Introduction
Streptococcus suis is a major pathogen
affecting pigs. It is endemic in countries involved in pig
husbandry; however, it may also lead to meningitis, endocarditis,
septicemia, arthritis, polyserositis, pneumonia and sudden death in
pigs. Occasionally, it may lead to serious zoonotic infections in
humans (1). The large-scale
outbreak of human S. suis (type) 2 infection, with the
feature of streptococcal toxic shock syndrome, in the Jiangsu and
Sichuan provinces of China (2)
indicated that S. suis remains a challenge for public
health.
Serotype 2 of S. suis (also termed SS2) is
considered to be the most virulent of the 33 established serotypes
of this pathogen. However, the primary factors contributing to its
virulence have yet to be fully elucidated. Previous studies on
virulence-associated factors of SS2 have focused primarily on the
bacterial capsular polysaccharide, muramidase-released protein,
extracellular protein factor and suilysin (3,4).
Over the last decade, a large number of putative virulence factors
associated with SS2 have been investigated (4), including fibronectin- and
fibrinogen-binding proteins (5),
opacity factor of S. suis (6), peptidoglycan (7), glutamine synthetase (8), di-peptidyl peptidase IV (9), inosine 5-monophosphate dehydrogenase
(10), trigger factor tig
gene (11), virulence-associated
gene A (12), Rgg transcription
regulator (13),
surface-associated subtilisin-like protease (14), catabolite control protein A
(15) and superoxide dismutase A
(16). Notably, an 89 K
pathogenicity island (PAI) (17)
and SalK/SalR (a two-component signal transduction system)
(18) have been identified as
requisites for the full virulence of ethnic Chinese isolates of
highly pathogenic SS2. However, the importance of these proteins in
the pathogenicity of SS2, and the pathogenesis of the infection
triggered by S. suis, remain to be elucidated.
To identify genes contributing to the virulence of
virulent strains, a previous study conducted suppression and
subtractive hybridization using a ZY458 virulent SS2 strain and a
13w avirulent SS2 strain (19). A
total of 42 genomic regions were identified as being present in the
virulent strain, but were absent in the avirulent one (19). Protein E gene (termed vapE)
is one of these 42 genes, although it is absent in the non-virulent
SS2 strain 1330. The objective of the present study was to
investigate the effects of the ∆458VapE mutation on the virulence
of SS2.
Materials and methods
Bacterial strains, plasmids and
primers
Bacterial strains and plasmids used in the present
study are listed in Table I. S.
suis 2 strains were cultured in brain-heart infusion (BHI)
broth (Difco; BD Biosciences, Franklin Lakes, NJ, USA) supplemented
with 5% (v/v) calf serum or BHI agar at 37°C. Escherichia
coli strain DH5α was used for cloning purposes and was cultured
in Luria broth (LB) or LB agar at 37°C (20). When recombinants were screened or
cultured, plates or broth were supplemented with appropriate
antibiotics at the following concentrations: i) Spectinomycin
(Spc), 100 µg/ml for S. suis 2 with plasmid pSET4s
and 50 µg/ml for DH5α with plasmid pSET4; ii) ampicillin
(Amp), 100 µg/ml for DH5α with plasmid pMD18-T; iii)
erythromycin (Ery), 8 µg/ml for S. suis 2 with
plasmid pAT18 and 150 µg/ml for DH5α with plasmid pAT18 (all
antibiotics from Sigma-Aldrich, St. Louis, MO, USA). DNA
extraction, cloning, transformation, and other molecular techniques
used in the present study were implemented following protocols
described previously (20). All
primer synthesis and DNA sequencing were outsourced to Invitrogen
(Thermo Fisher Scientific, Inc., Waltham, MA, USA).
 | Table IBacterial strains and plasmids used
in the current study. |
Table I
Bacterial strains and plasmids used
in the current study.
A, Plasmid
|
|---|
| Name | Description | Source |
|---|
| pMD18-T | A clone vector | Takara |
| pSET4s | Suicide vector,
shuttle vector between E. coli and S. suis (8,22) | Huazhong
Agricultural University, China |
| pAT18 | Shuttle vector
between E. coli and S. suis (21,29) | Huazhong
Agricultural University, China |
B, Bacterial strain
|
|---|
| Name | Description | Source |
|---|
| DH5α | Host cell for
maintaining the recombinant plasmids | Takara |
| ZY458 | S. suis 2
wild-type strain, diseased pig in Sichuan in 2005,
mrp+epf+sly+ | Our laboratory |
| 1330 | S. suis 2,
avirulence reference strain,
mrp−epf−sly− | Canada (30) |
| SP3 | S. suis 2,
Spanish strain, diseased pig,
mrp+epf+sly+ | Spain (31) |
| SP6 | S. suis 2,
Spanish strain, diseased pig,
mrp+epf+sly+ | Spain (31) |
| SP8 | S. suis 2,
carrier strain, healthy pig,
mrp−epf–sly− | Spain (31) |
| ZD89 | S. suis 2,
carrier strain, healthy pig,
mrp−epf–sly− | Veterinary
Institute of Harbin, China |
| B22 | S. suis 2,
carrier strain, healthy pig,
mrp−epf−sly− | Veterinary
Institute of Harbin, China |
| B3 | S. suis 2,
carrier strain, healthy pig,
mrp−epf–sly− | Veterinary
Institute of Harbin, China |
|
458ΔvapE | vapE
deletion mutant of S. suis 2 strain ZY458 | The present
study |
| 458ΔvapE
(pvapE) | Complemented strain
of 458ΔvapE, carrying the recombinant plasmid
pAT18::vapE | The present
study |
Construction of the vapE mutant
strain
The primers used in the current study are presented
in Table II. The PCR was
performed according to a standard protocol. Each reaction was
conducted using a 50 µl mixture containing 5 µl 10X
buffer, 50 pmol each primer, 2 mM each deoxynucleoside
triphosphate, 5 units Ex Taq polymerase (all obtained from Takara
Biotechnology Co., Ltd., Dalian, China) and 5 µl supernatant
of denatured bacteria. The PCR was performed with a Techgene
FTGENE2D thermocycler (Techne Ltd., Duxford, UK). In order to
generate a vapE gene-deleted mutant, a primer set was
designed with a 194 bp internal deletion in the vapE gene by
overlap extension polymerase chain reaction (OE PCR), using
denatured bacteria as the DNA template (20). Two pairs of primers (VapE-1/VapE-2
and VapE-3/VapE-4) were used to independently amplify the 674 and
743 bp fragments of vapE, including flanking sequences from
genomic DNA of the wild-type SS2 strain, ZY458. 16S rDNA was used
as the reference gene, the primers were as follows: Sense,
5′-AGAGTTTGATCCTGGCTCAG-3′ and antisense,
5′-ACGGCTACCTTGTTACGACTT-3′. Amplification was performed as
follows: i) Initial denaturation at 94°C for 3 min; ii) 30 cycles
of 94°C for 30 sec, 60°C for 30 sec and 72°C for 50 sec; and iii) a
final elongation step at 72°C for 10 min. Primers VapE-2 and VapE-3
contained 15 nt stretches complementary to each other; thus, the
PCR fragments were fused by OE PCR using primers VapE-1 and VapE-4.
The following PCR protocol was used: 3 min at 94°C, followed by 30
cycles at 94°C for 30 sec, 59°C for 30 sec and 72°C for 1 min 40
sec, followed by 72°C for 10 min. The resultant 1,417 bp PCR
product contained an internal in-frame deletion of 194 bp in the
vapE gene (from 1 to 194 nt). The PCR product was purified
by DNA Gel Extraction kit (Takara Biotechnology Co., Ltd.) Band
subsequently cloned into a pMD18-T vector using a pMD18-T Vector
Cloning kit (Takara Biotechnology Co., Ltd.) to generate the
pMD18-T::ΔvapE plasmid. The pMD18-T::ΔvapE plasmid was digested
with BamHI and EcoRI enzymes (Takara Biotechnology
Co., Ltd., Dalian, China), and the DNA fragment containing the
mutated vapE (ΔvapE) was then cloned into a pSET4s
thermosensitive suicide plasmid (21) to generate the pSET4s::ΔvapE suicide
plasmid. The resultant plasmid was confirmed by DNA sequencing and
transfected into a ZY458 SS2 strain to screen for deletion mutants
as described by Takamatsu et al (22). Subsequently, the SS2 strain ZY458
was electrotransfected with pSET4s::ΔvapE using the ECM 399
electroporation system (BTX Harvard Apparatus, Inc., Holliston, MA,
USA) at 2,000 V, and cultured at 28°C in the presence of Spc to
select the recombinant. The resultant ZY458 (pSET4s::ΔvapE) cells
were cultured in BHI broth with Spc at 28°C until the early
logarithmic growth phase, and then were shifted to 37°C and
incubated for an additional 10 h. Subsequently, the cultures were
diluted and spread onto BHI agar plates without antibiotic and
incubated overnight at 37°C. The cultures were screened for mutants
that had lost the vectors and had exchanged their wild-type allele
for a genetic segment containing the ΔvapE gene as a consequence of
homologous recombination via a double cross-over. A resultant
458ΔvapE mutant strain was verified by PCR amplification with the
primers VapE-5/VapE-R and further confirmed by DNA sequencing. The
PCR cycle protocol, performed on Techgene FTGENE2D thermocycler,
was as follows: i) Initial denaturation at 94° for 3 min; ii) 30
cycles of 94°C for 30 sec, 50°C for 30 sec and 72°C for 40 sec; and
iii) a final elongation step at 72°C for 10 min.
 | Table IIPrimers used for PCR amplification
and identification. |
Table II
Primers used for PCR amplification
and identification.
| Primer | Sequence
(5′-3′) | Restriction
site | Positiona (bp) |
|---|
| VapE-1 | GGATCCCACCAGCTTGCACATCGTC | BamHI | +850 to +868 |
| VapE-2 |
GCCTGTTCCACCTTTGATAGTTGCCC | | +194 to +220 |
| VapE-3 |
AAAGGTGGAACAGGCCTTCTTTCTATGGTC | | +195 to +209; −1 to
−15 |
| VapE-4 | GAATTCGAAAACCCCGAAATTTATCAAGTG | EcoRI | −721 to −744 |
| VapE-5 |
CCCTATCATTGATATAAATTCCCTC | | +301 to +325 |
| VapE-U |
TAGGTTTCCCCTTAAAGTGC | | +1059 to +1078 |
| VapE-D |
TACACCGCTAAACCTCTTTC | | −929 to −948 |
| VapE-F | GAATTCTCAGATTGTCATATTCACTAG | EcoRI | +1952 to +1972 |
| VapE-R | GGATCCATTGAGAAATACATGTTAG | BamHI | −302 to −320 |
Functional complemented vapE mutant
strain
To generate a functionally complemented strain of
458ΔvapE, the structural gene of vapE with its promoter
sequence (from 320 bp upstream of the start codon to 439 bp
downstream of the stop codon) was amplified using primers VapE-F
and VapE-R. PCR was performed on a Techgene FTGENE2D thermocycler
under the following conditions: i) Initiation and elongation, 3 min
at 94°; ii) 30 cycles of denaturation for 30 sec at 94°, annealing
for 30 sec at 50°C; and iii) elongation for 2 min at 72°C; and a
final elongation step at 72°C for 10 min. The PCR product (2,292
bp) was purified using the DNA Gel Extraction kit and subsequently
cloned into pMD18-T to generate the plasmid, pMD18-T::vapE. The
vapE fragment was then subcloned into shuttle vector pAT18. The
resultant plasmid was verified by DNA sequencing, and subsequently
used to electrotransfect the mutant strain 458ΔvapE using the ECM
399 electroporation system, as described above (BTX Harvard
Apparatus, Inc.), which was plated onto BHI agar supplemented with
Ery to screen for the complemented strain, 458ΔvapE (pvapE).
Bacterial growth curve
Wild-type strain S. suis 2 ZY458, mutant
strain 458ΔvapE and complemented strain 458ΔvapE (pvapE) were
separately inoculated into 100 ml BHI broth and incubated at 37°C.
Samples of culture were monitored by spectrophotometry using a T6
UV spectrophotometer (Beijing Purkinje General Instrument Co., Ltd,
Beijing, China). The absorbance was measured at 600 nm in a quartz
cuvette (Beijing Purkinje General Instrument Co., Ltd.) at 1 h
intervals. BHI broth minus the inoculation of bacteria served as a
blank.
Experimental infection of mice
The present study was approved by the Review Board
of the Academy of Military Medical Sciences (Changchun, China). All
animal experiments were conducted in accordance with the accepted
standards of the Animal Care and Use Committee of Academy of
Military Medical Sciences. The protocol was approved by the Animal
Care and Use Committee of Academy of Military Medical Sciences and
all efforts were made to minimize suffering. All experiments
involving mice were conducted in accordance with the Council for
International Organizations of Medical Sciences: International
Guiding Principles for Biomedical Research Involving Animals
(23). The bacterial cultures were
serially diluted in BHI broth and plated onto BHI agar plates in
order to determine the colony forming unit (CFU)/ml. The working
cultures for experimental infection were adjusted to a final
concentration of 1×109 CFU/ml.
Female BALB/c mice (age, 4 weeks; weight, 13–14 g)
were housed at 24±1°C and 60% relative humidity in a 12-h
light/dark cycle with access to food and water ad libitum.
They were randomly divided into four groups (12 mice/group), and
individual groups were injected intraperitoneally (i.p.) with 100
µl of the ZY458 wild-type strain, ZD89 carrier strain,
458ΔvapE mutant strain or 458ΔvapE (pvapE) complemented strain
cultures. The animals were monitored daily for 1 week for mortality
and clinical signs, including depression, swollen eyes, ruffled
hair, lethargy and nervous symptoms. Tissues of the heart, liver,
spleen, lung and kidney from infected mice (12 mice/group) were
harvested for detection of the SS2 bacteria by plating onto BHI
agar plates. Positive cultures were confirmed by PCR using VapE-F
and VapE-R primers (Table
II).
Bacterial colonization assay
To evaluate the pathogenicity of SS2, the capacity
of the bacteria to colonize the tissues of the heart, liver,
spleen, lung and kidney of infected mice was analyzed using a
colonization assay. Further BALB/c (13–14 g) mice were randomly
divided into three groups (nine mice/group) and injected i.p. with
1×108 CFU/mouse of one of the ZY458, ZD89 or 458ΔvapE
strains. One mouse from each group was euthanized by cervical
dislocation at 12, 24 and 36 h post-infection, and the tissues were
collected and ground with an electric pestle (Tiangen Biotech Co.,
Ltd., Beijing, China) in 0.9% NaCl (0.03 g tissue/ml). The
supernatants were diluted 10-fold and plated onto BHI agar plates.
Following an overnight incubation, the bacterial colonies were
counted and the data were expressed as CFU/g of tissue, as
described previously (24).
Distribution analysis of the vapE gene by
PCR
The primers VapE-F and VapE-R were designed on the
basis of the published sequence of the SS2 strain, 05ZYH33 (GenBank
accession no. NC_009442), to detect the vapE gene from the
genomic DNA of SS2 strains ZY458, SP3, SP6, 1330, ZD89, B22, B3 and
SP8 by PCR. 16S rDNA served as the reference gene. Each reaction
was conducted using a 25 µl mixture containing 2.5 µl
10X buffer, 25 pmol each primer, 2 mM each deoxynucleoside
triphosphate, 2.5 units Taq polymerase and 2.5µl supernatant
of denatured bacteria. The PCR was performed with Techgene FTGENE2D
thermocycler, under the following conditions: i) Initiation and
elongation for 3 min at 94°; ii) 30 cycles of denaturation for 30
sec at 94°, annealing for 30 sec at 50°C and elongation for 2 min
at 72°C; and iii) a final elongation step at 72°C for 10 min.
Amplicons were visualized by running at 100 V for 30 min on a 1%
agarose gel containing ethidium bromide (Takara Biotechnology Co.,
Ltd.). A DL2000 DNA ladder (Takara Biotechnology Co., Ltd.) was
used as a size marker.
Results
Generation of vapE mutant
A vapE deletion mutant strain, 458ΔvapE, was
generated by a homologous replacement method using ZY458 as the
parent strain. The vapE gene knock-out mutant strain was
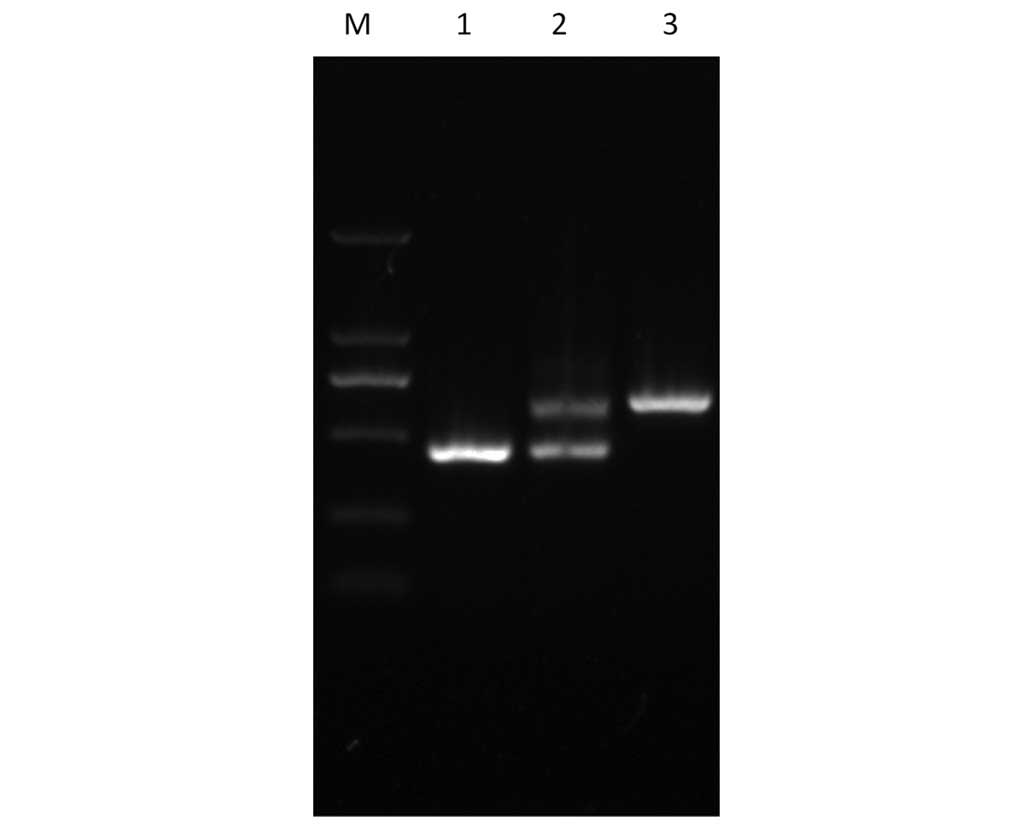
confirmed by PCR (Fig. 1).
Comparing the nucleotide sequences using basic local alignment
search tool (BLAST) searching revealed that a 194 bp segment of the
vapE gene (from 1 to 194 nt of the vapE coding
sequence) had been deleted without alteration of the remaining
sequence.
Generation of a functional complemented
ΔvapE mutant
The PCR-amplified structural gene of vapE was
cloned into a pAT18 shuttle vector. The resultant plasmid was
confirmed by PCR (Fig. 1) and
designated as pAT18::vapE, which was then electrotransfected into
458ΔvapE cells to produce the complemented strain, 458ΔvapE
(pvapE). A BLAST analysis of the nucleotide sequences in the
National Center for Biotechnology Information database with the
vapE gene confirmed that the amplified 2,292 bp fragment was
identical with the other strains.
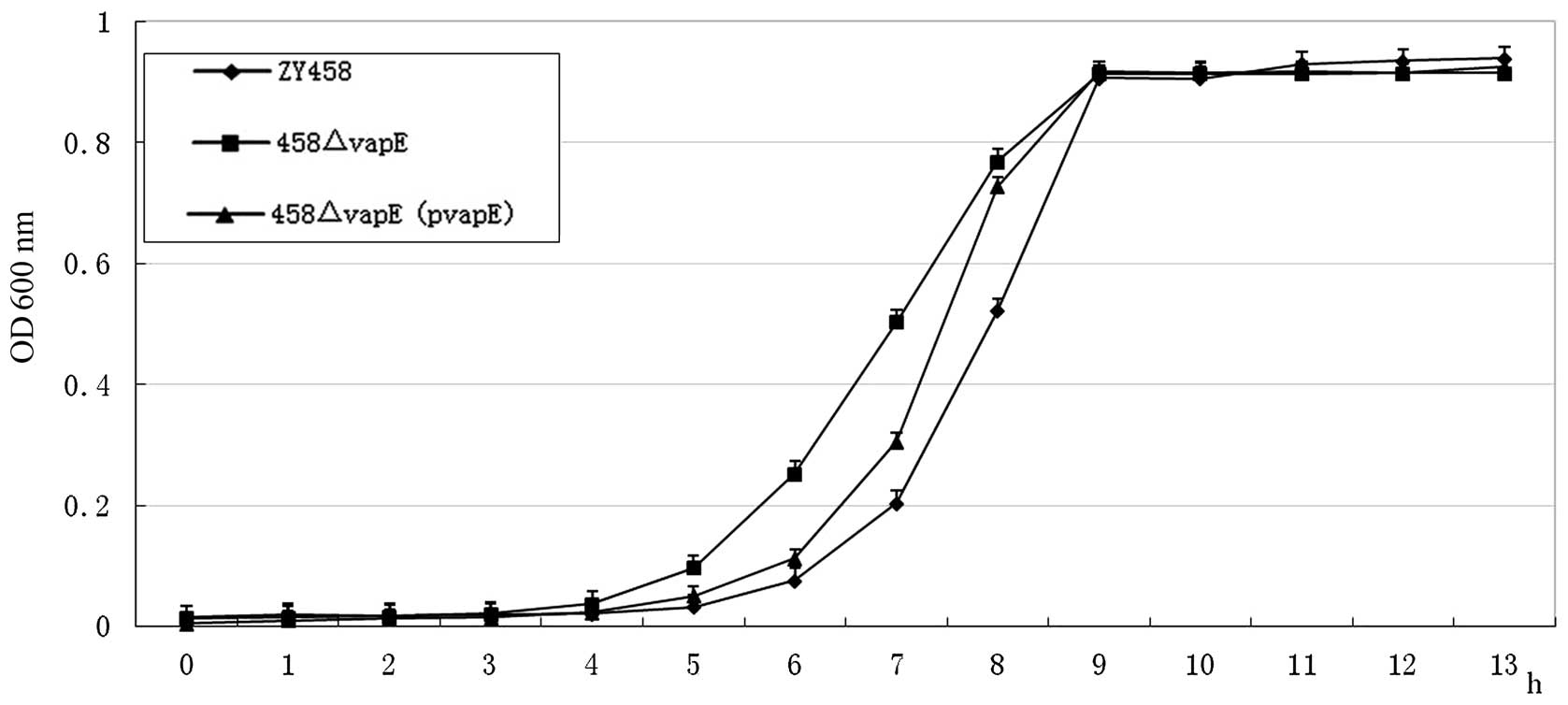
Bacterial growth rates
To determine whether the deletion of the vapE
gene leads to a defect in bacterial growth, the growth
characteristics of wild-type SS2 strain ZY458, the vapE
deletion mutant, 458ΔvapE, and the complemented strain, 458ΔvapE
(pvapE), were compared at 37°C in BHI broth. The optical density
values of the bacterial cultures at 600 nm were measured. The
growth curves indicated that the growth rates of the mutant strains
458ΔvapE and 458ΔvapE (pvapE) were similar to that of the wild-type
strain, ZY458 (Fig. 2).
The virulence of the ΔvapE mutant is
attenuated in mice
To further assess the effect of vapE deletion
on the pathogenesis of SS2, four groups of mice were infected with
one of the SS2 wild-type strain ZY458, the carrier strain, ZD89, or
the mutant strains, 458ΔvapE or 458ΔvapE (pvapE). The results
indicated that all mice infected with wild-type SS2 exhibited
severe clinical symptoms, including depression, apathy, fever,
anorexia, emaciation, swollen eyes and neural disorders, and died
within 2 days of infection (Table
III). Similarly, animals infected with the complemented strain
458ΔvapE(pvapE) developed severe clinical symptoms and 83.3% of the
mice died 2 days post-infection. By contrast, 100% of the mice
infected with mutant strain 458ΔvapE exhibited only mild clinical
symptoms in the first 2 days post-infection, and recovered fully
within a week. None of the mice injected with ZD89 developed any
clinical symptoms. Furthermore, SS2 bacteria were recovered from
the organs of mice infected with ZY458 and 458ΔvapE (pvapE).
However, no bacteria were detected in the heart, liver, spleen,
lung or kidney of any mice infected with 458ΔvapE or ZD89 over a
test period of 7 days post-infection (data not shown).
 | Table IIIVirulence of S. suis wild-type
and mutant strains evaluated in BALB/c micea. |
Table III
Virulence of S. suis wild-type
and mutant strains evaluated in BALB/c micea.
| Strain | Morbidity (%) | Mortality (%) | Percentage of mice
from which SS2 was isolated
|
|---|
| Heart | Liver | Spleen | Lung | Kidney |
|---|
| ZY458 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| ΔvapE | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| ΔvapE (pvapE) | 100.0 | 83.3 | 83.3 | 83.3 | 83.3 | 83.3 | 83.3 |
| Negative
(ZD89) | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
To further evaluate the virulence attenuation of
458ΔvapE, a bacterial colonization assay was performed. As
indicated in Table IV, a
reduction in clone-forming efficiency was observed in the infected
mice. Mice infected with ZY458 died within 36 h, in contrast, mice
infected with 458ΔvapE remained alive with
105–106 CFU/g of tissue.
 | Table IVBacterial colonization of the tissues
of mice infected with S. suis 2 (CFU/g tissue). |
Table IV
Bacterial colonization of the tissues
of mice infected with S. suis 2 (CFU/g tissue).
| Organ | ZY458 infection
| ΔvapE infection
| ZD89 infection
|
|---|
| 12 h | 24 h | 36 ha | 12 h | 24 h | 36 h | 12 h | 24 hb | 36 hb |
|---|
| Heart |
6.6×1010 |
2.0×108 | (−) |
1.3×1010 |
2.0×107 |
5.0×105 |
1.5×106 | (−) | (−) |
| Liver |
8.7×1010 |
1.3×109 | (−) |
3.0×1010 |
2.4×108 |
5.5×106 |
2.0×106 | (−) | (−) |
| Spleen |
2.8×1011 |
1.3×109 | (−) |
2.8×1010 |
5.5×108 |
1.2×106 |
3.0×106 | (−) | (−) |
| Lung |
1.1×1010 |
6.0×108 | (−) |
2.4×1010 |
4.2×107 |
5.5×105 |
7.0×106 | (−) | (−) |
| Kidney |
8.7×1010 |
1.5×108 | (−) |
1.7×1010 |
4.7×107 |
2.5×105 |
4.0×106 | (−) | (−) |
Distribution analysis of the vapE gene in
various S. suis 2 strains
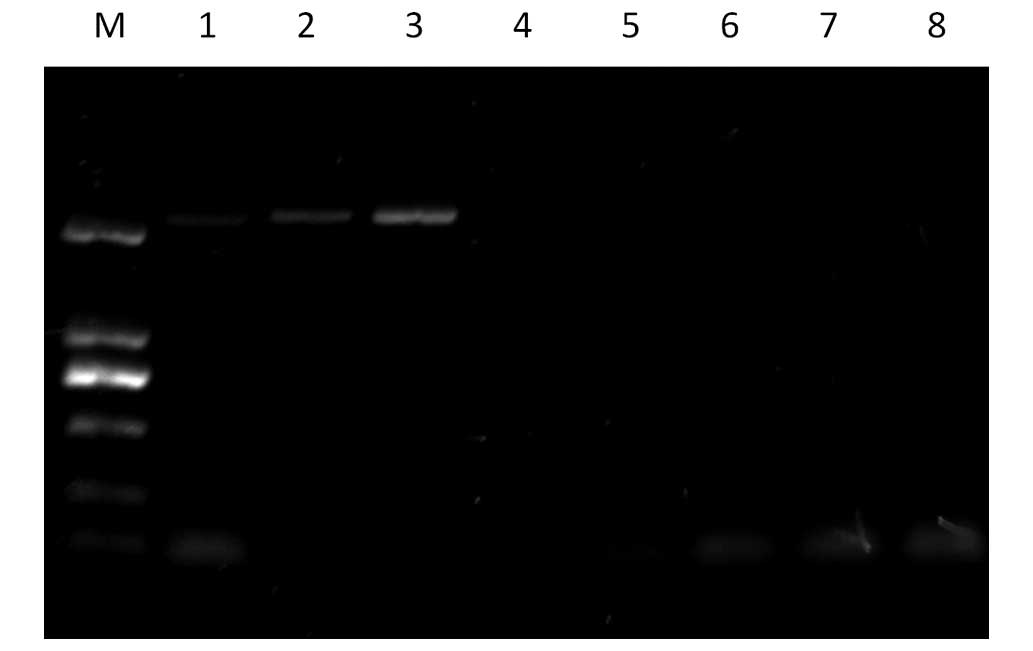
To determine whether the vapE gene exists
only in virulent strains of SS2, PCR amplification of the
vapE gene was performed using primers of VapE-F and VapE-R
(Table II). A 2,292 bp portion of
the target fragment was amplified from the virulent strains (ZY458,
SP3, SP6), but not from any of the avirulent (1330) and carrier
strains that were investigated (ZD89, B22, B3 and SP8) (Fig. 3).
Discussion
S. suis 2 is a major swine pathogen and a
zoonotic agent that leads to septicemia and meningitis in pigs and
humans (25). An improved
understanding of its pathogenesis is critical for developing
effective approaches to combat its severe infectivity. A 1,533 bp
vapE open reading frame sequence of the ZY458 virulent
strain was submitted to GenBank (accession no. JX270678). There was
a 100% identity match to the corresponding sequences of S.
suis P1/7 SSU1332 (GenBank accession no. AM946016), S.
suis A7 SSUA7_1349 (accession no. CP002570), S. suis
GZ1SSGZ1_1350 (accession no. CP000837), S. suis BM407
SSUBM407_1409 (accession no. FM252032), S. suis SC84
SSUSC84_1362 (accession no. FM252031), S. suis 98HAH33
SSU98_1525 (accession no. CP000408), S. suis 05ZYH33
SSU05_1514 (accession no. CP000407), and serotype 1/2 S.
suis SS12 SSU12_1401 (accession no. CP002640), serotype 14
S. suis JS14 SSUJS14_1484 (accession no. CP002465).
To investigate the role of the vapE gene in
the pathogenesis of SS2, a vapE in-frame deletion mutant of
wild-type strain ZY458 was generated using a gene knock-out, and
the impact of the vapE deletion on the virulence of S.
suis 2 was assessed in a mouse infection model. The current
findings indicated that mice infected with the ZY458 wild-type
strain or the 458ΔvapE (pvapE) complemented strain presented severe
clinical symptoms, including body weight loss, and died within 2
days of infection, suggesting that the complemented strain retained
the virulence of the wild-type strain. By contrast, mice inoculated
with the vapE deletion mutant developed only mild clinical
signs, with marginal weight loss in the first 2 days
post-infection, and they recovered fully within a week. This
indicates that deletion of the vapE gene in the ZY458
virulent strain leads to reduced pathogenicity of SS2 in mice.
Together with the molecular evidence that the vapE gene is
present only in virulent SS2 strains, these findings suggest that
the vapE gene is critical for the pathogenicity of S.
suis 2.
The exact function of the vapE gene in SS2
remains unclear; however, the vapE protein was predicted to be
cytoplasmic by the Cell-Ploc package (26). The corresponding gene products in
S. suis strains P1/7, BM407 and SC84 have been annotated as
'putative phage primase' in GenBank, and as 'virulence-associated
protein E' in other S. suis strains. In a previous study,
Wei et al (27) identified
various putative PAIs of S. suis 2, with the vapE
gene located in PAI4. Notably, this putative PAI only existed in
virulent S. suis 2 strains and was able to encode phage
integrases, certain hypothetical proteins, phage protein and tRNA.
On the basis of its original phage elements, vapE has been
suggested to be from a bacteriophage that integrated into the S.
suis 2 genome through a horizontal gene transfer (28). Its absence may reduce the
expression of other virulence-associated proteins in PAI4, thus
reducing the overall pathogenicity of S. suis 2. As a
result, animals exhibited mild clinical symptoms and recovered
fully after 2 days.
In conclusion, the results reported in the present
study clearly indicate that vapE is associated with the
pathogenicity of S. suis 2. Although its function requires
further investigation, this finding may contribute to the
understanding of the pathogenesis of S. suis 2 and may help
in the development of novel strategies against S. suis
infections. Further investigation will require research into the
transcriptome of S. suis ZY458 and 458ΔvapE.
Acknowledgments
We would like to thank Dr JF Fernández-Garayzábal
(Departamento Patología Animal I, Facultad de Veterinaria,
Universidad Complutense, Spain) and Dr Marcelo Gottschalk
(University of Montreal, Faculty of Veterinary Medicine, St.
Hyacinthe, QC, Canada) for kindly providing S. suis 2
strains and S. suis 2 avirulent reference strain 1330,
respectively. The present study was supported by grants from
National Natural Science Foundation of China (grant nos. 31172340
and 31101790/C1803).
References
|
1
|
Wertheim HF, Nghia HDT, Taylor W and
Schultsz C: Streptococcus suis: an emerging human pathogen. Clin
Infect Dis. 48:617–625. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yu H, Jing H, Chen Z, Zheng H, Zhu X, Wang
H, Wang S, Liu L, Zu R, Luo L, et al Streptococcus suis study
groups: Human Streptococcus suis outbreak, Sichuan, China. Emerg
Infect Dis. 12:914–920. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gottschalk M and Segura M: The
pathogenesis of the meningitis caused by Streptococcus suis: the
unresolved questions. Vet Microbiol. 76:259–272. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fittipaldi N, Segura M, Grenier D and
Gottschalk M: Virulence factors involved in the pathogenesis of the
infection caused by the swine pathogen and zoonotic agent
Streptococcus suis. Future Microbiol. 7:259–279. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
de Greeff A, Buys H, Verhaar R, Dijkstra
J, van Alphen L and Smith HE: Contribution of fibronectin-binding
protein to pathogenesis of Streptococcus suis serotype 2. Infect
Immun. 70:1319–1325. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Baums CG, Kaim U, Fulde M, Ramachandran G,
Goethe R and Valentin-Weigand P: Identification of a novel
virulence determinant with serum opacification activity in
Streptococcus suis. Infect Immun. 74:6154–6162. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fittipaldi N, Sekizaki T, Takamatsu D, de
la Cruz Domínguez-Punaro M, Harel J, Bui NK, Vollmer W and
Gottschalk M: Significant contribution of the pgdA gene to the
virulence of Streptococcus suis. Mol Microbiol. 70:1120–1135. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Si Y, Yuan F, Chang H, Liu X, Li H, Cai K,
Xu Z, Huang Q, Bei W and Chen H: Contribution of glutamine
synthetase to the virulence of Streptococcus suis serotype 2. Vet
Microbiol. 139:80–88. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ge J, Feng Y, Ji H, Zhang H, Zheng F, Wang
C, Yin Z, Pan X and Tang J: Inactivation of dipeptidyl peptidase IV
attenuates the virulence of Streptococcus suis serotype 2 that
causes streptococcal toxic shock syndrome. Curr Microbiol.
59:248–255. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang XH, He KW, Duan ZT, Zhou JM, Yu ZY,
Ni YX and Lu CP: Identification and characterization of inosine
5-monophosphate dehydrogenase in Streptococcus suis type 2. Microb
Pathog. 47:267–273. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu T, Zhao Z, Zhang L, Ma H, Lu K, Ren W,
Liu Z, Chang H, Bei W, Qiu Y and Chen H: Trigger factor of
Streptococcus suis is involved in stress tolerance and virulence.
Microb Pathog. 51:69–76. 2011. View Article : Google Scholar
|
|
12
|
Li P, Liu J, Zhu L, Qi C, Bei W, Cai X,
Sun Y and Feng S: VirA: A virulence-related gene of Streptococcus
suis serotype 2. Microb Pathog. 49:305–310. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zheng F, Ji H, Cao M, Wang C, Feng Y, Li
M, Pan X, Wang J, Qin Y, Hu F and Tang J: Contribution of the Rgg
transcription regulator to metabolism and virulence of
Streptococcus suis serotype 2. Infect Immun. 79:1319–1328. 2011.
View Article : Google Scholar :
|
|
14
|
Bonifait L and Grenier D: The SspA
subtilisin-like protease of Streptococcus suis triggers a
pro-inflammatory response in macrophages through a non-proteolytic
mechanism. BMC Microbiol. 11:472011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Willenborg J, Fulde M, de Greeff A, Rohde
M, Smith HE, Valentin-Weigand P and Goethe R: Role of glucose and
CcpA in capsule expression and virulence of Streptococcus suis.
Microbiology. 157:1823–1833. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tang Y, Zhang X, Wu W, Lu Z and Fang W:
Inactivation of the sodA gene of Streptococcus suis type 2 encoding
superoxide dismutase leads to reduced virulence to mice. Vet
Microbiol. 158:360–366. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen C, Tang J, Dong W, Wang C, Feng Y,
Wang J, Zheng F, Pan X, Liu D, Li M, et al: A glimpse of
streptococcal toxic shock syndrome from comparative genomics of S.
suis 2 Chinese isolates. PLoS One. 2:e3152007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li M, Wang C, Feng Y, Pan X, Cheng G, Wang
J, Ge J, Zheng F, Cao M, Dong Y, et al: SalK/SalR, a two-component
signal transduction system, is essential for full virulence of
highly invasive Streptococcus suis serotype 2. PLoS One.
3:e20802008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Qi C, Liu J, Zhu LW, Wang WD, Meng FQ and
Feng SZ: Genetic difference between Streptococcus suis serotype 2
virulent strain and avirulent strain by suppression subtractive
hybridization. Chin J Vet Sci. 588–593. 2009.In Chinese.
|
|
20
|
Sambrook J and Russell DW: Molecular
Cloning: A Laboratory Manual. 3rd edition. Cold Spring Harbor
Laboratory Press; New York: 2001
|
|
21
|
Takamatsu D, Osaki M and Sekizaki T:
Construction and characterization of Streptococcus suis-Escherichia
coli shuttle cloning vectors. Plasmid. 45:101–113. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Takamatsu D, Osaki M and Sekizaki T:
Thermosensitive suicide vectors for gene replacement in
Streptococcus suis. Plasmid. 46:140–148. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
No Authors Listed. CIOMS International
guiding principles for biomedical research involving animals.
Altern Lab Anim. 12:ii1985.PubMed/NCBI
|
|
24
|
Wang G, Lo LF and Maier RJ: The RecRO
pathway of DNA recombinational repair in Helicobacter pylori and
its role in bacterial survival in the host. DNA Repair (Amst).
10:373–379. 2011. View Article : Google Scholar
|
|
25
|
Gottschalk M, Segura M and Xu J:
Streptococcus suis infections in humans: The Chinese experience and
the situation in North America. Anim Health Res Rev. 8:29–45. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chou KC and Shen HB: Cell-PLoc: A package
of web servers for predicting subcellular localization of proteins
in various organisms. Nature Protoc. 3:153–162. 2008. View Article : Google Scholar
|
|
27
|
Wei W, Ding GH, Wang XQ, Sun JC, Tu K, Hao
P, Wang C, Cao ZW, Shi TL and Li YX: Comparative genome analysis of
Streptococcus suis. Chin Sci Bull. 51:808–818. 2006. View Article : Google Scholar
|
|
28
|
Juhas M, van der Meer JR, Gaillard M,
Harding RM, Hood DW and Crook DW: Genomic islands: Tools of
bacterial horizontal gene transfer and evolution. FEMS Microbiol
Rev. 33:376–393. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Trieu-Cuot P, Carlier C, Poyart-Salmeron C
and Courvalin P: Shuttle vectors containing a multiple cloning site
and a lacZ α gene for conjugal transfer of DNA from Escherichia
coli to gram-positive bacteria. Gene. 102:99–104. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Quessy S, Dubreuil JD, Caya M and Higgins
R: Discrimination of virulent and avirulent Streptococcus suis
capsular type 2 isolates from different geographical origins.
Infect Immun. 63:1975–1979. 1995.PubMed/NCBI
|
|
31
|
Vela AI, Goyache J, Tarradas C, Luque I,
Mateos A, Moreno MA, Borge C, Perea JA, Domínguez L and
Fernández-Garayzábal JF: Analysis of genetic diversity of
Streptococcus suis clinical isolates from pigs in Spain by
pulsed-field gel electrophoresis. J Clin Microbiol. 41:2498–2502.
2003. View Article : Google Scholar : PubMed/NCBI
|