Introduction
Polycystic ovary syndrome (PCOS) is the most common
endocrinopathy in reproductive-age women, affecting 8–18% (1). Insulin resistance (IR) is considered
to be the main pathological factor responsible for the hormonal
disturbances associated with the syndrome (2), and IR in PCOS patients confers a
substantial risk for developing type 2 diabetes and cardiovascular
diseases (3,4). Furthermore, IR has been reported to
be linked with reduced mitochondrial respiration (5). Reduced expression of nuclear-encoded
genes involved in oxidative phosphorylation (OXPHOS) has been
reported in skeletal muscle of patients with PCOS (6). In addition, analysis of oxygen
consumption in blood mononuclear cells (leukocytes) indicated that
in women with PCOS, mitochondrial complex I respiration is reduced
compared with that in age- and body mass index-matched control
subjects (7). Due to this central
role of mitochondrial impairment in PCOS, current studies focus on
mutations in the mitochondrial genome of patients with POCS.
With the purpose of elucidating the molecular basis
of PCOS, the present study performed a systematic and extensive
mutational screening for pathogenic mutations in the mitochondrial
genome of a patient with PCOS. Previous studies by our group showed
that mitochondrial OXPHOS complexes are hot spots for mutations
associated with PCOS, as well as several mitochondrial transfer
(mt-t)RNA mutations (8,9). These tRNA mutations included
tRNAGln T4395C, tRNACys G5821A,
tRNAAsp A7543G, tRNALys A8343G,
tRNAArg T10454C and tRNAGlu A14693G. These
mt-tRNA mutations were localized at highly conserved nucleotides,
which caused structural and functional alternations, and
consequently resulted a failure of mt-tRNA metabolism. The present
study reported on the clinical and molecular characterization of a
Han Chinese patient with PCOS-IR. A sequence analysis of the
mitochondrial genome showed the presence of ND5 T12338C and
tRNASer (UCN) C7492T mutations.
Subject and methods
Case subject
A female patient (age, 31 years) from the Hangzhou
area of Zhejiang Province (China) was referred to the Department of
Obstetrics and Gynecology (Hangzhou First People's Hospital,
Hangzhou, China) due to an irregular menstrual cycle and suspicion
of PCOS. Following obtainment of written informed consent from the
patient, blood samples were obtained and a clinical evaluation was
performed following protocols approved by the Ethics Committee of
Hangzhou First People's Hospital (Hangzhou, China). The patient
underwent a thorough physical examination, a laboratory assessment
of metabolic syndrome and routine electrocardiography. In addition,
200 age-matched control subjects (average age, 30 years) were
selected from a panel of unaffected women of Han Chinese ancestry
from the same region. A thorough analysis of the patient's personal
and the patient's family's medical history was performed, the
patient's symptoms were assessed and PCOS was diagnosed according
to the Rotterdam criteria (10):
i) Clinical signs of hyperandrogenism; ii) polycystic ovaries
symptom; iii) A ratio of luteinizing hormone (LH) levels vs.
follicle stimulating hormone (FSH) of >2.0, or total
testosterone levels of >2.64 nmol/l. In addition, a
comprehensive physical examination of the patient was performed to
confirm the absence of any other syndrome, including
hyperprolactinemia, thyroid and adrenal diseases, 21-hydroxylase
deficiency and androgen-secreting tumors. The patient had no family
history of PCOS.
Laboratory assessment
Serum levels of luteinizing hormone (LH),
follicle-stimulating hormone (FSH), estradiol, progesterone, total
testosterone (TT) and fasting insulin levels were measured by
electrochemiluminescence immunoas-says (Roche Diagnostics, Basel,
Switzerland). The plasma glucose levels were measured using a
Beckman glucose analyzer (Ramcon, Fullerton, CA, USA). Furthermore,
an oral glucose tolerance test was performed. In brief, a blood
sample was obtained from the antecubital vein at 0 and 120 min for
measurement of plasma glucose concentrations, with 0 min denoting
the level of fasting plasma glucose. Patients with a plasma glucose
concentration of <7.8 mmol/l at the 120-min time point were
categorized as having normal glucose tolerance, while those with
plasma glucose levels of 7.8-11.1 mmol/l were classified as having
impaired glucose tolerance, and those with glucose levels of
>11.1 mmol/l were indicated to have diabetes mellitus. The main
outcome measure of IR was calculated using the homeostatic model
assessment of IR (HOMA-IR), where a value of >3.16 denotes IR
according to the following formula:
Molecular and genetic analysis
Total DNA was isolated from peripheral blood
leukocytes of the patient using an Universal Genomic DNA Extraction
kit version 3.0 (Takara Bio Inc., Otsu, Japan). The entire mtDNA
was amplified in 24 overlapping fragments as previously described
(11), using 24 overlapping
primers provided by the BGI (Shenzhen, China). The PCR mixture
included 200 µM dNTPs, 2 µl buffer (10X), 0.2
µl Taq DNA polymerase and 15 mmol/l Mg2+ (Takara
Bio Inc., Otsu, Japan). After polymerase chain reaction
amplification, the fragments were purified and subsequently
analyzed by direct sequencing in an ABI 3730 automatic DNA
sequencer (Thermo Fisher Scientific, Inc., Waltham, MA, USA) using
the BigDye Terminator Cycle Sequencing kit (Sigma-Aldrich, St.
Louis, MO, USA). Sample sequences were compared with the revised
Cambridge Reference Sequence (GenBank accession no. NC_012920) from
MITOMAP, a human mitochondrial genome database (http://www.mitomap.org/MITOMAP) (12).
Results and Discussion
Laboratory examination of the patient showed that
the levels of TT were 2.89 nmol/l, and the LH/FSH ratio was 2.81.
In addition, the patient was insulin resistant. The clinical
laboratory parameters of the PCOS patient are listed in Table I.
 | Table IClinical and biochemical parameters of
the patient with polycystic ovary syndrome. |
Table I
Clinical and biochemical parameters of
the patient with polycystic ovary syndrome.
| Parameter | Value | Normal range |
|---|
| LH (IU/l) | 14.54 | 1.50–9.30 |
| FSH (IU/l) | 5.17 | 1.40–18.00 |
| Estradiol
(pmol/l) | 205.60 | 0.00–198.20 |
| Progesterone
(nmol/l) | 3.12 | 0.89–3.88 |
| Total testosterone
(nmol/l) | 2.89 | 8.36–28.70 |
| Prolactin
(µg/l) | 5.22 | 2.00–17.00 |
| OGTT (0 h)
(mmol/l) | 5.11 | 3.90–6.10 |
| OGTT (2 h)
(mmol/l) | 8.55 | – |
| Insulin (0 h)
(µU/ml) | 9.80 | 2.60–12.00 |
| HOMA-IR (x) | 3.72 | – |
Mutational screening of the mitochondrial genome
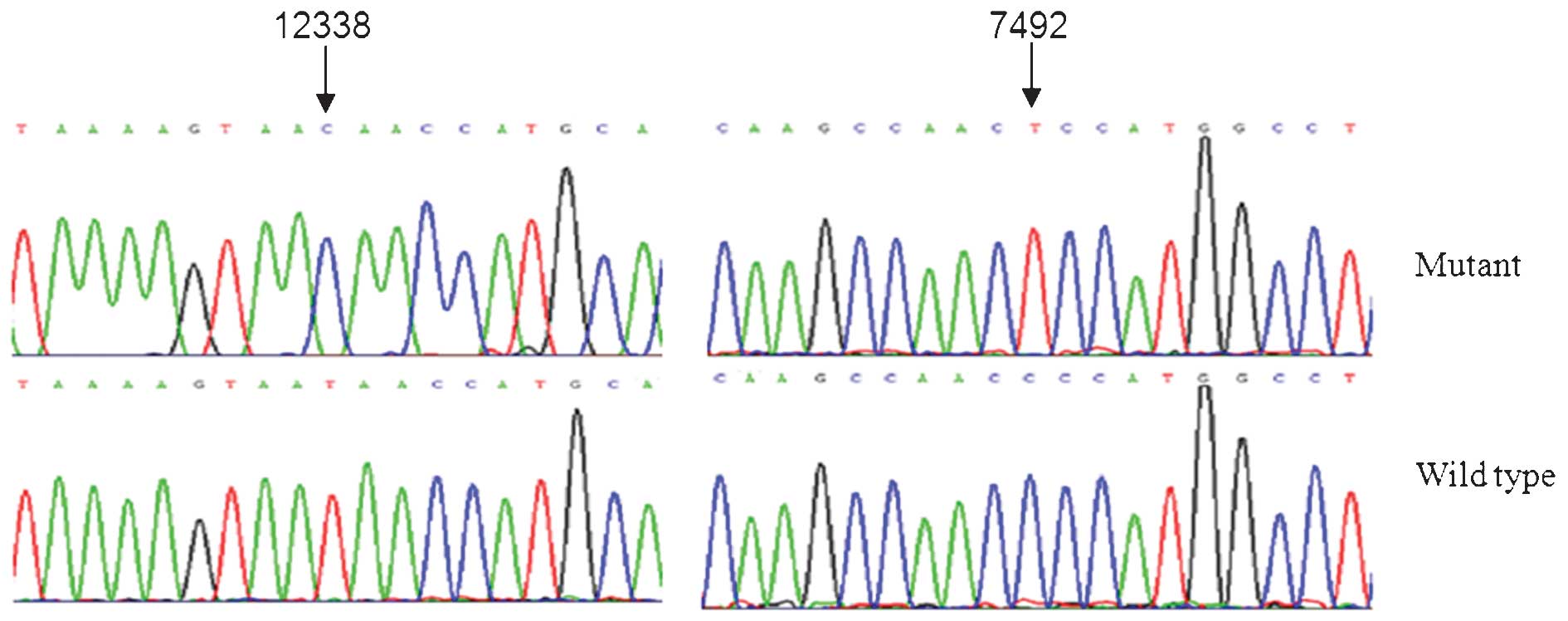
showed the presence of the homoplasmic ND5 T12338C and tRNASer
(UCN) C7492T mutations (Fig.
1), belonging to the human mitochondrial haplogroup F2
(13). Notably, the homoplasmic
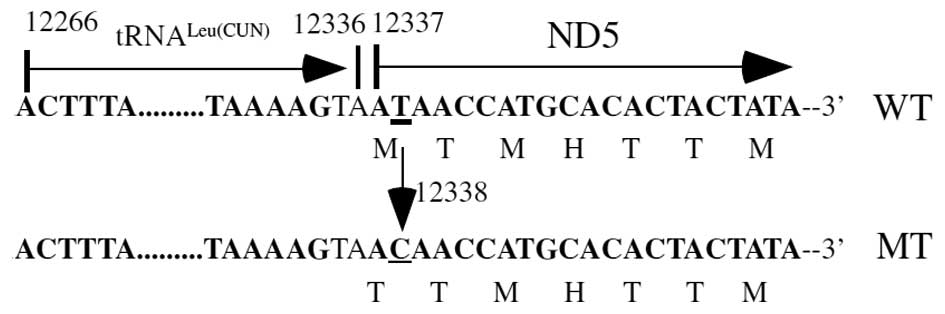
ND5 T12338C mutation was only detected in the patient with PCOS but
not in the healthy controls. This mutation is a result of a
replacement of the first amino acid, a translation-initiating
methionine, with a threonine in the ND5 polypeptide (Fig. 2). The encoding sequence of the
first methionine in the ND5 gene is extraordinarily conserved among
species and organelles ranging from bacteria to human mitochondria
(14). As a result, the truncated
ND5 protein was expected to be shortened by two amino acids, since
the ND5 T12338C mutation replaced the first amino acid from
methionine to threonine and, as methionine is the translational
initiation code, the mutant ND5 translates from the third
methionine. As reported by a previous study, the ND5 T12338C
mutation was also located in two nucleotides adjacent to the 3′ end
of the tRNALeu (CUN) gene (15). Consequently, this mutation altered
respiratory function, as well as the processing of RNA precursors,
thereby leading to a reduction in tRNALeu (CUN) levels.
The functional significance of the ND5 T12338C mutation in terms of
mitochondrial physiology was further supported by studies reporting
that the T12338C mutation may modulate the phenotypic expression of
the deafness-associated 12S rRNA A1555G mutation, and that it was
associated with essential hypertension and Leber's hereditary optic
neuropathy (16,17). The homoplasmic ND1 T3308C mutation
has been suggested to contribute to the higher penetrance of
hearing loss in a large African pedigree compared with that in
Japanese and French pedigrees carrying the tRNASer (UCN)
T7511C mutation (18,19). A significant reduction in
steady-state levels of ND1 mRNA and the adjacent tRNALeu
(UUR) observed in cybrids carrying the T3308C mutation was
likely due to an alteration in the processing of the H-strand
polycistronic RNA precursors or the destabilization of ND1 mRNA
(20). Thus, the ND5 T12338C
mutation, which is similar to the ND1 T3308C mutation (14,19),
may induce a reduction in ND5 mRNA expression levels, as well as
the steady state level of tRNALeu (CUN); therefore, this
mutation may cause mitochondrial dysfunction and, since
mitochondrial dysfunction is associated with the pathogenesis of
PCOS-IR (6,7), we propose the ND5 T12338C mutation is
associated with PCOS-IR.
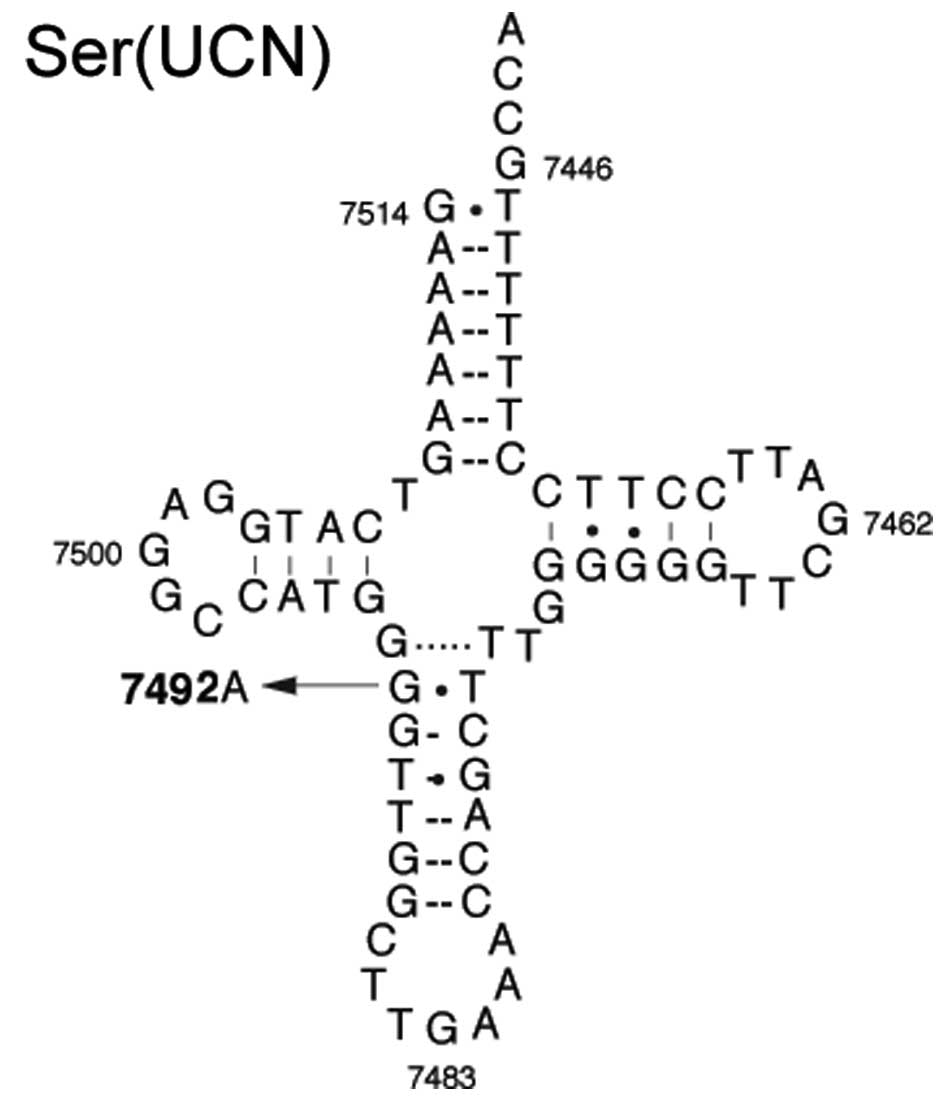
The homoplasmic C7492T mutation occurred at position
26 in the anticodon stem of the tRNASer (UCN)-encoding
gene (Fig. 3). Mutation at this
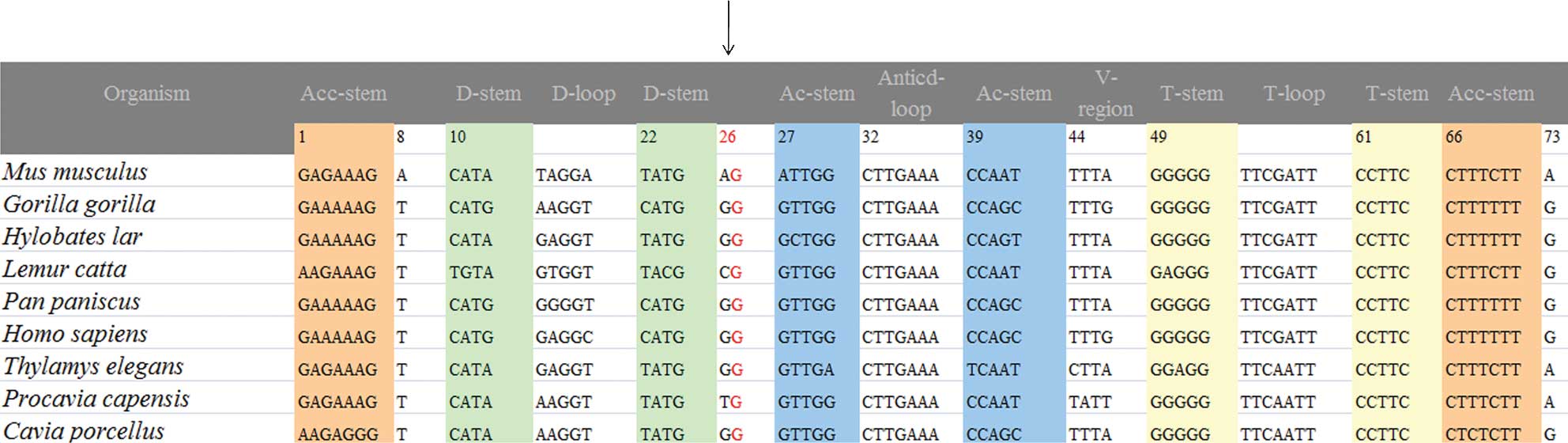
position is highly conserved among various species, which suggests
that it may be associated with the pathogenesis of PCOS-IR
(Fig. 4). In addition, the C7492T
mutation led to an alteration of base pairing (A26-U44), and
identical base pairing at the same position in tRNAIle
generated by the heteroplasmic T4295C mutation has been associated
with altered tRNA metabolism in chronic progressive external
ophthalmoplegia (21). Therefore,
it can be anticipated that the C7492T mutation may alter the
tertiary structure of tRNASer (UCN) and, since the
anticodon stem is critical for codon and anticodon interaction,
this mutation may reduce the steady state level of tRNASer
(UCN) as well as the aminoacylation ability. The resultant
shortage of tRNASer (UCN) may be responsible for defects
in mitochondrial protein synthesis. Consequently, the deficiency in
respiratory chain function will cause a reduction in ATP synthesis
and increase the generation of reactive oxygen species. Therefore,
this mutation may have an active role in the pathogenesis of
PCOS-IR. However, due to the complex molecular mechanisms of PCOS,
it is likely that the C7492T and T12338C mutations alone are
insufficient to produce the clinical phenotype; other factors,
including nuclear genes, epigenetic modifications and environmental
factors may contribute to the pathogenesis of PCOS.
Acknowledgments
The authors would like to thank the patient for
participating this study. This work was supported by grants from
the Ministry of Public Health of Zhejiang Province (grant no.
2013KYA158) and the Hangzhou Bureau of Science and Technology
(grant no. 20150633B16).
References
|
1
|
March WA, Moore VM, Willson KJ, Phillips
DI, Norman RJ and Davies MJ: The prevalence of polycystic ovary
syndrome in a community sample assessed under contrasting
diagnostic criteria. Hum Reprod. 25:544–551. 2010. View Article : Google Scholar
|
|
2
|
Dunaif A: Insulin resistance and the
polycystic ovary syndrome: Mechanism and implications for
pathogenesis. Endocr Rev. 18:774–800. 1997.PubMed/NCBI
|
|
3
|
Teede HJ, Hutchison SK and Zoungas S: The
management of insulin resistance in polycystic ovary syndrome.
Trends Endocrinol Metab. 18:273–279. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Moran LJ, Misso ML, Wild RA and Norman RJ:
Impaired glucose tolerance, type 2 diabetes and metabolic syndrome
in polycystic ovary syndrome: A systematic review and
meta-analysis. Hum Reprod Update. 16:347–363. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Petersen KF and Shulman GI: Etiology of
insulin resistance. Am J Med. 119(5 Suppl 1): S10–S16. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Skov V, Glintborg D, Knudsen S, Jensen T,
Kruse TA, Tan Q, Brusgaard K, Beck-Nielsen H and Højlund K: Reduced
expression of nuclear-encoded genes involved in mitochondrial
oxidative metabolism in skeletal muscle of insulin-resistant women
with polycystic ovary syndrome. Diabetes. 56:2349–2355. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Victor VM, Rocha M, Bañuls C,
Sanchez-Serrano M, Sola E, Gomez M and Hernandez-Mijares A:
Mitochondrial complex I impairment in leukocytes from polycystic
ovary syndrome patients with insulin resistance. J Clin Endocrinol
Metab. 94:3505–3512. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhuo G, Ding Y, Feng G, Yu L and Jiang Y:
Analysis of mitochondrial DNA sequence variants in patients with
polycystic ovary syndrome. Arch Gynecol Obstet. 286:653–659. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhuo G, Feng G, Leng J, Yu L and Jiang Y:
A 9-bp deletion homoplasmy in women with polycystic ovary syndrome
revealed by mitochondrial genome-mutation screen. Biochem Genet.
48:157–163. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rotterdam ESHRE/ASRM-Sponsored PCOS
Consensus Work-shop Group: Revised 2003 consensus on diagnostic
criteria and long-term health risks related to polycystic ovary
syndrome. Fertil Steril. 81:19–25. 2004. View Article : Google Scholar
|
|
11
|
Rieder MJ, Taylor SL, Tobe VO and
Nickerson DA: Automating the identification of DNA variations using
quality-based fluorescence re-sequencing: Analysis of the human
mitochondrial genome. Nucleic Acids Res. 26:967–973. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Brandon MC, Lott MT, Nguyen KC, Spolim S,
Navathe SB, Baldi P and Wallace DC: MITOMAP: A human mitochondrial
genome database-2004 update. Nucleic Acids Res. 33(Database issue):
D611–D613. 2005. View Article : Google Scholar :
|
|
13
|
Kong QP, Bandelt HJ, Sun C, Yao YG, Salas
A, Achilli A, Wang CY, Zhong L, Zhu CL, Wu SF, et al: Updating the
East Asian mtDNA phylogeny: A prerequisite for the identification
of pathogenic mutations. Hum Mol Genet. 15:2076–2086. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen B, Sun D, Yang L, Zhang C, Yang A,
Zhu Y, Zhao J, Chen Y, Guan M, Wang X, et al: Mitochondrial ND5
T12338C, tRNA(Cys) T5802C and tRNA(Thr) G15927A variants may have a
modifying role in the phenotypic manifestation of
deafness-associated 12S rRNA A1555 G mutation in three Han Chinese
pedigrees. Am J Med Genet A. 146A:1248–1258. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Andrews RM, Kubacka I, Chinerry PF,
Lightowlers RN, Turnbull DM and Howell N: Reanalysis and revision
of the Cambridge reference sequence for human mitochondrial DNA.
Nat Genet. 23:1471999. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Teng L, Zheng J, Leng J and Ding Y:
Clinical and molecular characterization of a Han Chinese family
with high penetrance of essential hypertension. Mitochondrial DNA.
23:461–465. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu XL, Zhou X, Zhou J, Zhao F, Zhang J,
Li C, Ji Y, Zhang Y, Wei QP, Sun YH, et al: Leber's hereditary
optic neuropathy is associated with the T12338C mutation in
mitochondrial ND5 gene in six Han Chinese families. Ophthalmology.
118:978–985. 2011. View Article : Google Scholar
|
|
18
|
Li R, Ishikawa K, Deng JH, Heman-Ackah S,
Tamagawa Y, Yang L, Bai Y, Ichimura K and Guan MX: Maternally
inherited nonsyndromic hearing loss is associated with the T7511C
mutation in the mitochondrial tRNASerUCN gene in a Japanese family.
Biochem Biophys Res Commun. 328:32–37. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li X, Fischel-Ghodsian N, Schwartz F, Yan
Q, Friedman RA and Guan MX: Biochemical characterization of the
mitochondrial tRNASer (UCN) T7511C mutation associated with
nonsyndromic deafness. Nucleic Acids Res. 32:867–877. 2004.
View Article : Google Scholar :
|
|
20
|
Rossmanith W, Tullo A, Potuschak T, Karwan
R and Sbisà E: Human mitochondrial tRNA processing. J Biol Chem.
270:12885–12891. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Silvestri G, Servidei S, Rana M, Ricci E,
Spinazzola A, Paris E and Tonali P: A novel mitochondrial DNA point
mutation in the tRNA (Ile) gene is associated with progressive
external ophtalmoplegia. Biochem Biophys Res Commun. 220:623–627.
1996. View Article : Google Scholar : PubMed/NCBI
|