Introduction
Tuberculosis is a disease caused by the
intracellular bacterium Mycobacterium tuberculosis, which
has a high mortality rate (1).
CD+ T lymphocytes have been reported to have a central
role in the defense against M. tuberculosis and T helper
1-type (Th1) T cell-mediated immunity is the primary response
during tuberculosis (2–4). A previous study reported that M.
tuberculosis infection in the absence of α-β T lymphocytes
caused mortality in mice within 48 days (5). Protective immunity against M.
tuberculosis is predominantly regulated by transcription
factors (TFs).
Numerous TFs have been reported to be involved in
the immune response to tuberculosis. Protective immunity against
M. tuberculosis infection was reported to be under
regulation of the TF signal transducer and activator of
transcription (Stat) 4 (6).
Nuclear factor of activated T cells p (NFATp) and tumor necrosis
factor (TNF) were suggested to regulate the inflammatory response,
while mice deficient for NFATp had an increased mortality rate
following the development of tuberculosis (7). BXH2 mice deficient for the
Irf8294c allele succumbed to the disease due to
uncontrolled growth of M. tuberculosis (8). Expression of spleen focus-forming
virus proviral integration 1 (Sfpi1) TF was demonstrated to be
increased following 28 days of M. tuberculosis infection
(9). Significant overlap between
the binding sites of interferon regulatory factor 8 (Irf8) and
Sfpi1 was observed using chromatin immunoprecipitation microarray
(ChIP-chip) analysis of tuberculosis infection (10). Th1 cells have a crucial role in
defense against tuberculosis. V-rel avian reticuloendotheliosis
viral oncogene homolog (Rel) B (RelB) TF is essential for
differentiation of Th1 cells and its deficiency was reported to
induce defects in the differentiation of these cells (11). In addition to RelB, Stat4 and Stat1
were demonstrated to be involved in the development of Th1 cells in
response to infection (12).
Therefore, investigating these individual TFs may elucidate
numerous underlying processes in tuberculosis; however, it may not
provide an overview of gene regulation during tuberculosis.
Construction of gene regulatory networks in
tuberculosis, which include genes and TFs, provide a novel
opportunity for understanding the dynamic of molecular processes
involved in the disease. Such networks have been constructed for
M. tuberculosis. A previous study reviewed and constructed a
gene regulatory network for M. tuberculosis genes involved
in persistence in order to provide insight into the molecular
mechanisms involved in persistency (13). In addition, a comprehensive network
of infection-associated processes in human macrophages following
M. tuberculosis infection was constructed (14) as well as a host intracellular
network for the regulation of M. tuberculosis survival
(15). However, there has not yet
been a regulatory network constructed for the TFs involved in the
early lung immune response to M. tuberculosis infection.
In the present study, a network was constructed of
the genes that were differentially expressed (DE) specifically in
the lung cells in response to tuberculosis. TF binding sites,
protein-protein interactions and expression data were integrated in
order to construct the gene regulatory network. Network analyses
using system biology tools were used to determine the most
prominent TFs involved in early lung immune responses to
tuberculosis.
Materials and methods
Microarray availability and
preprocessing
Raw data for early lung infection with M.
tuberculosis were obtained from the Gene Expression Omnibus
(GEO; http://www.ncbi.nlm.nih.gov/geo/) server database
(accession no. GSE23014). These data contributed by Kang et
al (16), contained time
course (0, 12, 15 and 21 days) expression data for infection of
C57BL/6 mice with the H37Rv strain of M. tuberculosis.
Microarray samples were divided into three groups for the
comparison and identification of DE genes during the early days of
lung response to tuberculosis: Group 1, data from 12 days post
infection was compared with that of day 0; group 2, comparison of
days 12 and 15 post infection; and group 3, comparison of day 15
with day 21 post infection.
A Robust Multi-array Averaging (RMA) algorithm was
used for normalization of raw data (17). DE genes were identified using a
two-sample Student’s t-test algorithm. These algorithms were each
performed using Flexarray software v1.6.2 (18). A fold change of 1.5 was set as the
threshold criteria for identifying DE genes. Group III data was
used for all subsequent analysis.
Functional clustering DE genes
In order to determine the enrichment process during
each comparison, the Databases for Annotation, Visualization and
Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/) bioinformatics tool
was used. Clusters with enrichment scores of >1.3 were regarded
to be meaningful clusters in the functional clustering analysis
(19).
TFs involved in regulating DE genes
In order to distinguish regulators of DE genes in
each comparison, data was submitted to the ChIP Enrichment Analysis
(ChEA; http://amp.pharm.mssm.edu/lib/chea.jsp) database
(20). This database analyzed
458,471 potential regulatory interactions based on ChEA from 221
publications. In addition, the TFactS database (http://www.tfacts.org/) was used, which integrates
data from experimentally validated TFs/targets regulatory
interactions. This database consists of data from 6,401 regulatory
interactions for 343 TFs which regulate 2,720 genes from the
following databases: PAZAR, Transcriptional Regulatory Element
Database, Nuclear Factor I Regulome Database and Transcription
Regulatory Regions Database, as well as literature. TFactS was used
to compare submitted DE gene data from the present study with
validated target genes available in its catalog in order to
identify the regulatory genes in the submitted datasets (21). TFs were limited based on their
P-value (<0.05) and altered expression (≥1.5 fold change).
TFs protein-protein interactions
The Biological General Repository for Interaction
Datasets (BioGRID; http://thebiogrid.org/) database was used to extract
valid protein-protein interactions information for DE TFs (22). In addition to BioGRID,
protein-protein interaction data was obtained from the Search Tool
for the Retrieval of Interacting Genes/Proteins (STRING; http://string-db.org/) database, which accommodates
physical (validated and predicted) and functional interactions.
Interactions with a confidence score >0.4 (medium confidence)
were incorporated into the networks (23).
The protein interactions obtained from BioGRID and
STRING were mapped to the expression data in order to identify only
meaningful interactions. These interactions contributed to the
construction of the TFs regulatory network and TFs protein-protein
interaction network.
Network construction and ontology
In order to construct regulatory networks, TF/target
interactions, TFs protein-protein interactions and expression data
were integrated and visualized using Cytoscape v3.0.2 (24). The enriched immune system processes
were investigated in the constructed network using the ClueGO v1.8
plugin for Cytoscape. A two-sided hypergeometric statistical test
and Bonferroni correction were used for P-value correction. In
addition, ClueGO was used to determine processes affected by the DE
TFs based on Kyoto Encyclopedia of Genes and Genomes (KEGG;
http://www.genome.jp/kegg/) pathway and
biological process analyses (25).
Centrality analysis of regulatory
network
Central genes in the regulatory network were
identified using a CentiScaPe v2.0 plugin for Cytoscape. Three
centrality indexes were used on a directed regulatory network:
Degree, betweenness and stress. Degree is considered to be the
simplest index for topological analysis; the number of adjacent
connected nodes to a given node (x) reveal the degree of this node.
In the directed network constructed, out-degree rather than
in-degree was considered. Nodes with a high degree are considered
the hubs of the network. In addition to degree, stress and
betweenness were used as centrality indexes to find hub TFs. These
two indexes provide complementary results from analysis of the
central genes (26).
Protein complexes and active modules
Protein-protein interactions obtained for DE TFs
from BioGRID and STRING were used to construct a protein
interaction network. This network was used to identify protein
complexes which may be involve in the response of lung cells to
early infection of tuberculosis. These protein complexes were
identified using MCODE v1.4.1 plugin for Cytoscape (27).
Integrated regulatory networks were composed of
expression data and interactions. Parts of this network
demonstrated increased activity compared with other parts based on
expression, these are referred to as active modules. These modules
were explored in Cytoscape using the JActiveModules v1.8 tool
(28). To identify active modules
in constructed regulatory networks using JActiveModules, the
expression values were converted to P-values. JActiveModules find
active modules using the loaded P-values.
Summary of methods
Overall, the most crucial DE TFs involve in early
lung immune response to tuberculosis were identified based on
several analyses, including presence in protein complexes,
contribution in active modules, centrality as well as role in
regulation of significant processes and tuberculosis-associated
genes. These analyses were divided into three parts: Involvement in
protein complexes and active modules; centrality analysis; and
regulation of the most enriched processes. DE TFs that were present
in ≥2 out of 3 of these analyses were assumed to be important
factors in early lung immune response to tuberculosis.
Results
Gene expression during different stages
of early lung responses to tuberculosis
Gene expression data obtained from early lung immune
response to tuberculosis was divided into three distinct groups. By
comparing the expression profiles at day 0 and 12 post infection
(Group I), 240 DE genes were identified, while 153 and 2,105 DE
genes were detected when the gene expression profiles were compared
between day 12 and 15 (Group II) and between day 15 and 21 (Group
III), respectively. As the number of DE genes obtained from group
III was higher than those of the two other groups, the third
comparison was selected to establish the regulatory network of TFs
in lungs during infection by M. tuberculosis.
Functional clustering analysis of DE genes obtained
from the comparison of day 15 and 21 post M. tuberculosis
infection resulted in the identification of 113 meaningful clusters
with an accepted enrichment score of >1.3. The presence of
immune system-associated terms, including those for signaling,
defense response, inflammatory response and T cell activation,
indicated the involvement of the immune system during the early
response of lung cells infected by M. tuberculosis.
Regulators of DE genes
ChEA and TFactS databases revealed the involvement
of 17 DE TFs in the regulation of early responses, including
eomesodermin, v-ets avian erythroblastosis virus E26 oncogene
homolog, enhancer of zeste homolog 2 (Ezh2), Irf8, jun B
proto-oncogene, kruppel-like factor 4 (Klf4),
myeloblastosis-related protein B (Mybl2), nuclear receptor
subfamily 3 group C member 1, Rel, RelB, Sfpi1, sex determining
region Y-box 17, Stat1, Stat2, Stat4, T-cell acute lymphocytic
leukemia 1 (Tal1) and thyrotroph embryonic factor. These DE TFs
contribute to the regulation of 1,253 out of 2,105 DE genes;
however, TFs were not identified for the remainder of the DE genes
submitted. Following protein-protein interaction analysis of these
TFs, a gene regulatory network was constructed, including all
seventeen TFs and 1,270 DE genes, which were revealed to be
connected by 4070 interactions.
TFs involved in protein complexes and
modules
In order to determine the involvement of TFs in
protein complexes, the MCODE tool for Cytoscape was used. A total
of 6 complexes were identified in early lung immune response to
tuberculosis with a score of ≥3. The top protein complex (score
30.516 based on the reference of MCODE plugin of Cytoscape)
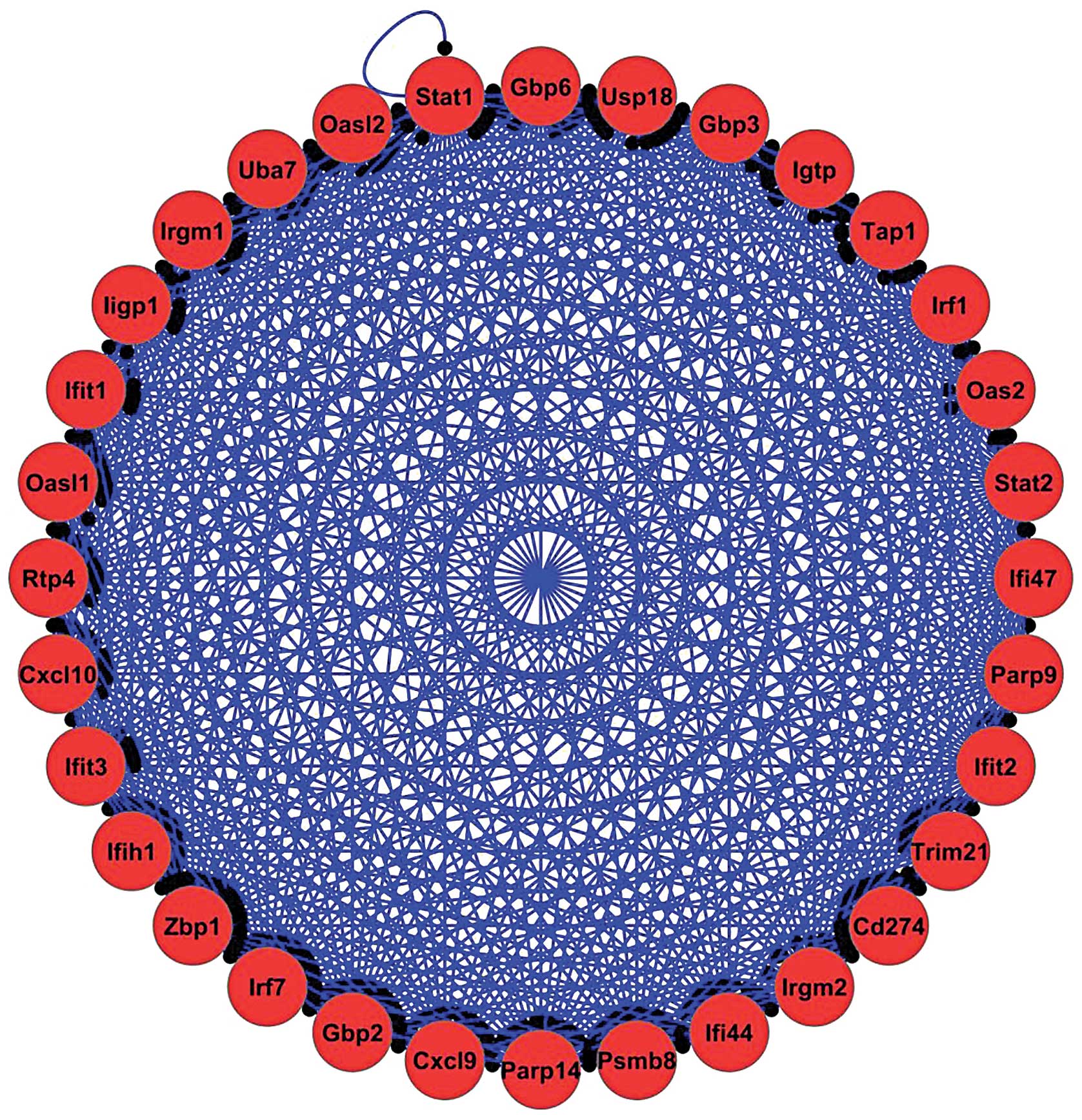
contained 32 DE genes with 474 linking interactions (Fig. 1). Biological process analysis of
this protein complex indicated that the innate immune response was
the top term, as determined using P-values and the number of DE
genes. The DE TFs Ezh2, Irf8, Klf4, Rel, RelB, Stat1 and Stat2 were
identified to be present in the protein complexes identifies.
The active modules of the regulatory network were
analyzed in order to provide an in-depth view of the activity of
the network’s components. TFs involved in these modules are likely
to be more important compared with other TFs. Based on the
biological process analysis of the top active modules, the
biological processes which were most affected by these TFs were as
follows: Response to cytokine stimulus, response to viruses and the
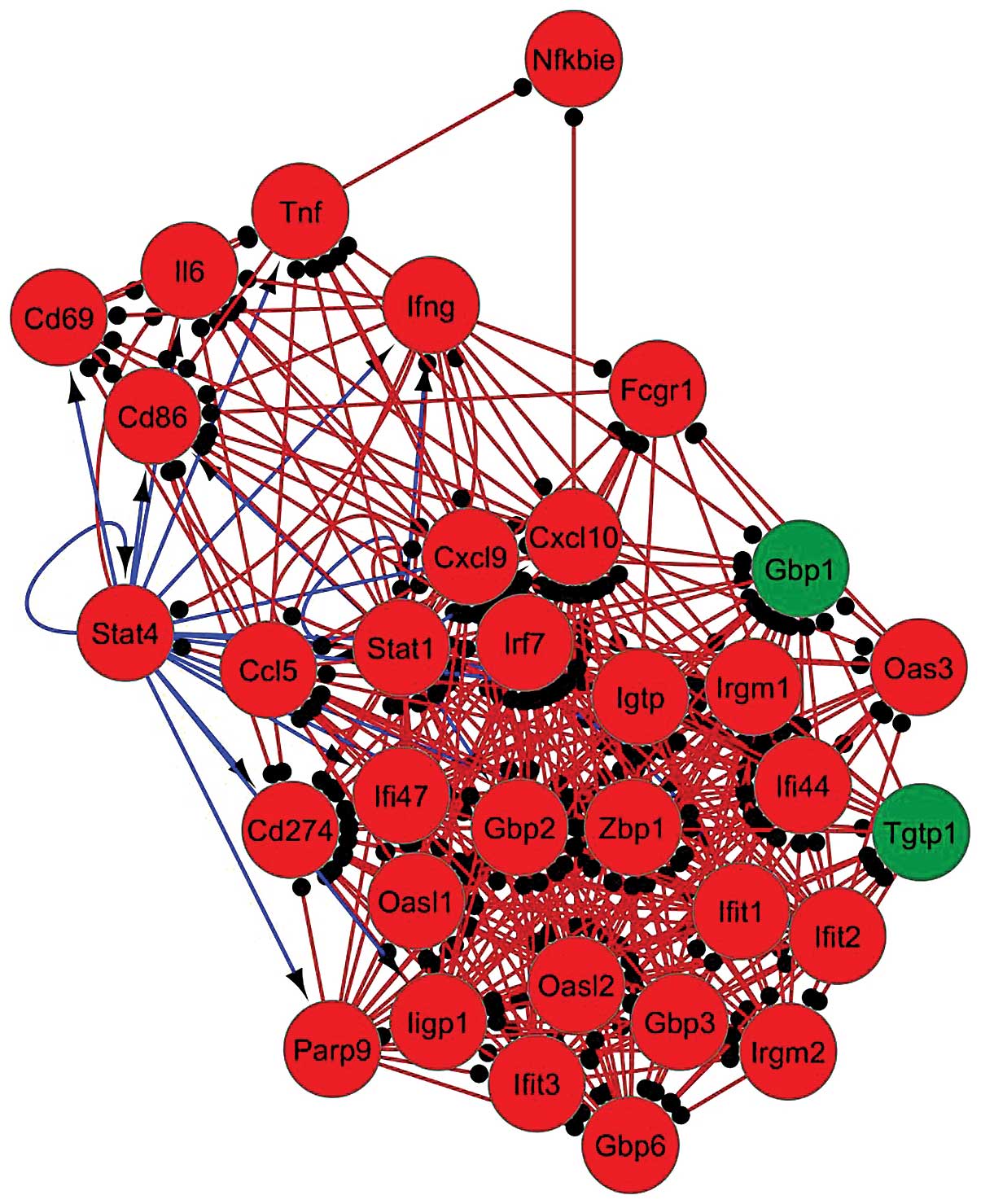
innate immune response. The top active module was composed of 33 DE
genes with 348 interactions (Fig.
2); DE genes in this module were primarily regulated by the TFs
Stat4 and Stat1. The top 5 modules were investigated for the
presence of DE TFs and the results revealed that the TFs Stat4,
Stat2, Stat1, Rel and RelB were detected in ≥1 of the these
modules. Of note, Stat4 and Stat1 were present in all of 5 modules.
These results therefore demonstrated the central role of Stat4 and
Stat1 in active modules in the early lung response to tuberculosis
infection. Overall, the TFs Ezh2, Irf8, Klf4, Rel, RelB, Stat1,
Stat2 and Stat4 were identified to be involved in protein complexes
and core active modules.
Central DE TFs in the regulatory
network
Centrality analysis using the CentiScaPe plugin in
Cytoscape highlighted the importance of TFs in the network topology
and regulation of the whole network (Table I). Three parameters were considered
in analyzing the network: Degree, betweenness and stress. According
to degree, Sfpi1, Klf4, Tal1, Ezh2 and Stat4 were the DE TFs with
highest interactions in the constructed gene regulatory network
during early infection of the lung with tuberculosis. Rankings for
stress and betweenness revealed that Sfpi1, Stat4, Ezh2, Mybl2 and
Irf8 were the top 5 central genes in gene regulatory network. The
overall ranking of the DE TFs, according to the mean ranking of the
three parameters, identified Sfpi1, Stat4, Ezh2, Mybl2 and Irf8 as
the top regulatory TFs in the constructed gene regulatory
network.
 | Table IRanking of TFs based on analyses of
three centrality indexes. |
Table I
Ranking of TFs based on analyses of
three centrality indexes.
| Symbol | TF name | Centrality indexes
| Mean | Rank |
|---|
| Degree | Stress | Betweenness |
|---|
| Sfpi1 | Spleen
focus-forming virus proviral integration 1 | 1 | 1 | 1 | 1.00 | 1 |
| Stat4 | Stat4 | 5 | 2 | 2 | 3.00 | 2 |
| Ezh2 | Enhancer of zeste
homolog 2 | 4 | 3 | 4 | 3.66 | 3 |
| Mybl2 |
Myeloblastosis-related protein B | 7 | 4 | 3 | 4.66 | 4 |
| Irf8 | Interferon
regulatory factor 8 | 8 | 5 | 5 | 6.00 | 5 |
| Sox17 | Sex determining
region Y-box 17 | 6 | 7 | 8 | 7.00 | 6 |
| Eomes | Eomesodermin | 10 | 8 | 6 | 8.00 | 7 |
| Stat1 | Stat1 | 12 | 6 | 7 | 8.33 | 8 |
| Klf4 | Kruppel-like factor
4 | 2 | 13 | 13 | 9.33 | 9 |
| Nr3c1 | Nuclear receptor
subfamily 3 group C member 1 | 11 | 9 | 9 | 9.66 | 10 |
| Stat2 | Stat2 | 13 | 11 | 11 | 11.66 | 11 |
| Junb | Jun B
proto-oncogene | 15 | 10 | 10 | 11.66 | 11 |
| Tal1 | T-cell acute
lymphocytic leukemia 1 | 3 | 17 | 17 | 12.33 | 12 |
| Rel | Rel | 14 | 12 | 12 | 12.66 | 13 |
| Erg | V-ets avian
erythroblastosis virus E26 oncogene homolog | 9 | 16 | 16 | 13.66 | 14 |
| RelB | RelB | 15 | 14 | 14 | 14.33 | 15 |
| Tef | Thyrotroph
embryonic factor | 16 | 15 | 15 | 15.33 | 16 |
TFs involvement in the regulation of
immune system processes
In order to determine the most important TFs in the
regulation of immune system processes, gene regulatory network
ontology was used to identify the affected immune processes and
their regulators (Table II).
According to the P-values of affected processes in immune system
analysis, the results demonstrated that the positive regulation of
T cell activation and proliferation were the most enriched process.
In the constructed regulatory gene network the most important
regulatory DE TFs (based on number of regulatory interactions) for
T cell activation included Sfpi1, Stat4. Tal1, Rel and Klf4.
Positive regulation of T cell proliferation contained 26 DE genes
that were primarily regulated by Sfpi1, Stat4, Tal1, Rel, Irf8,
Stat1, Klf4 and Mybl2 DE TFs in the network. The regulators
identified as the top five TFs involved in the positive regulation
of T cell proliferation term, some of which have the same number of
targets, for example Stat4 and Tal1 both regulate nine DE genes in
this term. In addition to these processes, DE genes associated with
tuberculosis were investigated based on KEGG pathway analysis of
the gene regulatory network. A total of 38 DE genes involved in
tuberculosis were found to be present in the regulatory network
constructed in the preset study. In addition, Sfpi1, Stat4, Tal1,
Irf8, Klf4 and Stat1 were determined to be the top 5 DE TFs
involved in regulation of Tuberculosis-associated DE genes in early
lung immune response to tuberculosis. Overall, 4 DE TFs were
identified (Sfpi1, Stat4, Tal1 and Klf4) which were actively
involved in the regulation of the following three processes:
Positive regulation of T cell activation, positive regulation of T
cell proliferation and tuberculosis-associated gene expression.
 | Table IIEnriched immune system processes in
the gene regulatory network for early lung response to tuberculosis
infection. |
Table II
Enriched immune system processes in
the gene regulatory network for early lung response to tuberculosis
infection.
| No. | Gene ontology
term | No. genes | % | P-value | Genes |
|---|
| 1 | Positive regulation
of T cell activation | 42 | 38.53 | 1.85E-04 | Ada, Aif1, Blm,
Ccl2, Ccl5, Ccr2, Cd28, Cd4, Cd5, Cd83, Coro1a, H2-Aa, H2-Ab1,
H2-DMa, Hes1, Ifng, Igfbp2, Ikzf1, Il12a, Il12b, Il12rb1, Il1b,
Il2, Il21, Il2ra, Il6, Il7r, Irf1, Itgal, Jak3, Lck, Malt1,
Nckap1l, Pdcd1lg2, Prkcq, Ptprc, Sash3, Spn, Thy1, Tnfsf11, Vcam1,
Zap70 |
| 2 | Positive regulation
of T cell proliferation | 26 | 43.33 | 4.08E-04 | Aif1, Blm, Ccl5,
Ccr2, Cd28, Coro1a, Hes1, Ifng, Igfbp2, Il12a, Il12b, Il12rb1,
Il1b, Il2, Il21, Il2ra, Itgal, Jak3, Nckap1l, Pdcd1lg2, Prkcq,
Ptprc, Sash3, Spn, Vcam1, Zap70 |
| 3 | Mature B cell
differentiation | 7 | 77.77 | 7.58E-04 | Ada, Bcl3,
Malt1, Plcg2, Pou2f2, Ptk2b, Tnfaip3 |
| 4 | Positive regulation
of isotype switching to IgG isotypes | 6 | 85.71 | 8.13E-04 | Cd28, Cd40,
Ifng, Il2, Ptprc, Tbx21 |
| 5 | Antigen processing
and presentation of exogenous peptide antigen via major
histocompatability complex class II | 9 | 64.28 | 1.00E-03 | Fcgr2b, H2-Aa,
H2-Ab1, H2-DMa, H2-DMb2, H2-Eb1, H2-Oa, Ifi30, Unc93b1 |
| 6 | Regulation of
isotype switching to IgG isotypes | 7 | 70.00 | 2.00E-03 | Cd28, Cd40,
Ifng, Il2, Il27ra, Ptprc, Tbx21 |
| 7 | Regulation of T
cell activation | 55 | 32.73 | 2.10E-03 | Ada, Aif1, Blm,
Ccl2, Ccl5, Ccr2, Cd274, Cd28, Cd4, Cd5, Cd83, Coro1a, Ctla4,
Ctnnb1, H2-Aa, H2-Ab1, H2-DMa, H2-Oa, Hes1, Ido1, Ifng, Igfbp2,
Ikzf1, Il12a, Il12b, Il12rb1, Il1b, Il2, Il21, Il2ra, Il6, Il7r,
Irf1, Itgal, Jak3, Lag3, Lat, Lck, Malt1, Nckap1l, Nfkbid, Nrarp,
Pdcd1lg2, Pde5a, Prkcq, Ptpn22, Ptpn6, Ptprc, Sash3, Sit1, Spn,
Thy1, Tnfsf11, Vcam1, Zap70 |
| 8 | Positive regulation
of lymphocyte activation | 49 | 32.66 | 3.70E-03 | Ada, Aif1, Blm,
Ccl2, Ccl5, Ccr2, Cd28, Cd4, Cd40, Cd5, Cd83, Cdkn1a, Coro1a,
H2-Aa, H2-Ab1, H2-DMa, Hes1, Ifng, Igfbp2, Ikzf1, Il12a, Il12b,
Il12rb1, Il15ra, Il1b, Il2, Il21, Il2ra, Il6, Il7r, Inpp5d, Irf1,
Itgal, Jak3, Lck, Malt1, Myd88, Nckap1l, Pdcd1lg2, Prkcq, Ptprc,
Sash3, Spn, Tbx21, Thy1, Tnfrsf4, Tnfsf11, Vcam1, Zap70 |
| 9 | Regulation of
lymphocyte proliferation | 45 | 33.33 | 4.50E-03 | Ada, Aif1, Blm,
Ccl5, Ccr2, Cd274, Cd28, Cd40, Cdkn1a, Coro1a, Ctla4, Ctnnb1,
Fcgr2b, Hes1, Ido1, Ifng, Igfbp2, Ikzf3, Il10, Il12a, Il12b,
Il12rb1, Il1b, Il2, Il21, Il2ra, Inpp5d, Irf1, Itgal, Jak3, Lst1,
Myd88, Nckap1l, Pdcd1lg2, Pde5a, Prkcq, Ptpn22, Ptpn6, Ptprc,
Sash3, Sox11, Spn, Tnfrsf4, Vcam1, Zap70 |
| 10 | Cellular response
to interferon-γ | 15 | 44.11 | 6.10E-03 | Aif1, Arg2,
Ass1, Ccl2, Ccl5, Gbp1, Gbp2, Gbp3, H2-Ab1, Il12b, Il12rb1, Irf1,
Jak2, Nos2, Stat1 |
In conclusion, according to the results of the
analyses of TF involvement in protein complexes and active modules,
centrality and the regulation of most enriched processes, it was
revealed that the top DE TFs involved in the regulation of the
early lung response to tuberculosis were Stat4, Irf8, Sfpi1, Ezh2
and Klf4.
Discussion
The present study identified possible novel TFs
involved in the regulation of early lung immune response to
tuberculosis in mice and their roles. Numerous TFs and their
regulated processes in this constructed regulatory network were
revealed to overlap with known regulators and affected processes
during tuberculosis.
Stat4 has a crucial role in the development of the
protective response against M. tuberculosis infection in
mice (6). CD4+ T cells
deficient for Stat4 were reported to be unable to differentiate
into Th1 cells during tuberculosis (29,30).
T cell receptors (TCR) induce activation of CD4+ T cells
when they encounter antigens presented by antigen presenting cells
(29). Activated T cells produce
interleukin (IL)-2 and express IL-12 receptors in response to this
signal. IL-12 receptors are activated on encountering IL-12, which
is produced and secreted by macrophages and CD8α+
dendritic cells; in addition, the activated IL-12 receptor
subsequently activates Stat4, which results in the initiation of
Th1 differentiation from CD4+ T cells during M.
tuberculosis infection (29).
During tuberculosis infection, Stat4 upregulation following IL-12
stimulation in bronchoalveolar cells was reported to lead to
increased expression of interferon γ (Infγ) (31).
Previous studies have suggested that Irf8 was
critical for the differentiation of myeloid cells and defense
against intracellular microbes (8,10). A
study by Marquis et al (8)
reported that uncontrolled M. tuberculosis growth occurred
in the spleen, liver and lungs of BXH2 mice with a defective
IRF8R294c allele and induced premature mortality
(8). Irf8 is a crucial regulator
of the immune responses mediated by Th1 cells and is involved in
regulation of toll-like signaling (32). A previous study reported that high
overlap between the binding sites of Irf8 and Spfi1 was observed in
tuberculosis infection, as determined using ChIP-chip data analysis
(10). Sfpi1 was demonstrated to
be involved in the development of mature macrophages as well as B
and T cells (33). This regulator
primarily affected the efficiency of T cell progenitors fate
commitment and/or their differentiation (34). In addition Sfpi1 was reported to
have a role in the generation of cytokine expression patterns in T
helper 2-type (Th2) cells (35).
Ezh2 is a methyltransferase component of polycomb
repressive complex (PRC2) (36).
Low expression of Ezh2 was reported in mature T cells; however,
following antigen recognition through the TCR and activation of T
cells Ezh2 expression was rapidly increased (36). Expression of cytokines during the
development of Th1 and Th2 cells was predominantly regulated by
PRC2 members (37). For example,
Infγ expression was downregulated in Th1 cells following
Ezh2 mRNA knock-down (37).
IL-4 and IL-13 are Th2-specific cytokines, the expression levels of
which were reported to be down-regulated in Th1 cells through the
methyltransferase activity of Ezh2 (38,39).
Mice with T cell knock-out of the Klf4 gene
were used to investigate its roles in the development and
differentiation of T cell. Klf4 was reported to be highly expressed
in mature T cells; however, it was demonstrated that upon T cell
activation, Klf4 expression was downregulated (40). Another study on the T cell
activation network revealed the role of Klf4 in the regulation of
transcription in tuberculosis (41).
Collectively, the present study aimed to identify
novel TFs and dissect their roles in tge concept of gene regulatory
network during early lung immune response to tuberculosis. This
analysis led to identification of 17 DE TFs involved in regulation
of numerous immunological and biological processes, including T
cell activation, T cell proliferation and tuberculosis-associated
gene expression. Constructed network analysis revealed Stat4, Irf8,
Sfpi1, Ezh2 and Klf4 as master regulators of early lung response to
tuberculosis. Identification of these master TFs extend the current
understanding of the underlying molecular mechanisms and may be
useful as candidate novel targets for novel tuberculosis
therapies.
References
|
1
|
World Health Organisation: Tuberculosis
Fact Sheet. World Health Organization; Geneva Switzerland: 2012
|
|
2
|
Leveton C, Barnass S, Champion B, Lucas S,
De Souza B, Nicol M, Banerjee D and Rook G: T-cell-mediated
protection of mice against virulent Mycobacterium tuberculosis.
Infect Immun. 57:390–395. 1989.PubMed/NCBI
|
|
3
|
Kaufmann SH: Immunity to intracellular
bacteria. Annu Rev Immunol. 11:129–163. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schluger NW and Rom WN: The host immune
response to tuberculosis. Am J Respir Crit Care Med. 157(3 Pt 1):
679–691. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mogues T, Goodrich ME, Ryan L, LaCourse R
and North RJ: The relative importance of T cell subsets in immunity
and immunopathology of airborne Mycobacterium tuberculosis
infection in mice. J Exp Med. 193:271–280. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sugawara I, Yamada H and Mizuno S:
Relative importance of STAT4 in murine tuberculosis. J Med
Microbiol. 52:29–34. 2003. View Article : Google Scholar
|
|
7
|
Via LE, Tsytsykova AV, Rajsbaum R, Falvo
JV and Goldfeld AE: The transcription factor NFATp plays a key role
in susceptibility to TB in mice. PLoS One. 7:e414272012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Marquis JF, LaCourse R, Ryan L, North RJ
and Gros P: Disseminated and rapidly fatal tuberculosis in mice
bearing a defective allele at IFN regulatory factor 8. J Immunol.
182:3008–3015. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Keller C, Hoffmann R, Lang R, Brandau S,
Hermann C and Ehlers S: Genetically determined susceptibility to
tuberculosis in mice causally involves accelerated and enhanced and
recruitment of granulocytes. Infect Immun. 74:4295–4309. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Marquis JF, Kapoustina O, Langlais D,
Ruddy R, Dufour CR, Kim BH, MacMicking JD, Giguère V and Gros P:
Interferon regulatory factor 8 regulates pathways for antigen
presentation in myeloid cells and during tuberculosis. PLoS Genet.
7:e10020972011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Corn RA, Hunter C, Liou HC, Siebenlist U
and Boothby MR: Opposing roles for RelB and Bcl-3 in regulation of
T-box expressed in T cells, GATA-3 and Th effector differentiation.
J Immunol. 175:2102–2110. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Szabo SJ, Sullivan BM, Peng SL and
Glimcher LH: Molecular mechanisms regulating Th1 immune responses.
Annu Rev Immunol. 21:713–758. 2003. View Article : Google Scholar
|
|
13
|
Wang X, Wang H and Xie J: Genes and
regulatory networks involved in persistence of Mycobacterium
tuberculosis. Sci China Life Sci. 54:300–310. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sambarey A, Prashanthi K and Chandra N:
Mining large-scale response networks reveals ‘topmost activities’
in Mycobacterium tuberculosis infection. Sci Rep. 3:23022013.
View Article : Google Scholar
|
|
15
|
Kumar D, Nath L, Kamal MA, Varshney A,
Jain A, Singh S and Rao KV: Genome-wide analysis of the host
intracellular network that regulates survival of Mycobacterium
tuberculosis. Cell. 140:731–743. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kang DD, Lin Y, Moreno JR, Randall TD and
Khader SA: Profiling early lung immune responses in the mouse model
of tuberculosis. PLoS One. 6:e161612011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Blazejczyk M, Miron M and Nadon R:
FlexArray: Statistical data analysis software for gene expression
microarrays, made with life scientists in mind. McGill University
and Génome Québec Innovation Centre; Montréal, QC: 2007
|
|
19
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lachmann A, Xu H, Krishnan J, Berger SI,
Mazloom AR and Ma’ayan A: ChEA: transcription factor regulation
inferred from integrating genome-wide ChIP-X experiments.
Bioinformatics. 26:2438–2444. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Essaghir A, Toffalini F, Knoops L, Kallin
A, van Helden J and Demoulin JB: Transcription factor regulation
can be accurately predicted from the presence of target gene
signatures in microarray gene expression data. Nucleic Acids Res.
38:e1202010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Stark C, Breitkreutz BJ, Reguly T, Boucher
L, Breitkreutz A and Tyers M: BioGRID: a general repository for
interaction datasets. Nucleic Acids Res. 34(Database Issue):
D535–539. 2006. View Article : Google Scholar :
|
|
23
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41(Database Issue): D808–D815. 2013. View Article : Google Scholar :
|
|
24
|
Saito R, Smoot ME, Ono K, Ruscheinski J,
Wang PL, Lotia S, Pico AR, Bader GD and Ideker T: A travel guide to
Cytoscape plugins. Nat Methods. 9:1069–1076. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bindea G, Mlcenik B, Hackl H, Charoentong
P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z and
Galon J: ClueGO: a Cytoscape plug-in to decipher functionally
grouped gene ontology and pathway annotation networks.
Bioinformatics. 25:1091–1093. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Scardoni G, Petterlini M and Laudanna C:
Analyzing biological network parameters with CentiScaPe.
Bioinformatics. 25:2857–2859. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ideker T, Ozier O, Schwikowski B and
Siegel AF: Discovering regulatory and signalling circuits in
molecular interaction networks. Bioinformatics. 18(Suppl 1):
233–240. 2002. View Article : Google Scholar
|
|
29
|
Rengarajan J, Szabo SJ and Glimcher LH:
Transcriptional regulation of Th1/Th2 polarization. Immunol Today.
21:479–483. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wurster AL, Tanaka T and Grusby MJ: The
biology of Stat4 and Stat6. Oncogene. 19:2577–2584. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Raju B, Hoshino Y, Belitskaya-Lévy I,
Dawson R, Ress S, Gold JA, Condos R, Pine R, Brown S, Nolan A, et
al: Gene expression profiles of bronchoalveolar cells in pulmonary
TB. Tuberculosis (Edinb). 88:39–51. 2008. View Article : Google Scholar
|
|
32
|
Zhao J, Kong HJ, Li H, Huang B, Yang M,
Zhu C, Bogunovic M, Zheng F, Mayer L, Ozato K, et al:
IRF-8/interferon (IFN) consensus sequence-binding protein is
involved in Toll-like receptor (TLR) signaling and contributes to
the cross-talk between TLR and IFN-gamma signaling pathways. J Biol
Chem. 281:10073–10080. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
McKercher SR, Torbett BE, Anderson KL,
Henkel GW, Vestal DJ, Baribault H, Klemsz M, Feeney AJ, Wu GE,
Paige CJ and Maki RA: Targeted disruption of the PU. 1 gene results
in multiple hematopoietic abnormalities. EMBO J. 15:5647–5658.
1996.PubMed/NCBI
|
|
34
|
Spain LM, Guerriero A, Kunjibettu S and
Scott EW: T cell development in PU. 1-deficient mice. J Immunol.
163:2681–2687. 1999.PubMed/NCBI
|
|
35
|
Chang HC, Han L, Jabeen R, Carotta S, Nutt
SL and Kaplan MH: PU. 1 regulates TCR expression by modulating
GATA-3 activity. J Immunol. 183:4887–4894. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
He S, Tong Q, Bishop DK and Zhang Y:
Histone methyltrans-ferase and histone methylation in inflammatory
T-cell responses. Immunotherapy. 5:989–1004. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jacob E, Hod-Dvorai R, Ben-Mordechai OL,
Boyko Y and Avni O: Dual function of polycomb group proteins in
differentiated murine T helper (CD4+) cells. J Mol Signal. 6:52011.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Koyanagi M, Baguet A, Martens J, Margueron
R, Jenuwein T and Bix M: EZH2 and histone 3 trimethyl lysine 27
associated with II4 and II13 gene silencing in Th1 cells. J Biol
Chem. 280:31470–31477. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lee GR, Kim ST, Spilianakis CG, Fields PE
and Flavell RA: T helper cell differentiation: regulation by cis
elements and epigenetics. Immunity. 24:369–379. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
An J, Golech S, Klaewsongkram J, Zhang Y,
Subedi K, Huston GE, Wood WH III, Wersto RP, Becker KG, Swain SL
and Weng N: Krüppel-like factor 4 (KLF4) directly regulates
proliferation in thymocyte development and IL-17 expression during
Th17 differentiation. FASEB J. 25:3634–3645. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Subbian S, O’Brien P, Kushner NL, Yang G,
Tsenova L, Peixoto B, Bandyopadhyay N, Bader JS, Karakousis PC,
Fallows D and Kaplan G: Molecular immunologic correlates of
spontaneous latency in a rabbit model of pulmonary tuberculosis.
Cell Commun Signal. 11:162013. View Article : Google Scholar : PubMed/NCBI
|