for the
normalization of serum miRNAs (37,38).
Therefore, in the present study, U6 was also included for RT-qPCR
analysis. Of the 11 genes examined, six miRNAs (hsa-miR-10a-5p,
hsa-miR-345-5p, hsa-miR-143-3p, hsa-miR-140-3p, hsa-miR-502-3p and
hsa-miR-141-3p) were identified with no or low expression (Ct
values >35) and were excluded for evaluation, while the
remaining five genes (hsa-miR-193a-5p, hsa-miR-16-5p, U6,
hsa-miR-191-5p and hsa-let-7d-3p), which were expressed in all
samples and had a mean Ct value of <35, were analyzed using
geNorm and NormFinder for stability. geNorm is a pairwise
comparison model, which calculates the optimal number of required
reference genes to obtain reliable results from RT-qPCR, while
NormFinder uses an analysis of variance-based model to estimate
intra- and intergroup variations in reference genes (39). In the present study, based on
NormFinder and geNorm analysis, hsa-miR-193a-5p was identified as
the most suitable gene with constant expression, and the
combination of hsa-miR-193a-5p and hsa-miR-16-5p was recommended as
the most suitable pair of reference genes for serum miRNA
investigations in BC. In addition, two methods were used in the
validation of hsa-miR-193a-5p and hsa-miR-16-5p, including the
analysis of expression levels in a large independent cohort and
validation with miR-148b-3p as a target miRNA in an additional
cohort. These results were consistent with a previous study, which
demonstrating upregulated levels of miR-148b-3p in serum from
patients with BC (22).
In conclusion, the results of the present study
demonstrated that normalization of miRNAs RT-qPCR data using a
combination of hsa-miR-193a-5p and hsa-miR-16-5p as reference genes
may produce reliable and accurate results for serum miRNA
investigations in BC. However, since the samples size was small, a
large sample size and multicenter trials are required to confirm
this conclusion.
Acknowledgments
The authors would like to thank Chengjun Zhou and
Junhui Zhen for their assistance in the histological evaluation of
samples.
References
|
1
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J and
Thun MJ: Cancer statistics, 2009. CA Cancer J Clin. 59:225–249.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kaufman DS, Shipley WU and Feldman AS:
Bladder cancer. Lancet. 374:239–249. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Botteman MF, Pashos CL, Redaelli A, Laskin
B and Hauser R: The health economics of bladder cancer: A
comprehensive review of the published literature.
Pharmacoeconomics. 21:1315–1330. 2003. View Article : Google Scholar
|
|
5
|
Malmström PU: Why has the survival of
patients with bladder cancer not improved? BJU Int. 101:267–269.
2008. View Article : Google Scholar
|
|
6
|
Falebita OA, Sweeney P and Lee G: Urine
cytology in the evaluation of urological malignancy revisited: is
it still necessary. Urol Int. 84:45–49. 2010. View Article : Google Scholar
|
|
7
|
Mengual L, Lozano JJ, Ingelmo-Torres M,
Gazquez C, Ribal MJ and Alcaraz A: Using microRNA profiling in
urine samples to develop a non-invasive test for bladder cancer.
Int J Cancer. 133:2631–2641. 2013.PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Heneghan HM, Miller N and Kerin MJ: MiRNAs
as biomarkers and therapeutic targets in cancer. Curr Opin
Pharmacol. 10:543–550. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mitchell PS, Parkin RK, Kroh EM, et al:
Circulating microRNAs as stable blood-based markers for cancer
detection. Proc Natl Acad Sci USA. 105:10513–10518. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hunter MP, Ismail N, Zhang X, et al:
Detection of microRNA expression in human peripheral blood
microvesicles. PLoS ONE. 3:e36942008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chim SS, Shing TK, Hung EC, Leung TY, Lau
TK, Chiu RW and Lo YM: Detection and characterization of placental
microRNAs in maternal plasma. Clin Chem. 54:482–490. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Genovesi LA, Anderson D, Carter KW, Giles
KM and Dallas PB: Identification of suitable endogenous control
genes for microRNA expression profiling of childhood
medulloblastoma and human neural stem cells. BMC Res Notes.
5:5072012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chang KH, Mestdagh P, Vandesompele J,
Kerin MJ and Miller N: MicroRNA expression profiling to identify
and validate reference genes for relative quantification in
colorectal cancer. BMC Cancer. 10:1732010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zheng G, Wang H, Zhang X, et al:
Identification and validation of reference genes for qPCR detection
of serum microRNAs in colorectal adenocarcinoma patients. PLoS ONE.
8:e830252013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cheng L, Montironi R, Davidson DD and
Lopez-Beltran A: Staging and reporting of urothelial carcinoma of
the urinary bladder. Mod Pathol. 22(Suppl 2): S70–S95. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Asaga S, Kuo C, Nguyen T, Terpenning M,
Giuliano AE and Hoon DS: Direct serum assay for microRNA-21
concentrations in early and advanced breast cancer. Clin Chem.
57:84–91. 2011. View Article : Google Scholar
|
|
20
|
Zheng G, Wang H, Zhang X, Yang Y, Wang L,
Du L, Li W, Li J, Qu A, Liu Y and Wang C: Identification and
validation of reference genes for qPCR detection of serum microRNAs
in colorectal adenocarcinoma patients. PLoS One. 8:e830252013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vandesompele J, De Preter K, Pattyn F,
Poppe B, Van Roy N, De Paepe A and Speleman F: Accurate
normalization of real-time quantitative RT-PCR data by geometric
averaging of multiple internal control genes. Genome Biol.
3:research0034. 2002, View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Adam L, Wszolek MF, Liu CG, Jing W, Diao
L, Zien A, Zhang JD, Jackson D and Dinney CP: Plasma microRNA
profiles for bladder cancer detection. Urol Oncol. 31:1701–1708.
2013. View Article : Google Scholar
|
|
23
|
Selth LA, Townley SL, Bert AG, Stricker
PD, Sutherland PD, Horvath LG, Goodall GJ, Butler LM and Tilley WD:
Circulating microRNAs predict biochemical recurrence in prostate
cancer patients. Br J Cancer. 109:641–650. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Köberle V, Kronenberger B, Pleli T, et al:
Serum microRNA-1 and microRNA-122 are prognostic markers in
patients with hepatocellular carcinoma. Eur J Cancer. 49:3442–3449.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang X, Liang M, Dittmar R and Wang L:
Extracellular microRNAs in urologic malignancies: Chances and
challenges. Int J Mol Sci. 14:14785–14799. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu R, Zhang C, Hu Z, et al: A
five-microRNA signature identified from genome-wide serum microRNA
expression profiling serves as a finger-print for gastric cancer
diagnosis. Eur J Cancer. 47:784–791. 2010. View Article : Google Scholar
|
|
27
|
Bediaga NG, Davies MP, Acha-Sagredo A, et
al: A microRNA-based prediction algorithm for diagnosis of
non-small lung cell carcinoma in minimal biopsy material. Br J
Cancer. 109:2404–2411. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bargaje R, Hariharan M, Scaria V and
Pillai B: Consensus miRNA expression profiles derived from
interplatform normalization of microarray data. RNA. 16:16–25.
2010. View Article : Google Scholar :
|
|
29
|
Chen X, Ba Y, Ma L, et al:
Characterization of microRNAs in serum: A novel class of biomarkers
for diagnosis of cancer and other diseases. Cell Res. 18:997–1006.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pritchard CC, Kroh E, Wood B, Arroyo JD,
Dougherty KJ, Miyaji MM, Tait JF and Tewari M: Blood cell origin of
circulating microRNAs: A cautionary note for cancer biomarker
studies. Cancer Prev Res (Phila). 5:492–497. 2012. View Article : Google Scholar
|
|
31
|
Scheffer AR, Holdenrieder S, Kristiansen
G, von Ruecker A, Müller SC and Ellinger J: Circulating microRNAs
in serum: novel biomarkers for patients with bladder cancer? World
J Urol. 32:353–358. 2014. View Article : Google Scholar
|
|
32
|
Zhu HT, Dong QZ, Wang G, Zhou HJ, Ren N,
Jia HL, Ye QH and Qin LX: Identification of suitable reference
genes for qRT-PCR analysis of circulating microRNAs in hepatitis B
virus-infected patients. Mol Biotechnol. 50:49–56. 2012. View Article : Google Scholar
|
|
33
|
Lardizábal MN, Nocito AL, Daniele SM,
Ornella LA, Palatnik JF and Veggi LM: Reference genes for real-time
PCR quantification of microRNAs and messenger RNAs in rat models of
hepatotoxicity. PLoS ONE. 7:e363232012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Song J, Bai Z, Han W, Zhang J, Meng H, Bi
J, Ma X, Han S and Zhang Z: Identification of suitable reference
genes for qPCR analysis of serum microRNA in gastric cancer
patients. Dig Dis Sci. 57:897–904. 2012. View Article : Google Scholar
|
|
35
|
Kreth S, Heyn J, Grau S, Kretzschmar HA,
Egensperger R and Kreth FW: Identification of valid endogenous
control genes for determining gene expression in human glioma.
Neurooncol. 12:570–579. 2010.
|
|
36
|
Hu Z, Dong J, Wang LE, et al: Serum
microRNA profiling and breast cancer risk: The use of miR-484/191
as endogenous controls. Carcinogenesis. 33:828–834. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Peltier HJ and Latham GJ: Normalization of
microRNA expression levels in quantitative RT-PCR assays:
Identification of suitable reference RNA targets in normal and
cancerous human solid tissues. RNA. 14:844–852. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang Y, Tang N, Hui T, Wang S, Zeng X, Li
H and Ma J: Identification of endogenous reference genes for
RT-qPCR analysis of plasma microRNAs levels in rats with
acetaminophen-induced hepatotoxicity. J Appl Toxicol. 33:1330–1336.
2013.PubMed/NCBI
|
|
39
|
Andersen CL, Jensen JL and Ørntoft TF:
Normalization of real-time quantitative reverse transcription-PCR
data: A model-based variance estimation approach to identify genes
suited for normalization, applied to bladder and colon cancer data
sets. Cancer Res. 64:5245–5250. 2004. View Article : Google Scholar : PubMed/NCBI
|