Introduction
Oral cancer is the eighth most common cause of
cancer-associated mortality, and >90% of oral malignancies are
oral squamous cell carcinomas (OSCC) (1). Due to the large numbers of patients,
OSCC has become an important health concern, and has gained
considerable public attention in numerous countries. Despite
improvements in treatment, the mortality rate of patients with OSCC
remains high, and it is estimated that half of patients with OSCC
will survive for only five years following diagnosis (1). With the development of biological
target therapies, treatment options for patients have significantly
expanded (2). Previously,
molecular markers of p53 have been used as treatment targets with
positive clinical outcomes, which prompted investigators to
identify efficient markers that would be able to rapidly predict
the presence of an OSCC tumour, and effectively improve patient
health (1).
A previous study demonstrated that inflammation, in
addition to acting as an innate immune response, may promote tumour
growth and progression (3).
Numerous solid tumour types exhibit inflammatory growth (3). During this process, inflammatory
cytokines and other mediators are induced by the tumour, acquiring
tumour-promoting activities (4).
Therefore, the inflammatory environment assists tumour development
and metastasis.
As an important regulator of inflammation,
macrophage migration inhibitory factor (MIF) is a pleiotropic
cytokine, which has a pro-inflammatory function and is involved in
immune response in the presence of stress, inflammation and
infection (5). Although the
signaling pathways activated by MIF remain to be fully elucidated,
previous studies (5,6) have suggested that MIF has a role in
disease-associated processes, particularly in neoplastic disorders
(6). It has become increasingly
evident that MIF influences biological mechanisms underlying tumour
growth and metastasis (6). MIF has
been reported to be overexpressed in numerous types of tumour and
may promote potential malignant activities in several ways:
Inhibition of apoptosis, promotion of angiogenesis and stimulation
of the cell cycle (5,6).
Previous reports have suggested the importance of
MIF in the development of OSCC. França et al (7) demonstrated that MIF-positive cells
were located in both the tumour parenchyma and inflammatory cells
in the OSCC tissue specimens. Dumitru et al (8) reported that high expression levels of
MIF are associated with higher lymph node metastasis, and reduced
survival of patients with head and neck cancer. In addition, the
investigators demonstrated that the effects of tumour-derived MIF
on neutrophils is a further mechanism by which MIF may modulate
neutrophil survival and enhance the migratory properties of OSCC
cells (8).
The aim of the present study was to examine whether
small interfering (si)RNA can be utilized to disrupt the biological
behavior of OSCC cells. Firstly, the expression levels of MIF in a
number of OSCC cell lines were investigated. Secondly, siRNA
targeting MIF were used to knock down the expression of MIF, and to
identify its effects on proliferation, migration and colony
formation in OSCC cells. Finally, the staining of MIF protein in
OSCC tissue samples from patients with OSCC was observed. The
present study aimed to determine the roles of MIF in the
progression of OSCC.
Materials and methods
Reagents
Dulbecco's modified Eagle's medium (DMEM), foetal
bovine serum (FBS), trypsin-EDTA and Invitrogen
Lipofectamine® 2000 were purchased from Thermo Fisher
Scientific, Inc. (Waltham, MA, USA). Primary monoclonal antibodies
against mouse anti-human MIF and α-tublin were obtained from Abcam
(Cambridge, MA, USA; cat. nos. ab55445 and ab15246), and primary
polyclonal antibodies against rat anti-human Twist1, matrix
metalloproteinase (MMP)-2 and MMP-9 were obtained from Santa Cruz
Biotechnology, Inc. (Dallas, TX, USA; cat. nos. sc134136, sc10736
and sc10737). The secondary antibodies, goat anti-mouse
immunoglobulin (Ig)G and goat anti-rabbit IgG, were supplied by
Bio-Rad Laboratories, Inc. (Hercules, CA, USA; cat. nos. STAR137P
and STAR121P).
Cell lines and culture conditions
Normal human epithelial cells (EP) were supplied by
the Queensland Institute of Medical Research (Brisbane, Australia).
Established Tca8113, SCC25 and HN5 human OSCC cell lines were
provided by Professor Qian Tao (Sun Yat-sen University, Guangzhou,
China), Professor Nickolas Saunders (Princess Alexandra Hospital,
Woolloongabba, Australia), and Professor Ming Wei (Griffith
University, Nathan, Australia), respectively, and were cultured in
a humidified atmosphere containing 5% CO2 and 95% air at
37°C. The Tca8113, SCC25 and HN5 cells were grown in DMEM,
supplemented with 10% FBS and 100 U/ml penicillin G and 100 mg/ml
streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C
in an incubator, containing 5% CO2 and 20%
O2.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
At the same time point, all cell lines were plated
into 6-well plates at a density of 1×106 cells/well.
Following overnight culture and when 90% of the cells attained
confluence, the total RNA was isolated from all cell lines using a
PureLink RNA Mini kit (Invitrogen; Thermo Fisher Scientific, Inc.).
RNA (1 µg) was reverse transcribed to cDNA using an iScript
cDNA Synthesis kit (Bio-Rad Laboratories, Inc.), according to the
manufacturer's instructions. Quantitative gene analysis was
performed for GAPDH and MIF using an EXPRESS SYBR®
GreenER™ qPCR Supermix Universal kit (Invitrogen; Thermo Fisher
Scientific, Inc.) and an icycler iQ5 Real-Time PCR system (Bio-Rad
Laboratories, Inc.). The primer sequences used to amplify the cDNA
were as follows: GAPDH, forward 5′-CTTAGAGGGACAAGTGGCG-3′ and
reverse 5′-ACGCTGAGCCAGTCAGTGTA-3′; MIF, forward
5′-TCGCGAGCTATAGAAGAATCA-3′ and reverse
5′-TGTTCAAGTCTTCGGAGTTTG-3′. Thermal cycling was performed at 95°C
for 2.5 min, followed by 45 cycles of amplification at 95°C for 10
sec, 58°C for 10 sec, 72°C for 25 sec and 72 cycles of elongation
at 60°C for 5 sec. The data were normalized against the internal
GAPDH control in order to obtain ΔCq. Finally, the fold-change of
the genes of interest relative to the untreated samples were
calculated using the 2−ΔΔCq method (9).
Western blot analysis
The total protein of all cell lines was extracted
using radioimmunoprecipitation lysis buffer (Thermo Fisher
Scientific, Inc.). The protein concentration was determined using a
Bicinchoninic Acid Protein Assay kit (Pierce Biotechnology, Inc.,
Rockford, IL, USA). A total of 40 µg protein was separated
by 10% SDS-PAGE (Bio-Rad Laboratories, Inc.). The proteins were
transferred onto polyvinylidene fluoride membranes (EMD Millipore,
Billerica, MA, USA) and were subsequently blocked with 5% non-fat
dry milk in Tris-buffered saline for 1 h at room temperature. The
membranes were incubated with the following primary antibodies: MIF
(1:2,000), Twist1 (1:200), MMP-2 (1:200), MMP-9 (1:200) and
α-tublin (1:3,000) overnight at 4°C, washed twice with
phosphate-buffered saline (PBS) and were subsequently incubated
with horseradish peroxidase-conjugated secondary antibodies
(Bio-Rad Laboratories, Inc.) for 1 h at room temperature. The
protein bands were subsequently detected with SuperSignal WestPico
Chemiluminescent Substrate (Thermo Fisher Scientific, Inc.) and
were visualized using a VersaDoc-MP Imaging system (Bio-Rad
Laboratories, Inc.).
Transient transfection of MIF siRNA
Tca8113, SCC25, HN5 OSCC cells were seeded into
six-well plates at a density of 1×104 cells/well.
Following 48 h incubation the cells reached 80% confluence and were
transiently transfected with MIF siRNA using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). The sequences for MIF siRNA (Invitrogen; Thermo
Fisher Scientific, Inc.) were as follows: Sense,
5′-ACAUCAACUAUUACGACAUGAACGCGGdTdT-3′ and anti-sense,
5′-CCGCGUUCAUGUCGUAAUAGUUGAUGUdTdT-3′; the sequences for negative
control (NC) were sense, 5′-GUUGCGCCCGCGAAUGAUAUUUAUAAUdTdT-3′ and
anti-sense, 5′-AUUAUAAAUAUCAUUCGCGGGCGCAACdTdT-3′. The
oligodeoxynucleotides for the NC were obtained following scrambling
of the siRNA oligodeoxynucleotide for MIF, and were determined to
not be associated with any mRNA sequence by BLAST (10). Cell transfection was performed,
according to the protocol of the transfection kit manufacturer.
Briefly, each sequence of MIF siRNA and 10 µl
Lipofectamine® 2000 was diluted in serum-free medium
(250 µl) at room temperature for 5 min, mixed together, and
incubated for 30 min at room temperature. The mixture was
subsequently administered to the Tca8113, SCC25 and HN5 cells, and
after 5 h of incubation, the medium was replaced with complete
medium.
Cell proliferation assay
At a density of 6×105 cells/well, OSCC
cells with or without MIF siRNA were seeded into 96-well plates.
Following incubation for 24, 48 and 72 h, cell proliferation was
detected using an MTT assay (Sigma-Aldrich, St. Louis, MO, USA).
The absorbance was measured at 590 nm using a Biomek Plate Reader
(Beckman Coulter, Gladesville, Australia), following the addition
of 20 µl MTT (5 mg/ml; Thermo Fisher Scientific, Inc.) for 4
h. All solutions were subsequently removed and 150 µl/well
dimethyl suphoxide was added to solubilize the formation crystals
produced from the MTT assay.
Transmigration assay
Transwell inserts (5 µm pores; Corning
Incorporated, Corning, NY, USA) were used for the transmigration
assay. A total of 300 µl Tca8113, SCC25 and HN5 cells
(3×105 cells/ml) with or without MIF siRNA, which were
resuspended in serum-free culture medium, were added in the upper
chamber of the transwell inserts, whereas 600 µl
completemedium was placed in the lower chamber. The chambers were
incubated for 10 h. The non-migrating cells were scraped off the
top of the Transwell, and the migrating cells were fixed and
stained with trypan blue (Sigma-Aldrich), and were observed under a
microscope (Olympus BX60, Olympus Corporation, Toyko, Japan). A
total of five fields under ×400 magnification were randomly
selected and counted by two independent observers.
Colony formation assay
To measure the rate of colony formation, following
transfection with MIF siRNA, Tca8113, SCC25 and HN5 OSCC cells were
evenly spread onto six-well plates at 500 cells/well and cultured
for 10 days at 37°C. Following the incubation period, the cells
were stained with trypan blue solution (Sigma-Aldrich) and their
images were captured using a digital camera (Olympus SH-2; Olympus
Corporation). All experiments were repeated in triplicate and
representative photos of the colonies were captured.
Immunohistochemistry
Formalin-fixed, paraffin embedded OSCC tissue
specimens from 20 patients (50–70 years old; male/female ratio,
1:1, no previous treatment) rectuited from Guanghua Dental Hospital
(Guangzhou, China) with OSCC were sectioned at 5 µm and
mounted onto poly-L-Lysine coated slides, deparaffinized in xylene
(Sigma-Aldrich) and rehydrated through graded ethanol. Informed
consent was obtained from each patient and ethical approval was
obtained from the Ethics Committee of the Hospital of Stomotology,
Sun Yat-Sen University (Guangzhou, China). The sections were
subsequently incubated with 3% H2O2 in
methanol for 30 min at room temperature. Following three washes
with PBS and blocking with normal goat serum (Nichirei Bioscience,
Tokyo, Japan) for 30 min, the sections were incubated overnight at
4°C with mouse monoclonal anti-human MIF primary antibody (1:500;
Thermo Fisher Scientific, Inc.), prior to being incubated with
secondary antibody for 30 min at room temperature following three
washes with PBS. Staining was performed using the
Histostain-Bulk-SP detection kit (Zymed; Thermo Fisher Scientific,
Inc.). The sections were counter-stained with Mayer's hematoxylin
(Sigma-Aldrich). Negative controls were prepared by substituting
the primary antibody with PBS.
Statistical analysis
Data analysis was performed using SAS version 8.1
(SAS Institute, Cary, CA, USA). A paired Student t-test was used to
compare two means, and one way analysis of variance was used to
compare >2 means. P<0.05 was considered to indicated a
statistically significant difference.
Results
mRNA and protein expression levels of MIF
are upregulated in OSCC cells
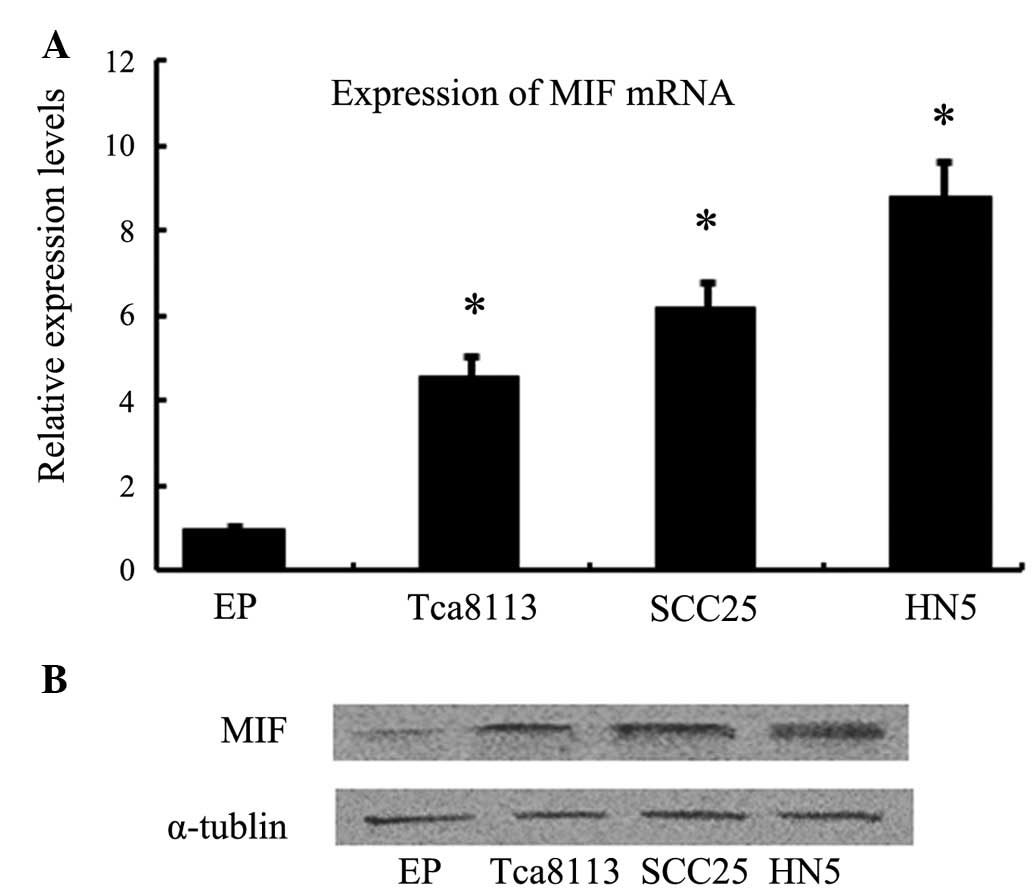
RT-qPCR and western blotting were used to detect
whether OSCC cells expressed MIF mRNA and protein. MIF mRNA and
protein were shown to be expressed in Tca8113, SCC25 and HN5 OSCC
cells, whereas normal epithelial cells exhibited low expression
levels (Fig. 1).
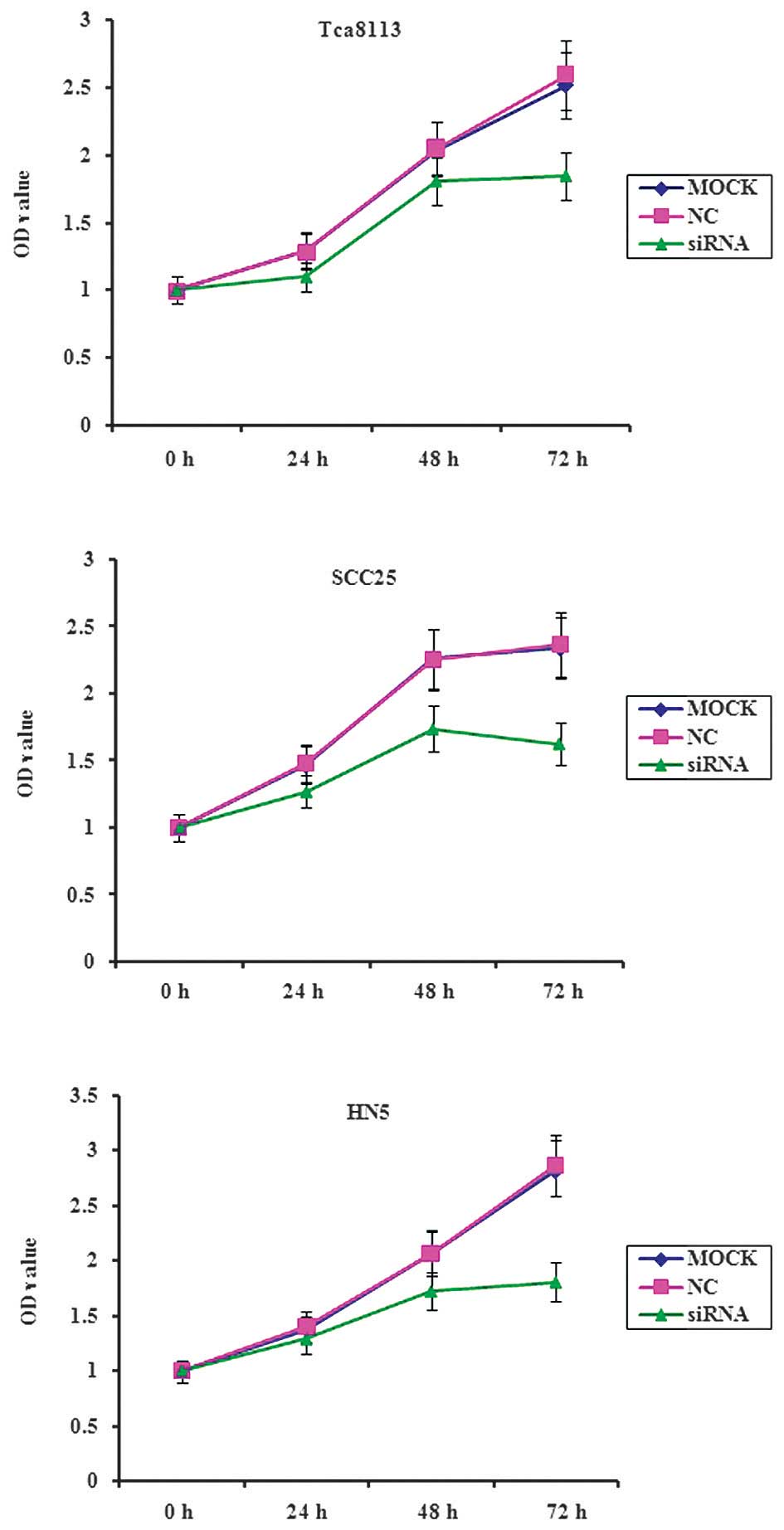
OSCC cell proliferation is inhibited by
MIF siRNA
An optimized experiment was performed to determine
the effective concentration and duration of transient transfection
of MIF siRNA. RT-qPCR was performed and demonstrated that the three
OSCC cell lines exhibited downregulation of MIF mRNA following
transfection with 100 nM MIF siRNA for 48 h. Furthermore, an MTT
assay determined that treatment with MIF siRNA significantly
decreased the proliferation of Tca8113, SCC25 and HN5 cells at each
time point within three days of culture (P<0.05; Fig. 2).
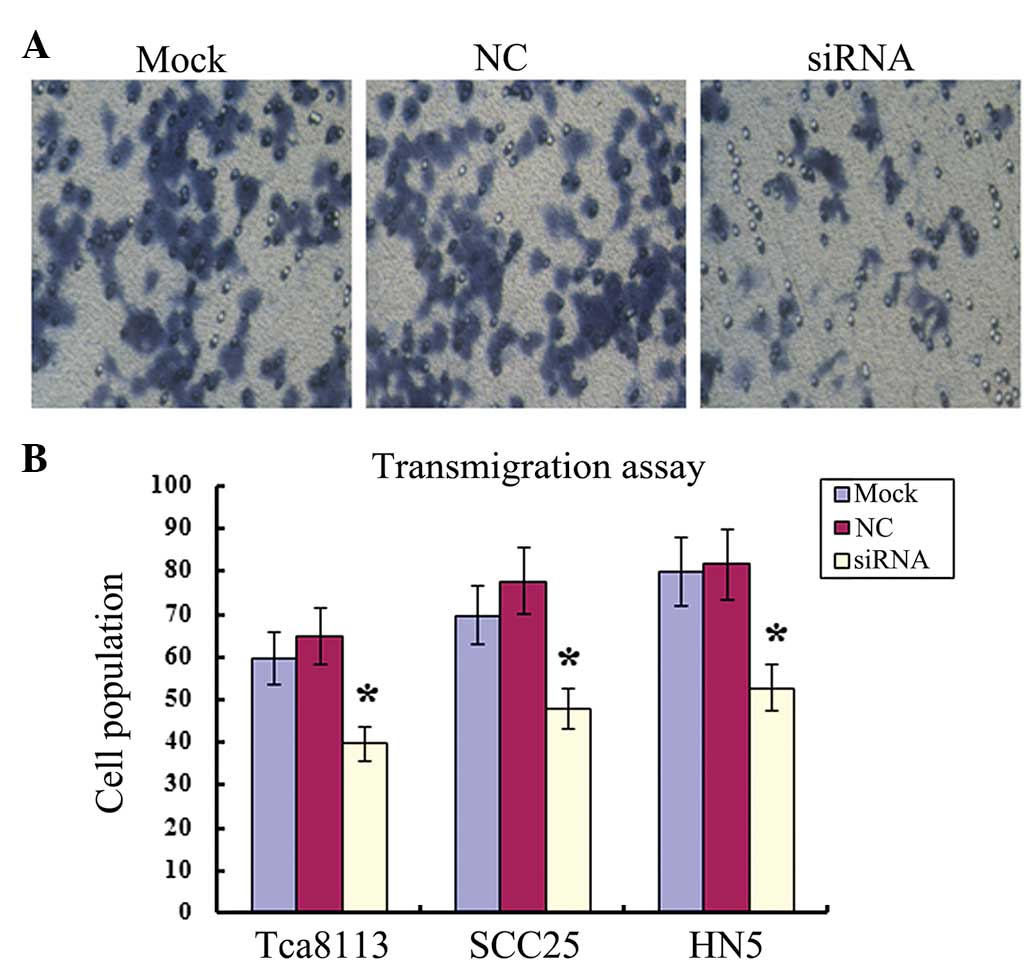
Migration of OSCC cells is reduced by MIF
siRNA
The migration ability of all OSCC cells was further
analyzed using a Transwell assay. The cell count of Tca8113, HN5
and SCC25 OSCC cells post-transfection with MIF siRNA, was
significantly decreased compared with the negative control or Mock
(no siRNA; P<0.05; Fig. 3).
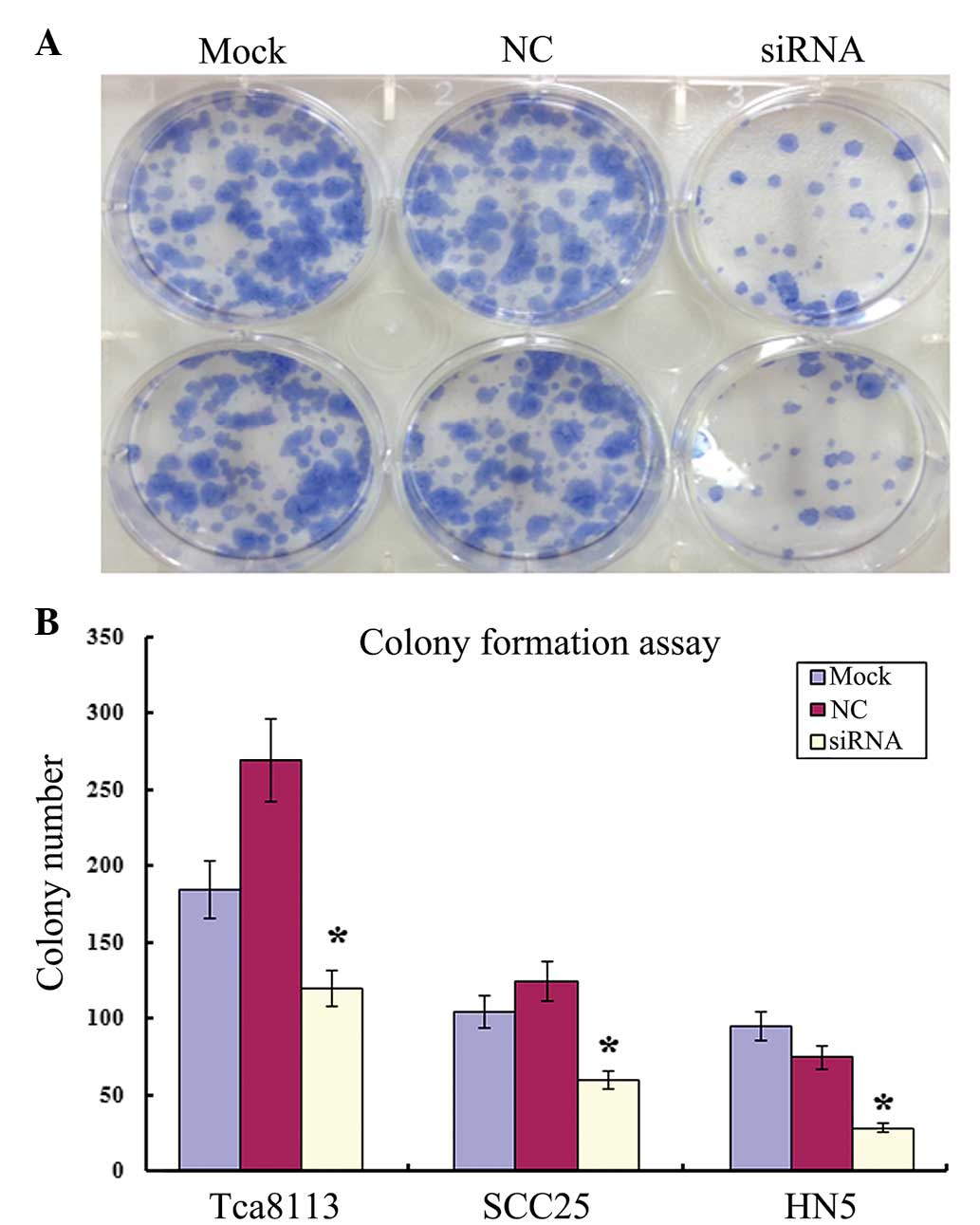
Colony formation of OSCC cells is
inhibited by MIF siRNA
Limited dilution was used to ensure that all OSCC
cells were at an equal number of 500 cells/well in each 6-well
plate. Single colony formation was inhibited in all OSCC lines
post-transfection with 100 nM MIF siRNA, compared with the negative
control and the Mock (Fig. 4).
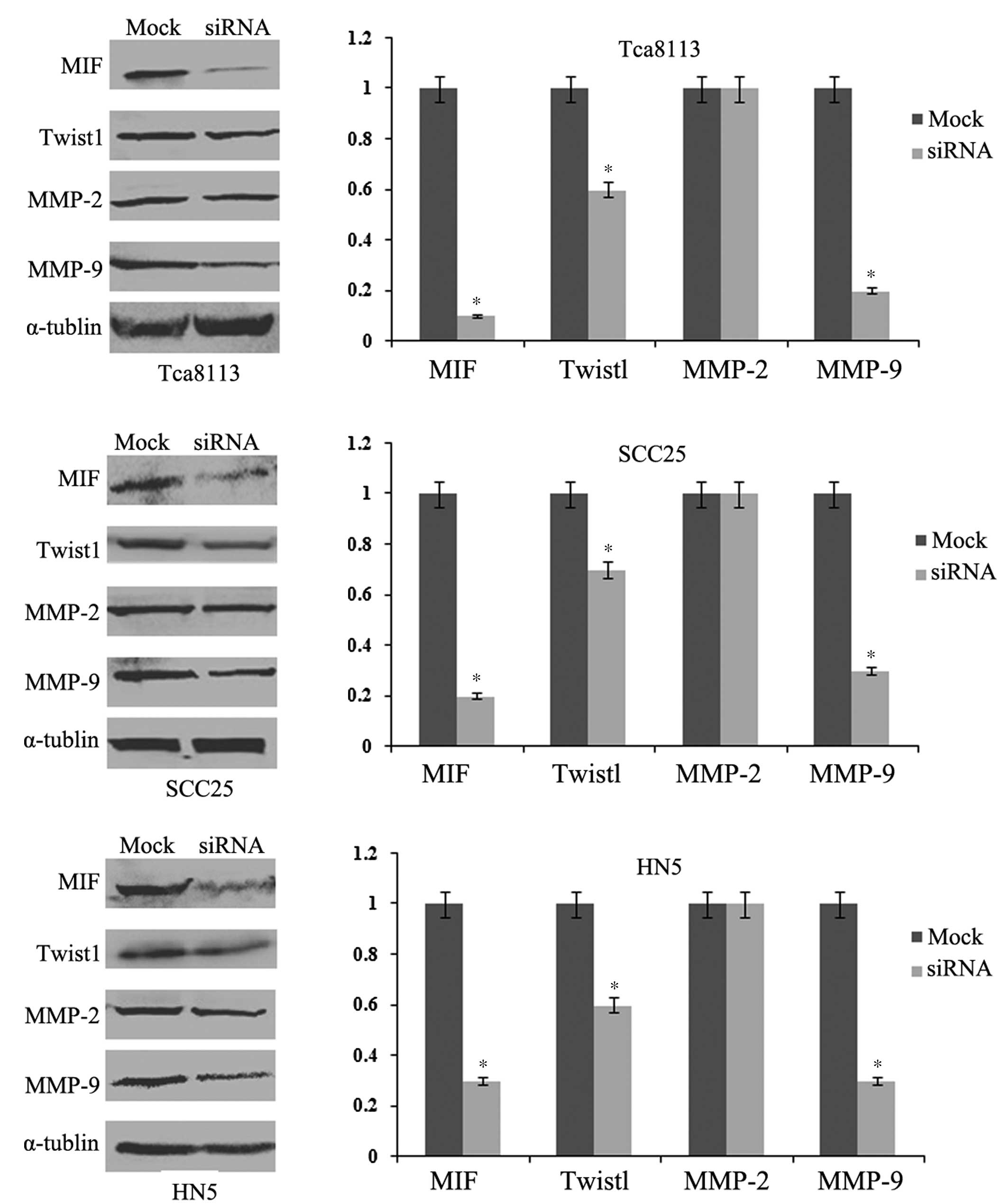
Pathway analysis of the
epithelial-mesenchymal transition (EMT) by western blotting
To explore the signaling pathways underlying the
functional changes of OSCC cells, western blotting was used to
detect the protein expression level changes in the molecular
markers of the EMT. Post-transfection with 100 nM MIF siRNA, the
protein expression levels of MIF in the Tca8113, SCC25 and HN5
cells were markedly downregulated (Fig. 5). The protein expression levels of
the transcriptional factor of the EMT, Twist1, decreased similarly
following the inhibition of MIF. MMP-2 and MMP-9 exhibited
different changes in protein expression levels, with MMP-2
remaining identical whereas MMP-9 decreased significantly.
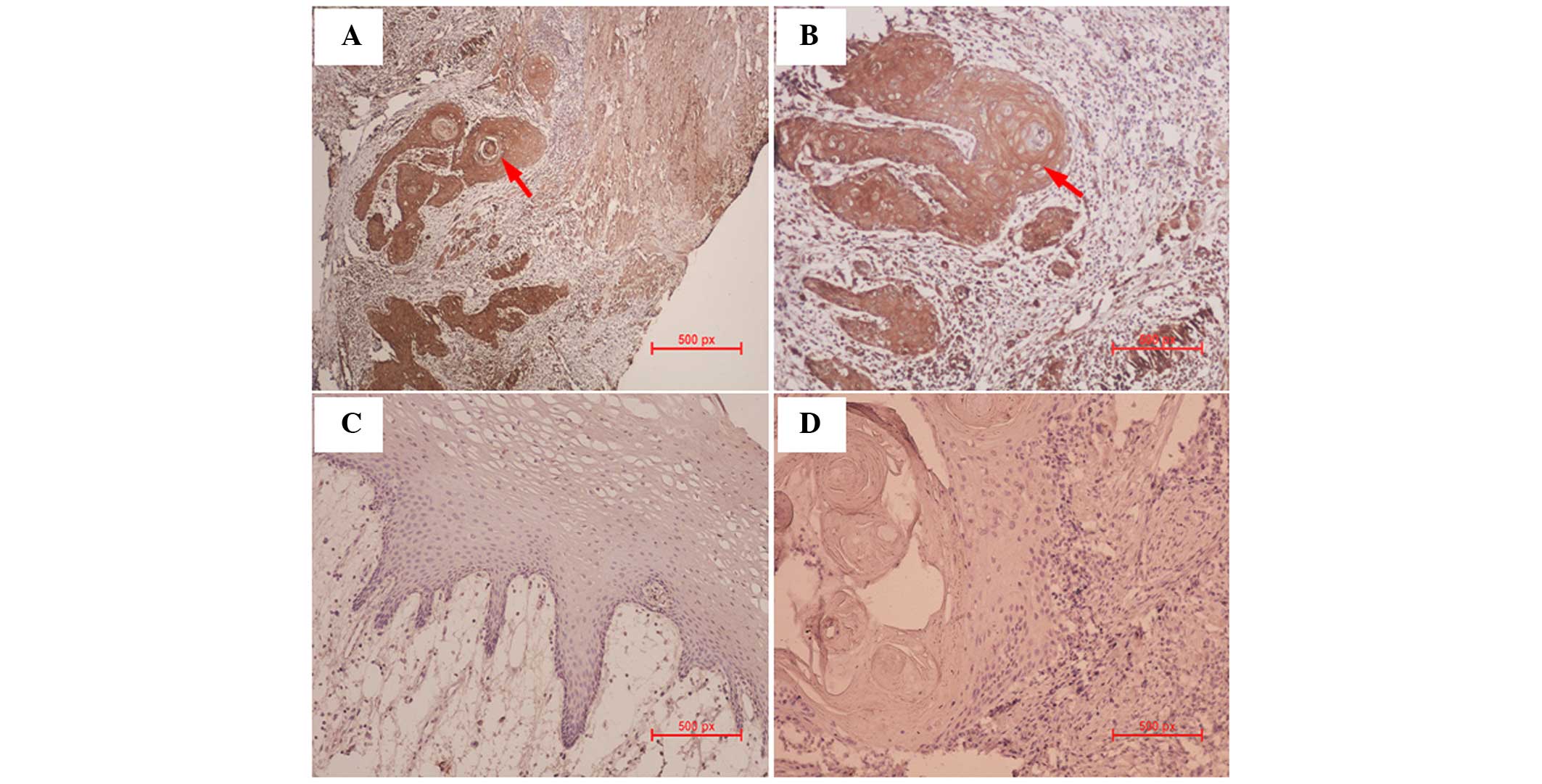
MIF is stained in the majority of OSCC
cells in the clinical tissue samples
Immunohistochemistry was performed to determine
whether OSCC tissue samples expressed MIF protein. In all clinical
samples, MIF was markedly stained in the OSCCs, in either the cell
membrane or cytoplasm, however, not in the nucleus (Fig. 6A and B). In normal oral mucosa, MIF
staining was less pronounced in the entire tissue (Fig. 6C). Negative control samples
exhibited no MIF staining (Fig.
6D).
Discussion
MIF was initially described as a factor inhibiting
macrophage migration (5).
Structural analysis demonstrated that the secondary structure of
MIF is similar to that of the major histocompatibility complex,
indicating its potential role in the immune system (11). Recent studies (5,6) have
reported that MIF was upregulated in various tumour cells. MIF may
promote malignant activities, increase cell migration and
invasiveness, and influence immune reactions to tumour growth
(6). OSCC usually spreads to
adjacent sites in the oral-maxillofacial region, often extending to
the jaw bones (1), and lymph node
metastases are common. However, molecular markers for OSCC with
prognostic and predictive significance remain to be identified.
The involvement of MIF in carcinogenesis and
autoimmune disorders make it a potential target for inhibition
(12). Certain molecules,
including Milatuzumab, inhibiting MIF action have been developed
and used in clinical trials (10).
Therefore, the present study aimed to determine whether MIF was a
therapeutic target candidate of OSCC. siRNA is effective for the
inhibition of specific gene expression, and its efficacy has been
demonstrated in previous in vitro and in vivo
studies. Meyer-Siegler et al (10) demonstrated that LNCaP and DU-145
prostate cancer cell lines exhibited increased mRNA expression of
MIF (10). Treatments aimed at
inhibiting MIF using siRNA or anti-MIF inhibitors significantly
decreased xenograft tumour volume and angiogenesis, providing a
novel therapeutic target for the treatment of androgen-independent
prostate cancer (10). The results
of the present study demonstrated that transient transfection with
MIF siRNA efficiently inhibited the production of MIF protein
within all OSCC cell lines. It also reduced the proliferation,
migration and colony formation abilities of OSCC cells, which
demonstrated the potential inhibitory value of MIF.
To further investigate the mechanisms underlying
these processes at the molecular level, several protein markers
were detected by western blotting. Since Twist1 is associated with
the EMT and tumour invasion, the protein expression levels of
Twist1 were quantified following transfection with MIF siRNA
(9). The results suggested that
the expression of Twsit1 was inhibited by MIF siRNA. The protein
expression levels of MMP-2 and MMP-9 were also quantified, and the
results demonstrated that the protein expression levels of MMP-9
decreased in all OSCCs, whereas those of MMP-2 revealed no change.
These results suggested that the EMT signaling pathways were
affected by MIF siRNA, which further elucidated the mechanisms
underlying the biological behavioral changes of OSCCs.
MIF acts as an upstream mediator of MMP family
members (13,14). Pakozdi et al (13) reported that MIF induced rheumatoid
arthritis synovial fibroblast expression of MMP-2 in a time and
concentration-dependent manner. The protein expression levels of
MMP-2 were significantly decreased in MIF gene-deficient compared
with wild-type mice joint homogenates. The authors further
demonstrated that MIF-induced upregulation of MMP-2 expression
required protein kinase C, c-jun N-terminal kinase and Src
signaling pathway activation. Kong et al (14) demonstrated that the expression
levels of MIF and MMP-9 were markedly upregulated in vulnerable
atheromatous plaques; suggesting that MIF may have a role in the
destabilization of human atherosclerotic plaques. They further
determined that MIF activated the MEK-ERK-MAP signaling pathway to
induce the expression of MMP-9 in murine macrophages. Activation of
this signaling pathway is necessary for the expression of MMP-9 and
activation in response to MIF stimulation (15). Although protein expression levels
of MMP-2 revealed no change in the present study, inhibition of MIF
protein efficiently downregulated the protein expression levels of
MMP-9.
In conclusion, the results of the present study
suggested that MIF may have an important role in the invasion,
migration and proliferation of OSCC. As determined by transfection
with MIF siRNA, the gene expression of MIF was effectively knocked
down in OSCC cells. MIF may mediate MMP-2/9 signaling pathways,
which correlate with the EMT, which suggested that MIF may serve as
a therapeutic target in the treatment of OSCC.
References
|
1
|
Quan J, Johnson NW, Zhou G, Parsons PG,
Boyle GM and Gao J: Potential molecular targets for inhibiting bone
invasion by oral squamous cell carcinoma: A review of mechanisms.
Cancer Metastasis Rev. 31:209–219. 2012. View Article : Google Scholar
|
|
2
|
Ziober BL, Mauk MG, Falls EM, Chen Z,
Ziober AF and Bau HH: Lab-on-a-chip for oral cancer screening and
diagnosis. Head Neck. 30:111–121. 2008. View Article : Google Scholar
|
|
3
|
Terzić J, Grivennikov S, Karin E and Karin
M: Inflammation and colon cancer. Gastroenterology.
138:2101–2114.e5. 2010. View Article : Google Scholar
|
|
4
|
Rendon BE, Willer SS, Zundel W and
Mitchell RA: Mechanisms of macrophage migration inhibitory factor
(MIF) dependent tumor microenvironmental adaptation. Exp Mol
Pathol. 86:180–185. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bach JP, Rinn B, Meyer B, Dodel R and
Bacher M: Role of MIF in inflammation and tumorigenesis. Oncology.
75:127–133. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nishihira J, Ishibashi T, Fukushima T, Sun
B, Sato Y and Todo S: Macrophage migration inhibitory factor (MIF):
Its potential role in tumor growth and tumor-associated
angiogenesis. Ann N Y Acad Sci. 995:171–182. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
França CM, Batista AC, Borra RC,
Ventiades-Flores JA, Mendonça EF, Deana AM, Mesquita-Ferrari RA, de
Natali Caly D, de Mello Rode S and Faria MR: Macrophage migration
inhibitory factor and oral cancer. J Oral Pathol Med. 42:368–373.
2013. View Article : Google Scholar
|
|
8
|
Dumitru CA, Gholaman H, Trellakis S,
Bruderek K, Dominas N, Gu X, Bankfalvi A, Whiteside TL, Lang S and
Brandau S: Tumor-derived macrophage migration inhibitory factor
modulates the biology of head and neck cancer cells via neutrophil
activation. Int J Cancer. 129:859–869. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Quan J, Elhousiny M, Johnson NW and Gao J:
Transforming growth factor-β1 treatment of oral cancer induces
epithelial-mesenchymal transition and promotes bone invasion via
enhanced activity of osteoclasts. Clin Exp Metastasis. 30:659–670.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Meyer-Siegler KL, Iczkowski KA, Leng L,
Bucala R and Vera PL: Inhibition of macrophage migration inhibitory
factor or its receptor (CD74) attenuates growth and invasion of
DU-145 prostate cancer cells. J Immunol. 177:8730–8739. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qi D, Hu X, Wu X, Merk M, Leng L, Bucala R
and Young LH: Cardiac macrophage migration inhibitory factor
inhibits JNK pathway activation and injury during
ischemia/reper-fusion. J Clin Invest. 119:3807–3816. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Greven D, Leng L and Bucala R: Autoimmune
diseases: MIF as a therapeutic target. Expert Opin Ther Targets.
14:253–264. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pakozdi A, Amin MA, Haas CS, Martinez RJ,
Haines GK III, Santos LL, Morand EF, David JR and Koch AE:
Macrophage migration inhibitory factor: A mediator of matrix
metallo-proteinase-2 production in rheumatoid arthritis. Arthritis
Res Ther. 8:R1322006. View
Article : Google Scholar
|
|
14
|
Kong YZ, Yu X, Tang JJ, Ouyang X, Huang
XR, Fingerle-Rowson G, Bacher M, Scher LA, Bucala R and Lan HY:
Macrophage migration inhibitory factor induces MMP-9 expression:
Implications for destabilization of human atherosclerotic plaques.
Atherosclerosis. 178:207–215. 2005. View Article : Google Scholar
|
|
15
|
Yu X, Lin SG, Huang XR, Bacher M, Leng L,
Bucala R and Lan HY: Macrophage migration inhibitory factor induces
MMP-9 expression in macrophages via the MEK-ERK MAP kinase pathway.
J Interferon Cytokine Res. 27:103–109. 2007. View Article : Google Scholar : PubMed/NCBI
|