Introduction
Inflammatory bowel disease (IBD), which is an
umbrella term for a collection of diseases that include Crohn's
disease (CD) and ulcerative colitis (UC), is characterized by
flares and periods of remission (1). The clinical course of IBD is quite
variable; however, ≥35% of patients receiving standardized care
experience relapse and 15% ultimately do not respond to medical
therapy (1). These varied outcomes
are likely due to genetic, microbial and immune heterogeneity, as
well as treatment strategies and lifestyle factors (2–4).
Disease relapses decrease the quality of life of affected
individuals. In the last few decades, studies have focused on the
identification of possible factors, including the neutrophil Fcγ
receptor I (CD64) index, anti-GM-CSF antibody (Ab) levels, serum
levels of ficolin-2, fecal calprotectin and Clostridium
difficile presence, that may correlate with disease activity
and predict disease relapse (5–10).
However, to the best of our knowledge, none of the previously
investigated factors were found to be reliable indicators of
disease activity. Once endoscopic remission has been achieved, it
is difficult for a physician to identify the optimum point for
ceasing or tapering IBD medication; previous studies demonstrated
that mucosal healing could not predict sustained remission in
patients with IBD following discontinuation of infliximab therapy
or thiopurine withdrawal (11–13).
At present, disease activity is assessed on the
basis of a clinical evaluation and endoscopic examination (11–13).
Initiating 'top down' biological treatment immediately following
surgery, rather than waiting for disease recurrence, has been shown
to provide the best rates of long-term prevention (14); thus indicating that underlying
mucosal inflammation may be present even following complete
endoscopic remission. A previous study demonstrated that some
patients exhibit endoscopic recurrence and yet require re-resection
within 1 year of the initial surgical resection of lesions
(13). Therefore, a reliable
method for the diagnosis of mucosal inflammation is required in
order to allow preventative medication to be initiated prior to the
development of irreversible intestinal damage (15). Such a diagnostic tool would enable
patients to be treated at the earliest signs of disease onset. At
present, endoscopy is the most sensitive method for detecting early
mucosal changes; a severe endoscopic recurrence at 1 year was shown
to predict a clinical relapse (16). However, in some cases, the severity
of an endoscopic lesion does not always match the clinical
manifestations; it was previously shown that certain patients with
mild IBD had a poor response to treatment and in some cases the
disease quickly relapsed during tapering or withdrawal of
medication despite evidence of endoscopic remission (15,16).
Therefore, it is possible that the remission assessed by clinical
and endoscopic evaluation was incomplete in these cases. Thus, a
clinical relapse is typically assessed based on the clinical
activity index (CAI) (17), or the
Crohn's Disease Activity Index (CDAI) (18), rather than on the endoscopic
findings.
The present study analyzed the gene expression
profiles of inflamed and unaffected colon mucosa from patients with
mild CD and UC, in order to establish a more sensitive method for
monitoring mucosal impairment.
Materials and methods
Patients
A total of 24 patients with IBD (13 men and 11
women; mean age, 41±15 years) visiting the Outpatient Clinic of
Zhongshan Hospital (Shanghai, China) between July 2013 and
September 2014 were enrolled in the present study. The disease
activity was assessed based on CDAI for patients with CD and CAI
for patients with UC, and the severity of the lesion was determined
based on endoscopic findings. The assessed lesions were limited to
the sigmoid colon and rectum of all patients. The group with mild
CD consisted of 8 patients with a CDAI between 150 and 220. The
mild UC group consisted of 16 patients with a CAI between 4 and 5.
A total of 5 patients with CD and 10 patients with UC were
undergoing treatment with 5-aminosalicylic acid prior to and during
the present study. As a control group, 9 healthy subjects,
including 5 men and 4 women (mean age, 46±10 years) without a
history of IBD or other known chronic diseases, were enrolled in
the present study. All colon biopsies were collected via a
colonoscopy and were immediately stored in RNAlater (Qiagen GmBH,
Mannheim, Germany). The present study was approved by the Ethical
Committee of Medical Research, Zhongshan Hospital, Fudan
University. Informed consent was obtained from all patients.
RNA extraction and purification
Colon biopsy specimens were homogenized and total
RNA was extracted using TRIzol® reagent (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) according to the manufacturer's
protocol. The RNA was checked for an RNA integrity number using an
Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA,
USA). Qualified total RNA was further purified using the RNeasy
micro kit (Qiagen GmBH) and RNase-Free DNase set (Qiagen GmBH).
Sample quality was assessed by a photospectrometer (Nano Drop
Technologies, Thermo Fisher Scientific, Inc.). The 260/280 ratios
in all samples were >1.8.
Microarray experiment
DNA microarrays (Affymetrix GeneChip Human
Transcriptome Array 2.0; Affymetrix, Santa Clara, CA, USA) were
performed according to the manufacturer's protocol. Briefly, total
RNA was amplified, labeled with biotin and purified using the
GeneChip® WT PLUS Reagent kit (cat. no. 902280;
Affymetrix). Hybridization to the array and washing was performed
with the GeneChip® Hybridization, Wash, and Stain kit
(cat. no. 900720; Affymetrix) in a Hybridization Oven 645 and a
Fluidics Station 450 (both Affymetrix). The arrays were scanned
using the GeneChip® Scanner 3000 (Affymetrix).
GeneChip® Command Console® software
(Affymetrix, Santa Clara, CA, US) was used to control the scanner
and summarize probe cell intensity data (CEL file generation) with
default settings and the raw data were normalized using Expression
Console software 1.4.1 (both Affymetrix).
The GeneChip Human Transcriptome Array 2.0 array
interrogates 44,699 well-annotated genes using >6 million
distinct probes. The array was designed based on the Homo
sapiens hg19 reference genome (http://hgdownload.soe.ucsc.edu/goldenPath/hg19/chromosomes/)
using the following databases: RefSeq (http://www.ncbi.nlm.nih.gov/refseq/), Ensembl
(http://www.ensembl.org/index.html),
UCSC Genome Browser (including known genes and lincRNA transcripts;
https://genome.ucsc.edu/), the Vertebrate Genome
Annotation (Vega) database (http://vega.sanger.ac.uk/index.html), the Mammalian
Gene Collection (v10; http://genecollections.nci.nih.gov/MGC/), NONCODE
(www.noncode.org/), the Long Noncoding RNA
Database (http://www.lncrnadb.org/) and the
Human lincRNA Catalog (http://www.broadinstitute.org/genome_bio/human_lincrnas/).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RNA (2 µg) extracted from colon biopsy
specimens from patients with UC was reverse-transcribed to cDNA
using the ReverTra Ace® qPCR RT kit (Toyobo Co., Ltd.,
Osaka, Japan), according to the manufacturer's protocol. Following
first strand cDNA synthesis, the mix was stored at −20°C in a
freezer prior to PCR analysis. mRNA expression levels were
determined by qPCR on an ABI 7500 system (Applied Biosystems;
Thermo Fisher Scientific, Inc.) using SYBR Premix Ex Taq
RT-PCR kit (Takara Bio Inc., Dalian, China), according to the
manufacturer's protocol. The cycling conditions were as follows:
95°C for 30 sec, followed by 40 cycles at 95°C for 5 sec and 60°C
for 34 sec. The following primers were used: Interferon (IFN)-γ
forward, 5′-TCGGTAACT GACTTGAATGTCCA-3′ and reverse,
5′-TCGCTTCCCTGTTTTAGCTGC-3′; interleukin (IL)-17 forward,
5′-AGCGCAACATGACAGTGAAG-3′ and reverse, 5′-GTGTAATTCCAGGGGGAGGT-3′;
and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) forward,
5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse,
5′-GGCTGTTGTCATACTTCTCATGG-3′. GAPDH was examined under identical
conditions as an internal control to demonstrate the equivalence of
the template. The expression levels of the transcripts were
evaluated using the 2−ΔΔCq method (19) on an ABI 7500 system (Applied
Biosystems; Thermo Fisher Scientific, Inc.).
Statistical analysis and
bioinformatics
Alterations in gene expression were defined using a
fold change cutoff of ≥±2. Heat maps were generated using ggplot
(http://ggplot.yhathq.com/). Cluster
analysis of differentially expressed genes was performed using
Affymetrix Transcriptome Analysis Console (TAC) software 2.0. Gene
Ontology (GO) analyses to assess the functions of genes were
performed using the Database for Annotation, Visualization and
Integrated Discovery (http://david.abcc.ncifcrf.gov/). Pathway analyses were
performed using GenMAPP (http://www.genmapp.org). The Student's t-test was
performed for RT-qPCR data analysis. P<0.05 was considered to
indicate a statistically significant difference.
Results
Large numbers of genes with altered
expression patterns are detected in the unaffected and inflamed
colon mucosa of patients with mild CD and UC
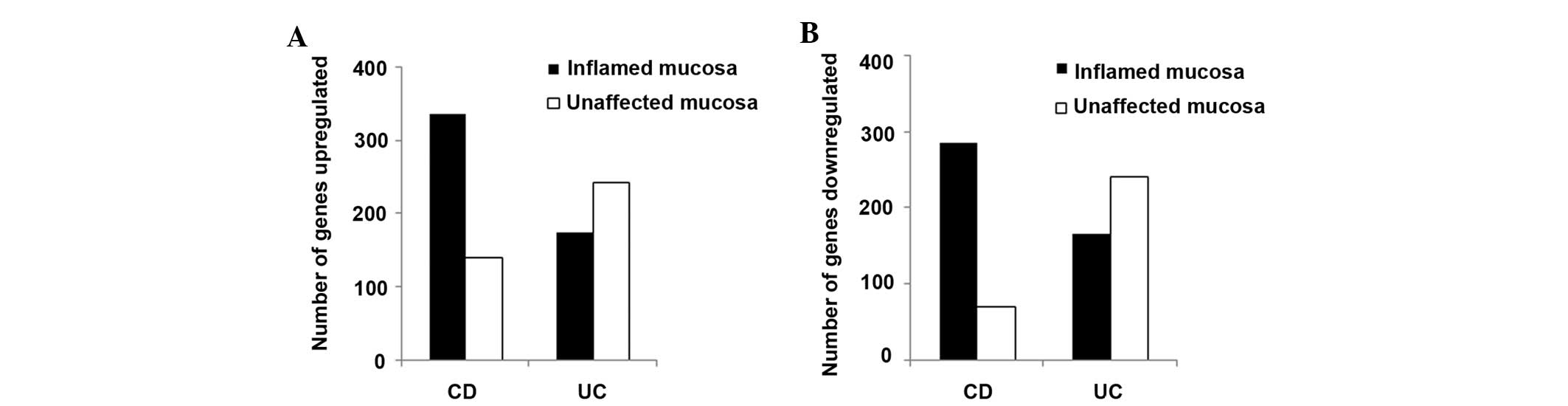
In inflamed colon biopsies, 620 genes (upregulated,
334; downregulated, 174) in patients with CD and 339 genes
(upregulated, 174; downregulated, 165) in patients with UC had
≥3-fold change in expression, as compared with the healthy
controls. In addition, a large number of genes with ≥3-fold change,
as compared with colon biopsies from normal controls, were
identified in the unaffected colon biopsies from patients with CD
(139 genes upregulated and 71 genes downregulated) and UC (243
genes upregulated and 240 genes downregulated) (Fig. 1). The number of genes with altered
expression levels was greater in the inflamed colon mucosa, as
compared with the unaffected colon mucosa, in patients with CD
(upregulated genes, 334 vs. 139, respectively; downregulated genes,
286 vs. 71, respectively). However, the number of genes with
altered expression levels was greater in the unaffected colon
mucosa, as compared with the inflamed colon mucosa, in patients
with UC (upregulated genes, 243 vs. 174, respectively;
downregulated genes, 240 vs. 165, respectively). These results
suggest that the abnormalities at the molecular level were not
limited to the endoscopic lesions derived from the colons of
patients with UC.
Gene expression patterns in the inflamed
and unaffected colon mucosa from patients with mild CD and UC are
similar
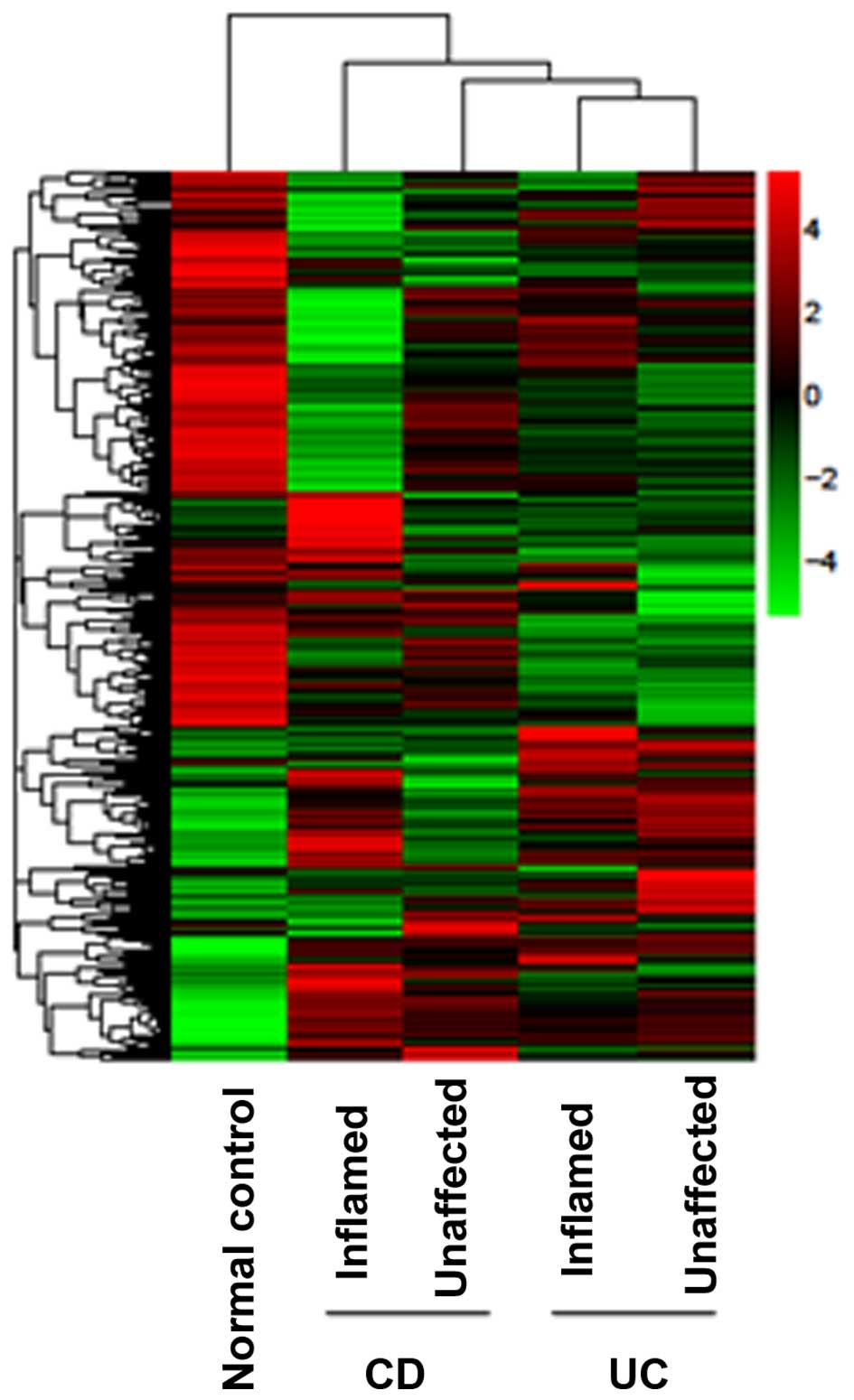
In order to further analyze the differences in the
gene expression patterns between the inflamed and unaffected colon
mucosa from patients with mild IBD, the intensity of genes with
altered expression from inflamed and unaffected colon biopsies was
investigated. The differentially expressed genes were clustered and
heat maps were generated. The intensity of the color on the heat
maps corresponds to the extent of gene expression alterations. The
heat map demonstrated that the expression profiles for the inflamed
and unaffected colon mucosa were overlapping, in particular for the
UC group (Fig. 2).
Molecular functions and pathways
associated with altered transcripts overlap in the inflamed and
unaffected colon mucosa from patients with mild IBD
GO analyses of the altered genes in the inflamed and
unaffected colon mucosas from patients with CD and UC were
conducted. The most prominent molecular functions of the altered
genes in the inflamed mucosa of patients with CD included roles in
the immune response, extracellular region and nucleus, whereas in
the unaffected mucosa of CD patients, they were the immune
response, antigen binding and complement activation. These results
suggested that there was an overlap in the molecular functions of
the altered transcripts between the inflamed and unaffected colon
mucosa of patients with CD.
The most prominent molecular functions of the
altered genes detected in the inflamed mucosa of patients with UC
included roles in the immune response, extracellular region and
extracellular space, whereas in the unaffected mucosa of patients
with CD, they were the immune response, extracellular region and
nucleus; thus suggesting that there was marked similarity and
overlap in the molecular functions of the transcripts between the
inflamed and unaffected colon mucosa in patients with UC (Table I).
 | Table IMolecular functions of the associated
genes with altered expression in inflamed and unaffected colon
mucosa from patients with inflammatory bowel disease. |
Table I
Molecular functions of the associated
genes with altered expression in inflamed and unaffected colon
mucosa from patients with inflammatory bowel disease.
| Name of molecular
function | Crohn's disease
| Ulcerative colitis
|
|---|
| Inflamed mucosa
P-value | Unaffected mucosa
P-value | Inflamed mucosa
P-value | Unaffected mucosa
P-value |
|---|
| Immune response
(GO:0006955) | 9.05E-13 | 2.54E-15 | 1.02E-16 | 6.56E-13 |
| Complement activation
(GO:0006958) | 1.41E-05 | 7.81E-11 | 7.92E-10 | 1.41E-05 |
| Antigen binding
(GO:0003823) | 1.85E-05 | 8.82E-13 | 6.25E-05 | 1.63E-04 |
| Extracellular matrix
(GO:0031012) | N/A | N/A | 1.54E-04 | 5.23E-04 |
| DNA binding
(GO:0003677) | N/A | N/A | 1.24E-04 | 1.04E-04 |
| Extracellular region
(GO:0005576) | 6.25E-08 | 4.67E-04 | 3.54E-14 | 2.98E-12 |
| Nucleus
(GO:0005634) | 4.00E-07 | N/A | 1.73E-04 | 1.34E-06 |
| Innate immune
response (GO:0045087) | N/A | 3.24E-05 | 2.33E-05 | N/A |
| Extracellular space
(GO:0005615) | 5.11E-06 | N/A | 7.33E-12 | N/A |
| Transcription,
DNA-dependent (GO:0006351) | 8.48E-05 | N/A | 5.00E-04 | 4.68E-06 |
| Chemokine activity
(GO:0008009) | 0.000144 | 0.00157 | 7.82E-05 | 0.00172 |
The pathways associated with these transcripts were
also investigated and the common pathways of the altered genes in
the inflamed and unaffected colon mucosa from the CD group
included: Staphylococcus aureus infection, mineral
absorption and protein digestion and absorption. Conversely, the
common pathways for the altered genes in the inflamed and
unaffected colon mucosa from the UC group included: S.
aureus infection, asthma, mineral absorption, protein digestion
and absorption, retinol metabolism and linoleic acid metabolism
(Table II). These results
suggested that there were similarities in the molecular functions
associated with the altered transcripts between the inflamed and
unaffected colon mucosa from CD and UC patients.
 | Table IIPathways of associated genes with
altered expression in inflamed and unaffected colon mucosa from
patients with inflammatory bowel disease. |
Table II
Pathways of associated genes with
altered expression in inflamed and unaffected colon mucosa from
patients with inflammatory bowel disease.
| Name of pathway | Crohn's disease
| Ulcerative colitis
|
|---|
| Inflamed mucosa
P-value | Unaffected mucosa
P-value | Inflamed mucosa
P-value | Unaffected mucosa
P-value |
|---|
| Staphylococcus
aureus infection (hsa05150) | 8.88E-07 | 0.004322 | 3.22E-08 | 4.64E-06 |
| Asthma
(hsa05310) | 2.34E-06 | N/A | 3.03E-05 | N/A |
| Mineral absorption
(hsa04978) | 6.93E-06 | 6.11E-09 | 0.000134 | 8.12E-06 |
| Olfactory
transduction (hsa04740) | 3.97E-05 | N/A | N/A | 0.000358 |
| Starch and sucrose
metabolism (hsa00500) | 8.49E-05 | N/A | N/A | 0.001897 |
| Protein digestion
and absorption (hsa04974) | 0.000136 | 0.000447 | 3.45E-08 | 1.28E-08 |
| Retinol metabolism
(hsa00830) | 3.97E-05 | N/A | 1.64E-06 | 6.25E-06 |
| Linoleic acid
metabolism (hsa00591) | N/A | 0.000152 | 4.58E-06 | 3.98E-05 |
| Chemokine signaling
pathway(hsa04062) | N/A | 0.000228 | N/A | N/A |
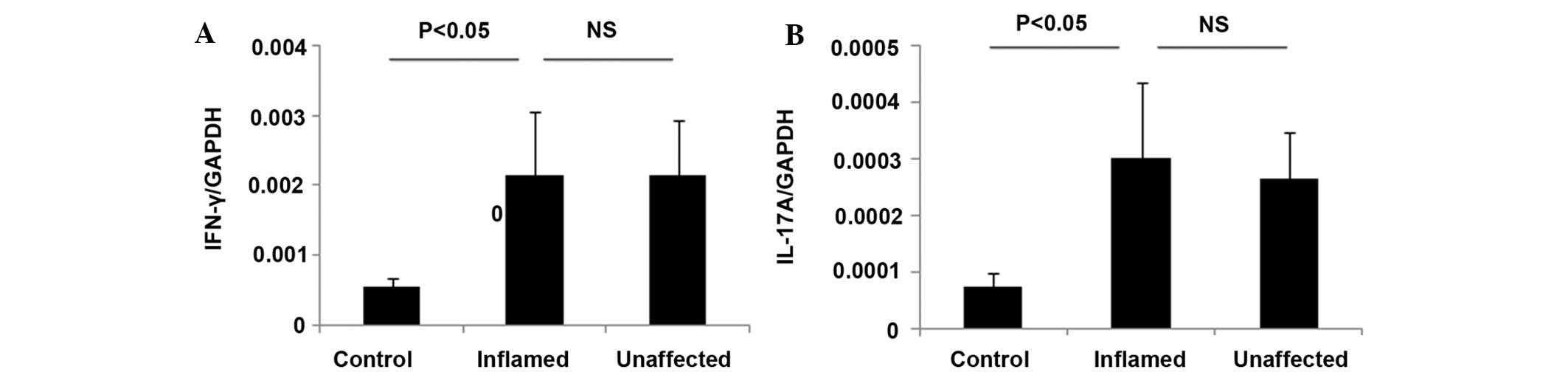
Inflammatory cytokines are elevated in
the inflamed and unaffected colon mucosa from patients with UC
Alterations in the expression levels of inflammatory
cytokines in the colon mucosa is a key pathological characteristic
of IBD, and it has been associated with disease activity and
mucosal inflammation (20–22). The present study investigated the
mRNA expression levels of IFN-γ and IL-17 in inflamed and
unaffected colon biopsies from patients with UC using RT-qPCR. The
CD patients were not analyzed as there were insufficient CD
patients for conducting RT-qPCR analyses. The levels of IFN-γ and
IL-17 were comparably elevated in the inflamed and unaffected colon
mucosa from patients with UC (Fig.
3), and were significantly different from the control
(P<0.05; Fig. 3).
Discussion
The cause of relapses in patients with CD and UC are
largely unknown, which may be due to the lack of a sensitive method
for monitoring mucosal inflammation before an endoscopic
abnormality becomes apparent (2–4). The
incidence of endoscopic recurrence in patients with CD at 1 year
following curative resection is as high as 75% (20). Subjective symptoms are not reliable
indicators of disease activity, whereas endoscopy is an objective
tool (16). However, the present
study demonstrated that endoscopic observations may be insufficient
for detecting an underlying impairment of the gut mucosa, since
pathological molecular alterations occurred in the inflamed and
unaffected colon mucosa of patients with mild IBD.
The present study demonstrated that large numbers of
genes were abnormally expressed (either upregulated or
downregulated) in patients with mild IBD, as compared with normal
controls. These results indicated that the evaluation of gene
expression profiles may be considered a sensitive and reliable
method for detecting mucosal inflammation. Furthermore, the present
study demonstrated that there were similarities in the gene
expression alterations and inflammatory cytokine expression levels
between the inflamed and unaffected mucosa from patients with IBD,
in particular in patients with UC. This finding suggested that
mucosal inflammation may not be limited to endoscopic lesions in
the gut, and that molecular abnormalities may better diagnose an
impairment of intestinal homeostasis and subclinical intestinal
inflammation. Consistent with these results, a previous study
demonstrated that there were marked overlaps in the gene expression
alterations between the small bowel proximal to the pouch and the
pouch itself in patients with UC following restorative
proctocolectomy (23).
The molecular functions of the altered genes
associated with IBD in the present study fell into three main
categories: Inflammation, nutrition absorption and cell structure.
Therefore, the altered expression of inflammation-associated genes
may be the initial abnormality in the pathogenesis of IBD, which is
supported by the finding that IBD is a disease caused by an
unbalanced immune response to intestinal commensal organisms
(24–26). However, it is also possible that
abnormal cell structure may trigger the impairment of barrier
function prior to the induction of an unbalanced immune response to
the gut microbiota (27). The
precise associations among inflammation, nutrition absorption and
cell structure require further investigation.
It was demonstrated that, as well as nutrition
digestion, absorption and metabolism, S. aureus infection
was overrepresented in the inflamed and unaffected colon mucosa
from CD and UC patients. A case of a hospitalized CD patient with
an intestinal methicillin-resistant S. aureus infection has
been previously reported (28). In
addition, a previous study demonstrated that S aureus
infection increased and was associated with the risk of mortality
in hospitalized patients with IBD (29). However, none of the patients
enrolled in the present study were hospitalized nor did they
exhibit clinical manifestations of a S. aureus infection.
Therefore, the significance of the S. aureus infection
pathway as a tool for predicting the occurrence of a relapse in
patients with IBD requires further investigation. In addition, it
may be useful to screen patients with IBD for S. aureus
infections.
The management of IBDs often depends on the disease
severity (2); however, the
assessment of disease activity may be critical for the design of an
optimal therapeutic strategy in order to prevent the recurrence of
IBDs. The stopping or tapering of medication for IBD is usually
objective and dependant on the experience of the individual
physician. The present study demonstrated that gene expression
profile analyses may be considered a sensitive and reliable method
for the detection of subclinical mucosal inflammation in recurrent
IBD patients with mild symptoms. Abnormal gene expression profiles
in patients with normal endoscopic findings may predict the
occurrence of relapses after treatment has been stopped or tapered.
In addition, for such patients, the maintenance of treatment may be
considered. Therefore, the results of the present study may be of
value in clinical practice regarding the diagnosis and development
of therapeutic strategies for patients with IBDs. One limitation of
the present study was the relatively small size of the cohort due
to economical reasons. Therefore, the results require verification
using larger groups of patients.
In conclusion, the present study analyzed the gene
expression profiles of inflamed and unaffected colon mucosa from
patients with mild CD and UC. Marked alterations in the gene
expression profiles of these patients were identified, and there
was marked similarity and overlap in the gene expression
alterations that occurred between the inflamed and unaffected
mucosa of patients with UC. The results of the present study
suggested that gene expression profiling may be considered a more
sensitive tool, as compared with an endoscopic evaluation, for the
detection of a colon mucosa impairment in patients with IBDs, in
particular in patients with UC.
Acknowledgments
The authors would like to thank Shanghai South Gene
Technology (Shanghai, China) for assistance with the microarray
study and bioinformatics analysis. The present study was supported
by the National Natural Science Foundation of China (grant no.
81370510) and the Shanghai Committee of Science and Technology
(grant no. 13140902000).
References
|
1
|
Orlando A, Mocciaro F, Renna S, Scimeca D,
Rispo A, Lia Scribano M, Testa A, Aratari A, Bossa F, Tambasco R,
et al: Early post-operative endoscopic recurrence in Crohn's
disease patients: data from an Italian Group for the study of
inflammatory bowel disease (IG-IBD) study on a large prospective
multicenter cohort. J Crohns Colitis. 8:1217–1221. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pineton de Chambrun GP and Sandborn WJ:
IBD in 2011: Advances in IBD management-towards a tailored
approach. Nat Rev Gastroenterol Hepatol. 9:70–72. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Martin TD, Chan SS and Hart AR:
environmental factors in the relapse and recurrence of inflammatory
bowel disease: A review of the literature. Dig Dis Sci.
60:1396–1405. 2015. View Article : Google Scholar
|
|
4
|
Spooren CE, Pierik MJ, Zeegers MP, Feskens
EJ, Masclee AA and Jonkers DM: Review article: The association of
diet with onset and relapse in patients with inflammatory bowel
disease. Aliment Pharmacol Ther. 38:1172–1187. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schaffer T and Schoepfer AM: Serum
ficolin-2 correlates worse than fecal calprotectin and CRP with
endoscopic Crohn's disease activity. J Crohns Colitis. 8:1125–1132.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Minar P, Haberman Y, Jurickova I, Wen T,
Rothenberg ME, Kim MO, Saeed SA, Baldassano RN, Stephens M,
Markowitz J, et al: Utility of neutrophil Fcγ receptor I (CD64)
index as a biomarker for mucosal inflammation in pediatric Crohn's
disease. Inflamm Bowel Dis. 20:1037–1048. 2014.PubMed/NCBI
|
|
7
|
Däbritz J, Bonkowski E, Chalk C, Trapnell
BC, Langhorst J, Denson LA and Foell D: Granulocyte macrophage
colony-stimulating factor auto-antibodies and disease relapse in
inflammatory bowel disease. Am J Gastroenterol. 108:1901–1910.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Naismith GD, Smith LA, Barry SJ, Munro JI,
Laird S, Rankin K, Morris AJ, Winter JW and Gaya DR: A prospective
evaluation of the predictive value of faecal calprotectin in
quiescent Crohn's disease. J Crohns Colitis. 8:1022–1029. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Foell D, Wittkowski H and Roth J:
Monitoring disease activity by stool analyses: From occult blood to
molecular markers of intestinal inflammation and damage. Gut.
58:859–868. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Masclee GM, Penders J, Jonkers DM, Wolffs
PF and Pierik MJ: Is clostridium difficile associated with relapse
of inflammatory bowel disease? Results from a retrospective and
prospective cohort study in the Netherlands. Inflamm Bowel Dis.
19:2125–2131. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sorrentino D, Nash P, Viladomiu M,
Hontecillas R and Bassaganya-Riera J: Stopping anti-TNF agents in
patients with Crohn's disease in remission: Is it a feasible
long-term strategy? Inflamm Bowel Dis. 20:7570–766. 2014.
|
|
12
|
Dai C, Liu WX, Jiang M and Sun MJ: Mucosal
healing did not predict sustained clinical remission in patients
with ibd after discontinuation of one-year infliximab therapy. PLoS
One. 9:e1107972014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kennedy NA, Kalla R, Warner B, Gambles CJ,
Musy R, Reynolds S, Dattani R, Nayee H, Felwick R, Harris R, et al:
Thiopurine withdrawal during sustained clinical remission in
inflammatory bowel disease: Relapse and recapture rates, with
predictive factors in 237 patients. Aliment Pharmacol Ther.
40:1313–1323. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Regueiro M, Kip KE, Baidoo L, Swoger JM
and Schraut W: Postoperative therapy with infliximab prevents
long-term Crohn's disease recurrence. Clin Gastroenterol Hepatol.
12:1494–1502. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Farkas K, Lakatos PL, Nagy F, Szepes Z,
Miheller P, Papp M, Palatka K, Bálint A, Bor R, Wittmann T and
Molnár T: Predictors of relapse in patients with ulcerative colitis
in remission after one-year of infliximab therapy. Scand J
Gastroenterol. 48:1394–1398. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Keyashian K: Does endoscopic assessment of
mucosal healing affect IBD management? Dig Dis Sci. 59:2351–2353.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mańkowska-Wierzbicka D, Swora-Cwynar E,
Poniedziałek B, Adamski Z, Dobrowolska A and Karczewski J:
Usefulness of selected laboratory markers in ulcerative colitis.
Eur Cytokine Netw. Oct 13–2015.Epub ahead of print.
|
|
18
|
Monteleone G, Di Sabatino A, Ardizzone S,
Pallone F, Usiskin K, Zhan X, Rossiter G and Neurath MF: Impact of
patient characteristics on the clinical efficacy of mongersen
(GED-0301), an oral Smad7 antisense oligonucleotide, in active
Crohn's disease. Aliment Pharmacol Ther. Jan 13–2016.Epub ahead of
print. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
20
|
Neurath MF: Cytokines in inflammatory
bowel disease. Nat Rev Immunol. 14:329–342. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hundorfean G, Neurath MF and Mudter J:
Functional relevance of T helper 17 (Th17) cells and the IL-17
cytokine family in inflammatory bowel disease. Inflamm Bowel Dis.
18:180–186. 2012. View Article : Google Scholar
|
|
22
|
Aine B, Grainne L and Kate K:
Transcriptional profiling of the colonic mucosa and epithelial
cells of patients with acutely active ulcerative colitis. Inflamm
Bowel Dis. (Suppl 1): S10–S11. 2014. View Article : Google Scholar
|
|
23
|
Yanai H, Ben-Shachar S, Baram L, Elad H,
Gitstein G, Brazowski E, Tulchinsky H, Pasmanik-Chor M and Dotan I:
Gene expression alterations in ulcerative colitis patients after
restorative proctocolectomy extend to the small bowel proximal to
the pouch. Gut. 64:756–764. 2015. View Article : Google Scholar
|
|
24
|
Abraham C and Medzhitov R: Interactions
between the host innate immune system and microbes in inflammatory
bowel disease. Gastroenterology. 140:1729–1737. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
MacDonald TT, Biancheri P, Sarra M and
Monteleone G: What's the next best cytokine target in IBD? Inflamm
Bowel Dis. 18:2180–2189. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cader MZ and Kaser A: Recent advances in
inflammatory bowel disease: Mucosal immune cells in intestinal
inflammation. Gut. 62:1653–1664. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Henderson P, van Limbergen JE, Schwarze J
and Wilson DC: Function of the intestinal epithelium and its
dysregulation in inflammatory bowel disease. Inflamm Bowel Dis.
17:382–395. 2011. View Article : Google Scholar
|
|
28
|
Nguyen GC, Patel H and Chong RY: Increased
prevalence of and associated mortality with methicillin-resistant
Staphylococcus aureus among hospitalized IBD patients. Am J
Gastroenterol. 105:371–377. 2010. View Article : Google Scholar
|
|
29
|
Bryant RV, Winer S, Travis SP and Riddell
RH: Systematic review: Histological remission in inflammatory bowel
disease. Is 'complete' remission the new treatment paradigm? An
IOIBD initiative. J Crohns Colitis. 8:1582–1597. 2014. View Article : Google Scholar : PubMed/NCBI
|