Introduction
Kidney cancer is one of the most common types of
malignancy in developed countries, and is increasing in developing
countries (1,2). Kidney cancer is more prevalent in
males, and its estimated incidence and mortality rates in males are
~111,100 and 43,000 per year in developed countries (1). Renal cell carcinoma (RCC) accounts
for ~90% of all renal tumors and 3.7% of all types of cancer in
adults (3,4). Clear cell RCC, which accounts for 80%
of patients with RCC, is the most aggressive histological type of
RCC, which has the highest rate of metastasis and the poorest
prognosis (5). RCC is
characterized by a lack of early-warning signs, protean clinical
manifestations, and resistance to radiotherapy and chemotherapy
(6). Developments in current
understanding of the underlying molecular biology of renal cell
carcinoma have led to systemic treatments, which have markedly
improved patient outcomes (7).
Therefore, it is necessary and worthwhile to identify novel
biomarkers for RCC to improve diagnosis and treatment.
MicroRNAs (miRNAs), which regulate ~50% of human
genes by binding to the 3′-untranslated regions (UTRs), are
important in a wide range of biological and pathological processes,
including cell differentiation, migration, growth, proliferation,
apoptosis and metabolism (7–10).
In tumorigenesis, miRNAs can function as oncogenes and tumor
suppressors, and are predominantly dependent on their target genes
(11). The aberrant expression of
miRNAs has been reported between malignant and normal renal
tissues, including between four histological subtypes of RCC, which
suggest that miRNAs may provide a useful tool for diagnostic and
prognostic improvements, and for the identification of predictive
biomarkers (12). Specific miRNAs
implicated in the pathogenesis of RCC include miRNA-23b, which
targets the tumor suppressor gene, proline oxidase, which is
unregulated in RCC, and functions as an oncogene by promoting
proliferation and suppressing apoptosis. Therefore, decreasing the
expression of miR-23b may be an effective method to inhibit kidney
tumor growth (13).
Previously, studies have found that miR-130b is
dysregulated in several types of human cancer, including chronic
myeloid leukemia (14), T-cell
leukemia (15), melanoma (16), head and neck cancer (17), thyroid cancer (18), pituitary cancer (19), ovarian cancer (20), endometrial cancer (21), colorectal cancer (22), gastric cancer (23), esophageal cancer (24), bladder cancer (25), prostate cancer (26), pancreatic cancer (27) and non-small cell lung cancer
(28). Previous microarray chip
studies have shown that the expression of miR-130b is significantly
higher in RCC tissues, compared with adjacent normal tissues
(29,30). However, the expression of miR-130b
in RCC has not been validated by quantitative polymerase chain
reaction (qPCR) analysis, and the function of miR-130b in RCC
requires further investigation. The aim of the present study was to
validate the upregulation of miR-130b, and examine the effects of
miR-130b on cellular migration, proliferation and apoptosis in RCC.
The present study indicated that miR-130b is a potential biomarker
for early diagnosis and a therapeutic target for the treatment of
RCC.
Materials and methods
Cell lines and transfection
Human RCC cell lines (786-O and ACHN) and the 293T
normal human embryo kidney cell line, were obtained from Guangdong
and Shenzhen Key Laboratory of Male Reproductive Medicine and
Genetics (Shenzhen, China), and were cultured in Dulbecco's
modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), containing 10% fetal bovine serum (Gibco; Thermo
Fisher Scientific, Inc.) and 1% antibiotics (100 µ/ml
penicillin and 100 mg/ml streptomycin sulfates; Gibco). All cells
were incubated at 37°C in a humidified chamber containing 5%
CO2. The 786-O, ACHN and 293T cells (4×105
cells/well) were plated into 6-well plates (BD Biosciences,
Franklin Lakes, NJ, USA) with three replicate wells, respectively.
Total RNA, including miRNA, was extracted using an miRneasy Mini
kit (Qiagen, Valencia, CA, USA) following harvesting of cells with
trypsin.
In order to downregulate the expression of miR-130b
in the cells, an miRNA oligonucleotide was chemically synthesized
by GenePharma Company, Ltd. (Shanghai, China). The sequences were
as follows: hsa-miR-130b inhibitor, 5′-AUGCCCUUUCAUCAUUGCACUG-3′;
and hsa-miR-130b inhibitor negative control (NC),
5′-CAGUACUUUUGUGUAGUACAA-3′.
Clinical specimens
Paired clinical specimens (size, 0.5×0.5×0.5
cm3) of fresh RCC and adjacent normal tissues (located
2.0 cm outside of the visible RCC lesions), were obtained from 42
patients with RCC from the Department of Urology, Peking University
Shenzhen Hospital (Shenzhen, China) between September 2012 and
November 2014. Written informed consent was obtained from all
patients, and the collection and use of the samples included in the
present study were reviewed and approved by the ethics committee of
the Peking University Shenzhen Hospital (Shenzhen, China). Once
dissected, all fresh samples were immersed in RNAlater (Qiagen),
frozen in liquid nitrogen and then stored at −80°C. The
clinicopathological information of the patients is presented in
Table I. Total RNA, including
miRNA, was extracted from all tissue specimens using an miRneasy
Mini kit (Qiagen, Valencia, CA, USA).
 | Table IClinicopathological characteristics
of patients with renal cell carcinoma. |
Table I
Clinicopathological characteristics
of patients with renal cell carcinoma.
| Characteristic | Cases (n) |
|---|
| Mean age (range;
years) | 53 (29–72) |
| Gender | |
| Male/female | 28/14 |
| Histological
type | |
| Clear
cell/papillary | 36/6 |
| pT-stage | |
| T1/T2/T3+T4 | 24/16/2 |
| Fuhrman grade | |
| I/II/III/IV | 14/18/7/3 |
| AJCC clinical
stages | |
| I/II/III+IV | 24/15/3 |
RNA purification and
reverse-transcription qPCR (RT-qPCR)
Tissue samples were homogenized in 1 ml TRIzol
reagent (Invitrogen; Thermo Fisher Scientific) per 100 mg of tissue
using a power homogenizer (Tiangen Biochemical Science and
Technology Co., Ltd., Beijing, China). Subsequently, homogenates
were incubated for 5 min at room temperature to permit the complete
dissociation of nucleoprotein complexes. Subsequent to vigorous
manual agitation for 15 sec, samples were centrifuges at 12,000 × g
for 15 min at 4°C. The total RNA, which was extracted from the
tissues and cells, was purified using an RNeasy® Maxi
kit (Qiagen), according to the manufacturer's protocol. The RNA
samples with 260/280 ratios of 1.8–2.0 were used for further
experiments. A total of 1 µg total RNA was reverse
transcribed into cDNA using an miScript II RT kit (Qiagen).
The expression levels of miR-130b were analyzed
using an miScriptSYBR®green PCR kit (Qiagen) using the
Roche light-cycler 480 Real-Time PCR system (Roche Diagnostics
GmbH, Mannheim, Germany). The 20-µl reaction mixture
contained 10 µl 2X QuantiTect SYBR Green PCR Master mix, 2
µl 10X miScript Universal Primer, 0.4 µl specific
miRNA primer, 1 µl cDNA template and RNase-free water. The
thermocycling steps were 95°C for 15 min, followed by 40 cycles of
94°C 15 sec, 55°C for 30 sec and 72°C for 30 sec. U6 was used as an
endogenous control to normalize the data. The mRNA expression
levels were presented as the fold difference relative to U6, which
was based on the relative quantification equation
(2−ΔΔCq): ΔΔCq = (meanCqtumor -
meanCqcontrol) − (meanCqnormal -
meanCqcontrol) (32).
The sequence of the miR-130b forward primer was
5′-CAGTGCAATGATGAAAGGGCAT-3′ and the reverse primer was universal
primer, provided in the miScriptSYBR®green PCR kit
(Qiagen). The primer sequences of U6 were forward
5′-CTCGCTTCGGCAGCACA-3′ and revers 5′-ACGCTTCACGAATTTGCGT-3′.
Wound healing assay
A wound healing assay was performed to evaluate the
migratory ability of the 786-O and ACHN cells in vitro. The
786-O or ACHN cells (~3×105/dish) were seeded into
12-well dishes and cultured for 24 h, prior to being transfected
with 100 pmol of either the miR-130b inhibitor or the negative
control using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific). At 6 h post-transfection, a sterile 200
µl pipette tip was used to make a scratch through the cell
layer. The cells were then rinsed with phosphate-buffered saline
(PBS) and cultured in serum-free DMEM at 37°C. Images of the
scratches were captured using a digital camera system, 0, 24 and 48
h following the introduction of the scratches. The relative
migratory distances (%) were measured using an MIAS-2000 computer
image analysis system (Leica Microsystems GmbH, Wetzlar, Germany),
and the experiments were performed in triplicate and repeated at
least three times.
MTT assay
An MTT assay was used to analyze the cell
proliferation in the cell groups. The 786-O or ACHN cells(~5,000
cells) were plated into each well of a 96-well, plate with five
replicate wells for each condition. Each well was transfected with
5 pmol miR-130b inhibitor or negative control, and measurements
were obtained at 0, 24, 48 or 72 h post-transfection. Blank control
wells were also included, which contained DMEM only. Prior to the
measurement, 20 µl MTT (5 mg/ml; Sigma-Aldrich, St. Louis,
MO, USA) was added to each well, and the 96-well plates were
incubated at 37°C in a humidified chamber containing 5%
CO2 for 6 h. Following incubation, the MTT medium
mixtures were discarded and 120 µl dimethyl sulphoxide
(DMSO; Sigma-Aldrich) was added. Following agitation for 30 min at
room temperature, the absorbance was measured using an ELISA
microplate reader (Model 680; Bio-Rad Laboratories, Inc., Hercules,
CA, USA) at a wavelength of 490 nm (with 630 nm as the reference
wavelength).
Cell apoptosis assay
Enumeration of the rate of apoptosis of the cells
was performed by staining with fluorescein isothiocyanate
(FITC)-conjugated Annexin V and propidium iodide (PI), obtained
from Invitrogen (Thermo Fisher Scientific, Inc.) using flow
cytometry (Epics Xl-4, Beckman Coulter, Brea, CA, USA). The 786-O
or ACHN cells (~3×105) were plated in each well of
6-well plates for the cell apoptosis assay. The cells were
transfected with 200 pmol of either miR-130b inhibitor or the
negative control for 6 h. At 48 h post-transfection, the cells,
including floating cells, were harvested, washed twice with 4°C PBS
and resuspended in 100 µl 1X binding buffer [10 mM
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 140 mM NaCl,
2.5 mM CaCl2, pH 7.4; Invitrogen] at a concentration of
at least 3×106 cells per ml. This suspension (100
µl) was stained with 5 µl of Annexin V-FITC and 5
µl PI for 15 min at room temperature in the dark. Following
the addition of 400 µl binding buffer to each tube, the
cells were analyzed using flow cytometry. Each experiment was
performed at least three times.
Statistical analysis
All data are presented as the mean ± standard
deviation of three independent experiments. Statistical analyses
were performed using the SPSS 19.0 statistical software package
(IBM SPSS, Armonk, NY, USA). Statistical significance was
determined using Student's t-test. For comparison of the expression
levels of miR-130b in matched tumor and adjacent normal samples, a
paired t-test was used. P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression of miR-130b is upregulated in
RCC tissues and cells
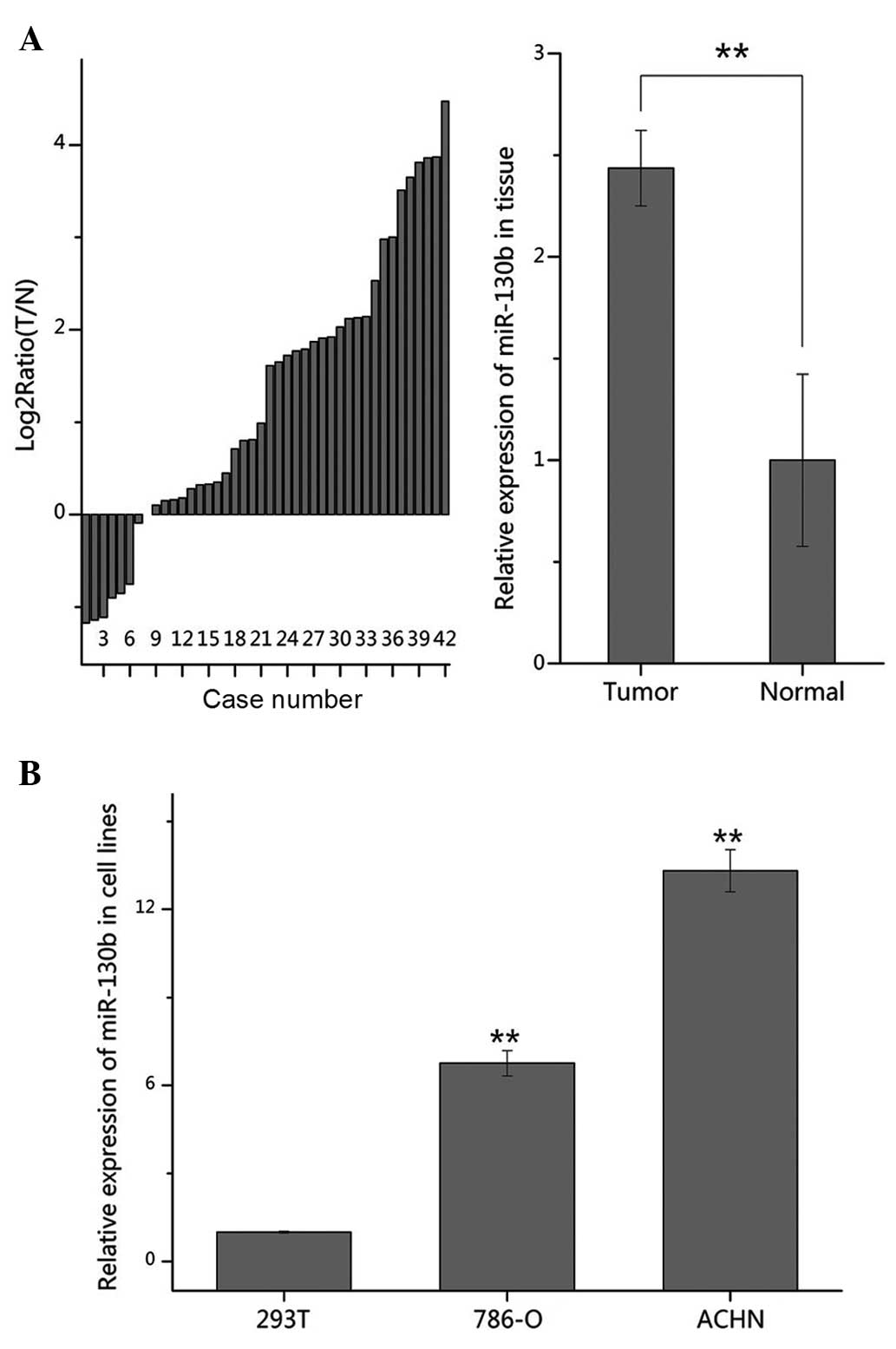
To confirm the expression of miR-130b in RCC, the
present study used RT-qPCR to quantify and compare the expression
levels of miR-13b between the 42 RCC and paired adjacent normal
tissue samples, and between the 786-O and ACHN RCC cell lines and
293T cell line. As shown in Fig.
1A, the expression of miR-130b was significantly increased
(35/42) in the tumor tissues, compared with the normal tissue
samples (P<0.01). It was also revealed that the expression
levels of miR-130b were significantly upregulated in the 786-O and
ACHN cells, compared with the 293T cells (Fig. 1B). These results suggested that
miR-130b acts as an oncogene in RCC.
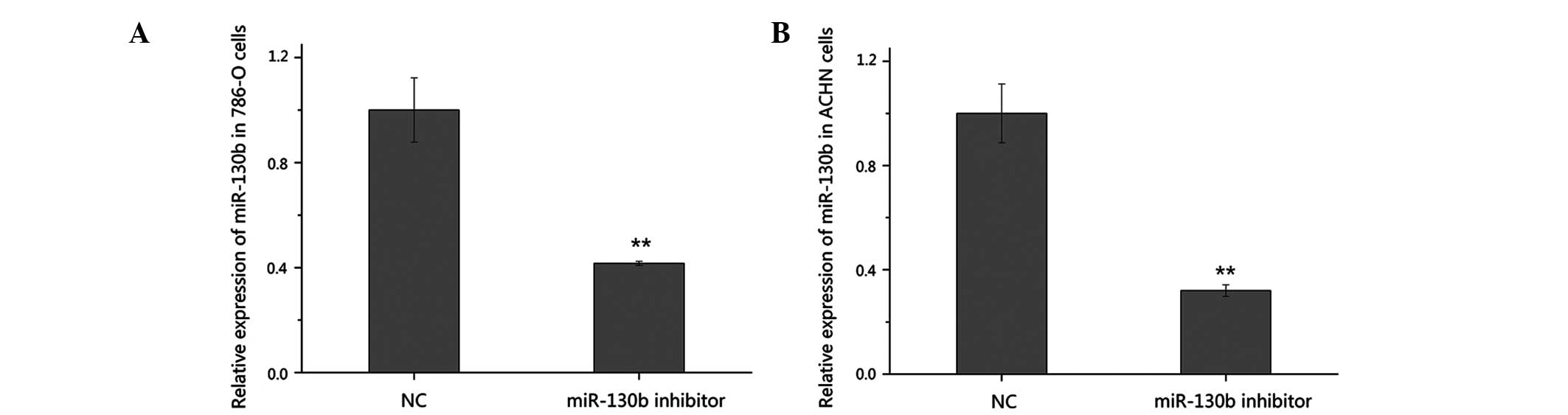
Validation of the suppression of miR-130b
by RT-qPCR
To further investigate the function of miR-130b in
RCC, the high expression level of miR-130b was suppressed in the
present study using a chemically synthesized miR-130b inhibitor.
The silencing efficiency of the miR-130b inhibitor was validated
using RT-qPCR 48 h post-transfection. As shown in Fig. 2, the expression of miR-130b
decreased by 58.4% in the 786-O cells and by 68.0% in the ACHN
cells transfected with the inhibitor, compared with the cells
transfected with the negative control.
A reduction in the expression of miR-130b
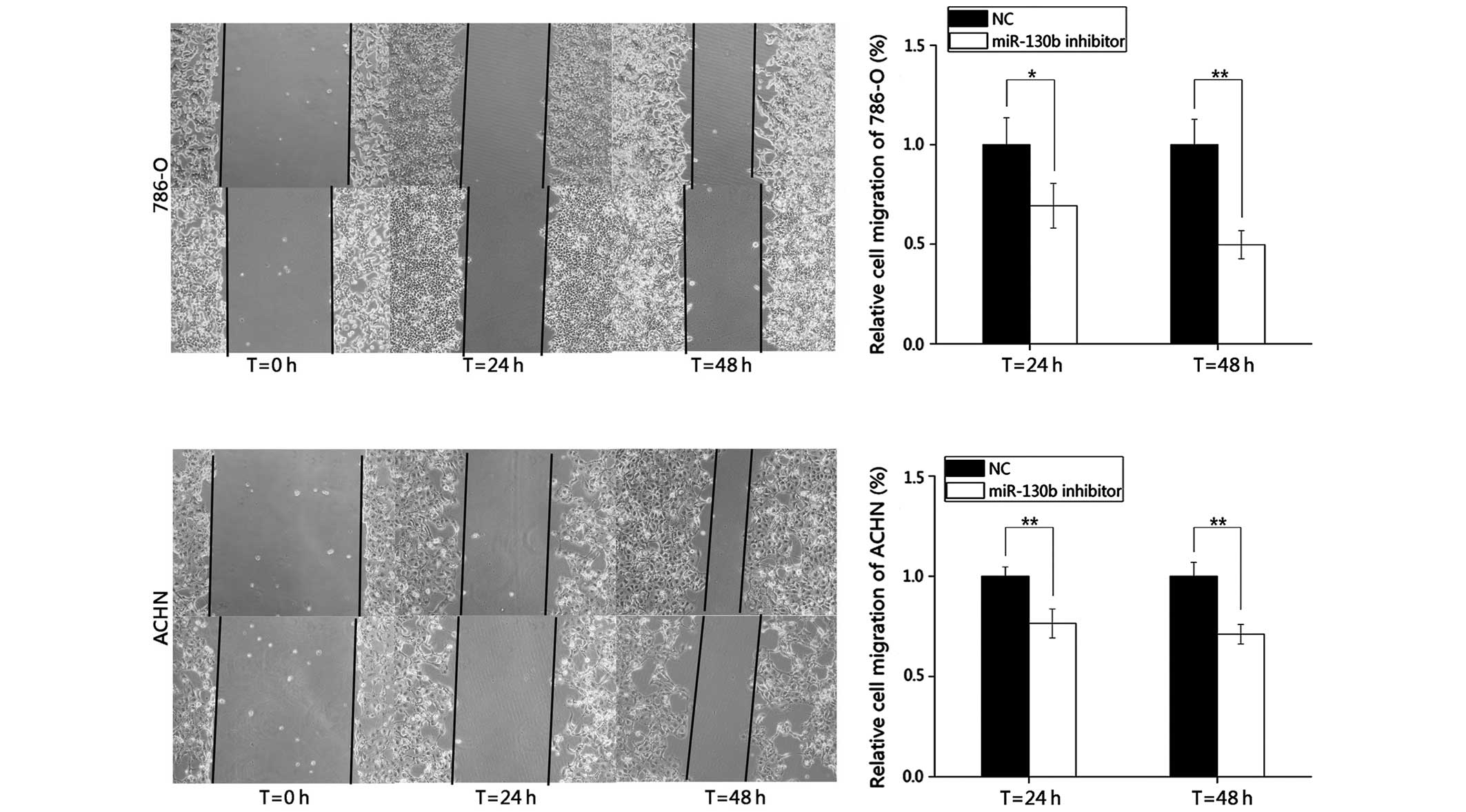
inhibits RCC cell migration
In the present study, wound healing assays were
performed to observe the function of miR-130b in cell migration.
Images of the scratch wounds were captured 0, 24 and 48 h following
transfection using a digital camera system (Fig. 3). The results demonstrated that the
migratory distances of the cells transfected with the miR-130b
inhibitor after 24 and 48 h were markedly inhibited by 30.66 and
50.29% in the 786-O cells, and by 23.54 and 28.92% in the ACHN
cells, respectively, compared with the negative control group.
These results indicated that the reduction in the expression of
miR-130b inhibited the migration of the RCC cells.
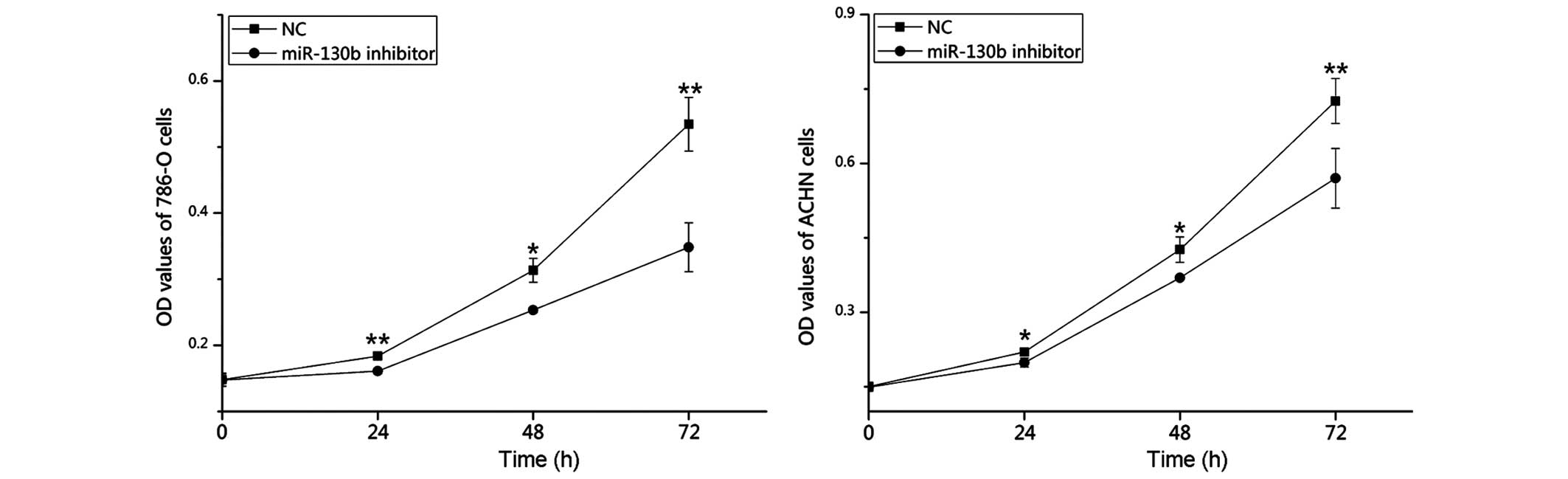
Downregulation of miR-130b suppresses RCC
cell proliferation
The potential effect of miR-130b on the
proliferation of RCC cells was determined using MTT assays. The
miR-130b inhibitor and negative control groups were measured at 0,
24, 48 and 72 h post-transfection. The statistical analyses of the
optical density values demonstrated that the proliferation of the
786-O cells was inhibited at 24, 48 and 72 h by 12.34 (P<0.01),
19.15 (P<0.05) and 34.81% (P<0.01), respectively. In
addition, the proliferation of the ACHN cells decreased by 9.56
(P<0.05), 13.80 (P<0.05) and 23.08% (P<0.01) at 24, 48 and
72 h post-transfection, respectively. These results suggested that
the miR-130b inhibitor reduced the growth of the 786-O and ACHN
cells, compared with the negative control inhibitor (Fig. 4).
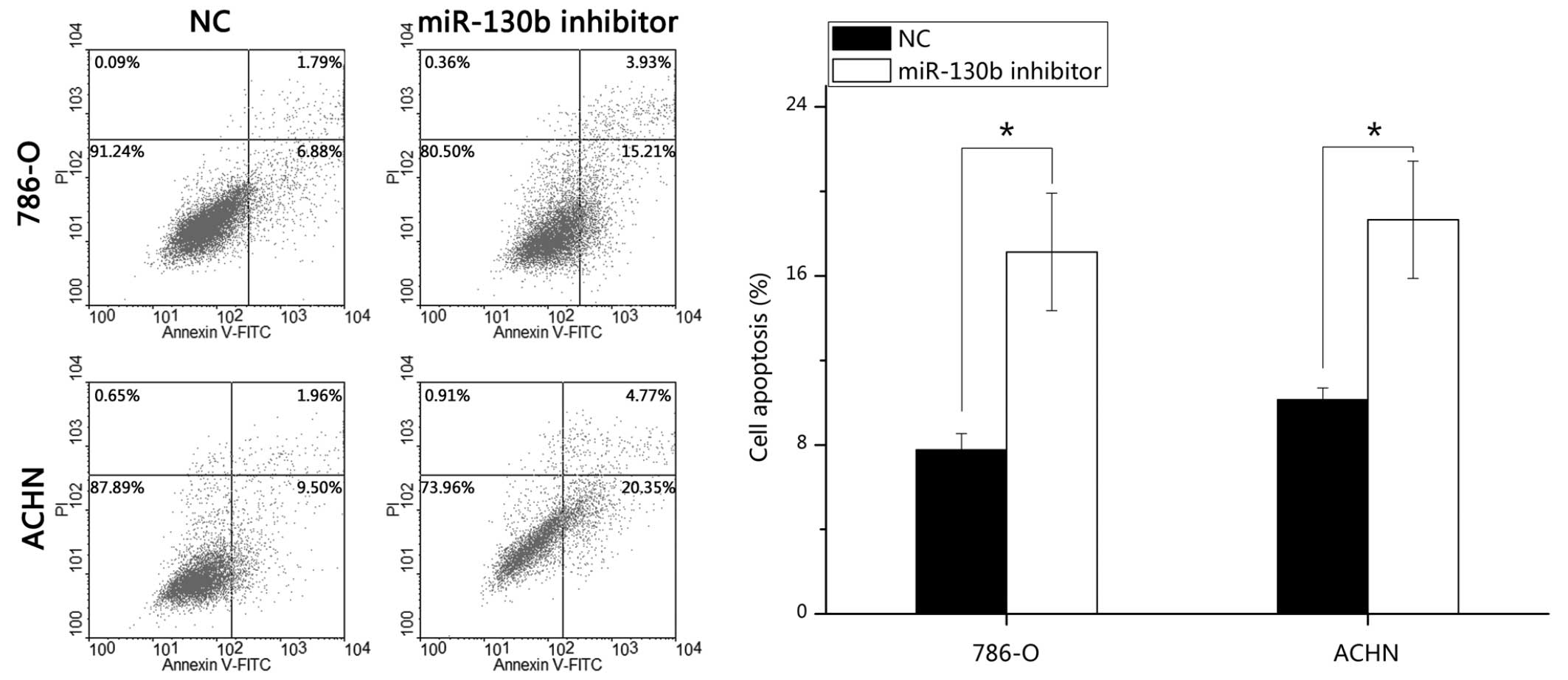
miR-130b inhibitor induces RCC cell
apoptosis
To determine the potential role of miR-130b on the
apoptosis of RCC cells, FITC-conjugated Annexin V and PI assays
were performed. Following transfection of the cells with the
miR-130b inhibitor or negative control for 48 h, the 786-O and ACHN
cells were harvested, stained and analyzed. The results
demonstrated that the average early apoptotic rates of the 786-O
cells transfected with the miR-130b inhibitor and the negative
control were 17.13 and 7.77%, respectively (P<0.05), and the
average apoptotic rates of the ACHN cells were 18.65 and 10.13%,
respectively (P<0.05). These data indicated that the miR-130b
inhibitor promoted RCC cell apoptosis (Fig. 5).
Discussion
The development of cancer involves the
overexpression of oncogenes and silencing of tumor suppressor
genes. The well-known dysregulated signal transduction pathways in
RCC tumorigenesis, including the anti-oncogene, von Hippel Lindau
(VHL) and oncogene, vascular endothelial growth factor (VEGF)
(13). miRNAs are a class of small
non-coding RNA, which exert their function by targeting specific
genes, including hypoxia-inducible factor, mammalian target of
rapamycin, VEGF and VHL, which are key molecules involved in the
initiation and development of clear cell RCC, through translation
inhibition or the induction of mRNA degradation (32,33).
For example, miR-141 and 200c, which are downregulated in RCC,
affect epithelial-mesenchymal transition (EMT) via zinc finger
homeobox 1b-mediated repression of E-cadherin (34). In addition, the increased
expression of miR-29b, due to ectopic expression of VHL, inhibits
the protein expression of TIs11B, a known negative regulator of
VEGF (35), suggesting that
miR-29b possesses oncogenic activity. By targeting different
important genes, affecting cellular proliferation, differentiation,
migration and apoptosis, miRNAs have been shown to be crucial in
carcinogenesis and cancer progression.
miR-130b has been reported to be dysregulated in
several types of cancer (14–28),
in which the expression of miR-130b is either upregulated or
downregulated. miR-130b is either upregulated and function as an
oncogene by targeting tumor suppressor genes, or downregulated and
identified as an anti-oncogene by decreasing oncogenes.
Chronic myeloid leukaemia (CML) is a
myeloproliferative disorder, which is characterized by the
expression of the oncoprotein, Bcr-Abl kinase. miRNA-130a/b are
regulated by BCR-ABL and downregulate the expression of the tumor
suppressive gene, CCN3 (36).
miR-130b is consistently upregulated in human T-cell leukemia virus
1-transformed cells, targeting the tumor suppressor protein, tumor
protein 53-induced nuclear protein 1 (15). Leone et al reported that the
levels of miR-130b are markedly reduced in pituitary adenomas, and
that the overexpression of miR-130b inhibits cell proliferation,
arresting cells in the G1 and G2 phases of the cell cycle by
targeting cyclin A2 (19). In
endometrial cancer, the overexpression of miR-130b and loss of
DICER1 induced abnormal expression of EMT-associated genes, which
constitute a loop regulation of the miR-130b-DICER1-EMT axis
(21). In colorectal cancer,
Colangelo et al (22)
demonstrated that the upregulation of miR-130b exhibits clinical
relevance, as it is linked to advanced cases of colorectal cancer,
poor patient prognosis, and molecular features of enhanced EMT and
angiogenesis by directly targeting peroxisome
proliferator-activated receptor γ (PPARγ) in vitro and in
vivo. From another perspective, miR-130b significantly
decreases cell migration and invasion by downregulating integrin β1
(37). In gastric cancer, the
overexpression of miR-130b increases cell viability, reduces cell
death and decreases the expression of B cell lymphoma-2-interacting
mediator of cell death in transforming growth factor-β mediated
apoptosis, subsequent to the downregulation in the protein
expression of Runt-related transcription factor 3 (23). However, miR-130b is significantly
downregulated and exerts a suppressive effect in prostate and
pancreatic cancer metastasis through the downregulation of matrix
metalloproteinase 2 and signal transducer and activator of
transcription 3, respectively (26,27).
However, the expression and function of miR-130b in
RCC have not been reported previously. Previous miRNA profiling
studies have shown the upregulation of miR-130b in RCC (29,30).
In the present study, the increased expression of miR-130b as
validated in RCC tissues and 786-O and ACHN RCC cell lines,
compared with adjacent normal tissues and 293T cell lines, using
RT-qPCR. In addition, the downregulation of miR-130b through
synthesized miR-130b inhibitor was found to weaken cellular
migration and proliferation, and induce apoptosis in the 786-O and
ACHN cells, which was shown in the wound-healing, MTT and apoptosis
assays, respectively. These results suggested that miR-130b may be
characterized as an oncogene in RCC by regulating cell migration,
proliferation and apoptosis.
It appears contradictory that miR-130b was
identified as an oncogene in certain types of cancer, but as a
tumor suppressor gene in others. This inconsistency may be
explained by imperfect binding of the miRNA to the 3′-UTR in
mammals (8). Due to imperfect
complementarity, a single miRNA can potentially regulate hundreds
of genes, including oncogenes and tumor suppressor genes, and a
specific gene can be regulated by several miRNAs. In addition, the
expression pattern of genes and miRNAs is tissue-, organ- and
time-specific. Therefore, miR-130b is upregulated in CML,
endometrial cancer, colorectal cancer and gastric cancer, but
reduced in pituitary adenoma, prostate and pancreatic cancer. Even
in the same type of cancer, by targeting different genes, a
specific miRNA can function differently. For example, in colorectal
cancer, miR-130b decreases cell migration and invasion by
decreasing intergrin β1 (37), but
promotes cell proliferation, EMT and angiogenesis by targeting
PPARγ (22).
To the best of our knowledge, the results of the
present study provide novel insight into the roles and possible
mechanisms underlying the effects of miR-130b in the occurrence and
development of RCC. miR-130b was significantly upregulated in human
RCC tissues and cell lines, and was observed to function as an
oncogene by affecting cell migration, proliferation and apoptosis
in RCC cell lines. In addition, the data obtained in the present
study suggest that miR-130b may be a promising biomarker for early
diagnosis, and a therapeutic target for the treatment of RCC.
Further investigations are required to define the roles and target
genes of miR-130b in RCC and other types of cancer.
Acknowledgments
The present study was supported by the National
Natural Science Foundation of China (no. 81101922), the Science and
Technology Development Fund Project of Shenzhen (nos.
CYJ20130402114702124 and JCYJ20150403091443329) and the fund of
Guangdong Key Medical Subject.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Zhang S, Zhao P, Zeng H,
Zou X and He J: Annual report on status of cancer in China, 2010.
Chin J Cancer Res. 26:48–58. 2014.PubMed/NCBI
|
|
3
|
Tavani A and La Vecchia C: Epidemiology of
renal-cell carcinoma. J Nephrol. 10:93–106. 1997.PubMed/NCBI
|
|
4
|
National Cancer Institute: Surveillance,
Epidemiology and End Results Program. SEER Stat Fact Sheets: Kidney
and Renal Pelvis Cancer. http://seer.cancer.gov/statfacts/html/kidrp.html.
Accessed December 12, 2015.
|
|
5
|
Rini BI, Campbell SC and Escudier B: Renal
cell carcinoma. Lancet. 373:1119–1132. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Motzer RJ, Bander NH and Nanus DM:
Renal-cell carcinoma. N Engl J Med. 335:865–875. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huntzinger E and Izaurralde E: Gene
silencing by microRNAs: Contributions of translational repression
and mRNA decay. Nat Rev Genet. 12:99–110. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Krol J, Loedige I and Filipowicz W: The
widespread regulation of microRNA biogenesis, function and decay.
Nat Rev Genet. 11:597–610. 2010.PubMed/NCBI
|
|
10
|
Carthew RW and Sontheimer EJ: Origins and
Mechanisms of miRNAs and siRNAs. Cell. 136:642–655. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fendler A, Stephan C, Yousef GM and Jung
K: MicroRNAs as regulators of signal transduction in urological
tumors. Clin Chem. 57:954–968. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rydzanicz M, Wrzesiński T, Bluyssen HA and
Wesoły J: Genomics and epigenomics of clear cell renal cell
carcinoma: Recent developments and potential applications. Cancer
Lett. 341:111–126. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu W, Zabirnyk O, Wang H, Shiao YH,
Nickerson ML, Khalil S, Anderson LM, Perantoni AO and Phang JM:
MiR-23b targets proline oxidase, a novel tumor suppressor protein
in renal cancer. Oncogene. 29:4914–4924. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ferreira AF, Moura LG, Tojal I, Ambrósio
L, Pinto-Simões B, Hamerschlak N, Calin GA, Ivan C, Covas DT,
Kashima S and Castro FA: ApoptomiRs expression modulated by BCR-ABL
is linked to CML progression and imatinib resistance. Blood Cells
Mol Dis. 53:47–55. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yeung ML, Yasunaga J, Bennasser Y, Dusetti
N, Harris D, Ahmad N, Matsuoka M and Jeang KT: Roles for microRNAs,
miR-93 and miR-130b and tumor protein 53-induced nuclear protein 1
tumor suppressor in cell growth dysregulation by human T-cell
lymphotrophic virus 1. Cancer Res. 68:8976–8985. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sand M, Skrygan M, Sand D, Georgas D,
Gambichler T, Hahn SA, Altmeyer P and Bechara FG: Comparative
microarray analysis of microRNA expression profiles in primary
cutaneous malignant melanoma, cutaneous malignant melanoma
metastases and benign melanocytic nevi. Cell Tissue Res. 351:85–98.
2013. View Article : Google Scholar
|
|
17
|
Chen Z, Jin Y, Yu D, Wang A, Mahjabeen I,
Wang C, Liu X and Zhou X: Down-regulation of the microRNA-99 family
members in head and neck squamous cell carcinoma. Oral Oncol.
48:686–691. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dettmer MS, Perren A, Moch H, Komminoth P,
Nikiforov YE and Nikiforova MN: MicroRNA profile of poorly
differentiated thyroid carcinomas: New diagnostic and prognostic
insights. J Mol Endocrinol. 52:181–189. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Leone V, Langella C, D'Angelo D, Mussnich
P, Wierinckx A, Terracciano L, Raverot G, Lachuer J, Rotondi S,
Jaffrain-Rea ML, et al: Mir-23b and miR-130b expression is
downregulated in pituitary adenomas. Mol Cell Endocrinol. 390:1–7.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang L, Zhu MJ, Ren AM, Wu HF, Han WM, Tan
RY and Tu RQ: A ten-microRNA signature identified from a
genome-wide microRNA expression profiling in human epithelial
ovarian cancer. PLoS One. 9:e964722014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li BL, Lu C, Lu W, Yang TT, Qu J, Hong X
and Wan XP: MiR-130b is an EMT-related microRNA that targets DICER1
for aggression in endometrial cancer. Med Oncol. 30:4842013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Colangelo T, Fucci A, Votino C, et al:
MicroRNA-130b promotes tumor development and is associated with
poor prognosis in colorectal cancer. Neoplasia. 15:1086–1099. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lai KW, Koh KX, Loh M, Tada K, Subramaniam
MM, Lim XY, Vaithilingam A, Salto-Tellez M, Iacopetta B, Ito Y and
Soong R; Singapore Gastric Cancer Consortium: MicroRNA-130b
regulates the tumour suppressor RUNX3 in gastric cancer. Eur J
Cancer. 46:1456–1463. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhao BS, Liu SG, Wang TY, Ji YH, Qi B, Tao
YP, Li HC and Wu XN: Screening of microRNA in patients with
esophageal cancer at same tumor node metastasis stage with
different prognoses. Asian Pac J Cancer Prev. 14:139–143. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ratert N, Meyer HA, Jung M, Lioudmer P,
Mollenkopf HJ, Wagner I, Miller K, Kilic E, Erbersdobler A, Weikert
S and Jung K: MiRNA profiling identifies candidate mirnas for
bladder cancer diagnosis and clinical outcome. J Mol Diagn.
15:695–705. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen Q, Zhao X, Zhang H, Yuan H, Zhu M,
Sun Q, Lai X, Wang Y, Huang J, Yan J and Yu J: MiR-130b suppresses
prostate cancer metastasis through down-regulation of MMP2. Mol
Carcinog. 23:222042014.
|
|
27
|
Zhao G, Zhang JG, Shi Y, Qin Q, Liu Y,
Wang B, Tian K, Deng SC, Li X, Zhu S, et al: MiR-130b is a
prognostic marker and inhibits cell proliferation and invasion in
pancreatic cancer through targeting STAT3. PLoS One. 8:e738032013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mitra R, Edmonds MD, Sun J, Zhao M, Yu H,
Eischen CM and Zhao Z: Reproducible combinatorial regulatory
networks elucidate novel oncogenic microRNAs in non-small cell lung
cancer. RNA. 20:1356–1368. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wulfken LM, Moritz R, Ohlmann C,
Holdenrieder S, Jung V, Becker F, Herrmann E, Walgenbach-Brünagel
G, von Ruecker A, Müller SC and Ellinger J: MicroRNAs in renal cell
carcinoma: Diagnostic implications of serum miR-1233 levels. PLoS
One. 6:e257872011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wu X, Weng L, Li X, Guo C, Pal SK, Jin JM,
Li Y, Nelson RA, Mu B, Onami SH, et al: Identification of a
4-microRNA signature for clear cell renal cell carcinoma metastasis
and prognosis. PLoS One. 7:e356612012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
32
|
Maher ER: Genomics and epigenomics of
renal cell carcinoma. Semin Cancer Biol. 23:10–17. 2013. View Article : Google Scholar
|
|
33
|
Chow TF, Mankaruos M, Scorilas A, Youssef
Y, Girgis A, Mossad S, Metias S, Rofael Y, Honey RJ, Stewart R, et
al: The miR-17-92 cluster is over expressed in and has an oncogenic
effect on renal cell carcinoma. J Urol. 183:743–751. 2010.
View Article : Google Scholar
|
|
34
|
Nakada C, Matsuura K, Tsukamoto Y,
Tanigawa M, Yoshimoto T, Narimatsu T, Nguyen LT, Hijiya N, Uchida
T, Sato F, et al: Genome-wide microRNA expression profiling in
renal cell carcinoma: Significant down-regulation of miR-141 and
miR-200c. J Pathol. 216:418–427. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sinha S, Dutta S, Datta K, Ghosh AK and
Mukhopadhyay D: Von Hippel-Lindau gene product modulates TIS11B
expression in renal cell carcinoma: Impact on vascular endothelial
growth factor expression in hypoxia. J Biol Chem. 284:32610–32618.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Suresh S, McCallum L, Lu W, Lazar N,
Perbal B and Irvine AE: MicroRNAs 130a/b are regulated by BCR-ABL
and down-regulate expression of CCN3 in CML. J Cell Commun Signal.
5:183–191. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhao Y, Miao G, Li Y, Isaji T, Gu J, Li J
and Qi R: MicroRNA-130b suppresses migration and invasion of
colorectal cancer cells through downregulation of integrin β1
[corrected]. PLoS One. 9:e879382014. View Article : Google Scholar
|