Introduction
Bone homeostasis is a dynamic process, which
maintains bone skeletal mass and quality, and protects organs and
hematopoiesis by bone remodeling throughout life (1,2).
Bone remodeling is a physiological process that involves a
functional balance between osteoclast-regulated bone resorption and
osteoblast-induced bone formation (3,4).
However, an imbalance in bone remodeling, induced by various
diseases, aging, smoking, increasing inflammation factors or
estrogen deficiency following menstruation, leads to adult skeletal
defects, including osteoporosis, rheumatoid arthritis, Paget's
disease and osteolysis associated with periodontal disease
(5). These diseases are
characterized by the excessive activity of osteoclasts (5).
Osteoclasts are bone-resorbing multinuclear cells,
which originate from hematopoietic stem cells of the
monocyte/macrophage lineage (6).
The differentiation of osteoclasts is regulated by the two
cytokines, receptor activator of nuclear factor (NF) κ-B ligand
(RANKL) and macrophage colony-stimulating factor (M-CSF) (7). M-CSF provides survival signals and
induces the expression of the RANK receptor in osteoclast and
precursor cells (1). RANKL is
expressed in osteoblasts and binds to the RANK receptor expressed
in osteoclast precursor cells (1,8).
This binding leads to the activation of key signaling pathways in
osteoclast differentiation through activated osteoclasia and the
regulation of factors assisting bone resorption (7). RANKL is a member of the tumor
necrosis factor (TNF) superfamily, which is expressed in
osteoblasts (4,9). Binding of RANKL to its receptor,
RANK, on osteoclast precursors results in the recruitment of TNF
receptor-associated factor (TRAF) family proteins, including TRAF6
(10). TRAF6 induces not only
mitogen-activated protein kinases (MAPKs), but also activates NF-κB
(2). This signaling subsequently
activates the c-Fos transcription factor and nuclear factor of
activated T cell cytoplasmic 1 (NFATc1) (10,11).
These transcription factors regulate the expression of several
osteoclast-specific genes (3).
To identify compounds, which inhibit osteoclast
differentiation, the present study used centipedegrass extract
(CGE) in a bone marrow culture system. During the process, it was
investigated whether osteoclast formation was suppressed by
Eremochloa ophiuroides CGE. E. ophiuroides is native
to China and Southeast Asia, and has become one of the most popular
lawn grasses in South America (12,13).
In a previous study, maysin was identified as a component of E.
ophiuroides centipedegrass by liquid chromatography-mass
spectrometry (12). CGE also
contains several C-glycosyl flavones and phenolic constituents,
including luteolin, orientin, isoorientin, rhamnosylisoorientin,
derhamnoslymaysin and luteoin-6-C-boivinopyranose (12,14).
A previous study reported that the methanolic extracts from the
leaves of E. ophiuroides centipedegrass exhibit anticancer
and anti-adipogenic activities in several cell types (12,15).
However, there are no reports on the inhibitory
effects of CGE on osteoclastogenesis. In the present study, the
anti-osteoclastogenic effects and underlying signaling pathway of
CGE in RANKL-stimulated primary precursor cells without cytotoxity
were investigated. The results demonstrated for the first time, to
the best of our knowledge, that CGE significantly suppresses
RANKL-induced osteoclast differentiation by modulating
osteoclast-specific genes, transcription factors and signaling
molecules (16). Thus, these
results provided evidence that CGE may represent an
anti-osteoclastogenic agent for use in the treatment of bone loss
diseases.
Materials and methods
Materials and antibodies
Cell culture medium and fetal bovine serum (FBS)
were obtained from Invitrogen (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). RANKL and M-CSF were purchased from PeproTech
(Rocky Hill, NJ, USA). The Leukocyte Acid Phosphatase Assay kit and
3-(4,5-dimethyl-thiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Preparation of the CGE
The dried leaves of E. ophiuroides
(centipedegrass; 5 kg) were ground in a Wiley mill (Weiber, India)
and passed through a 420-μm sieve. The final ground sample
(1 kg) was extracted three times with 100 liters of 80% methanol
(MeOH; v/v) for 24 h with constant agitation at an ambient
temperature in the dark. The extracts were filtered using No. 2
filter paper (Advantech Japan Co., Ltd., Tokyo, Japan) and
concentrated in vacuo. The MeOH extracts were then
fractionated with n-hexane and ethyl acetate (EA)(Daejung Chemicals
& Metals Co., Ltd., Siheung, Korea). The EA extracts were
concentrated in vacuo using an EYELA N-1001 rotary
evaporator (Tokyo Rikakikai Co., Ltd., Tokyo, Japan) and the dried
compounds were dissolved in MeOH. The dissolved MeOH extracts were
diluted in 20% MeOH and chromatographed on a Toyopearl HW-40C resin
(Tosoh, Tokyo, Japan) column using 70% MeOH (elution volume, 700
ml). The fraction was evaporated and freeze-dried, and the dried
extracts were reconstituted in dimethyl sulfoxide (DMSO; final
concentration, 1 mg/ml; Sigma-Aldrich) for cell treatment. CGE was
analyzed using an HPLC-MS (Agilent 1260 series; Agilent
Technologies, Santa Clara, CA, USA) in a positive ion mode. The CGE
was injected (10 μl) and chromatographed on a YMC-Pack ODS-A
(150×4.6 mm; l.D. S-5 μl; 12 nm) with a linear gradient
between buffer A (0.1% formic acid; Wako Pure Chemical Industries,
Ltd., Osaka, Japan) and buffer B (100% MeOH). The mobile phase was
programed as follows: 100% buffer A at 10 min, 50% buffer A at 30
min and 0% buffer A at 60 min. The calibration curves were
generated using high performance liquid chromotography (HPLC) with
standards, including rhamnosylisoorientin, derhamnoxylmaysin,
maysin, eremonetin (all kindly donated by Tae Hoon Kim, Daegu
University, South Korea), chlorogenic acid, orientin, isoorientin,
luteolin and icariin (used as a standard compound) (all
Sigma-Aldrich; data not shown). The flow rate was 0.5 ml/min and
ultraviolet detection was performed at 360 nm with a 1260 Infinity
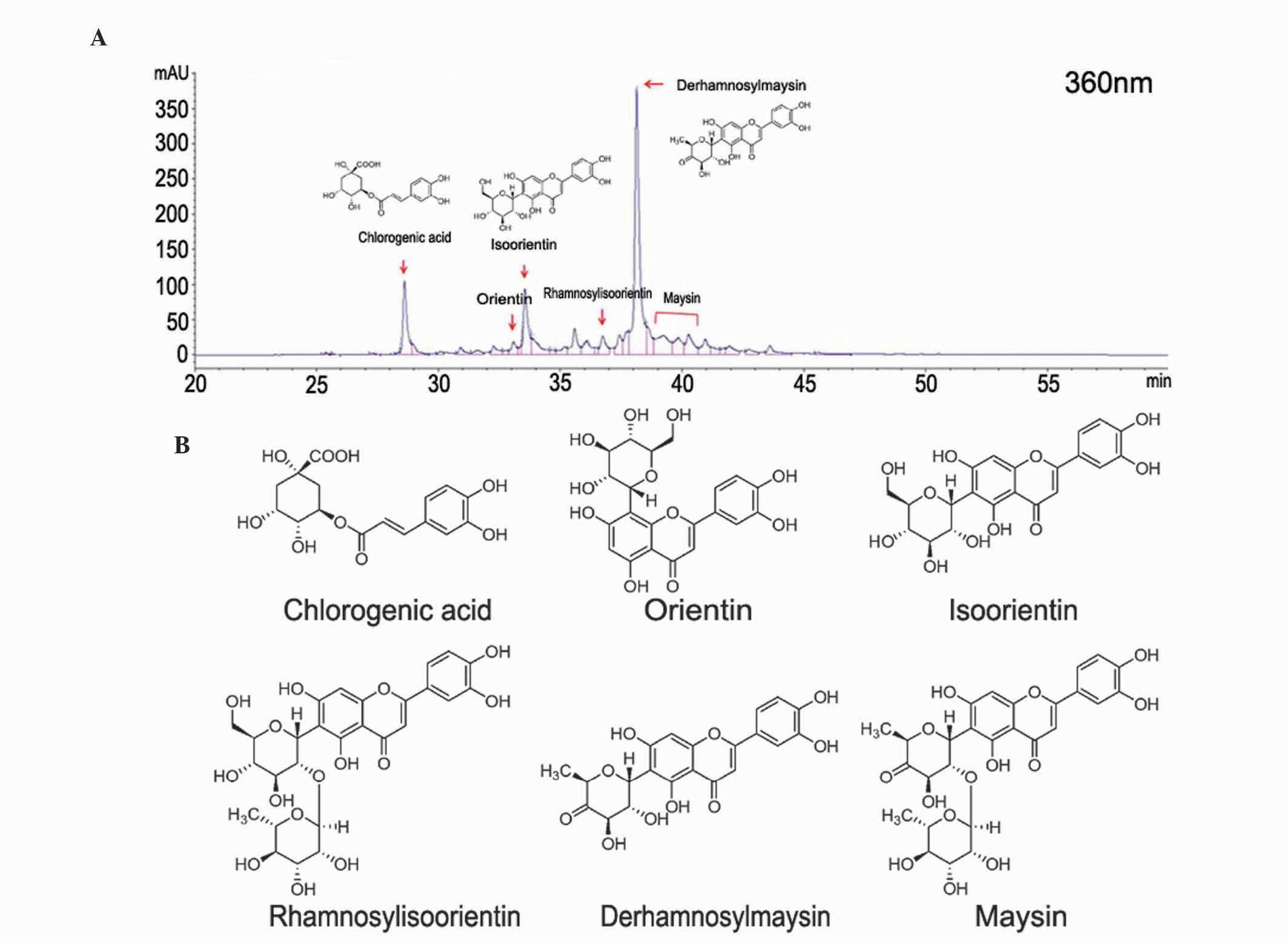
Diode Array Detector (Agilent Technologies, Santa Clara, CA, USA).
The CGE contained predominantly chlorogenic acid, at ~20% [43.4
μg/mg dry weight(D.W)], derahnosylmaysin at ~65% (108.4
μg/mg D.W), maysin at ~5% (193.6 μg/mg D.W), as shown
in Fig. 1.
Isolation of bone marrow-derived
macrophages (BMMs) and cell culture
A total of 5 male ICR mice (5-week-old; body weight,
30–32 g) were obtained from SLC, Inc. (Kotoh-cho, Japan). The mice
were maintained under a 12-h light/dark cycle, with access to food
and water ad libitum and allowed to acclimate to their
environment for 1 week prior to beginning experiments. To isolate
the BMMs, marrow cells were cultured from the femurs and tibias of
5 mice in α-minimal essential medium (α-MEM) containing 10% FBS
with 100 U/ml penicillin and 100 μg/ml streptomysin (Gibco;
Thermo Fisher Scientific, Inc.) in the presence of M-CSF (30 ng/ml)
at 37°C in 5% CO2. After 3 days, the nonadherent cells
were removed by washing with fresh medium, and the adherent cells
were used as BMMs. The BMMs were further cultured for 3 days in an
osteoclastogenic medium (α-MEM containing 10% FBS, 30 ng/ml M-CSF
and 100 ng/ml RANKL). The cells were then cytochemically stained
for tartrate-resistant acid phosphatase (TRAP), an osteoclast
marker protein.
TRAP staining
The BMMs were replated and then further cultured in
medium containing M-CSF (30 ng/ml) and RANKL (100 ng/ml) for 3
days. The cells were washed with phosphate-buffered saline (PBS)
and fixed 3.7% formaldehyde (Sigma-Aldrich) for 10 min. Following
washing with PBS, the cells were incubated with 0.1% Triton X-100
(Sigma-Aldrich) for 1 min. The cells were then washed and incubated
for between 40 min and 1 h at 37°C in the dark with a mixture of
Fast Garnet GBC, sodium nitrite, naphthol AS-BI phosphoric acid,
acetate and tatrate from the Leukocyte Acid Phosphatase Assay kit
(Sigma-Aldrich), according to the manufacturer's instructions. The
cells were washed with distilled water, and TRAP-positive
multinucleated cells containing three or more nuclei were counted
under an Olympus IX71 microscope (Olympus Corporation, Tokyo,
Japan).
Cell viability
The cells (1×104 cells/well) were seeded
in a 96-well plate and incubated overnight in media supplemented
with 10% FBS. Various concentrations of CGE (5, 10, 20, 40 or 80
μg/ml) were then added to the cells. The cells were
incubated at 37°C in 5% CO2 for 24 or 48 h, washed with
PBS and were then treated with medium containing 100 μg/ml
3-(4,5-dimeth-ylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
for 3 h at 37°C. The cells were then washed with PBS and dissolved
in 200 ml DMSO. The resulting intracellular purple formazan was
quantified from the absorbance at 540 nm using an Infinite M200
microplate reader (Tecan Group, Ltd., Männedorf, Switzerland).
Reverse transcription-polymerase chain
reaction (RT-PCR)
Total RNA was isolated from the cultured cells using
TRIzol reagent (Molecular Research Center, Inc., Cincinnati, OH,
USA) and cDNA synthesis was performed using Maxime RT Premix
(iNtron Biotechnology, Inc., Seongnam, Korea), according to the
manufacturer's protocol. The PCR primers were purchased from Neo
Probe (Daejeon, Korea). The cDNA was amplified in a reaction mix
made up to 20 μl, containing 1 μl cDNA, Maxime PCR
Premix kit (iNtron Biotechnology, Inc.), distilled water and 10
pmol of forward and of reverse primers, using the following primer
sets:
c-Fos, forward 5′-CTGGTGCAGCCCACT CTG GTC-3′ and
reverse 5′-CTTTCAGCAGATTGGCAATCTC-3′; NFATc1, forward
5′-CAACGCCCTGACCACCGATAG-3 and reverse 5′-GGCTGCCTTCCGTCTCATAGT-3′;
osteoclast-associated immunoglobulin-like receptor (OSCAR), forward
5′-CTGCTGGTAACGGATCAGCTCCCCAGA-3′ and reverse
5′-CCAAGGAGCCAGAACCTTCGAAACT-3′; matrix metalloproteinase (MMP)-9,
forward 5′-CGTCGTGATCCCCACTTACT-3′ and reverse
5′-AGAGTACTGCTTGCCCAGGA-3′; cathepsin K, forward
5′-AGGCGGCTATATGACCACTG-3′ and reverse 5′-CCGAGCCAAGAGAGCATATC-3′;
dendritic cell-specific transmembrane protein (DC-STAMP), forward
5′-GCAAGGAACCCAAGGAGTCG-3′ and reverse 5′-CAGTTGGCCCAGAAAGAGGG-3′;
and GAPDH, forward 5′-GGTGAAGGTCGGTGTGAACG-3′ and reverse
5′-CTCGCTCCTGGAAGATGGTG-3′.
Following initial denaturation at 95°C for 2 min,
PCR was performed for various cycle numbers, at varying annealing
temperatures, as follows: c-Fos, 58°C, 32 cycles; NFATc1, 59°C, 32
cycles; OSCAR, 59°C, 32 cycles; MMP-9, 58°C, 33 cycles; cathespin
K, 59°C, 32 cycles; and DC-STAMP, 60°C; 31 cycles. The reaction
products were separated on 1% agarose gels (Lonza Group, Basel,
Switzerland) and stained with GelRed (Biotium, Hayward, CA, USA).
These cycles also included denaturation steps at 95°C for 30 sec
and elongation steps at 72°C for 45 sec. The relative levels of
c-Fos, NFATc1, OSCAR, MMP-9, cathepsin K and DC-STAMP were
normalized to GAPDH. To quantify expression levels, band intensity
was compared using Gel-Pro version 3.0 software (Exon Intron Inc.,
Loganville, PA, USA).
Immunoblot analysis
Following washing with cold PBS, cells were
harvested and lysed using lysis buffer (Rockland, Inc.,
Gilbertsville, PA, USA). Following centrifugation at 18,300 × g for
20 min at 4°C, the supernatants were used as cell extract. Equal
concentrations of cell extract (40 mg/ml), determined using a
bicinchoninic acid assay (Thermo Fisher Scientific, Inc.), were
separated using SDS-PAGE on 8–12% gels and then transferred onto
polyvinylidene diflioride membranes (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). The membranes were blocked with 5% skim milk in
Tris-buffered saline (TBS) containing 0.1% Tween-20 (TBST) at room
temperature for 1 h, and then incubated overnight at 4°C with
primary antibodies diluted in 5% skim milk in TBST. These were:
Rabbit anti-c-Fos (dilution, 1:1,000; cat no. 2250; Cell Signaling
Technology, Inc., Danvers, MA, USA); mouse anti-NFATc1 (dilution,
1:1,000; cat. no. sc-7294; Santa Cruz Biotechnology, Inc., Dallas,
TX, USA); and rabbit anti-GAPDH (dilution, 1:2,000; cat. no. 2118;
Cell Signaling Technology, Inc.). The membranes were washed three
times in TBST for 30 min, with shaking. The membranes were then
incubated with the secondary antibodies horseradish peroxidase
(HRP)-conjugated horse anti-mouse IgG (dilution, 1:2,000; cat. no.
7076; NFAT1-incubated membrane) or HRP-conjugated goat anti-rabbit
IgG (dilution, 1:2,000; cat. no. 7074; all other membranes) (both
Cell Signaling Technology, Inc.) for 1 h at room temperature.
Following this, the membranes were washed twice in TBST for 10 min,
with shaking. These immunocomplexes were visualized on X-ray film
(GE Healthcare Life Sciences, Chalfont, UK) using Enhanced
Chemiluminescence Prime Western Blotting Detection reagent (GE
Healthcare Life Sciences), according to the manufacturer's
protocol. To quantify expression levels, band intensity was
compared using Gel-Pro software.
Lipopolysaccharide (LPS)-induced bone
resorption
A total of 21 ICR mice (6-week-old) were divided
into three groups (n=7 per group). The control group mice were
injected twice with PBS, instead of LPS, at day 0 and day 4 (PBS
group). The remaining mice received an intraperitoneal injection of
LPS (5 mg/kg body weight) at day 0 and day 4. Half of the
LPS-treated mice were injected orally with CGE 1 day prior to LPS
injection, and every day following LPS injection (LPS+CGE group).
The other half of the LPS-treated mice received vehicle (10 mM
KOH), instead of CGE (LPS group). All mice were sacrificed by
cervical dislocation 8 days following LPS injection. The left
femurs of the mice were scanned using a high-resolution micro-CT
scanner (Skyscan 1076; Bruker, Kontich, Belgium). Bone trabecular
parameter analyses were performed with the micro-CT data using the
software provided by Skyscan. All animal experiments were performed
in accordance with the principles of the Care and Use of Laboratory
Animals (17), and approved by the
Institutional Animal Care and Use Committee of Korea Atomic Energy
Research Institute (Jeongeup Si, Korea).
Statistical analysis
All experimental data are expressed as the mean ±
standard error of the mean. All experiments were repeated at least
three times, unless otherwise indicated. Statistical differences
between the control and experimental groups were analyzed using
Student's t-test with SPSS version 9.0 software (SPSS Inc.,
Chicago, IL, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
CGE inhibits osteoclast
differentiation
To determine the effects of CGE on
osteoclastogenesis, the effect of CGE on osteoclast formation was
examined using the bone marrow cells. Osteoclasts were generated
from mouse BMMs in the presence of CGE in osteoclastogenesis. RANKL
(100 ng/ml) induced TRAP-positive multinucleated osteoclast
differentiation in the BMMs, however CGE reduced the formation and
numbers of TRAP-positive multinucleated cells generated, with
76.4±10.3, 62.7±7.9, 69.5±6.8, 44.6±3.0 and 10.7±3.37% inhibition
at 5, 10, 20, 40 and 80 μg/ml, respectively (Fig. 2A and B). Therefore, CGE reduced the
formation and numbers of TRAP-positive multinucleated cells in a
concentration-dependent manner. CGE inhibited the negative effect
on osteoclastogenesis. The present study also measured the effects
of CGE in the BMMs using an MTT assay to exclude the possibility
that the inhibition was due to cytotoxicity. CGE demonstrated no
cytotoxic effects at the concentrations found to effectively
inhibit osteoclast formation (Fig.
2C). This result, suggesting that osteoclastogenesis was
suppressed by CGE, did not affect cell growth rate or induce toxic
effects in the BMMs.
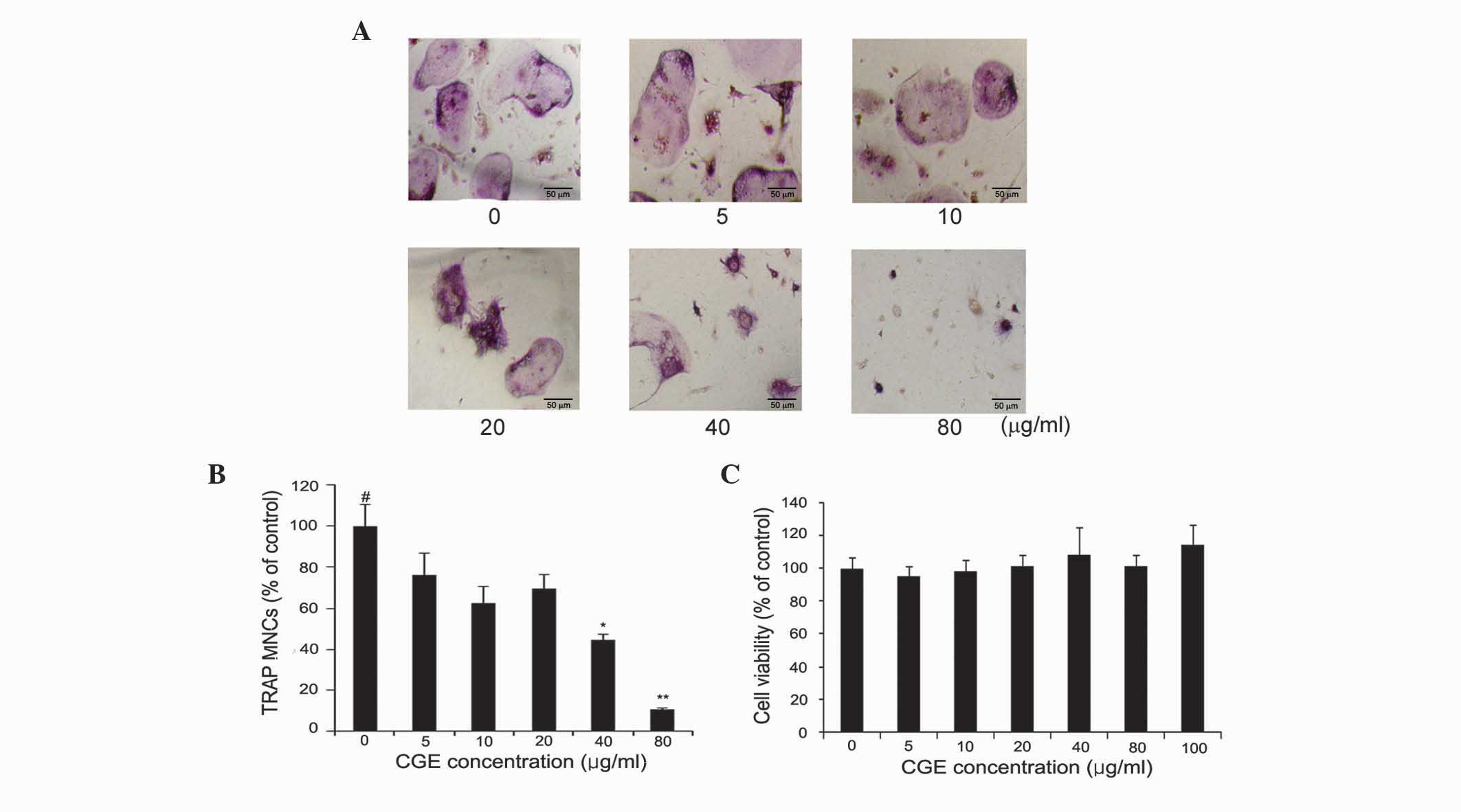 | Figure 2Effect of CGE on RANKL-mediated
osteoclast differentiation. (A) BMMs were cultured for 3 days with
the indicated concentrations of CGE in the presence of M-CSF (30
ng/ml) and RANKL (100 ng/ml). After 3 days, the cells were fixed
and stained for TRAP. (B) TRAP-positive MNCs were counted. (C)
Cytotoxicity of CGE on BMMs. The effect of CGE on cell viability
was measured using a
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay.
The results are expressed as the mean ± standard error of the mean.
**P<0.01 and *P<0.05 vs.
vehicle-treated cells (#). Magnification, ×100. MNCs,
multinucleated cells. CGEs, centipedegrass extracts; RANKL,
receptor activator of nuclear factor κ-B ligand; BMMs, bone
marrow-derived macrophages; M-CSF, macrophage colony-stimulating
factor, TRAP, tartrate-resistant acid phosphatase. |
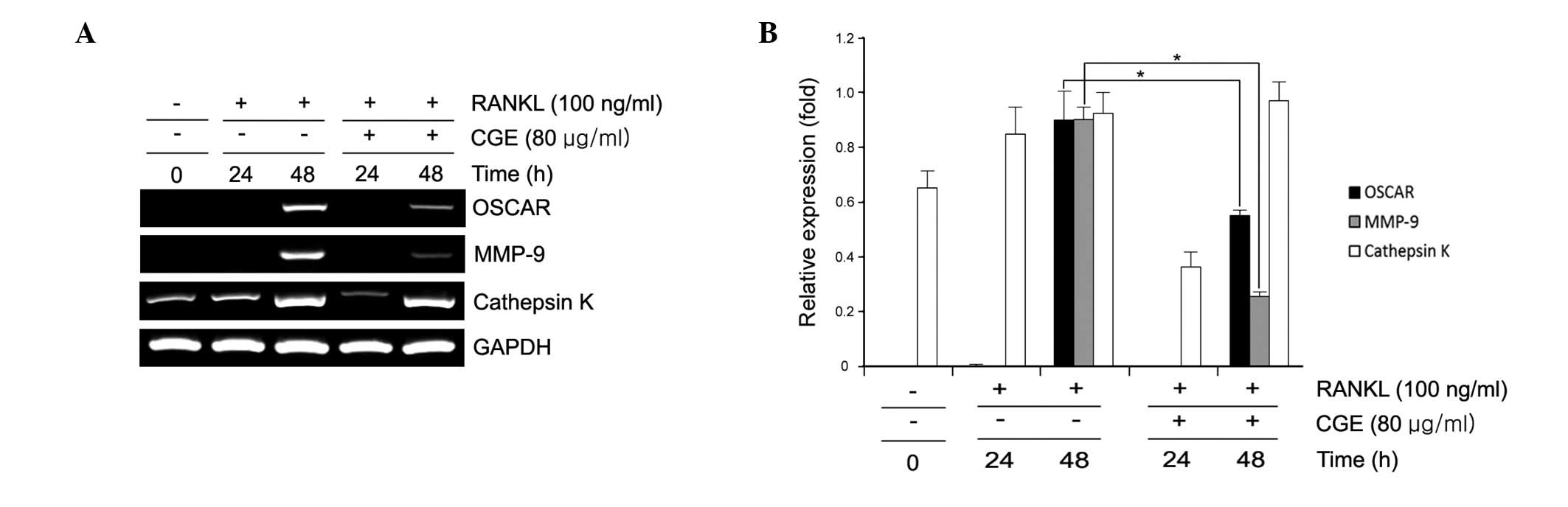
Regulation of osteoclastogenic-specific
genes in RANKL-induced BMMs by CGE
Osteoclast differentiation is associated with the
upregulation of specific genes in response to RANKL (7). In the present study, RANKL
significantly induced the expression of MMP-9, OSCAR and cathepsin
K in the BMMs. However, CGE decreased the expression of these
osteoclastogenesis-associated genes in a time-dependent manner
(Fig. 3). CGE did not affect the
expression of the housekeeping gene, GAPDH. These data showed that
CGE had a specific effect on the regulation of certain genes
induced during osteoclast differentiation. This raises the
possibility that CGE may inhibit osteoclast differentiation through
the inhibition of the RANKL-induced expression of c-Fos and
NFATc1.
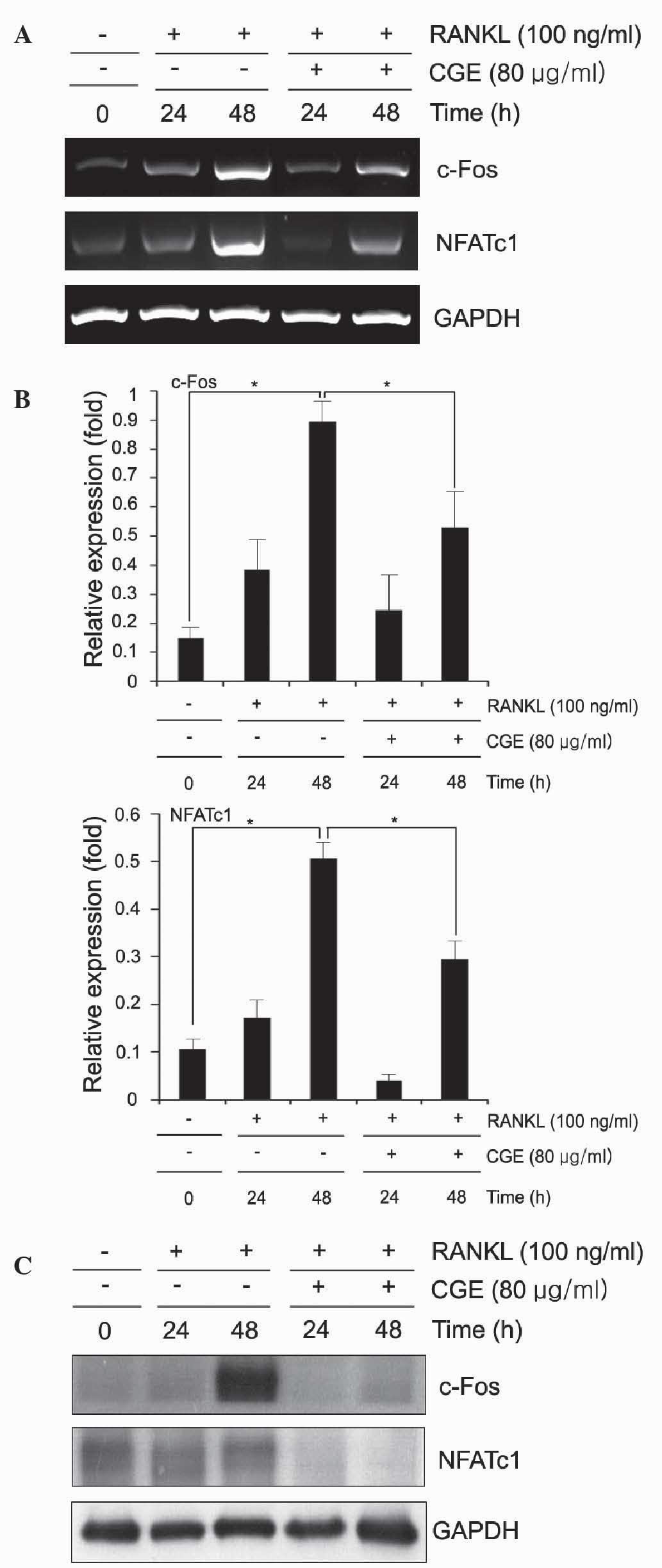
CGE inhibits the expression of NFATc1 and
c-Fos in RANKL-induced osteoclastogenesis
Osteoclast differentiation is regulated by the
induction of various genes in response to RANKL and RANK binding.
NFATc1 has been shown to be upregulated following RANKL
stimulation, and is important for osteoclast differentiation
(4). Therefore, the present study
examined the effect of CGE on the expression levels of NFATc1 and
c-Fos. The osteoclast precursors were pretreated with CGE and then
stimulated with RANKL at various time points. The results revealed
that the mRNA levels of c-Fos and NFATc1 were increased in response
to RANKL, however the expression levels of c-Fos and NFATc1 were
significantly inhibited by CGE (Fig.
4A and B). To compare with the result s of the RT-PCR analyses,
the present study examined the protein levels using immunobloting.
The protein expression levels of NFATc1 and c-Fos l increased in
response to RANKL, however the expression levels of c-Fos and
NFATc1 decreased following pretreatment with CGE (Fig. 4C). These results suggested that CGE
inhibited oateoclastogenesis by reducing the expression levels of
c-Fos and NFATc1 induced by RANKL.
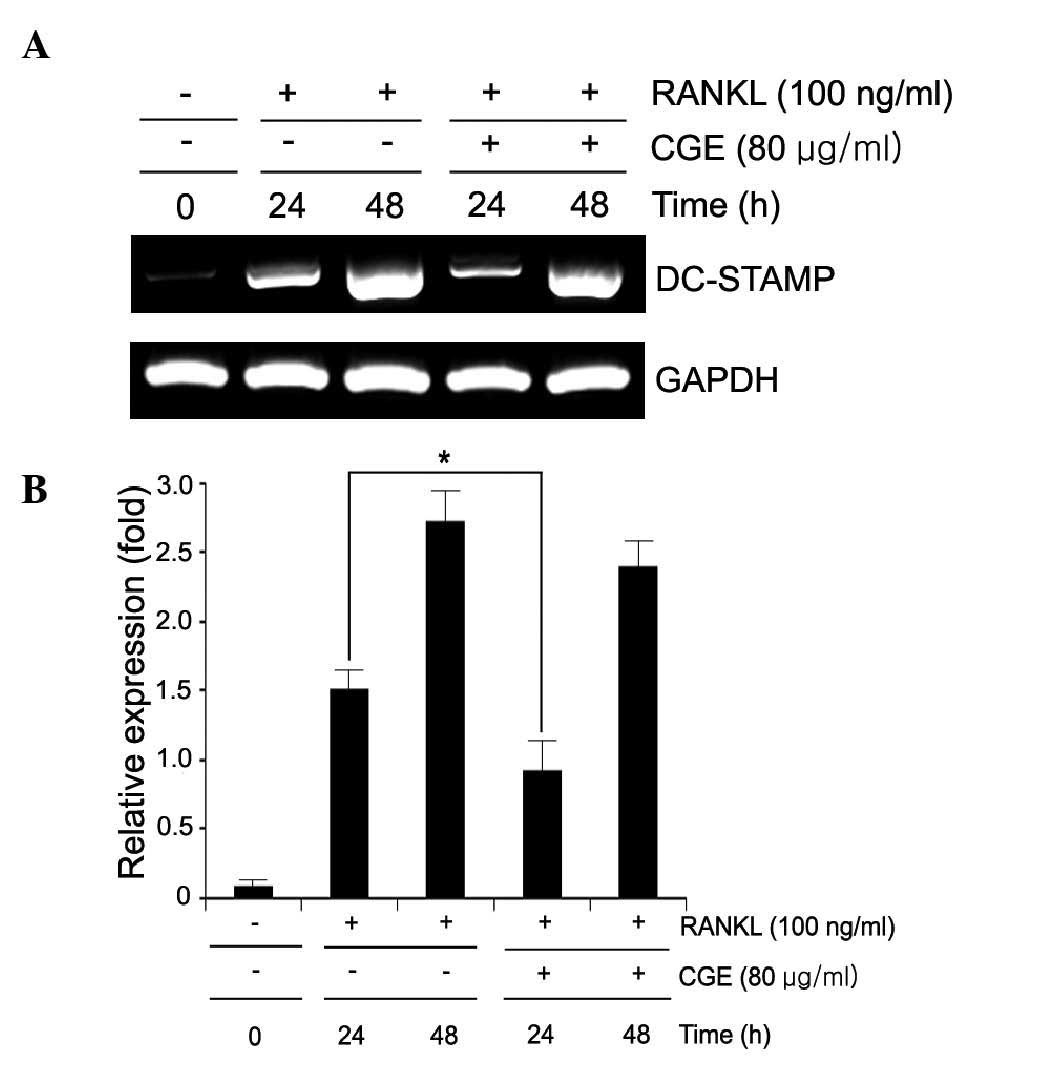
CGE suppresses osteoclast cell-cell
fusion through down- regulation of the expression of DC-STAMP
DC-STAMP is an important regulator of osteoclast
cell fusion (15,16). In the present study, the number of
fused TRAP-positive multi-nuclear osteoclasts was increased by
RANKL however, CGE suppressed RANKL-induced osteoclast fusion
(Fig. 2A). It order to determine
the effect of CGE on the expression of DC-STAMP, RT-PCR analysis
was performed. It was revealed that CGE attenuated the mRNA
expression of the fusion marker, DC-STAMP, confirming the
inhibitory effect of CGE on RANKL-mediated osteoclast fusion
(Fig. 5A and B).
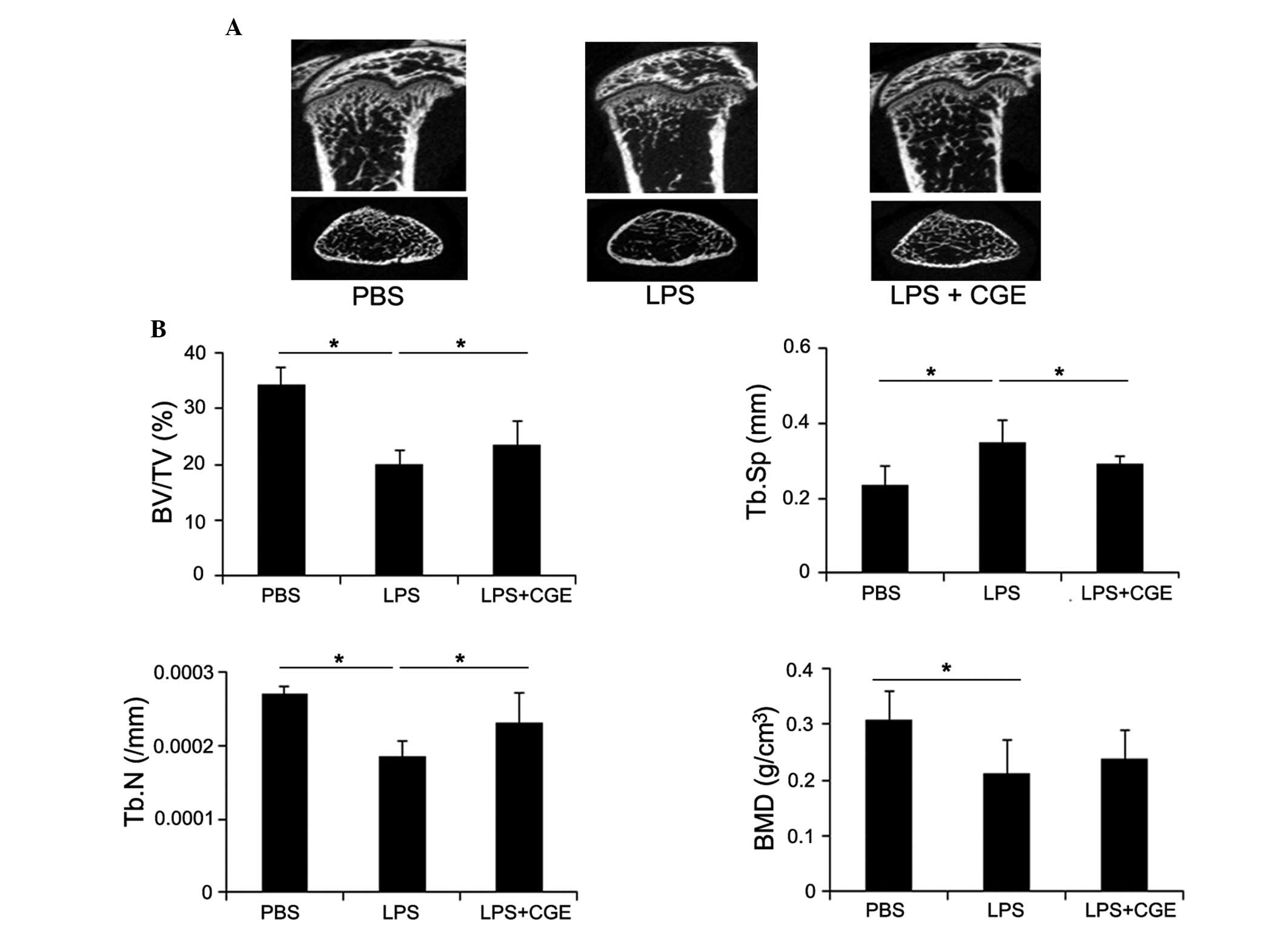
CGE on in vivo LPS-induced bone
resorption
The present study investigated the effects of CGE on
bone erosion in vivo. For this experiment, an animal model
of endotoxin-induced bone resorption was used (1,18,19).
The mice were challenged with LPS and treated either with or
without CGE. The femurs of the mice were collected 8 days following
LPS injection (day 0), and the collected femurs were scanned using
micro-CT. The LPS-injection mice exhibited profound decreases in
trabecular and cortical bone densities of the femur. The micro-CT
analyses also revealed that LPS-induced bone loss was reduced in
the femurs of the CGE-treated LPS-mediated mice (Fig. 6A). To enable more detailed analyses
of the effect of CGE in the bone microstructure, the femoral
radiographic results were analyzed to assess the trabecular bone
microstructure using measurement software. The calculation of the
microstructure indices of trabecular bone density revealed the
volume of trabecular bone (TB) per unit of total bone mass (TV).
The BV/TV was decreased following LPS induction, and this reduction
was observed, to a lesser extent, in the CGE-treated group
(Fig. 6B). The LPS-induced
reduction in trabecular number and bone mineral density were also
attenuated, whereas trabecular separation was increased by CGE
(Fig. 6B).
Discussion
Osteoclasts are dynamic cells, which are capable of
resorbing bone matrix (2). The
excessive formation of this cell type causes bone-destructive
diseases, including osteoporosis, periodontits and rheumatoid
arthritis (5).
Bone resorption is affected by various factors,
which govern osteoclast number and activity (5). The major signals of osteoclast
differentiation are RANKL and the RANK binding pathway. This
binding activates TRAF6, which induces MAPKs, including ERK, JNK
and p38, and also activates NF-κB (2). This signaling subsequently activates
the transcription factors, c-Fos, PU.1 and NFATc1, all of which are
required for osteoclastogenesis (10). These transcription factors undergo
nuclear translocation and regulate the expression of several
osteoclast-specific genes, including cathepsin K, TRAP, OSCAR,
MMP-9 and NFATc1 (3). c-Fos causes
the expression of NFATc1 during RANKL-induced osteoclast
differentiation. In addition, NFATc1 can induce osteoclast
differentiation in the absence of RANKL. Thus, c-Fos and NFATc1 are
essential factors in osteoclast differentiation (20). Therefore, the inhibition of
osteoclast formation and/or its activation may have therapeutic
applications for pathological bone diseases (2,5).
In the present study, it was demonstrated that CGE
inhibited osteoclast generation from primary BMMs. The data
suggested that CGE reduced osteoclast formation, compared to the
RANKL-stimulated control groups (Fig.
2A and B), and that these results were not a result of
cytotoxic effects of CGE (Fig.
2C). The interaction of RANKL and RANK results in the
activation of various signaling cascades during osteoclast
differentiation and activation. This signaling pathway involved
three well known MAPKs, including JNK, ERK and p38, and the
activation of signaling molecules induces transcription factors,
including NF-κB, NFATc1 and activator-protein-1 (AP-1), which are
essential for osteoclast differentiation (1,2,7). In
the present study, CGE did not inhibit the activation of MAPKs or
NF-κB in response to RANKL (data not shown). Therefore, it is
unlikely that CGE directly regulated the activation of MAPKs/NF-κB
downstream signaling. The transcription factor, NFATc1, is also
critical in osteoclastogenesis RANKL-RANK binding activation
through intracellular TRAF6 and the c-Fos signaling pathway
(1). This is a master regulator of
osteoclast differentiation, which auto-amplifies and affects the
expression of osteoclast-specific genes, including TRAP, calcitonin
receptor, OSCAR, cathepsin K and MMP-9 (1). The present study suggested that CGE
suppressed the RANKL-induced activation of c-Fos and NFATc1
(Fig. 4). Additionally, the mRNA
levels of osteoclast markers, including OSCAR, MMP-9 and cathepsin
K, were inhibited by CGE (Fig. 3).
Multinucleated giant cells are formed by the fusion of osteoclasts,
and it has been reported that DC-STAM and the d2 isoform of
vacuolar (H+) ATPase (V-ATPase), V(0) domain (Atp6v0d2),
are key regulators of osteoclast cell-cell fusion, which are
regulated by NFATc1 (3). The
present study investigated how CGE regulates osteoclast maturation,
and examined whether CGE stimulation regulated the expression of
DC-STAMP (3,21,22).
The RANKL-induced BMMs significantly increased the expression of
DC-STAMP, however, the expression of this marker was inhibited by
CGE treatment, in a time-dependent manner (Fig. 5A and B). These results indicated
that the osteoclast differentiation induced by RANKL required the
osteoclastogenesis-associated genes regulated by NFATc1. However,
CGE exposure in the BMMs suppressed the expression levels of
osteoclastogenesis-associated genes, including MMP-9, OSCAR,
cathepsin K and DC-STAMP, as well as c-Fos and NFATc1.
The present demonstrated the inhibitory effects of
CGE on osteoclastogenesis in primary precusor cells. The present
study also suggested the molecular mechanism underlying this
inhibition, involving transcription factors, including NFATc1 and
c-Fos, which act in genes involved in osteoclast differentiation.
CGE inhibited the expression of DC-STAMP, decreasing osteoclast
cell-cell fusion (Fig. 7). In
addition, CGE decreased bone loss and attenuated associated
parameters in the LPS-induced in vivo model (Fig. 6). The results suggested that CGE
may offer potential for used in the development of a therapeutic
drug for the treatment of bone-resorbing diseases, including
osteoporosis, rheumatoid arthritis and advanced periodontal
disease.
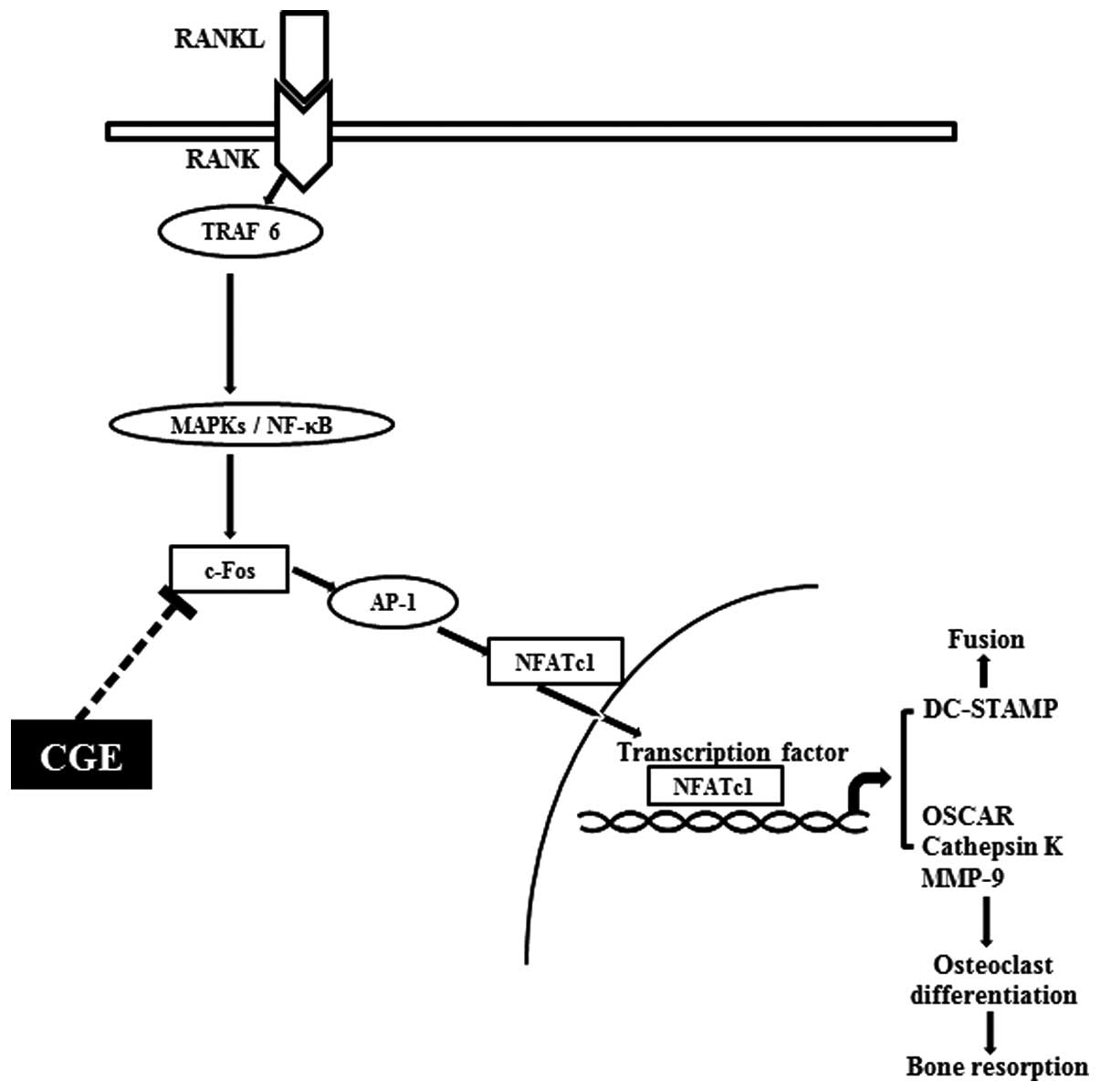 | Figure 7Schematic diagram of the inhibitory
function of CGE on osteoclastogenesis in RANK-RANKL signaling. The
binding of RANKL to RANK led to the activation of downstream
molecules, including NF-κB and MAPK. However, these molecules were
not affected by CGE. CGE reduced the induction of c-Fos and NFATc1,
leading to decreased osteoclastogenesis-associated gene expression,
and osteoclast formation and function. CGE, centipedegrass extract;
RANKL, receptor activator of nuclear factor κ-B ligand; MAPK,
mitogen-activated protein kinase; NF-κB, nuclear factor-κB; TRAF6,
TNF receptor-associated factor 6; AP-1, activator protein-1;
NFATc1, nuclear factor of activated T cell cytoplasmic; DC-STAMP
dendritic cell-specific transmembrane protein; OSCAR;
osteoclast-associated immunoglobulin-like receptor; MMP, matrix
metalloproteinase. |
Acknowledgments
This study was supported by the National Research
Foundation of Korea, funded by the Korean Ministry of Science, ICT
and Future Planning.
References
|
1
|
Kwak HB, Lee BK, Oh J, Yeon JT, Choi SW,
Cho HJ, Lee MS, Kim JJ, Bae JM, Kim SH and Kim HS: Inhibition of
osteoclast differentiation and bone resorption by rotenone, through
down-regulation of RANKL-induced c-Fos and NFATc1 expression. Bone.
46:724–731. 2010. View Article : Google Scholar
|
|
2
|
Kim HJ, Yoon KA, Lee MK, Kim SH, Lee IK
and Kim SY: A novel small molecule, NecroX-7, inhibits osteoclast
differentiation by suppressing NF-κB activity and c-Fos expression.
Life Sci. 91:928–934. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Choi J, Choi SY, Lee SY, Lee JY, Kim HS,
Lee SY and Lee NK: Caffeine enhances osteoclast differentiation and
maturation through p38 MAP kinase/Mitf and DC-STAMP/CtsK and TRAP
pathway. Cell Signal. 25:1222–1227. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ghayor C, Correro RM, Lange K,
Karfeld-Sulzwe LS, Grätz KW and Weber FE: Inhibition of osteoclast
differentiation and bone resorption by N-Methylpyrrolidone. J Biol
Chem. 286:24458–24466. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kim HH, Kim JH, Kwak HB, Huang H, Han SH,
Ha H, Lee SW, Woo ER and Lee ZH: Inhibition of osteoclast
differentiation and bone resorption by tanshinone IIA isolated from
Salvia miltiorrhiza Bunge. Biochem Pharmacol. 67:1647–1656. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim BG, Kwak HB, Choi EY, Kim HS, Kim MH,
Kim SH, Choi MK, Chun CH, Oh J and Kim JJ: Amorphigenin inhibits
osteoclast differentiation by suppressing c-Fos and nuclear factor
of activated T cells. Anat Cell Biol. 43:310–316. 2010. View Article : Google Scholar
|
|
7
|
Fumimoto R, Sakai E, Yamaguchi Y, Sakamoto
H, Fukuma Y, Nishishita K, Okamoto K and Tsukuba T: The coffee
diterpene kahweol prevents osteoclastogenesis via impairment of
NFATc1 Expression and blocking of Erk phosphorylation. J Pharmacol
Sci. 118:479–486. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Park YR, Eun JS, Choi HJ, Nepal M, Kim DK,
Seo SY, Li R, Moon WS, Cho NP, Cho SD, et al: Hexane-Soluble
fraction of the common fig, Ficus carica, inhibits osteoclast
differentiation in murine bone marrow-derived macrophages and RAW
264.7 Cells. Korean J Physiol Pharmacol. 13:417–424. 2009.
View Article : Google Scholar
|
|
9
|
Wada T, Nakashima T, Hiroshi N and
Penninger JM: RANKL-RANK signaling in osteoclastogenesis and bone
disease. Trends Mol Med. 12:17–25. 2006. View Article : Google Scholar
|
|
10
|
Mochizuki A, Takami M, Miyamoto Y,
Nakamaki T, Tomoyasu S, Kadono Y, Tanaka S, Inoue T and Kamijo R:
Cell adhesion signaling regulates RANK expression in osteoclast
precursors. PLoS One. 7:e487952012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao Q, Wang X, Liu Y, He A and Jia R:
NFATc1: Functions in osteoclasts. Int J Biochem Cell Biol.
42:576–579. 2010. View Article : Google Scholar
|
|
12
|
Badaboina S, Bai HW, Park CH, Jang DM,
Choi BY and Chung BY: Molecular mechanism of apoptosis induction in
skin cancer cells by the centipedegrass extract. BMC complement
Altern Med. 13:3502013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Barampuram S, Chung BY, Lee SS, An BC, Lee
EM and Cho JY: Development of an embryogenic callus induction
method for centipede grass (Eremochloa ophiuroides Munro) and
subsequent plant regeneration. In Vitro Cell Dev Bio Plant.
45:155–161. 2009. View Article : Google Scholar
|
|
14
|
Wiseman BR, Gueldner RC, Lynch RE and
Severson RF: Biochemical activity of centipedegrass against fall
armyworm larvae. J Chem Ecol. 16:2677–2690. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Park HJ, Chung BY, Lee MK, Song Y, Lee SS,
Chu GM, Kang SN, Song YM, Kim GS and Cho JH: Centipede grass exerts
anti-adipogenic activity through inhibition of C/EBPβ, C/EBPα, and
PPARγ expression and the AKT signaling pathway in 3T3-L1
adipocytes. BMC Complement Altern Med. 12:2302012. View Article : Google Scholar
|
|
16
|
Choi HJ, Park YR, Nepal M, Choi BY, Cho
NP, Choi SH, Heo SR, Kim HS, Yang MS and Soh Y: Inhibition of
osteoclastogenic differentiation by Ikarisoside A in RAW 264.7
cells via JNK and NF-kappaB signaling pathways. Eur J Pharmacol.
636:28–35. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
National Research Council (US) Committee
for the Update of the Guide for the Care and Use of Laboratory
Animals: Guide for the Care and Use of Laboratory Animals. 8th
edition. National Academies Press (US); Washington, DC, USA;
2011
|
|
18
|
Han KY, Yang D, Chang EJ, Lee Y, Huang H,
Sung SH, Lee ZH, Kim YC and Kim HH: Inhibition of osteoclast
differentiation and bone resorption by sauchinone. Biochem
Pharmacol. 74:911–923. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nepal M, Choi HJ, Choi BY, Yang MS, Chae
JI, Li L and Soh Y: Hispidulin attenuates bone resorption and
osteoclastogenesis via the RANKL-induced NF-κB and NFATc1 pathways.
Eur J Pharmacol. 715:96–104. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kwak HB, Yang D, Ha H, Lee JH, Kim HN, Woo
ER, Lee S, Kim HH and Lee ZH: Tanshinone IIA inhibits osteoclast
differentiation through down-regulation of c-Fos and NFATc1. Exp
Mol Med. 38:256–264. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yagi M, Miyamoto T, Sawatani Y, Iwamoto K,
Hosogane N, Fujita N, Morita K, Ninomiya K, Suzuki T, Miyamoto K,
et al: DC-STAMP is essential for cell-cell fusion in osteoclasts
and foreign body giant cells. J Exp Med. 202:345–351. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yagi M, Miyamoto T, Toyama Y and Suda T:
Role of DC-STAMP in cellular fusion of osteoclasts and macrophage
giant cells. J Bone Miner Metab. 24:355–358. 2006. View Article : Google Scholar : PubMed/NCBI
|