Introduction
Streptococcus suis type 2 (S. suis 2)
is an important zoonotic pathogen with a global distribution. This
strain can cause acute septicemia, meningitis, arthritis and
endocarditis with potentially fatal outcomes. Infections can lead
to severe toxic symptoms with high mortality in humans (1). Since this bacterium can be
transmitted in a variety of ways, it is a serious threat to the pig
industry and its workers. Therefore, this strain has aroused
significant concern in this community due to economic losses in the
meat industry and threats to human health (2,3).
Carbon catabolite repression (CCR) is important for
microbial catabolism, and catabolite control protein A (ccpA) is a
key regulatory factor for CCR and global carbon metabolism.
CcpA-mediated catabolite repression is necessary for the adaptation
of Gram-positive bacteria to changes in the environment (4). It is currently thought that
ccpA-mediated CCR is present in a variety of Gram-positive bacteria
with low GC content. CcpA regulates gene expression in important
metabolic pathways through specific functional domains and impacts
bacterial virulence through the metabolic regulatory functions of
ccpA (5,6).
As an important member of the
LacI/GalR transcriptional regulator family, ccpA
controls numerous metabolic processes in Gram-positive bacteria and
has important roles in the biofilm formation by S. suis
(7). Previous studies have found
that the envelope of S. suis was affected by the deletion of
ccpA, creating strains similar to those without an envelope.
Furthermore, the anti-phagocyte capabilities of strains without the
ccpA protein were significantly reduced as compared with those of
parental strains, indicating that ccpA has an important regulatory
role in cellular metabolism and structural features of S.
suis (8). Since ccpA has
pleiotropic regulatory functions, ccpA knockout affects numerous
physiological processes. Therefore, it is necessary to study the
association between ccpA structure and function. Investigating the
effect of ccpA on protein regulation is important for understanding
this protein's functional domains and active sites.
In the present study, isobaric tag for relative and
absolute quantification (iTRAQ)-based proteomics technology was
used to analyze the regulatory effect of the ccpA gene on the
biological functions of various genes and to provide gene
expression levels for further study of this gene in S. suis
pathogenicity.
Materials and methods
Strains and culture methods
S. suis and the ccpA-mutant strains from our
laboratory (9) were cultured in
Todd-Hewitt broth (Oxoid Ltd., Basingstoke, UK) at 37°C under 5%
CO2. Cells in the exponential growth phase were
used.
Sample preparation
After 200 µg cells were collected by
centrifugation, SDT lysis buffer [4% SDS, 100 mM Tris-HCl pH 8.0,
100 mM dithiothreitol (DTT)] purchased from Bio-Rad Laboratories,
Inc. (Hercules, CA, USA) was added and samples were then mixed by
vortexing and placed in a boiling water bath for 10 min. The
samples were then ultrasonically disrupted and heated again for 5
min in a boiling water bath. The supernatant was removed and
protein quantification was performed using the bicinchoninic acid
method.
Enzyme digestion and peptide
labeling
From each sample, 200 µg protein was taken
and DTT was added to a final concentration of 100 mM. The mixtures
were heated in a boiling water bath for 3 min and then cooled to
room temperature. The products were combined with 200 µl UA
buffer (Bio-Rad Laboratories, Inc.; 8 M Urea and 150 mM Tris HCl,
pH 8.0) and mixed. The mixtures were transferred into
ultrafiltration centrifuge tubes with a 10-kDa cutoff point and
centrifuged at 14,000 xg for 15 min. The precipitates were
re-suspended in 200 µl UA buffer and centrifuged at 14,000
xg for 15 min. The filtrates were discarded and the precipitates
were re-suspended in 100 µl Iodoacetamide (Bio-Rad
Laboratories, Inc.; 50 mM in UA) in the dark for 30 min and then
centrifuged at 14,000 xg for 10 min. The precipitates were
re-suspended in 100 µl UA buffer and centrifuged at 14,000
xg for 10 min. This process was repeated twice. The precipitates
were re-suspended in 100 µl dissolution buffer and
centrifuged at 14,000 x g for 10 min, which was also repeated
twice. The precipitates were re-suspended in 40 µl trypsin
buffer (Promega, Madison, WI, USA; 5 µg trypsin in 40
µl dissolution buffer), agitated at 600 rpm for 1 min and
incubated at 37°C for 16-18 h. The products were centrifuged at
14,000 xg for 10 min using fresh collection tubes, the filtrates
were collected, and peptide quantification was performed by
measuring the optical density at 280 nm. After enzymolysis, peptide
labeling was performed using an iTRAQ 8-plex Multiplex kit (Ab
Sciex, Framingham, MA, USA) according to the manufacturer's
instructions. For native S. suis, iTRAQ reagents 113, 114
and 115 were used, and for mutant strains, reagents 116, 117 and
118 were used.
Peptide fractionation with strong cation
exchange (SCX) chromatography
SCX chromatography was performed using an AKTA
Purifier 100 (GE Healthcare, Little Chalfont, UK) with a 4.6×100 mm
polysulfoethyl column (5 µm; 200 Å) (PolyLC Inc., Columbia,
MD, USA). Buffer A contained 10 mM KH2PO4 and
25% acetonitrile (ACN; pH 3.0; Sigma-Aldrich, St. Louis, MO, USA).
Buffer B contained 10 mM KH2PO4 pH 3.0, 500
mM KCl and 25% ACN. Samples were collected and lyophilized prior to
desalination in a C18 Cartridge (Sigma-Aldrich).
Mass spectrometry (MS)
An Easy nLC system (Thermo Fisher Scientific,
Waltham, MA, USA) run at a nanoliter flow rate was used for liquid
separation of each sample. Buffer A was 0.1% formic acid in water
and Buffer B was 0.1% formic acid in 84% ACN. Chromatographic
columns were equilibrated with 95% Buffer A. Samples were loaded
into a Thermo scientific EASY-Spray column (2 cmx100 µm 5
µm-C18) using an auto sampler and then separated on a Thermo
scientific EASY column (75 µmx100 mm 3 µm-C18) at a
flow rate of 250 nl/min. MS was performed using a Q-Ex active mass
spectrometer (Thermo Fisher Scientific) with an analysis time of
120 min, a positive ion detection method, a precursor ion scanning
range of 300–1,800 m/z, a first-order mass spectrum resolution of
70,000 at m/z 200, an AGC target of 3e6, a first order maximum
injection time (IT) of 10 msec, one scan range and a dynamic
exclusion of 40.0 sec. The mass-charge ratios of the peptides and
peptide fragments were obtained as follows: Ten-fragments spectra
(MS2 scan) were collected after each full scan, the MS2 Activation
Type was HCD, the isolation window was 2 m/z, the second-order MS
resolution was 17,500 at m/z 200 with one microscan. The
second-order Maximum IT was 60 msec, the normalized collision
energy was 30 eV and the under fill ratio was 0.1%.
Data analysis
The raw data of the MS analysis were derived from
RAW files. Database searches and quantitative analyses were
performed using the software Mascot 2.2 of Proteome Discoverer 1.4
(Thermo Fisher Scientific). For protein-abundance ratios measured
using iTRAQ, a 1.2-fold change was set as the threshold and a
two-tailed P-value <0.05 to identify significant changes. The
database was downloaded from the National Center of Biotechnology
Information (NCBI) on 2013-08-09, and the
NCBI_Streptococcus_suis. Fasta results contained 89,409
sequences (http://www.ncbi.nlm.nih.gov/protein?term=txid1307
[Organism]). Mascot 2.2 was used for the library search. The
localized sequence alignment software NCBI Basic Local Alignment
Search Tool (BLAST 2.2.28+-win32. ext; http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to
perform sequence alignment between the identified proteins and
protein sequences in the NCBInr database. Using the similarity
principle, functional information from homologous proteins was used
to create protein function annotation. The mapping function of
BLAST2GO (https://www.blast2go.com/; version
2.7.0) was used to extract gene ontology (GO) function entries
correlated with the aligned sequences for all differentially
expressed proteins. The Go Slim was annotated for the function of
the target protein by GO for generic.obo. The KEGG (Kyoto
Encyclopedia of Genes and Genomes) Automatic Annotation Server
(KAAS; http://www.genome.jp/tools/kaas/) was used to align
target-protein and Streptococcal sequences in the KEGG GENES
database. KEGG Orthology (KO) identifiers of homologous/similar
proteins were assigned to the relevant KEGG pathway. Carbon
metabolism pathway analysis was performed with the Pathway Builder
Tool 2.0 (Protein Lounge, San Diego, CA, USA).
Statistical analyses
All statistical analyses were performed using the
Student's t-test by Perseus 1.3 (http://141.61.102.17/perseus_doku/doku.php?id=start).
Differences with a P-value of 0.05 or less were considered
statistically significant.
Results
iTRAQ analysis results
A total of 1,167 proteins were identified from S.
suis and ccpA mutant strains from the original mass
spectrometry data, 55 of which were differentially expressed.
The mapping function of Blast2GO (Version 2.7.0) was
used to extract the GO function entries correlated with the aligned
sequences for all of the differentially expressed proteins. As a
result, 269 GO function entries associated with the sequences of 45
differentially expressed proteins (81.8%) were extracted. In the
functional annotation process, the sequences of a total of 37
differentially expressed proteins were annotated with 87 GO
function entries. The final statistical results after supplementary
annotation showed that a total of 47 proteins were annotated with
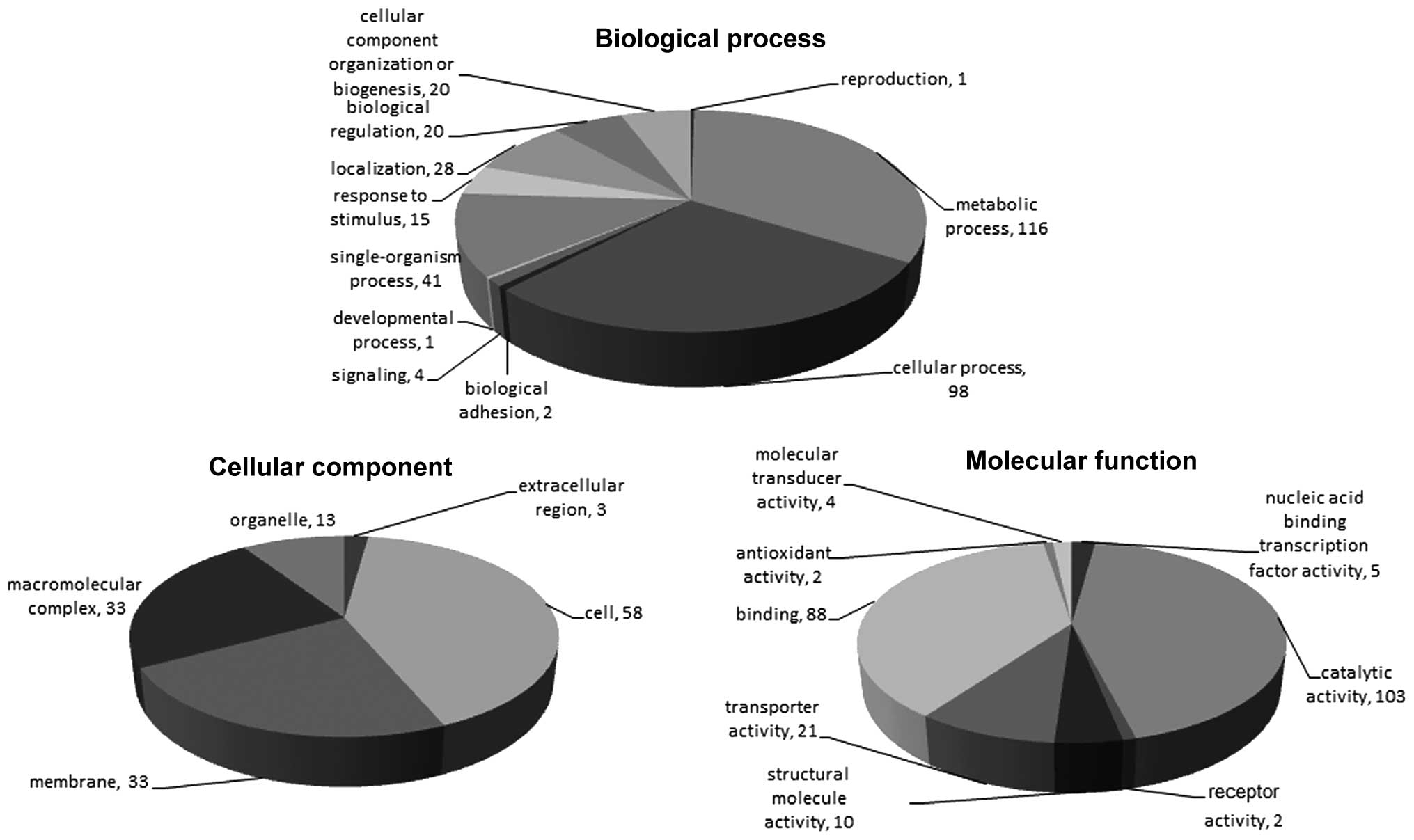
188 GO function entries (Fig.
1).
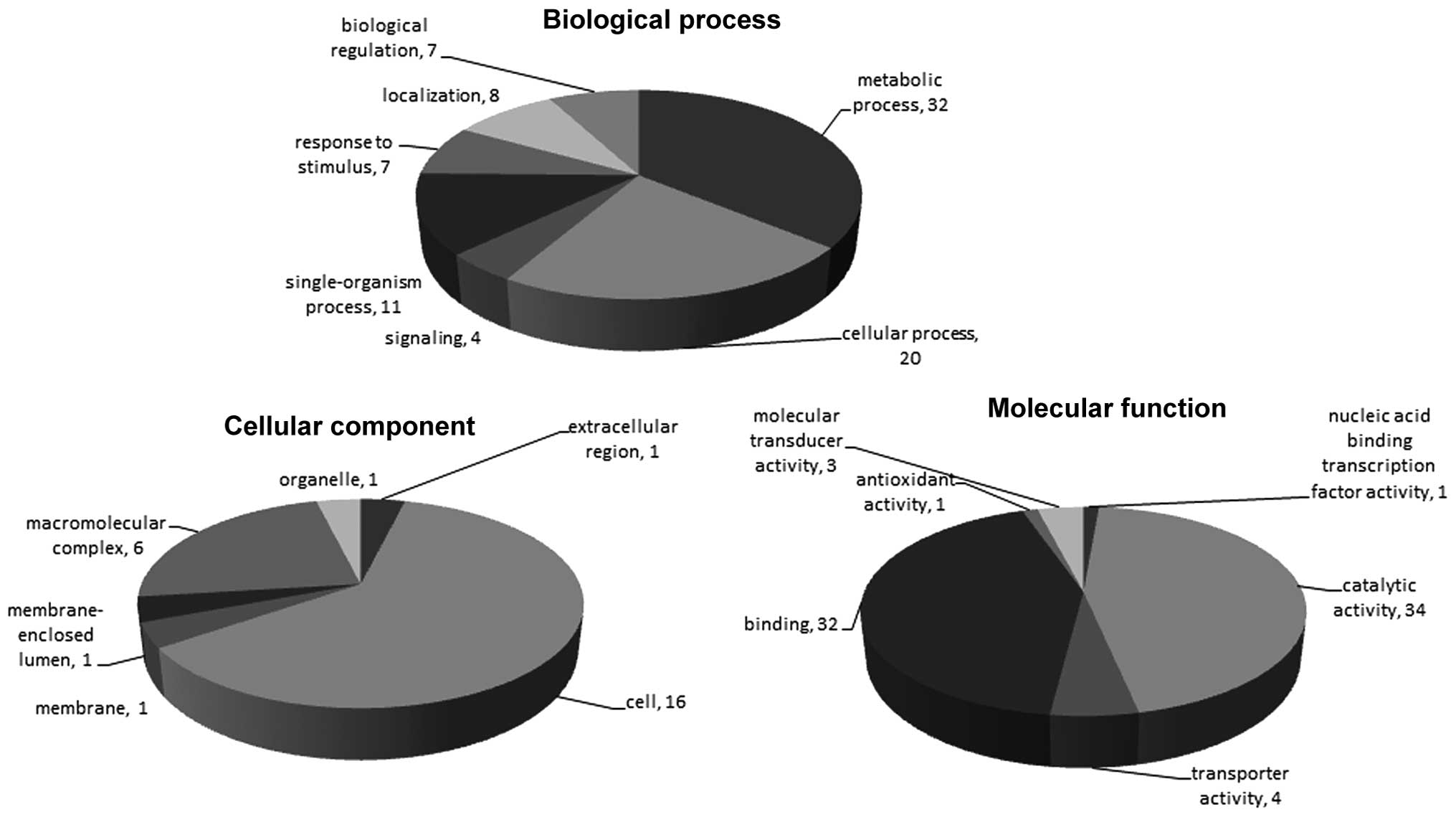
GO for generic.obo was used to annotate the
functions of the target proteins, and a total of 47 protein
sequences were annotated by 194 GO Slim function entries (Fig. 2).
KAAS was used to align target-protein and
Streptococcal sequences from the KEGG GENES database, and the KO
identifiers of homologous/similar proteins were assigned to the
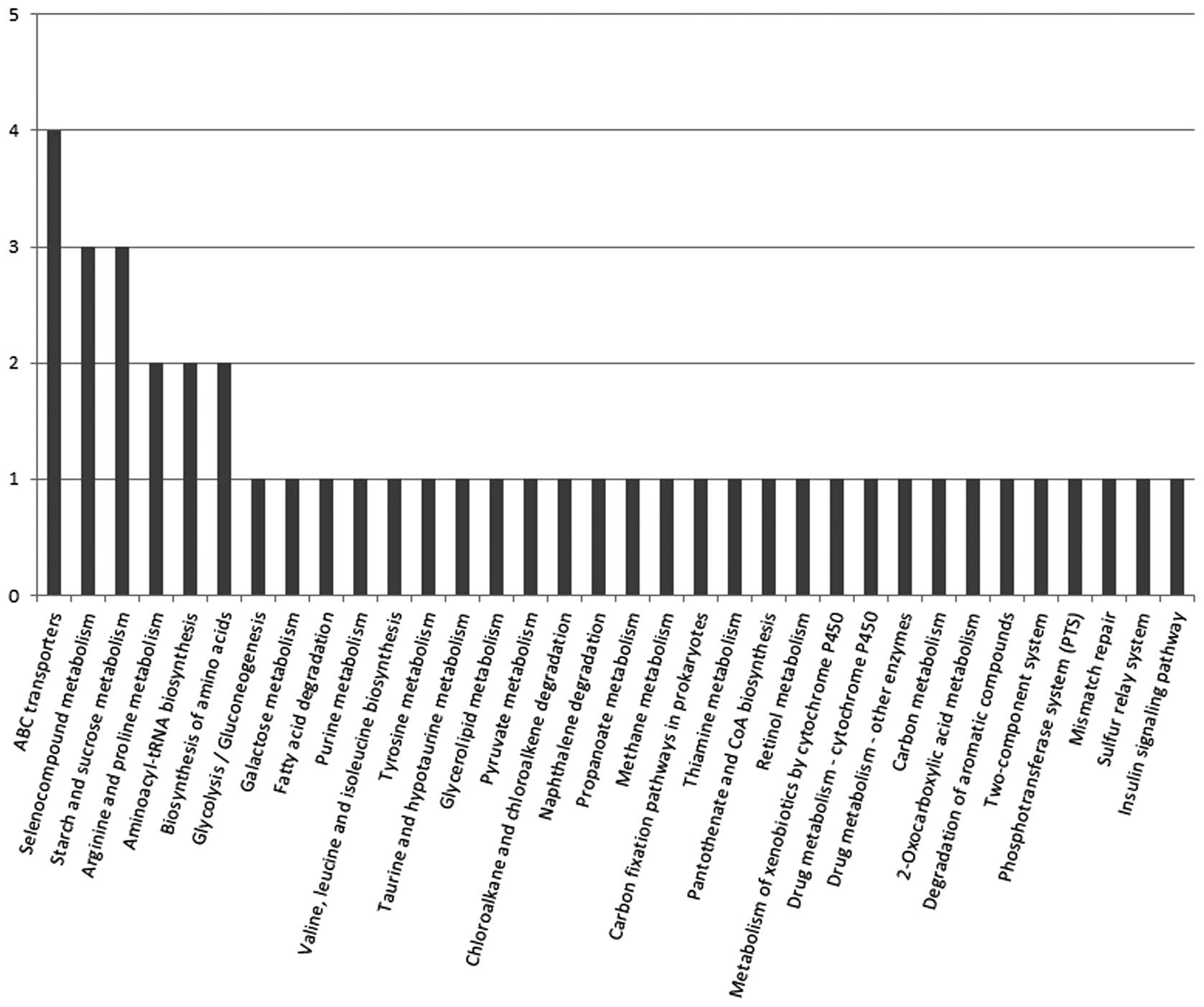
relevant KEGG pathways. A total of 34 KEGG signal/metabolic
pathways associated with the sequences of 21 differential proteins
were extracted (Fig. 3).
Regulatory function of proteins or
enzymes in carbon metabolic pathways
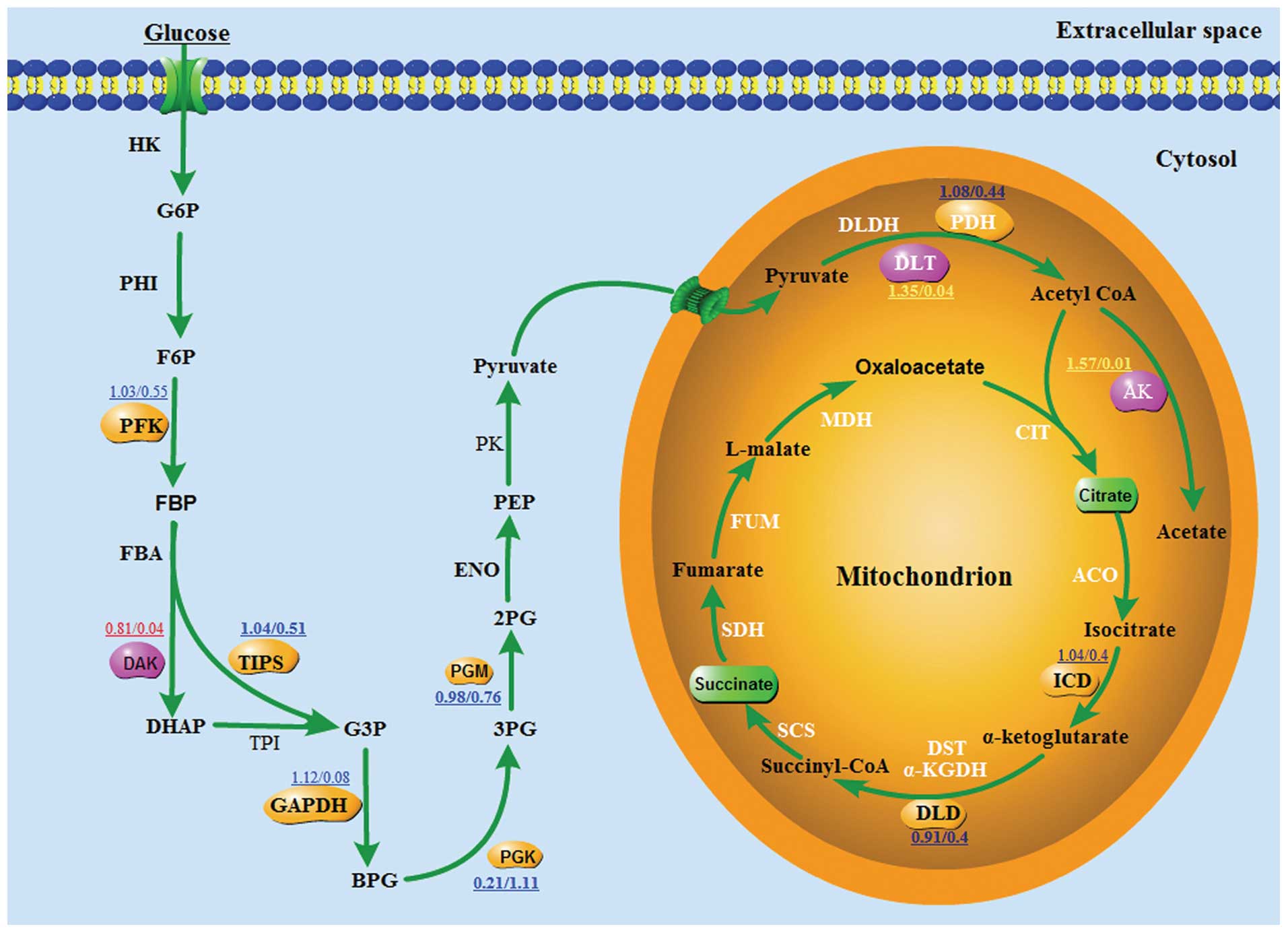
The data of the metabolic pathway analyses indicated
that dihydrolipoyl transacetylase and dihydroxyacetone kinase and
acetate kinase exhibited differential effects on the carbon
metabolism between the native S. suis type 2 and ccpA mutant
strains. These differential effects on the carbon metabolic
pathways between the native and the ccpA mutant strain indicate
that ccpA is directly or indirectly involved in the carbon
metabolism of bacteria. In particular, ccpA was indicated to have
certain effects on the cellular metabolism of bacteria (Fig. 4).
Discussion
Two large-scale outbreaks of S. suis
infection occurred in Jiangsu Province and Sichuan Province of
China in 1998 and 2005, respectively. These two public health
events attracted worldwide attention and became the focus of
domestic and international studies (10,11).
Although the exact pathological mechanisms of Streptococcus
suis has remained elusive, a number of potential virulence
factors have been uncovered (12).
CcpA is not only involved in the regulation of
carbon and nitrogen metabolism in bacteria, but also in specific
physiological processes, including sporification, solvent
production and virulence-gene expression in various microorganisms
(13). In pathogens such as
Staphylococcus aureus, ccpA directly regulates and activates
the expression of virulence genes (12), while in Clostridium
perfringens, it may regulate sporification and the expression
of virulence genes, thereby affecting cellular function (14).
This gene may regulate the metabolic processes of
bacteria at different levels and affect bacterial virulence,
thereby exerting differential effects to cause pathological changes
in infected organisms (6). CcpA
deletion can reduce the activity of glycolytic enzymes, including
enolase and other metabolic enzymes. In infected hosts, certain
enzymes are associated with pathogenic virulence and specific
metabolic requirements. Therefore, the virulence of a ccpA-deletion
strain is obviously lower than that of a wild-type parent strain
(15,16). However, ccpA directly activates the
expression of virulence genes in certain pathogens, such as
Streptococcus pyogenes (17, 18). Thus, it is necessary to conduct
in-depth studies of the structure, metabolic regulation and
immunogenicity of ccpA, a regulator with multiple effects.
In the present study, differentially expressed
proteins or peptides that primarily impact bacterial functions,
including catalytic activity, metabolic processes, ion binding and
nucleotide binding, were identified using iTRAQ technology.
Bioinformatic analysis showed that ccpA markedly influenced protein
metabolism in the bacterium as well as the processes of growth,
metabolism and infection. Metabolism, signal transduction and
pathway regulation for these processes were similar to the
observations of the metabolomics analysis of the ccpA gene of S.
suis performed in the present study. The metabolomics analysis
of the present study found that ccpA mutation caused changes in
metabolites, including glutamate, guanine, uridine and inosine,
which are primarily involved in the metabolism of amino acids,
nucleic acids, fats and certain small-molecule organic acids. These
results have been verified by a further metabolomics study by our
group (19). A comparison of the
data from the two studies indicated that ccpA affected overall
pathogenicity, metabolites and virulence through different pathways
(9,19). The altered functions of various
metabolic proteins, including peptidoglycan glycosyltransferase,
enolase, sortase, GAPDH, 6-phosphogluconate dehydrogenase and
glutamine synthetase may have affected the virulence of the
bacteria, which was also reported by a previous study (20).
These results indicated that ccpA serves similar
roles in S. suis and Streptococcus pneumoniae. CcpA
regulates carbohydrate metabolism in bacteria by various
mechanisms, thereby affecting the function and virulence of
bacteria (8,21). The regulation of bacterial proteins
by ccpA influences transcriptional regulation and signaling-pathway
activation via different pathways, thereby impacting the
phagocytosis of bacteria and bacterial virulence (22,23).
The differentially expressed proteins identified in the present
study included proteins associated with metabolism and molecular
transport. This demonstrated that ccpA regulates the expression of
intracellular products and various associated proteins by affecting
metabolism, molecular transport and transcription, hence
influencing the metabolic processes and virulence of bacteria.
These changes were similar to regulatory processes in Gram-positive
bacteria, including Streptococcus pneumonia and
Clostridium difficile (24,25).
In the present study, iTRAQ-based proteomic
technology has proven a useful tool for the functional study of the
ccpA protein in S. suis. It was shown that ccpA influences
important metabolic and regulatory pathways of this bacterium.
These effects are mainly associated with glucose metabolism, amino
acid metabolism, nucleic acid synthesis and adenosine triphosphate
binding cassette transporters. The study of the specific impact of
ccpA on metabolism, the mechanism of regulatory function through
functional domain transformation and the transformation from
multiple, complex effects to a single, simple effect are the focus
of future research.
Acknowledgments
The iTRAQ experiments and analysis were performed by
Shanghai Applied Protein Technology (Shanghai, China). The
metabolic pathway analysis was performed by Shanghai Omics Space
Biotech Co., Ltd (Shanghai, China). The present study was funded by
the National Natural Science Foundation of China (grant nos.
31101790/C1803 and 31172340/C180501).
References
|
1
|
Arends JP and Zanen HC: Meningitis caused
by Streptococcus suis in humans. Rev Infect Dis. 10:131–137. 1988.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gottschalk M, Xu J, Calzas C and Segura M:
Streptococcus suis: A new emerging or an old neglected zoonotic
pathogen? Future Microbiol. 5:371–391. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang J, Zhu J, Ren H, et al: Rapid visual
detection of highly pathogenic Streptococcus suis serotype 2
isolates by use of loop-mediated isothermal amplification. J Clin
Microbiol. 51:3250–3256. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kaufman GE and Yother J: CcpA-dependent
and-independent control of beta-galactosidase expression in
Streptococcus pneumoniae occurs via regulation of an upstream
phosphotransferase system-encoding operon. J Bacteriol.
189:5183–5192. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hondorp ER and McIver KS: The Mga
virulence regulon: Infection where the grass is greener. Mol
Microbiol. 66:1056–1065. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Poncet S, Milohanic E, Mazé A, et al:
Correlations between carbon metabolism and virulence in bacteria.
Contrib Microbiol. 16:88–102. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sumby P, Barbian KD, Gardner DJ, et al:
Extracellular deoxyribonuclease made by group A Streptococcus
assists pathogenesis by enhancing evasion of the innate immune
response. Proc Natl Acad Sci USA. 102:1679–1684. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Willenborg J, Fulde M, de Greeff A, et al:
Role of glucose and CcpA in capsule expression and virulence of
Streptococcus suis. Microbiology. 157:1823–1833. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lang X, Wan Z, Pan Y and Wang X and Wang
X, Bu Z, Qian J, Zeng H and Wang X: Investigation into the role of
catabolite control protein A in the metabolic regulation of
Streptococcus suis serotype 2 using gene expression profile
analysis. Exp Ther Med. 10:127–132. 2015.PubMed/NCBI
|
|
10
|
Tang J, Wang C, Feng Y, et al:
Streptococcal toxic shock syndrome caused by Streptococcus suis
serotype 2. PLoS Med. 3:e1512006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sriskandan S and Slater JD: Invasive
disease and toxic shock due to zoonotic Streptococcus suis: An
emerging infection in the East? PLoS Med. 3:e1872006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Seidl K, Stucki M, Ruegg M, et al:
Staphylococcus aureus CcpA affects virulence determinant production
and antibiotic resistance. Antimicrob Agents Chemother.
50:1183–1194. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Warner JB and Lolkema JS: CcpA-dependent
carbon catabolite repression in bacteria. Microbiol Mol Biol Rev.
67:475–490. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Varga J, Stirewalt VL and Melville SB: The
CcpA protein is necessary for efficient sporulation and enterotoxin
gene (cpe) regulation in Clostridium perfringens. J Bacteriol.
186:5221–5229. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang Y, Dang Y, Wang X, et al: Comparative
proteomic analyses of Streptococcus suis serotype 2 cell
wall-associated proteins. Curr Microbiol. 62:578–588. 2011.
View Article : Google Scholar
|
|
16
|
Titgemeyer F and Hillen W: Global control
of sugar metabolism: A gram-positive solution. Antonie Van
Leeuwenhoek. 82:59–71. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Deutscher J, Herro R, Bourand A, Mijakovic
I and Poncet S: P-Ser-HPr-a link between carbon metabolism and the
virulence of some pathogenic bacteria. Biochim Biophys Acta.
1754:118–125. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cho K and Caparon M: Patterns of virulence
gene expression differ between biofilm and tissue communities of
Streptococcus pyogenes. Mol Microbiol. 57:1545–1556. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lang X, Wan Z, Bu Z and Wang X and Wang X,
Zhu L, Wan J, Sun Y and Wang X: Catabolite control protein A is an
important regulator of metabolism in Streptococcus suis type 2.
Biomed Rep. 2:709–712. 2014.PubMed/NCBI
|
|
20
|
Meijerink M, Ferrando ML, Lammers G,
Taverne N, Smith HE and Wells JM: Immunomodulatory effects of
Streptococcus suis capsule type on human dendritic cell responses,
phagocytosis and intracellular survival. PLoS One. 7:e358492012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Iyer R, Baliga NS and Camilli A:
Catabolite control protein A (CcpA) contributes to virulence and
regulation of sugar metabolism in Streptococcus pneumoniae. J
Bacteriol. 187:8340–8349. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Seshasayee AS, Bertone P, Fraser GM and
Luscombe NM: Transcriptional regulatory networks in bacteria: from
input signals to output responses. Curr Opin Microbiol. 9:511–519.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Segura M, Gottschalk M and Olivier M:
Encapsulated Streptococcus suis inhibits activation of signaling
pathways involved in phagocytosis. Infect Immun. 72:5322–5330.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kaufman GE and Yother J: CcpA-dependent
and -independent control of beta-galactosidase expression in
Streptococcus pneumoniae occurs via regulation of an upstream
phosphotransferase system-encoding operon. J Bacteriol.
189:5183–5192. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Karlsson S, Burman LG and Åkerlund T:
Induction of toxins in Clostridium difficile is associated with
dramatic changes of its metabolism. Microbiology. 154:3430–3436.
2008. View Article : Google Scholar : PubMed/NCBI
|