Introduction
Intrahepatic cholangiocarcinoma (ICC) is a rare type
of primary liver cancer that originates from cholangiocytes, and is
ranked as one of the top five causes of cancer-associated
mortality. Furthermore, the 5-year survival rate of patients with
ICC is poor, predominantly due to high recurrence and metastasis
(1,2); therefore, further investigation
regarding potential diagnostic and therapeutic targets may aid in
reducing the mortality rate of ICC.
Vascular endothelial growth factor (VEGF)-C is an
important member of the VEGF family, which has been reported to be
associated with the progression of ICC (3). High expression of VEGF-C is
significantly correlated with vascular invasion, lymph node
metastasis, and the presence of positive surgical margins, as well
as poor survival rate (4).
However, the regulatory mechanisms of VEGF-C in ICC remain to be
elucidated.
MicroRNAs (miRs) are a class of non-coding RNAs,
18–25 nucleotides in length. It has been well established that miRs
directly bind to the 3′-untranslated region (UTR) of their target
mRNAs, leading to mRNA degradation or inhibition of protein
translation (5). By regulating the
expression of their target genes, miRs are involved in the
regulation of cell survival, proliferation, differentiation and
migration (6). Furthermore, miRs
have important roles in the development and progression of various
types of human cancer, including ICC (7,8). A
previous study demonstrated that low expression of miR-124,
regulated by the hepatitis C virus core protein, was able to
enhance the migration and invasion of ICC cells (9). In addition, miR-200c has been shown
to activate epithelial-mesenchymal transition via direct inhibition
of neural cell adhesion molecule 1 in ICC (10).
Reduced expression of miR-101 has been detected in
numerous types of human cancer, and is associated with the invasion
and progression of malignancies (11). Zhang et al (12) demonstrated that miR-101 was
markedly downregulated in cholangiocarcinoma specimens and cell
lines, as compared with in non-cancerous biliary epithelial cells,
suggesting that downregulation of miR-101 may be involved in the
development of cholangiocarcinoma. However, whether miR-101
participates in the progression of ICC, as well as the underlying
molecular mechanism, remain to be elucidated.
The present study aimed to explore the molecular
mechanisms by which miR-101 and VEGF-C mediate the migration and
invasion of ICC cells.
Materials and methods
Tissue specimen collection
The present study was approved by the Ethical
Committee of The Third Xiangya Hospital of Central South University
(Changsha, China). A total of 15 ICC tissue samples and matched
adjacent normal tissue samples were obtained from the Department of
Hepatobiliary and Pancreatic Surgery, the Third Xiangya Hospital of
Central South University. The patients providing the samples
consisted of 11 males and 4 females with an average age 53.5 years
old (41–64 years old). ICC was diagnosed by doctors from the
Department of Pathology and tissues were obtained by surgical
resection. The patients were recruited between June 2011 and
January 2012. All patients provided their written consent for the
present study. All tissues were immediately snap-frozen in liquid
nitrogen following surgical removal, and stored at −70°C until
further use.
Cell culture
The ICC-9810 human ICC cell line and normal human
intrahepatic biliary epithelial cells (HIBEC) were obtained from
the Institute of Cell Biology at the Chinese Academy of Sciences
(Shanghai, China). The cells were cultured in Dulbecco's modified
Eagle's medium (DMEM; Invitrogen Life Technologies, Carlsbad, CA,
USA) supplemented with 10% fetal bovine serum (FBS; Invitrogen Life
Technologies) at 37°C in a humidified incubator containing 5%
CO2.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
For mRNA detection, total RNA was extracted from
cells and tissues using TRIzol® reagent (Life
Technologies, Carlsbad, CA, USA), according to the manufacturer's
instructions. The tissues were homogenized with liquid nitrogen in
a mortar (Ziyi Company, Shanghai, China). RevertAid First-Strand
cDNA Synthesis kit (Fermentas, Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) was used to reverse transcribe RNA into cDNA (1
µg used). Subsequently, iQ™ SYBR Green Supermix (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) was used to perform qPCR
with the ABI 7500 thermocycler (Applied Biosystems Life
Technologies, Foster City, CA, USA). The cycling conditions were as
follows: 95°C for 5 min, followed by 40 cycles of 95°C for 30 sec
and 60°C for 30 sec. The sequences of the specific primers from
Shanghai Shenggong Co., Ltd. (Shanghai, China) were as follows:
VEGF-C, forward 5′-GGCTGGCAACATAACAGAGAA-3′, reverse
5′-CCCCACATCTATACACACCTCC-3′; and GAPDH, forward
5′-ACAACTTTGGTATCGTGGAAGG-3′, and reverse
5′-GCCATCACGCCACAGTTTC-3′. GAPDH was used as an internal reference.
Relative expression was analyzed using the 2−ΔΔCt
method.
For miR detection, total RNA was extracted using
TRIzol® reagent, according to the manufacturer's
instructions. miRNA Reverse Transcription kit (Life Technologies)
was used to convert RNA into cDNA (1 µg used). Subsequently,
qPCR was conducted using the miRNA QRT-PCR Detection kit
(GeneCopoeia, Rockville, MD, USA) and the ABI 7500 ther-mocycler.
The cycling conditions were as follows: 95°C for 5 min, followed by
40 cycles of 95°C for 30 sec and 60°C for 30 sec. U6 was used as an
internal reference. Relative expression was analyzed using the
2−ΔΔCt method.
Western blotting
Tissues and cells were solubilized in cold
radioimmunoprecipitation lysis buffer (Invitrogen Life
Technologies). The tissues were homogenized with liquid nitrogen in
a mortar (Ziyi Company, Shanghai, China). Proteins were separated
by 10% SDS-PAGE (Beyotime Institute of Biotechnology, Shanghai,
China), and transferred onto a polyvinylidene difluoride (PVDF;
Invitrogen Life Technologies) membrane. The PVDF membrane was then
incubated with phosphate-buffered saline containing 5% milk at 4°C
overnight. Subsequently, the PVDF membrane was incubated with
monoclonal mouse anti-human VEGF-C (ab63221; 1:100) and monoclonal
mouse anti-human GAPDH (ab8245; 1:100) primary antibodies (Abcam,
Cambridge, UK) at room temperature for 3 h. The membrane was then
incubated with rabbit anti-mouse secondary IgG antibodies (Abcam)
at room temperature for 1 h. An Enhanced Chemiluminescence kit
(Pierce Biotechnology, Inc., Rockford, IL, USA) was then used to
perform chemiluminescent detection. The relative protein expression
was analyzed using Image-Pro plus software 6.0 (Media Cybernetics,
Inc., Rockville, MD, USA) and was presented as the density ratio
versus GAPDH.
Transfection
Lipofectamine® 2000 (Life Technologies)
was used to perform cell transfection, according to the
manufacturer's instructions. For miR-101 and VEGF-C functional
analysis, ICC-9810 cells (5×106 cells) were transfected
for 48 h at 37°C with 100 mM each of scrambled miR as a negative
control (NC), miR-101 mimics, miR-101 inhibitor (Life
Technologies), VEGF-C-small interfering (si)RNA, or VEGF-C plasmid
(Nlunbio, Changsha, China), respectively. Non-transfected ICC-9810
cells were used as control.
Bioinformatics analysis
Bioinformatics analysis was conducted to predict the
putative targets of miR-101 using Targetscan online software
(www.targetscan.org).
Dual luciferase reporter assay
A mutant (MUT) 3′-UTR of VEGF-C was generated using
the Directed Mutagenesis kit (Stratagene, Agilent Technologies,
Inc., Santa Clara, CA, USA), according to the manufacturer's
instructions. The wild type (WT) or MUT 3′-UTR of VEGF-C (Nlunbio)
was inserted into the psiCHECK™2 vector (Promega Corporation,
Madison, WI, USA). Subsequently, ICC-9810 cells (5×106
cells) were transfected with psiCHECK™2-WT VEGF-C 3′-UTR or
psiCHECK™2-MUT VEGF-C 3′-UTR vector, with or without miR-101 mimics
(100 nM), and then incubated at 37°C in an atmosphere containing 5%
CO2 for 48 h. The Dual-Luciferase Reporter Assay system
(Promega Corporation) was then used to determine the luciferase
activities on an LD400 luminometer (Beckman Coulter, Inc., Brea,
CA, USA). Renilla luciferase activity was normalized to
firefly luciferase activity.
Cell migration and invasion assays
Cell migration and invasion assays were performed
using Transwell chambers (BD Biosciences, Franklin Lakes, NJ, USA).
For the invasion assay, the Transwell chambers were pre-coated with
Matrigel (BD Biosciences), whereas the chambers used for the
migration assay were not. A cell suspension containing
5×105 cells/ml was prepared in serum-free media, and 300
µl cell suspension was added into the upper chamber. A total
of 500 µl DMEM supplemented with 10% FBS was added to the
lower chamber. The cells were incubated for 24 h at 37°C. A
cotton-tipped swab was then used to carefully wipe away the cells
that did not migrate or invade through the pores. The filters were
fixed in 90% alcohol and stained with crystal violet
(Sigma-Aldrich, St. Louis, MO, USA). Cell number was determined in
five fields randomly selected under an inverted microscope
(IX73-A21PH; Olympus Corporation, Tokyo, Japan).
Statistical analysis
SPSS 17.0 software (SPSS Inc., Chicago, IL, USA) was
used to perform statistical analysis. All data are expressed as the
mean ± standard deviation. Differences were analyzed using one-way
analysis of variance. P<0.05 was considered to indicate a
statistically significant difference.
Results
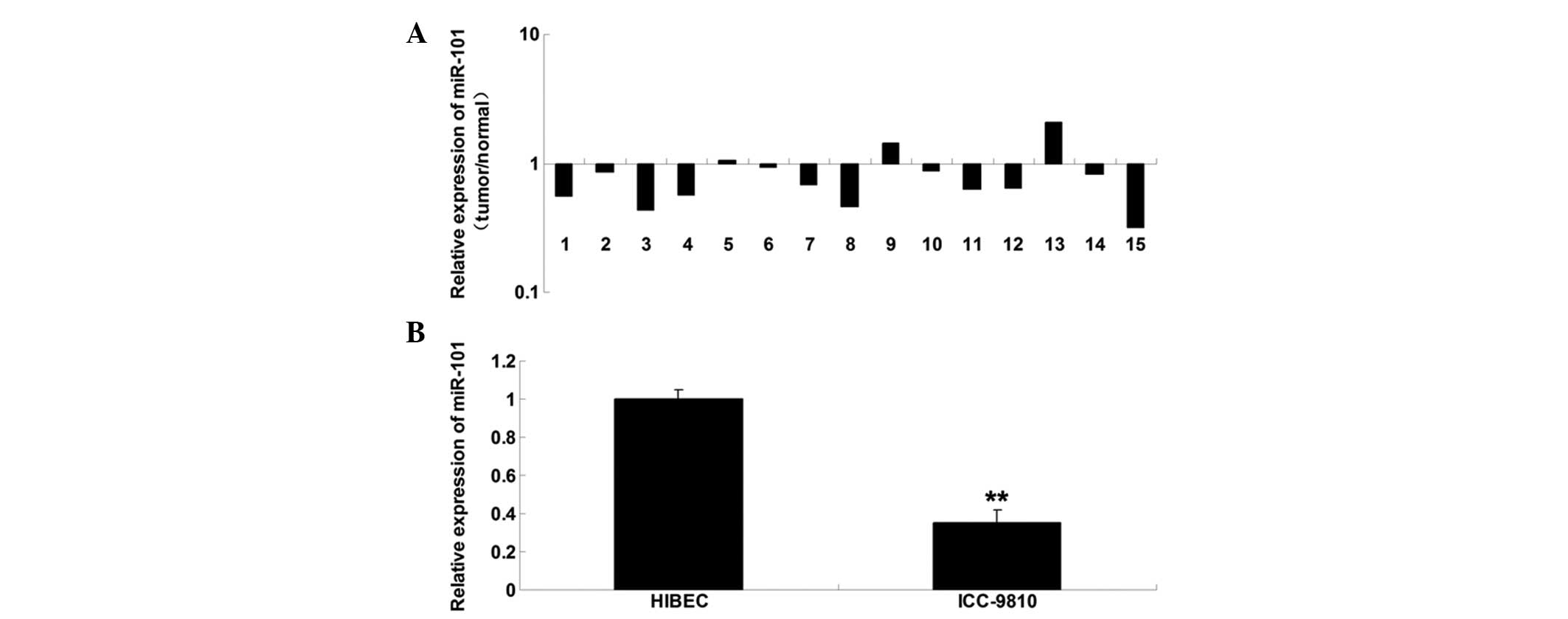
miR-101 is downregulated in ICC tissue
samples and cell lines
To determine the role of miR-101 in ICC, RT-qPCR was
conducted to examine the expression levels of miR-101 in ICC tissue
samples. miR-101 was markedly downregulated in the ICC tissue, as
compared with in the matched adjacent normal tissue (Fig. 1A). Furthermore, the miR-101
expression levels were determined in ICC cells; miR-101 was also
downregulated in the ICC-9810 human ICC cell line, as compared with
in the normal HIBECs (Fig. 1B).
These findings suggest that deregulation of miR-101 may be
associated with the development of ICC.
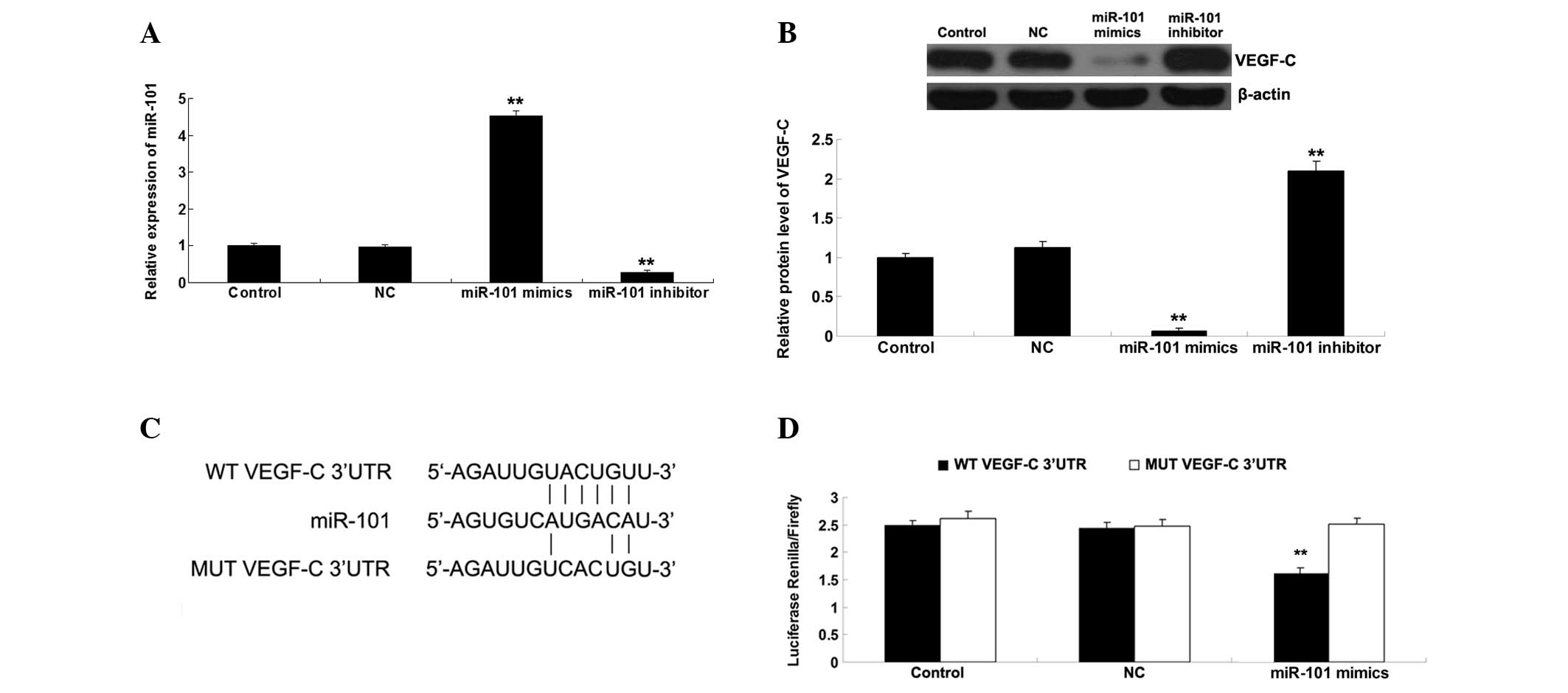
VEGF-C is a target gene of miR-101
Bioinformatics analysis suggested that VEGF-C is a
potential target gene of miR-101. Therefore, the present study
investigated whether miR-101 could mediate the expression of VEGF-C
in ICC-9810 cells. As presented in Fig. 2A, the miR-101 levels in ICC-9810
cells transfected with miR-101 mimics were significantly
upregulated compared with the control group. By contrast, the
miR-101 levels in ICC-9810 cells transfected with the miR-101
inhibitor were markedly reduced compared with the control group. In
addition, transfection with NC miR did not affect the miR-101 in
ICC-9810 cells. These above data indicate that the transfection
efficiency was satisfactory. Subsequently, the protein expression
levels of VEGF-C were determined using western blotting. As shown
in Fig. 2B, upregulation of
miR-101 markedly decreased the protein expression levels of VEGF-C,
whereas downregulation of miR-101 significantly increased the
protein expression levels of VEGF-C in the ICC-9810 cells. To
further verify whether miR-101 may directly bind to seed sequences
in the VEGF-C 3′-UTR in ICC-9810 cells, WT and MUT VEGF-C 3′-UTRs
were generated (Fig. 2C) and a
luciferase reporter assay was performed. Luciferase activity was
significantly reduced in the cells co-transfected with the WT
VEGF-C 3′UTR and miR-101 mimics; however, there was no difference
in luciferase activity in the cells co-transfected with the MUT
VEGF-C 3′UTR and miR-101 mimics, as compared with the control group
(Fig. 2D). These results indicate
that VEGF-C is a direct target of miR-101, and the protein
expression levels of VEGF-C are negatively regulated by miR-101 in
ICC-9810 cells.
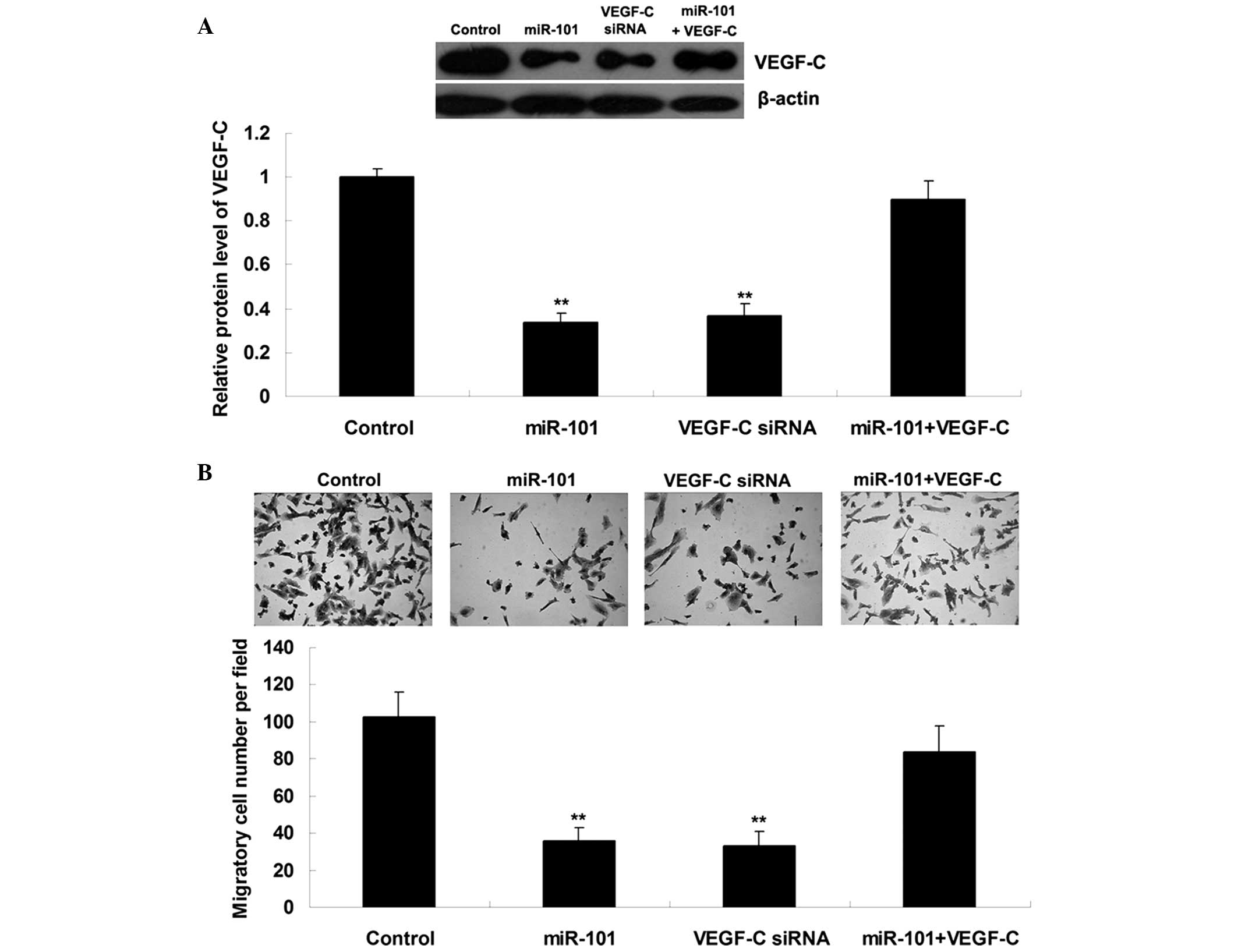
miR-101 suppresses ICC cell migration by
targeting VEGF-C
Since VEGF-C has been suggested to be involved in
ICC metastasis, the present study further examined the roles of
miR-101 and VEGF-C in the regulation of ICC cell migration.
ICC-9810 cells were transfected with miR-101 mimics, VEGF-C siRNA,
or co-transfected with miR-101 mimics and VEGF-C plasmid. As shown
in Fig. 3A, transfection
efficiency was satisfactory. Upregulation of miR-101 or knockdown
of VEGF-C markedly suppressed the migration of ICC-9810 cells;
however, the suppressive effects of miR-101 upregulation on
ICC-9810 cell migration were significantly attenuated following
overexpression of VEGF-C (Fig.
3B). These results suggest that miR-101 may inhibit ICC-9810
cell migration by directly targeting VEGF-C.
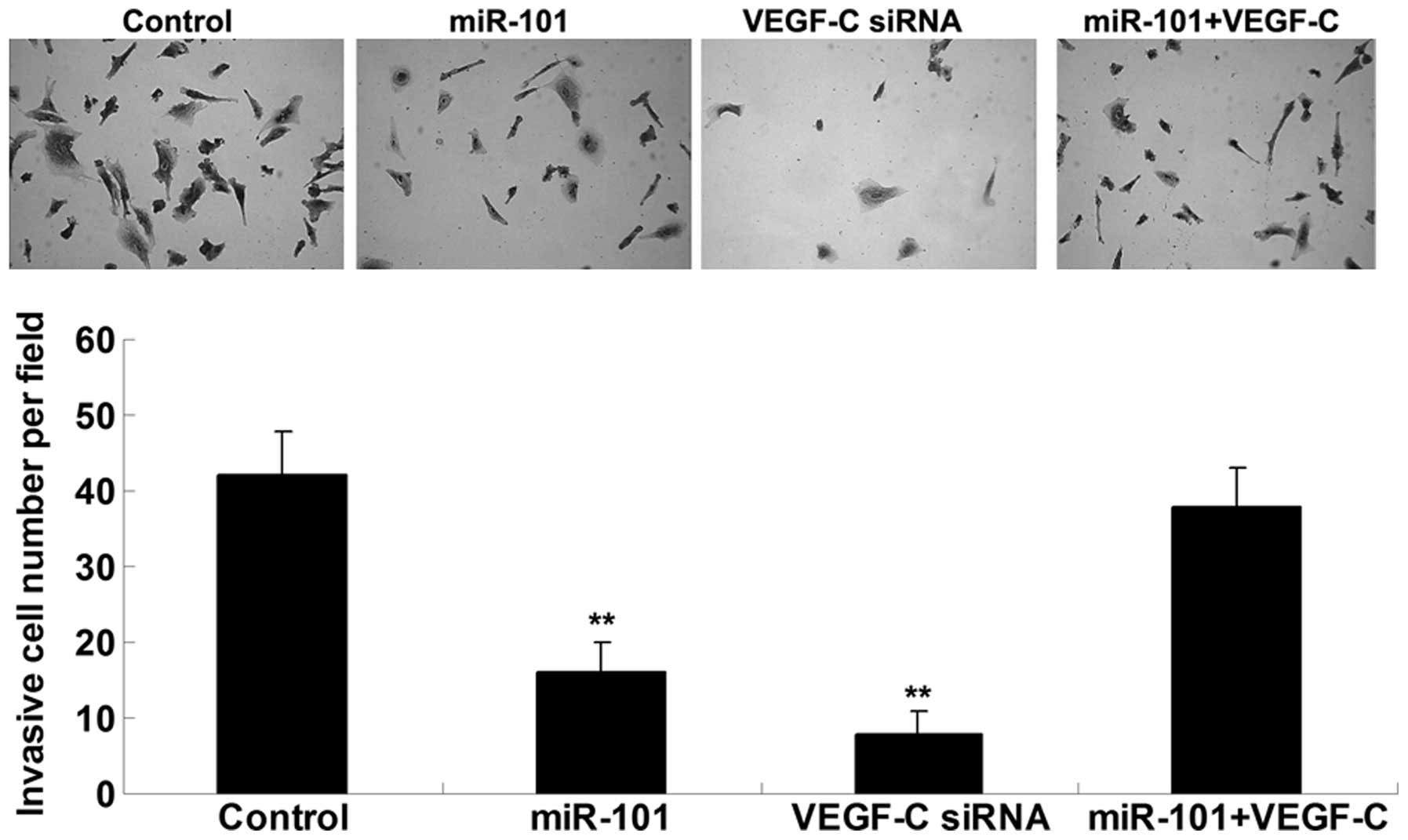
miR-101 suppresses ICC cell invasion by
targeting VEGF-C
The present study also examined the effects of
miR-101 and VEGF-C on ICC cell invasion. Concordant with the cell
migration results, upregulation of miR-101 or knockdown of VEGF-C
significantly inhibited ICC-9810 cell invasion; however, the
suppressive effects of miR-101 overexpression on ICC-9810 cell
invasion were reversed following upregulation of VEGF-C (Fig. 4). These results indicate that
miR-101 may inhibit ICC-9810 cell invasion via direct inhibition of
VEGF-C.
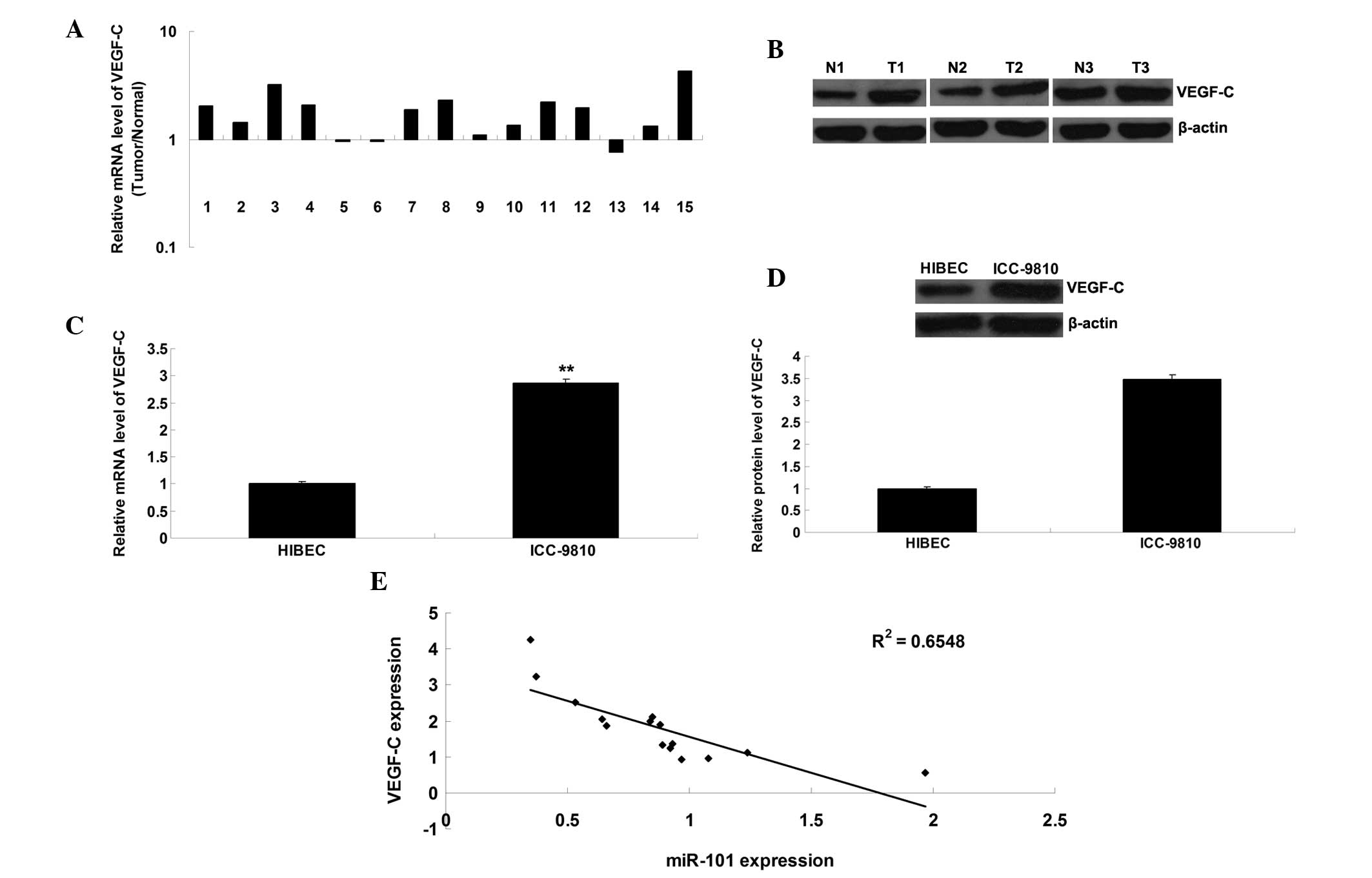
VEGF-C expression is markedly upregulated
in ICC tissue samples and cells
The present study further detected the mRNA and
protein expression levels of VEGF-C in ICC tissue samples and cell
lines. As shown in Fig. 5A and B,
the mRNA and protein expression levels of VEGF-C were frequently
upregulated in ICC tissue, as compared with in the matched adjacent
normal tissue. Furthermore, the mRNA and protein expression levels
of VEGF-C were upregulated in the ICC-9810 cells, as compared with
in the normal HIBECs (Fig. 5C and
D). In addition, an inverse correlation was detected between
miR-101 and VEGF-C expression in the ICC tissue (Fig. 5E).
Discussion
miR-101 has previously been demonstrated to act as a
tumor suppressor in numerous types of malignant tumors (11,13).
The results of the present study demonstrated that miR-101 was
significantly downregulated in ICC tissue samples and cell lines.
In addition, a novel target gene of miR-101 was identified, VEGF-C,
the expression of which was negatively regulated by miR-101 in
ICC-9810 cells. These results suggested that miR-101 was able to
inhibit the migration and invasion of ICC-9810 cells, at least in
part, via direct suppression of VEGF-C protein expression. In
addition, VEGF-C was shown to be upregulated in ICC tissue samples
and cells, and its expression was inversely correlated with miR-101
expression in ICC tissue.
Increasing evidence has demonstrated that miR-101
acts as a tumor suppressor in numerous types of human malignancies
(11). The expression of miR-101
has previously been shown to be frequently reduced in various types
of cancer, including non-small cell lung cancer cells, gastric
cancer, ovarian cancer, endometrial cancer and cervical cancer
(14–19). Yan et al (20) demonstrated that miR-101 was able to
inhibit lung tumorigenesis via suppression of DNA methlytransferase
3a-dependent DNA methylation. Wang et al (21) reported that miR-101 may promote the
apoptosis of breast cancer cells by targeting Janus kinase 2. In
addition, Zhang et al (12)
demonstrated that miR-101 was notably downregulated in
cholangiocarcinoma tissue samples and cell lines, thus suggesting
that miR-101 may participate in cholangiocarcinoma progression.
However, the detailed role of miR-101 in ICC has yet to be
investigated. The present study is the first, to the best of our
knowledge, to detect a marked downregulation of miR-101 expression
in ICC tissue and cell lines, suggesting that miR-101 may also be
involved in ICC progression.
A previous study demonstrated that miR-101 has a
suppressive role in the regulation of cancer metastasis (22). Zhang et al (23) reported that miR-101 was able to
inhibit the metastasis of osteosarcoma cells via downregulation of
enhancer of zeste homolog 2 expression (23). Wang et al (24) demonstrated that miR-101 may
suppress the migration and invasion of thyroid cancer cells by
directly targeting Rac1. The present study demonstrated that
restoration of miR-101 expression markedly inhibited the migration
and invasion of ICC-9810 cells, thus suggesting that miR-101 may
also exert an inhibitory effect on ICC metastasis.
VEGF-C is an important member of the VEGF family,
which has been demonstrated to participate in angiogen-esis
(25). VEGF-C has a role in the
regulation of proliferation and motility of endothelial cells, and
can also affect vascular permeability (26). The present study identified VEGF-C
as a novel target gene of miR-101, and the expression of VEGF-C was
shown to be negatively regulated by miR-101. VEGF-C has previously
been shown to be involved in the development and progression of
various types of human cancer (27). In addition, VEGF-C is associated
with lymph node metastasis in numerous types of human malignancy,
and knockdown of VEGF-C expression notably inhibits cancer
metastases (27).
VEGF-C is also associated with the progression of
ICC (4,28). Park et al (4) examined the expression of VEGF-C in
surgical specimens from 36 patients with ICC, and determined that
VEGF-C was frequently upregulated in ICC tissue. Furthermore,
VEGF-C expression was significantly correlated with lymph node
metastasis, the presence of positive surgical margins and poor
survival rate, thus suggesting that strong VEGF-C expression may be
an independent factor that indicates poor prognosis (4). Shi et al (28) demonstrated that Dickkopf-related
protein 1 enhanced tumor cell invasion and promoted the lymph node
metastasis of ICC via upregulation of VEGF-C. The present study
reported that siRNA-mediated VEGF-C downregulation markedly
inhibited the migration and invasion of ICC-9810 cells, whereas
upregulation of VEGF-C attenuated the suppressive effects of
miR-101 overexpression on ICC-9810 cell migration and invasion.
These findings indicated that VEGF-C may be involved in
miR-101-mediated migration and invasion of ICC cells.
In conclusion, the results of the present study
suggest that miR-101 has an inhibitory role in the regulation of
ICC cell migration and invasion, at least in part, via direct
inhibition of VEGF-C expression. Therefore, miR-101/VEGF-C
signaling may serve as a potential diagnostic and therapeutic
target for ICC.
References
|
1
|
Farges O and Fuks D: Clinical presentation
and management of intrahepatic cholangiocarcinoma. Gastroenterol
Clin Biol. 34:191–199. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Luh F, Kuei A, Fann P, Chu P and Yen Y:
Intrahepatic cholangiocarcinoma and hepatitis: Case study and
literature review. Anticancer Res. 29:3239–3243. 2009.PubMed/NCBI
|
|
3
|
Aishima S, Nishihara Y, Iguchi T, Taguchi
K, Taketomi A, Maehara Y and Tsuneyoshi M: Lymphatic spread is
related to VEGF-C expression and D2-40-positive myofibroblasts in
intrahepatic cholangiocarcinoma. Mod Pathol. 21:256–264. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Park BK, Paik YH, Park JY, Park KH, Bang
S, Park SW, Chung JB, Park YN and Song SY: The clinicopathologic
significance of the expression of vascular endothelial growth
factor-C in intrahepatic cholangiocarcinoma. Am J Clin Oncol.
29:138–142. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human MicroRNA targets. PLoS Biol.
2:e3632004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen L, Yan HX, Yang W, Hu L, Yu LX, Liu
Q, Li L, Huang DD, Ding J, Shen F, et al: The role of microRNA
expression pattern in human intrahepatic cholangiocarcinoma. J
Hepatol. 50:358–369. 2009. View Article : Google Scholar
|
|
9
|
Zeng B, Li Z, Chen R, Guo N, Zhou J, Zhou
Q, Lin Q, Cheng D, Liao Q, Zheng L and Gong Y: Epigenetic
regulation of miR-124 by hepatitis C virus core protein promotes
migration and invasion of intrahepatic cholangiocarcinoma cells by
targeting SMYD3. FEBS Lett. 586:3271–3278. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Oishi N, Kumar MR, Roessler S, Ji J,
Forgues M, Budhu A, Zhao X, Andersen JB, Ye QH, Jia HL, et al:
Transcriptomic profiling reveals hepatic stem-like gene signatures
and interplay of miR-200c and epithelial-mesenchymal transition in
intrahepatic cholangiocarcinoma. Hepatology. 56:1792–1803. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gui T and Shen K: miRNA-101: A potential
target for tumor therapy. Cancer Epidemiol. 36:537–540. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang J, Han C, Zhu H, Song K and Wu T:
miR-101 inhibits cholangiocarcinoma angiogenesis through targeting
vascular endothelial growth factor (VEGF). Am J Pathol.
182:1629–1639. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lei Q, Shen F, Wu J, Zhang W, Wang J and
Zhang L: MiR-101, downregulated in retinoblastoma, functions as a
tumor suppressor in human retinoblastoma cells by targeting EZH2.
Oncol Rep. 32:261–269. 2014.PubMed/NCBI
|
|
14
|
Zhang JG, Guo JF, Liu DL, Liu Q and Wang
JJ: MicroRNA-101 exerts tumor-suppressive functions in non-small
cell lung cancer through directly targeting enhancer of zeste
homolog 2. J Thorac Oncol. 6:671–678. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang HJ, Ruan HJ, He XJ, Ma YY, Jiang XT,
Xia YJ, Ye ZY and Tao HQ: MicroRNA-101 is down-regulated in gastric
cancer and involved in cell migration and invasion. Eur J Cancer.
46:2295–2303. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cui J, Eldredge JB, Xu Y and Puett D:
MicroRNA expression and regulation in human ovarian carcinoma cells
by luteinizing hormone. PLoS One. 6:e217302011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hiroki E, Akahira J, Suzuki F, Nagase S,
Ito K, Suzuki T, Sasano H and Yaegashi N: Changes in microRNA
expression levels correlate with clinicopathological features and
prognoses in endometrial serous adenocarcinomas. Cancer Sci.
101:241–249. 2010. View Article : Google Scholar
|
|
18
|
Lin C, Huang F, Zhang YJ, Tuokan T and
Kuerban G: Roles of MiR-101 and its target gene Cox-2 in early
diagnosis of cervical cancer in Uygur women. Asian Pac J Cancer
Prev. 15:45–48. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pang Y, Young CY and Yuan H: MicroRNAs and
prostate cancer. Acta Biochim Biophys Sin (Shanghai). 42:363–369.
2010. View Article : Google Scholar
|
|
20
|
Yan F, Shen N, Pang J, Xie D, Deng B,
Molina JR, Yang P and Liu S: Restoration of miR-101 suppresses lung
tumorigenesis through inhibition of DNMT3a-dependent DNA
methylation. Cell Death Dis. 5:e14132014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang L, Li L, Guo R, Li X, Lu Y, Guan X,
Gitau SC, Wang L, Xu C, Yang B and Shan H: miR-101 promotes breast
cancer cell apoptosis by targeting Janus kinase 2. Cell Physiol
Biochem. 34:413–422. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
He XP, Shao Y, Li XL, Xu W, Chen GS, Sun
HH, Xu HC, Xu X, Tang D, Zheng XF, et al: Downregulation of miR-101
in gastric cancer correlates with cyclooxygenase-2 overexpression
and tumor growth. FEBS J. 279:4201–4212. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang K, Zhang Y, Ren K, Zhao G, Yan K and
Ma B: MicroRNA-101 inhibits the metastasis of osteosarcoma cells by
downregulation of EZH2 expression. Oncol Rep. 32:2143–2149.
2014.PubMed/NCBI
|
|
24
|
Wang C, Lu S, Jiang J, Jia X, Dong X and
Bu P: Hsa-microRNA-101 suppresses migration and invasion by
targeting Rac1 in thyroid cancer cells. Oncol Lett. 8:1815–1821.
2014.PubMed/NCBI
|
|
25
|
Hu J, Cheng Y, Li Y, Jin Z, Pan Y, Liu G,
Fu S, Zhang Y, Feng K and Feng Y: microRNA-128 plays a critical
role in human non-small cell lung cancer tumourigenesis,
angiogenesis and lymphangiogenesis by directly targeting vascular
endothelial growth factor-C. Eur J Cancer. 50:2336–2350. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Olofsson B, Jeltsch M, Eriksson U and
Alitalo K: Current biology of VEGF-B and VEGF-C. Curr Opin
Biotechnol. 10:528–535. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Su JL, Yen CJ, Chen PS, Chuang SE, Hong
CC, Kuo IH, Chen HY, Hung MC and Kuo ML: The role of the
VEGF-C/VEGFR-3 axis in cancer progression. Br J Cancer. 96:541–545.
2007. View Article : Google Scholar
|
|
28
|
Shi RY, Yang XR, Shen QJ, Yang LX, Xu Y,
Qiu SJ, Sun YF, Zhang X, Wang Z, Zhu K, et al: High expression of
Dickkopf-related protein 1 is related to lymphatic metastasis and
indicates poor prognosis in intrahepatic cholangiocarcinoma
patients after surgery. Cancer. 119:993–1003. 2013. View Article : Google Scholar
|