Introduction
Fetal macrosomia has been inconsistently defined as
a birth weight of >4,000, 4,500 or 5,000 g, regardless of
gestational age (1,2). In China, the most widely used
definition of macrosomia is a birth weight ≥4,000 g. Macrosomia can
lead to adverse birth outcomes for the newborn, including perinatal
asphyxia, neonatal mortality and shoulder dystocia (1,3). In
addition, mothers delivering macrosomic fetuses are at an increased
risk of experiencing prolonged labor, C-section, abnormal
hemorrhage and perineal laceration (4,5).
Numerous studies have indicated that high birth weight is
associated with long-term health risks for the newborn, including
an increased risk of obesity, diabetes and certain types of cancer
(6–9). In the past two to three decades, the
incidence of macrosomia in developed countries has increased by
15–25% (2). In China, the
macrosomia occurrence rate increased from 6.5% in 2006 (10) to 7.3% in 2011 (11). However, the mechanisms responsible
for fetal macrosomia remains poorly understood.
A large body of research has indicated that
epigenetic alterations in placental tissues are associated with
adverse pregnancy outcomes and fetal programming (12–14).
MicroRNAs (miRNAs) are short noncoding RNA molecules (~22
nucleotides in length) and are important in post-transcriptional
gene regulation through their binding with hundreds of different
mRNAs (15). A wide range of
biological activities are affected by miRNAs, including cellular
proliferation, differentiation, apoptosis and the processes of
numerous diseases, including cancer, obesity, insulin resistance
and diabetes (16,17). Previous investigations have
demonstrated that aberrant expression of a number of miRNAs in the
placenta was associated with low birth weight and preeclampsia
(18,19). Maccani et al (20) reported that decreased levels of
placental miR-16 and -21 were associated with poor fetal growth. In
addition, miR-143 was implicated as a crucial regulator of
adipogenesis (21). Since evidence
suggested that miR-16, -21 and -143 are important in fetal
development, the current study sought to further characterize the
association of these miRNAs with macrosomia.
The present study examined the expression of
miRs-16, -21 and -143 in the placenta of macrosomic and normal
pregnancies and demonstrated miRNA dysregulation in macrosomic
placenta.
Materials and methods
Study population
Subjects were recruited at Yuying Children's
Hospital of Wenzhou Medical University (Wenzhou, China). Macrosomia
was defined as a neonate with a birth weight ≥4,000 g. Normal birth
weight was defined as 2,500–3,999 g. Samples from macrosomia cases
and controls were collected from women between the ages of 18 and
42 years with healthy pregnancies (i.e. pregnancies without
hypertension, hepatitis, heart disease, psychological disorders,
gestational diabetes or impaired glucose tolerance), whose infants
were full-term (≥37 weeks) and viable, with no known genetic
disorders. Maternal weight gain during pregnancy was divided into
three levels, including low, moderate and high, according to
recommendations from the Institute of Medicine (22). A total of 67 macrosomic and 64
control pregnancies were selected. All subjects provided written
informed consent to participate in the current study, and the
research protocol was approved by the Ethics Committee of Wenzhou
Medical University.
Placenta collection
A placental biopsy of ~1 g was excised from the
maternal side of each placenta, 2 cm from the umbilical cord
insertion site and free of maternal decidua, within 15 min of
delivery of the placenta. Biopsies were cut into small sections and
immediately placed in RNAlater (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). Samples were incubated at 4°C overnight and
stored at −80°C until RNA extraction was performed.
miRNA extraction and quantitative
detection
Total RNA was extracted from placental tissue using
a MicroRNA Isolation kit (BioTek China, Beijing, China), according
to the manufacturer's instructions. Extracted RNA was quantified by
measuring absorbance at 260 and 280 nm using the Nano-200
Micro-Spectrophotometer (Allsheng Instruments, Co., Ltd., Hangzhou,
China). Reverse transcription (RT) of miRNA (200 ng) was performed
using the ReverTra Ace® qPCR RT kit (Toyobo Co., Ltd.,
Osaka, Japan) with Bulge-Loop™ hsa-miR-16-5p (miRQ0000069-1-2),
hsa-miR-21-5p (miRQ0000076-1-2), hsa-miR-143-3p (miRQ0000435-1-2)
and U6 small nuclear (sn) RNA (MQP-0202) stem-loop RT-primers
(Guangzhou RiboBio Co., Ltd., Guangzhou, China), according to the
manufacturer's protocol. U6 snRNA was used as an internal control.
Reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) assay was performed using Thunderbird SYBR qPCR Mix
(Toyobo Co., Ltd.) and the CFX96 Touch Real-Time PCR Detection
System (Bio-Rad Laboratories, Inc., Hercules, CA, USA), in
accordance with the manufacturer's instructions. Cycling conditions
were as follows: 95°C for 3 min, followed by 30 cycles of 95°C for
10 sec, 60°C for 20 sec, and 70°C for 10 sec. To check PCR product
homogeneity, the temperature was elevated from 65 to 99°C in
0.1°C/sec increments to obtain a melting curve. miRNA expression
levels were calculated using the 2−ΔΔCq method (23).
Bioinformatic prediction of miRNA targets
and pathway analysis
miRTarBase (mirtarbase.mbc.nctu.edu.tw, release 4.5) was used to
identify target genes (24). DAVID
(Database for Annotation, Visualization and Integrated Discovery;
david.abcc.ncifcrf.gov) was used to
identify pathways involving these target genes (25).
Statistical analysis
Basic characteristics of the macrosomia and control
groups were compared using unpaired t-tests for continuous
variables and reported as the mean ± standard deviation.
Categorical variables were compared using χ2 tests. The
miRNA expression data were not normally distributed, therefore,
relative expression was presented as the median (interquartile
range) and the Mann-Whitney U test was used to analyze differences
in miRNA expression levels between the macrosomia and control
groups. miRNA expression levels were divided by quartile, using the
lowest quartile as the reference group. Logistic regression models
were used to determine the risk factors of giving birth to a
newborn with macrosomia. Spearman's correlation coefficients were
used to examine the relationship between miR-21 and -143
expression. All statistical analyses were conducted using SPSS
software, version 14.0 (SPSS, Inc., Chicago, IL, USA). All P-values
reported were two-tailed, and P<0.05 was considered to indicate
a statistically significant difference.
Results
Subject characteristics
Demographic data are presented in Table I. A total of 131 mothers and their
neonates were recruited for the current study. The average birth
weight of neonates with macrosomia was 4,303.4±219.5 g, while the
average weight of control neonates was 3,398.0±362.2 g. Maternal
weight gain during pregnancy was significantly higher in mothers
with macrosomic neonates compared with the control group
(P<0.001), and neonates with macrosomia were more likely to be
male (P=0.004). There were no significant differences in
gestational age, maternal weight prior to pregnancy, maternal body
mass index (BMI) prior to pregnancy, or maternal age between the
macrosomia and control groups.
 | Table ICharacteristics of the study
sample. |
Table I
Characteristics of the study
sample.
| Characteristic | Controls
(n=64) | Macrosomia cases
(n=67) | P-value |
|---|
| Birthweight, g | 3,398.0±362.2 | 4,303.4±219.5 | <0.001 |
| Gestational age,
weeks | 39.1±1.0 | 39.4±1.0 | 0.094 |
| Maternal weight
prior to pregnancy, kg | 52.3±7.0 | 54.5±7.2 | 0.110 |
| Height, cm | 159.9±3.6 | 160.5±4.5 | 0.445 |
| Maternal BMI prior
to pregnancy, kg/m2 | 20.5±2.7 | 21.2±2.8 | 0.195 |
| Maternal weight
gain during pregnancy, kg | 15.4±5.7 | 20.1±5.7 | <0.001 |
| Maternal age,
years | 27.8±4.4 | 29.0±3.8 | 0.097 |
| Tobacco during
pregnancy, n (%) | | | |
| No | 64 (100) | 67 (100) | |
| Yes | 0 | 0 | |
| Infant gender, n
(%) | | | 0.004 |
| Male | 30 (46.9) | 48 (71.6) | |
| Female | 34 (53.1) | 19 (28.4) | |
| Delivery method, n
(%) | | | <0.001 |
| Vaginal | 55 (85.9) | 32 (47.8) | |
| C-section | 9 (14.1) | 35 (52.2) | |
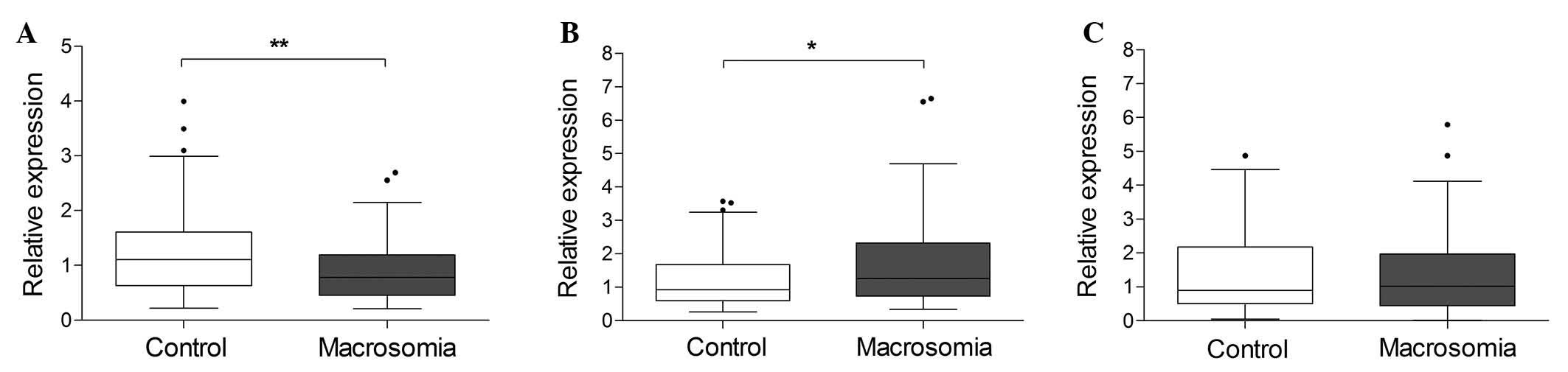
miRNA expression in macrosomic and
control placenta
To determine whether miR-16, -21 or -143 were
differentially expressed in macrosomic and control placenta, their
expression was measured using RT-qPCR. The Mann-Whitney U-test
demonstrated that miR-143 was expressed at lower levels in
macrosomic placenta (Fig. 1A;
P=0.003), whereas miR-21 expression was significantly higher in
macrosomic placenta (Fig. 1B; P=
0.043) compared with the control samples. However, no significant
difference in miR-16 expression levels was detected between the two
groups (Fig. 1C; P=0.955).
Association of miRNAs with
macrosomia
The present study examined whether increased miR-21
expression and decreased miR-143 expression were associated with
the likelihood of macrosomia using logistic regression models, and
adjusted for potential confounders, including maternal age, weight
prior to pregnancy, weight gain during pregnancy, gestational age,
infant gender and delivery method. miRNA expression levels were
divided by quartile (Table II).
As presented in Table II, the
likelihood of macrosomia increased with miR-21 expression quartile
as follows: Q2, OR=6.67 (95% CI, 1.39–32.05); Q3, OR=4.10 (95% CI,
0.88–19.11); and Q4, OR=16.19 (95% CI, 2.46–106.68). Conversely,
higher miR-143 quartiles were associated with protection against
macrosomia as follows: Q2, OR=0.22 (95% CI, 0.049–0.98); Q3,
OR=0.11 (95% CI, 0.024–0.55); and Q4 OR=0.16 (95% CI, 0.032–0.79).
High maternal weight gain during pregnancy and male gender were
also demonstrated to be risk factors for macrosomia.
 | Table IIResults of logistic regression models
investigating the association between individual miRNA expression
and macrosomiaa. |
Table II
Results of logistic regression models
investigating the association between individual miRNA expression
and macrosomiaa.
A, miR-21
|
|---|
| Effect | n (%) | χ2 | OR (95% CI) | P-value |
|---|
| miR-21 relative
expression quartiles | | | | 0.025 |
| Q1, ≤0.67 | 28 (25) | | 1.00 | |
| Q2, 0.68–1.10 | 27 (24) | 5.61 | 6.67
(1.39–32.05) | 0.018 |
| Q3, 1.11–1.95 | 33 (30) | 3.22 | 4.10
(0.88–19.11) | 0.073 |
| Q4, >1.95 | 23 (21) | 8.38 | 16.19
(2.46–106.68) | 0.004 |
| Maternal weight
gain during pregnancyb | | | | 0.025 |
| Low | 16 (16) | | 1.00 | |
| Moderate | 34 (34) | 5.87 | 3.20
(0.481–21.25) | 0.229 |
| High | 55 (50) | 3.48 | 9.78
(1.55–61.90) | 0.015 |
| Infant gender | | | | 0.002 |
| Female | 46 (41) | | 1.00 | |
| Male | 65 (59) | 9.29 | 6.36
(1.94–20.90) | 0.002 |
B, miR-143
|
|---|
| Effect | n (%) | χ2 | OR (95% CI) | P-value |
|---|
| miR-143 relative
expression quartiles | | | | 0.042 |
| Q1, ≤0.52 | 27 (24) | | 1.00 | |
| Q2, 0.53–0.90 | 29 (26) | 3.92 | 0.22
(0.049–0.98) | 0.048 |
| Q3, 0.91–1.35 | 29 (26) | 7.33 | 0.11
(0.024–0.55) | 0.007 |
| Q4, >1.35 | 27 (24) | 5.08 | 0.16
(0.032–0.79) | 0.024 |
| Maternal weight
gain during pregnancyb | | | | 0.010 |
| Low | 18 (16) | | 1.00 | |
| Moderate | 38 (34) | 6.60 | 3.76
(0.488–28.95) | 0.204 |
| High | 56 (50) | 5.07 | 14.43
(1.88–110.49) | 0.010 |
| Infant gender | | | | 0.015 |
| Female | 46 (41) | | 1.00 | |
| Male | 66 (59) | 5.91 | 4.05
(1.31–12.52) | 0.015 |
Interaction between miR-21 and -143
Low miR-143 and high miR-21 expression levels were
associated with an increased risk of developing macrosomia.
Therefore, the current study analyzed the interaction between the
expression of the two miRNAs and macrosomia risk (Table III). Logistic regression analysis
demonstrated no significant difference in the risk of developing
macrosomia among samples with high miR-143 and low miR-21, high
miR-143 and high miR-21 or low miR-143 and low miR-21 levels.
However, low placental miR-143 combined with high miR-21 levels
were associated with increased likelihood of macrosomia (OR=26.47,
95% CI, 2.91-240.79). This suggested an interaction between miR-21
and -143 expression in macrosomia risk.
 | Table IIIInteraction between miR-143 and
miR-21 expression and macrosomiaa. |
Table III
Interaction between miR-143 and
miR-21 expression and macrosomiaa.
| Effect | n (%) | χ2 | OR (95% CI) | P-value |
|---|
| miR-143 | | | | 0.005 |
| High | 54 (48.6) | | 1.00 | |
| Low | 57 (51.4) | 7.74 | 5.87
(1.69–20.44) | 0.005 |
| miR-21 | | | | 0.018 |
| Low | 57 (51.4) | | 1.00 | |
| High | 54 (48.6) | 5.64 | 4.59
(1.31–16.15) | 0.018 |
| miR-143 and
miR-21 | | | | 0.032b |
| High miR-143, low
miR-21 | 17 (15.3) | | 1.00 | |
| High miR-143, high
miR-21 | 37 (33.4) | 1.92 | 3.57
(0.59–21.63) | 0.166 |
| Low miR-143, low
miR-21 | 40 (36.0) | 2.65 | 4.53
(0.74–27.95) | 0.103 |
| Low miR-143, high
miR-21 | 17 (15.3) | 8.46 |
26.47(2.91–240.79) | 0.004 |
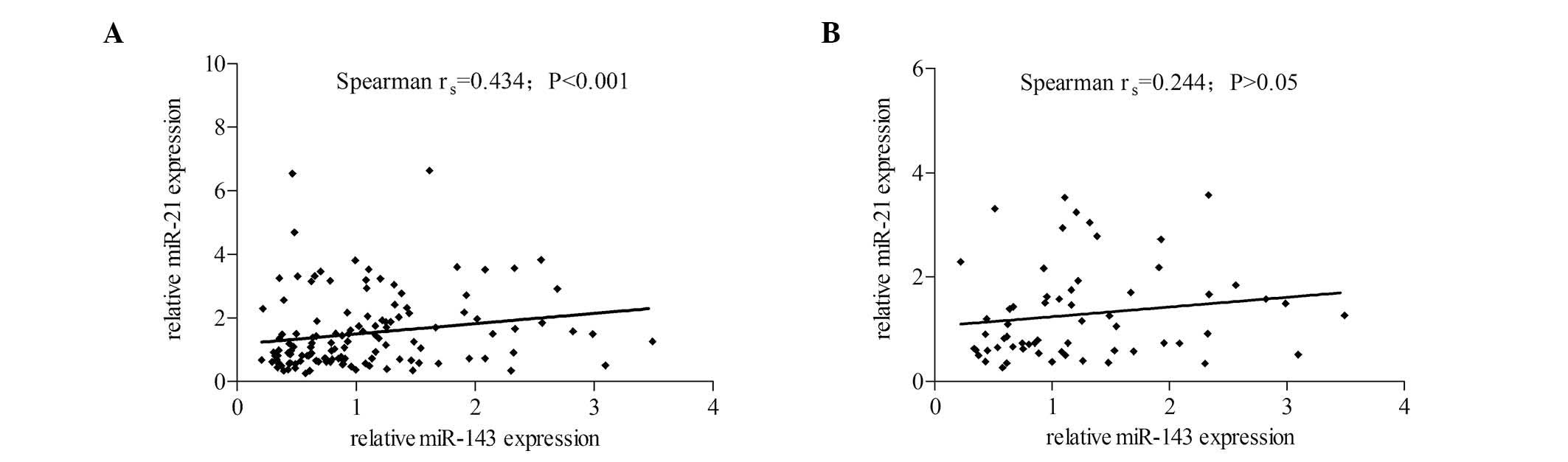
Correlation between miR-143 and miR-21
expression
As low miR-143 and high miR-21 levels were
demonstrated to be associated with macrosomia risk, the current
study analyzed whether miR-143 and -21 levels were inversely
correlated in macrosomic placenta. However, the microRNAs were
positively correlated in macrosomia samples (Spearman
rs= 0.43; P<0.001; Fig.
2A) Levels of miR-143 and miR-21 were not correlated in the
control group (P>0.05; Fig.
2B).
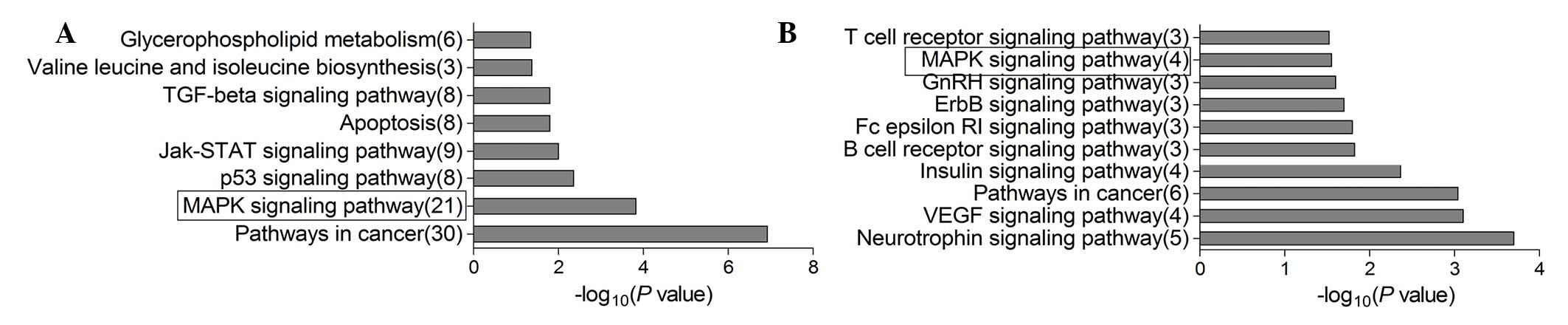
Pathway analysis
To understand the potential mechanisms by which
miR-143 and -21 function in shaping macrosomia risk, the present
study identified their potential target genes using miRTarBase, a
database that provides experimentally validated miRNA-target
interactions. The database suggested 489 target genes for miR-21
and 20 for miR-143 (Table IV).
Potential signaling pathways associated with the target genes,
generated by DAVID, are presented in Fig. 3. Identified target genes for miR-21
and -143 included those associated with the mitogen-activated
protein kinase (MAPK) signaling pathway, an important pathway in
cellular proliferation, growth and apoptosis.
 | Table IVRepresentative target genes of miR-21
and miR-143. |
Table IV
Representative target genes of miR-21
and miR-143.
miR-21 target gene
| miR-143 target gene
|
|---|
| Gene ID | Official full
name | Gene ID | Official full
name |
|---|
| PLPP1 | Phospholipid
phosphatase 1 | KRAS | Kirsten rat sarcoma
viral oncogene homolog |
| PLD2 | Phospholipase
D2 | KLF4 | Kruppel-like factor
4 |
| BCAT1 | Branched chain
amino-acid transaminase 1 | MYO6 | Myosin VI |
| LARS | Leucyl-tRNA
synthetase | DNMT3A | DNA
(cytosine-5-)-methyltransferase 3α |
| PDHA2 | Pyruvate
dehydrogenase (lipoamide) α2 | FNDC3B | Fibronectin type
III domain containing 3B |
| TGFB1 | Transforming growth
factor β1 | MAPK7 | Mitogen-activated
protein kinase 7 |
| TGFBR2 | Transforming growth
factor β receptor II | COX2 | Cytochrome c
oxidase subunit II |
| SP1 | Sp1 transcription
factor | COL1A1 | Collagen, type I,
α1 |
| MYC | v-myc avian
myelocytomatosis viral oncogene homolog | HRAS | Harvey rat sarcoma
viral oncogene homolog |
| FAS | Fas cell surface
death receptor | FSCN1 | Fascin
actin-bundling protein 1 |
| IL1B | Interleukin 1β | HK2 | Hexokinase 2 |
| BCL2 | B-cell CLL/lymphoma
2 | SERPINE1 | Serpin peptidase
inhibitor, clade E, member 1 |
| STAT3 | Signal transducer
and activator of transcription 3 | FHIT | Fragile histidine
triad |
| PIAS3 | Protein inhibitor
of activated STAT3 | MACC1 | Metastasis
associated in colon cancer 1 |
| AKT2 | v-akt murine
thymoma viral oncogene homolog 2 | PTGS2 |
Prostaglandin-endoperoxide synthase 2 |
| PTEN | Phosphatase and
tensin homolog | JAG1 | Jagged 1 |
| CDK6 | Cyclin-dependent
kinase 6 | AKT1 | v-akt murine
thymoma viral oncogene homolog 1 |
| MAPK3 | Mitogen-activated
protein kinase 3 | MDM2 | MDM2 proto-oncogene
E3 ubiquitin protein ligase |
| WNT5A | Wingless-type MMTV
integration site family member 5A | BCL2 | B-cell CLL/lymphoma
2 |
| EGFR | Epidermal growth
factor receptor | MMP13 | Matrix
metallopeptidase 13 |
Discussion
As the mediator between mother and fetus, the
placenta secretes various hormones, delivers nutrients to the
fetus, facilitates gas exchange and serves as a barrier to protect
the fetus from adverse environmental conditions, thus, is crucial
for fetal development (26).
Previous studies have demonstrated that aberrant miRNA expression
in the placenta was associated with preeclampsia (19), intrauterine growth retardation and
large birth size (18). The
results of the current study indicated that dysregulation of miR-21
and -143 expression was associated with macrosomia, and that there
was an association between the two miRNAs with regards to risk of
macrosomia. Collectively, the evidence suggested that aberrant
placental miRNA expression serves a prominent role in abnormal
fetal development.
miR-21 is established as an oncogene that is
important during tumor progression (27). In previous studies, the
upregulation of miR-21 in esophageal squamous cell carcinoma
induced cell proliferation and invasion (28), and anti-miR-21 inhibited cell
growth in breast tumor tissue (29). In addition, aberrant expression of
miR-21 has been demonstrated to affect apoptosis and metastasis in
glioma and colorectal cancer cells (30,31).
Placental and cancer cells share numerous similar features,
including proliferative, invasive and migratory capacity, and
shared epigenetic mechanisms for regulating trophoblast invasion
(32,33). Thus, the present study hypothesized
that miR-21 may affect placental function by influencing cellular
processes such as proliferation, thereby affecting fetal
development. The data demonstrated that placental miR-21 expression
was upregulated in macrosomia cases, which is consistent with a
previous investigation (34).
Maccani et al (20)
reported that low placental miR-21 expression was a risk factor for
small for gestational age (SGA). Similarly, a study in mice
demonstrated that miR-21 inhibition reduced obesity through the
reduction of adipocyte size (35).
Other reports have indicated that miR-21 regulated adipogenesis in
human primary adipocytes and that its expression was positively
correlated with BMI (36). miR-21
was also reported to regulate adipogenic differentiation and
adipose proliferation (37,38).
Therefore, the present study hypothesized that the upregulation of
placental miR-21 leads to macrosomia by promoting cell
differentiation and proliferation. However, further experimental
evidence is required to evaluate this.
To date, the role of miR-143 in the placenta has
remained unknown. To the best of our knowledge, the current study
was the first to demonstrate that placental miR-143 was
downregulated in macrosomia. Among the miRNAs associated with human
obesity, miR-143 was the first reported to regulate adipocyte
differentiation (39). Chen et
al (21) observed that
decreased miR-143 expression promoted adipogenic growth and
differentiation of adipose tissue-derived stromal cells by
targeting MAPK kinase 5. Compared with control placenta, placenta
from macrosomic pregnancies has greater capacity to support growth
and differentiation, as it must provide more space and transport
more nutrients to a larger fetus (40). Therefore, it can be explained why
miR-143 expression was reduced in macrosomic placenta. Other
research has reported miR-143 to be a pivotal regulator of cellular
activities, targeting multiple mRNAs involved in cell
proliferation, survival and apoptosis. For example, overexpression
of miR-143 repressed cell proliferation in melanoma and gastric
cancer by targeting syndecan-1 (41) and cyclooxygenase-2 (42), which are also expressed in the
placenta (43,44). This may partially explain the
downregulation of miR-143 in macrosomic placenta demonstrated in
the current study. Consistent with the present results, another
investigation demonstrated that miR-143 was downregulated in
maternal plasma in macrosomic pregnancies (45). However, an association between
miR-143 in maternal plasma and placenta remains unclear.
Another notable observation of the current study was
the opposing changes to the miR-21 and -143 levels in association
with macrosomia. Similarly, Keller et al (36) reported that the expression of
miR-21 was positively correlated with BMI. Expression of miR-21 was
higher in the adipocyte tissue of adults with BMI>30 than in
that of adults with lower BMIs, whereas miR-143 expression was
reduced adults with lower BMIs. The same trend was also been
demonstrated in colorectal carcinoma and colorectal liver
metastasis tissues, which exhibit overexpression of miR-21 and
underexpression of miR-143 relative to control tissues (46). These results suggest a functional
correlation between miR-21 and -143 expression, as does the
relationship identified by the logistic regression analysis in the
current study.
According to DAVID analysis (Fig. 3), miR-21 and -143 were associated
with MAPK signaling, which is a pivotal pathway in cell
proliferation and differentiation. miR-143 has been previously
reported to target extracellular signal-regulated kinase 5 (ERK5),
a member of the MAPK family that promotes cell proliferation and
growth. Overexpression of miR-143 reduced ERK5 expression and
activation, thus inhibiting cell proliferation and growth in human
colon carcinoma (47). By
contrast, miR-21 promoted proliferation of Eca109 cells and
inhibited apoptosis by activating the ERK1/2/MAPK pathway (48). Together, these results predict an
inverse correlation between levels of miR-143 and -21.
Unexpectedly, the current study demonstrated a positive correlation
between the two miRNAs in macrosomic but not in control placenta,
suggesting that a more complex mechanism co-regulates miR-21 and
-143. One such plausible mechanism is proposed as follows: A
restrictive correlation between miR-21 and -143 in the regulation
of fetal weight. Essentially, the two miRNAs independently and
inversely regulate growth in fetuses of normal weight, but in
macrosomia the two have a mutually restraining relationship, that
is, with the risk factor miR-21 increased, the protective factor
miR-143 was also increased, to protect the fetus from overgrowth.
This may be a homeostatic mechanism to maintain normal growth in
the fetus.
Previous studies have demonstrated that miR-16
targets genes in the pathways regulating numerous cellular
processes (49,50). Low levels of placental miR-16
expression are associated with SGA (20). However, the current study did not
identify significant differences in placental miR-16 expression
between macrosomic and control placenta. In addition, the present
study demonstrated that male infants were at a greater risk of
developing macrosomia than females. This result is consistent with
a number previous reports (11,51).
One explanation for this result is that male embryos have a higher
rate of cell division and metabolism than female embryos (52).
It is noted that this was a hospital-based
case-control study. All subjects were recruited from a large,
comprehensive hospital that serves all people in the Wenzhou
region. Thus, our data are likely to be representative of the
population in the Wenzhou region. Regardless, due to the small
sample size, the confidence intervals were relatively large.
In summary, the present study demonstrated that low
miR-143 expression and high miR-21 expression are risk factors for
macrosomia. Furthermore, there is an association between the two
miRNAs and macrosomia. These results provide evidence for the
effect of aberrant placental miRNA expression in macrosomia and
offer valuable insight into the mechanisms underlying macrosomia.
Future studies are required to determine the functional mechanism
linking placental miRNA expression to macrosomia and may provide
novel information on the potential use of miRNAs as a marker of
abnormal fetal growth.
Acknowledgments
The National Natural Science Foundation of China
(no. 81072378), and the Natural Science Funds of Zhejiang (no.
Y2101185). We gratefully acknowledge the placenta samples provided
by the Department of Obstetrics, Yuying Children's Hospital of
Wenzhou Medical University. We also thank the residents of the
Wenzhou region for their support for our epidemiological study.
References
|
1
|
Boulet SL, Alexander GR, Salihu HM and
Pass M: Macrosomic births in the united states: Determinants,
outcomes, and proposed grades of risk. Am J Obstet Gynecol.
188:1372–1378. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Henriksen T: The macrosomic fetus: A
challenge in current obstetrics. Acta Obstet Gynecol Scand.
87:134–145. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang X, Decker A, Platt RW and Kramer MS:
How big is too big? The perinatal consequences of fetal macrosomia.
Am J Obstet Gynecol. 198. pp. 517 e1–6. 2008, View Article : Google Scholar
|
|
4
|
Alsammani MA and Ahmed SR: Fetal and
maternal outcomes in pregnancies complicated with fetal macrosomia.
N Am J Med Sci. 4:283–286. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Stotland NE, Caughey AB, Breed EM and
Escobar GJ: Risk factors and obstetric complications associated
with macrosomia. Int J Gynaecol Obstet. 87:220–226. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ahlgren M, Wohlfahrt J, Olsen LW, Sørensen
TI and Melbye M: Birth weight and risk of cancer. Cancer.
110:412–419. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Boney CM, Verma A, Tucker R and Vohr BR:
Metabolic syndrome in childhood: Association with birth weight,
maternal obesity, and gestational diabetes mellitus. Pediatrics.
115:e290–e296. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Harder T, Plagemann A and Harder A: Birth
weight and risk of neuroblastoma: A meta-analysis. Int J Epidemiol.
39:746–756. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ognjanovic S, Carozza SE, Chow EJ, Fox EE,
Horel S, McLaughlin CC, Mueller BA, Puumala S, Reynolds P, Von
Behren J and Spector L: Birth characteristics and the risk of
childhood rhabdomyosarcoma based on histological subtype. Br J
Cancer. 102:227–231. 2010. View Article : Google Scholar :
|
|
10
|
Yu DM, Zhai FY, Zhao LY, Liu AD, Yu WT,
Jia FM, Zhang JG and Li J: Incidence of fetal macrosomia and
influencing factors in China in 2006. Chin J Child Health Care.
16:11–13. 2008.
|
|
11
|
Li G, Kong L, Li Z, Zhang L, Fan L, Zou L,
Chen Y, Ruan Y, Wang X and Zhang W: Prevalence of macrosomia and
its risk factors in china: A multicentre survey based on birth data
involving 101,723 singleton term infants. Paediatr Perinat
Epidemiol. 28:345–350. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Desgagné V, Hivert MF, St-Pierre J, Guay
SP, Baillargeon JP, Perron P, Gaudet D, Brisson D and Bouchard L:
Epigenetic dysregulation of the IGF system in placenta of newborns
exposed to maternal impaired glucose tolerance. Epigenomics.
6:193–207. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Prickett AR and Oakey RJ: A survey of
tissue-specific genomic imprinting in mammals. Mol Genet Genomics.
287:621–630. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xu X, Yang X, Liu Z, Wu K, Liu Z, Lin C,
Wang Y and Yan H: Placental leptin gene methylation and macrosomia
during normal pregnancy. Mol Med Rep. 9:1013–1018. 2014.PubMed/NCBI
|
|
15
|
Arora S, Rana R, Chhabra A, Jaiswal A and
Rani V: miRNA-transcription factor interactions: A combinatorial
regulation of gene expression. Mol Genet Genomics. 288:77–87. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Alvarez-Garcia I and Miska EA: MicroRNA
functions in animal development and human disease. Development.
132:4653–4662. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Amiel J, de Pontual L and Henrion-Caude A:
miRNA, development and disease. Adv Genet. 80:1–36. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang D, Na Q, Song WW and Song GY: Altered
expression of miR-518b and miR-519a in the placenta is associated
with low fetal birth weight. Am J Perinatol. 31:729–734. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu P, Zhao Y, Liu M, Wang Y, Wang H, Li
YX, Zhu X, Yao Y, Wang H, Qiao J, et al: Variations of microRNAs in
human placentas and plasma from preeclamptic pregnancy.
Hypertension. 63:1276–1284. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Maccani MA, Padbury JF and Marsit CJ:
miR-16 and miR-21 expression in the placenta is associated with
fetal growth. PLoS One. 6:e212102011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen L, Hou J, Ye L, Chen Y, Cui J, Tian
W, Li C and Liu L: MicroRNA-143 regulates adipogenesis by
modulating the MAP2K5-ERK5 signaling. Sci Rep.
4:38192014.PubMed/NCBI
|
|
22
|
Institute of Medicine: Nutrition During
Pregnancy. National Academies Press; Washington D.C.: 1990
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
24
|
Hsu SD, Tseng YT, Shrestha S, Lin YL,
Khaleel A, Chou CH, Chu CF, Huang HY, Lin CM, Ho SY, et al:
miRTarBase update 2014: An information resource for experimentally
validated miRNA-target interactions. Nucleic Acids Res. 42:D78–D85.
2014. View Article : Google Scholar :
|
|
25
|
Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar
|
|
26
|
Hogg K, Price EM, Hanna CW and Robinson
WP: Prenatal and perinatal environmental influences on the human
fetal and placental epigenome. Clin Pharmacol Ther. 92:716–726.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Selcuklu SD, Donoghue MT and Spillane C:
miR-21 as a key regulator of oncogenic processes. Biochem Soc
Trans. 37:918–925. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mori Y, Ishiguro H, Kuwabara Y, Kimura M,
Mitsui A, Ogawa R, Katada T, Harata K, Tanaka T, Shiozaki M and
Fujii Y: MicroRNA-21 induces cell proliferation and invasion in
esophageal squamous cell carcinoma. Mol Med Rep. 2:235–239.
2009.PubMed/NCBI
|
|
29
|
Si ML, Zhu S, Wu H, Lu Z, Wu F and Mo YY:
miR-21-mediated tumor growth. Oncogene. 26:2799–2803. 2007.
View Article : Google Scholar
|
|
30
|
Quintavalle C, Donnarumma E, Iaboni M,
Roscigno G, Garofalo M, Romano G, Fiore D, De Marinis P, Croce CM
and Condorelli G: Effect of miR-21 and miR-30b/c on TRAIL-induced
apoptosis in glioma cells. Oncogene. 32:4001–4008. 2013. View Article : Google Scholar
|
|
31
|
Asangani IA, Rasheed SA, Nikolova DA,
Leupold JH, Colburn NH, Post S and Allgayer H: MicroRNA-21 (miR-21)
post-transcriptionally downregulates tumor suppressor Pdcd4 and
stimulates invasion, intravasation and metastasis in colorectal
cancer. Oncogene. 27:2128–2136. 2008. View Article : Google Scholar
|
|
32
|
Ferretti C, Bruni L, Dangles-Marie V,
Pecking AP and Bellet D: Molecular circuits shared by placental and
cancer cells, and their implications in the proliferative, invasive
and migratory capacities of trophoblasts. Hum Reprod Update.
13:121–141. 2007. View Article : Google Scholar
|
|
33
|
Perry JK, Lins RJ, Lobie PE and Mitchell
MD: Regulation of invasive growth: Similar epigenetic mechanisms
underpin tumour progression and implantation in human pregnancy.
Clin Sci (Lond). 118:451–457. 2010. View Article : Google Scholar
|
|
34
|
Jiang H, Wu W, Zhang M, Li J, Peng Y, Miao
TT, Zhu H and Xu G: Aberrant upregulation of miR-21 in placental
tissues of macrosomia. J Perinatol. 34:658–663. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Seeger T, Fischer A, Muhly-Reinholz M,
Zeiher AM and Dimmeler S: Long-term inhibition of miR-21 leads to
reduction of obesity in db/db mice. Obesity (Silver Spring).
22:2352–2360. 2014. View Article : Google Scholar
|
|
36
|
Keller P, Gburcik V, Petrovic N, Gallagher
IJ, Nedergaard J, Cannon B and Timmons JA: Gene-chip studies of
adipogenesis-regulated microRNAs in mouse primary adipocytes and
human obesity. BMC Endocr Disord. 11:72011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kim YJ, Hwang SH, Cho HH, Shin KK, Bae YC
and Jung JS: MicroRNA 21 regulates the proliferation of human
adipose tissue-derived mesenchymal stem cells and high-fat
diet-induced obesity alters microRNA 21 expression in white adipose
tissues. J Cell Physiol. 227:183–193. 2012. View Article : Google Scholar
|
|
38
|
Kim YJ, Hwang SJ, Bae YC and Jung JS:
MiR-21 regulates adipogenic differentiation through the modulation
of TGF-beta signaling in mesenchymal stem cells derived from human
adipose tissue. Stem Cells. 27:3093–3102. 2009.PubMed/NCBI
|
|
39
|
Esau C, Kang X, Peralta E, Hanson E,
Marcusson EG, Ravichandran LV, Sun Y, Koo S, Perera RJ, Jain R, et
al: MicroRNA-143 regulates adipocyte differentiation. J Biol Chem.
279:52361–52365. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Larqué E, Ruiz-Palacios M and Koletzko B:
Placental regulation of fetal nutrient supply. Curr Opin Clin Nutr
Metab Care. 16:292–297. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li R, Zhang L, Jia L, Duan Y, Li Y, Wang
J, Bao L and Sha N: MicroRNA-143 targets Syndecan-1 to repress cell
growth in melanoma. PLoS One. 9:e948552014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wu XL, Cheng B, Li PY, Huang HJ, Zhao Q,
Dan ZL, Tian DA and Zhang P: MicroRNA-143 suppresses gastric cancer
cell growth and induces apoptosis by targeting COX-2. World J
Gastroenterol. 19:7758–7765. 2013. View Article : Google Scholar
|
|
43
|
Hanna N, Bonifacio L, Reddy P, Hanna I,
Weinberger B, Murphy S, Laskin D and Sharma D: IFN-gamma-mediated
inhibition of COX-2 expression in the placenta from term and
preterm labor pregnancies. Am J Reprod Immunol. 51:311–318. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Szabo S, Xu Y, Romero R, Fule T, Karaszi
K, Bhatti G, Varkonyi T, Varkonyi I, Krenacs T, Dong Z, et al:
Changes of placental syndecan-1 expression in preeclampsia and
HELLP syndrome. Virchows Archiv. 463:445–458. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ge Q, Zhu Y, Li H, Tian F, Xie X and Bai
Y: Differential expression of circulating miRNAs in maternal plasma
in pregnancies with fetal macrosomia. Int J Mol Med. 35:81–91.
2015.
|
|
46
|
Kulda V, Pesta M, Topolcan O, Liska V,
Treska V, Sutnar A, Rupert K, Ludvikova M, Babuska V, Holubec L Jr
and Cerny R: Relevance of miR-21 and miR-143 expression in tissue
samples of colorectal carcinoma and its liver metastases. Cancer
Genet Cytogenet. 200:154–160. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Borralho PM, Simões AE, Gomes SE, Lima RT,
Carvalho T, Ferreira DM, Vasconcelos MH, Castro RE and Rodrigues
CM: miR-143 overexpression impairs growth of human colon carcinoma
xenografts in mice with induction of apoptosis and inhibition of
proliferation. PLoS One. 6:e237872011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Liu F, Zheng S, Liu T, Liu Q, Liang M, Li
X, Sheyhidin I, Lu X and Liu W: MicroRNA-21 promotes the
proliferation and inhibits apoptosis in Eca109 via activating
ERK1/2/MAPK pathway. Mol Cell Biochem. 381:115–125. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang Y, Fan H, Zhao G, Liu D, Du L, Wang
Z, Hu Y and Hou Y: miR-16 inhibits the proliferation and
angiogenesis-regulating potential of mesenchymal stem cells in
severe pre-eclampsia. FEBS J. 279:4510–4524. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zuo W, Wang ZZ and Xue J: Artesunate
induces apoptosis of bladder cancer cells by miR-16 regulation of
COX-2 expression. Int J Mol Sci. 15:14298–14312. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Koyanagi A, Zhang J, Dagvadorj A, Hirayama
F, Shibuya K, Souza JP and Gülmezoglu AM: Macrosomia in 23
developing countries: An analysis of a multicountry,
facility-based, cross-sectional survey. Lancet. 381:476–483. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Mittwoch U: Blastocysts prepare for the
race to be male. Human Reprod. 8:1550–1555. 1993.
|