Introduction
Hepatocellular carcinoma (HCC) is the predominant
histologic subtype of primary liver cancer, accounting for 70–85%
of cases in the majority of countries, and is a leading cause of
cancer-associated mortality each year worldwide (1,2).
Although a number of advances in surgical techniques and medical
care have been achieved over the last two decades, long-term
survival of HCC remains low due to the high rate of recurrence and
metastasis (3,4). Furthermore, the majority of HCC
patients present with advanced metastatic disease at the initial
diagnosis. HCC develops via a complex process associated with
multi-step genetic and epigenetic changes (5,6).
Thus, it is key to develop novel approaches to aid early diagnosis,
prognosis determination, and therapeutic strategies to treat
HCC.
Non-coding RNAs (ncRNAs) are sub-divided into two
major classes according to size, small ncRNAs are <200 nt in
length and long ncRNAs are >200 nt in length (7). In recent years, microRNAs have been
identified as oncogenes or tumor suppressor genes due to the
effects exerted on cancer cells via post-transcriptional regulation
of protein expression (8–10). By contrast, lncRNAs were previously
hypothesized to be transcriptional noise. However, lncRNAs are key
in important functions at numerous levels, including X chromosome
inactivation, chromatin remodeling, and transcriptional repression
(11–13).
With the development of deep sequencing
technologies, lncRNAs have increasingly been associated with
various human diseases, particularly in types of cancer (14–16).
LncRNAs exert important regulatory effects in the programming of
cellular functions and may be potential oncogenes or tumor
suppressor RNAs (17). Prostate
cancer-associated ncRNA transcripts 1 (PCAT-1) is a long non-coding
RNA that was originally identified as a biomarker for prostate
cancer (18). High expression
levels of PCAT-1 were also observed to be involved in the
progression of colorectal cancer (19), and upregulation of PCAT-1 has also
been reported to be associated with advanced clinical stage and
poor prognosis in esophageal squamous carcinoma (20). However, the role of PCAT-1 in HCC
remains to be elucidated.
In the present study, PCAT-1 was demonstrated to be
crucial in the regulation of proliferation, migration and apoptosis
of HCC cells. To the best of our knowledge, this is the first study
that directly illustrates the role of PCAT-1 in HCC. Thus, PCAT-1
may serve as a potential prognostic biomarker and therapeutic
target in HCC.
Materials and methods
Patient samples
HCC tissue samples and adjacent morphologically
normal tissue samples were collected from 82 patients enrolled in
the present study who underwent surgery between August 2009 and May
2013 at the Second Affiliated Hospital, Harbin Medical University
(Harbin, China). None of the patients had received percutaneous
ablation, chemoembolization or radiotherapy prior to the surgery.
The diagnosis was based on detailed examination of sections stained
with hematoxylin and eosin and was confirmed in all cases by the
Department of Pathology of The Second Affiliated Hospital. The
present study was approved by the Institutional Review Board of The
Second Affiliated Hospital and written informed consent was
obtained from all patients. Specimens were obtained immediately
following surgical resection and stored at −80°C for further
analysis.
Cell culture
The HepG2 and Bel-7402 hepatocellular carcinoma cell
line and the L02 normal liver epithelial cell line were purchased
from the Institute of Biochemistry and Cell Biology, Chinese
Academy of Science (Shanghai, China). All cell lines was cultured
in Invitrogen Dulbecco's modified Eagle's medium (DMEM; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10%
fetal bovine serum (FBS; Invitrogen; Thermo Fisher Scientific,
Inc.) at 37°C in a 5% CO2 atmosphere.
Cell transfection
The cells were cultured in a 6-well or 24-well
plates for 24 h and then transfected with overexpression plasmid
vectors or small hairpin (sh)RNAs. All transfections were performed
using Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. PCAT-1
shRNA and negative control shRNA were obtained from Shanghai
GenePharma Co., Ltd. (Shanghai, China) with the sequence
5′-GAGAAAGCAUCUGUACCCUUACAA U-3′. The overexpression plasmid vector
for PCAT-1, PCAT-1-pcDNA3.1 + vector, was supplied and synthesized
by Invitrogen (Thermo Fisher Scientific, Inc.). The pcDNA3.1 +
empty vector was used as a negative control.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the tissue samples or
the transfected cells using TRIzol (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. RNA was
reverse transcribed using SuperScript First Strand cDNA system
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocols. The temperature protocol was as follows:
65°C for 5 min, 4°C for 1 min, 50°C for 50 min and finally 85°C for
5 min. Thirty-six cycles of qPCR were performed at 98°C for 2 min,
98°C for 10 sec, 60°C for 30 sec, and 72°C for 30 sec; the final
stage was performed at 72°C for 5 min. The primer sequences were as
follows: Sense, 5′-AATGGCATG AACCTGGGAGGCG-3′ and antisense,
5′-GGCTTTGGGAAGTGCTTTGGAG-3′ for PCAT-1; and sense,
5′-ACATCAAGAAGGTGGTGAAGCAGG -3′ and antisense,
5′-CGTCAAAGGTGGAGGAGTGGGT-3′ for GAPDH (Invitrogen; Thermo Fisher
Scientific, Inc.. qPCR was performed using the SYBR®
Premix Ex Taq™ II PCR kit (Takara Bio, Inc., Otsu, Japan)
according to the manufacturer's protocols on the ABI
Prism® 7000 Real-Time PCR system (Applied Biosystems;
Thermo Fisher Scientific, Inc.). Data was collected and analyzed by
SDS 2.3 software (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The expression level of PCAT-1 was normalized using the Cq
of the housekeeping gene GAPDH. The relative quantitative value was
expressed using the 2−ΔΔCq method (21). Each experiment was performed in
triplicate and repeated three times.
Cell Counting Kit-8 (CCK-8) assay
Cell proliferation was detected using the CCK-8
assay (Dojindo Molecular Technologies, Kumamoto, Japan), according
to the manufacturer's protocols. Cells were seeded in 96-well
plates and cultured in normal medium (DMEM + FBS) for 24 h prior to
transfection with PCAT-1 shRNA or negative control shRNA and
PCAT-1-pcDNA3.1 + vector or pcDNA3.1 + empty vector. At 0, 24, 48,
and 72 h after transfection, the absorbance of each well at a
wavelength of 450 nm was measured by a microplate reader (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). All experiments were
performed three times.
Cell migration assay
Following washing with phosphate-buffered saline
(Sigma-Aldrich, St. Louis, MO, USA), HepG2 or Bel-7402 cells were
detached with trypsin (Sigma-Aldrich), and resuspended in
serum-free medium (DMEM). The cell suspensions (200 µl;
4×105 cells/ml) was added to the upper chamber with a
non-coated membrane (EMD Millipore, Billerica, MA, USA) for the
Transwell migration assays (Costar; Corning Life Sciences,
Cambridge, MA, USA). Culture medium containing 10% FBS was added to
the bottom wells of the chambers. Following incubation for 24 h,
the cells were removed using cotton swabs, fixed with 100% methanol
(Sigma-Aldrich) and stained with 0.2% Crystal Violet
(Sigma-Aldrich). Images were acquired using a Leica DMI 6000B
microscope (magnification, ×100; Leica Microsystems Inc., Richmond
Hill, ON, Canada).. The assays were repeated at least three
times.
Caspase-3 ELISA assay
HepG2 and Bel-7402 cells were transfected with
PCAT-1 shRNA or negative control shRNA, and PCAT-1-pcDNA3.1 +
vector or pcDNA3.1 + empty vector. After 48 h transfection,
apoptosis was determined by calculating the activity of caspase-3
using the Caspase-3 ELISA activity assay kit (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocols.
Optical density (OD) values were measured by using a microplate
reader (Model 550; Bio-Rad Laboratories, Inc.). Data were presented
as the ratios between the OD values of PCAT-1 shRNA-transfected
cells or those of PCAT-1-pcDNA3.1 + vector-transfected cells and
their respective controls. All experiments were performed in
triplicate.
Statistical analysis
Data are presented as the mean ± standard deviation.
The difference in expression levels of PCAT-1 between HCC and
matched healthy samples were analyzed using paired samples t-test.
The differences between PCAT-1 shRNA or negative control
shRNA-transfected cells and PCAT-1-pcDNA3.1 + vector or pcDNA3.1 +
empty vector-transfected cells in the CCK-8 assay was analyzed
using analysis of variance. The independent samples t-test was used
to analyze other data. Statistical analyses were performed using
SPSS 17.0 (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered
to indicate a statistically significant difference.
Results
PCAT-1 expression levels were increased
in hepatocellular carcinoma tissues and cell lines compared with
controls
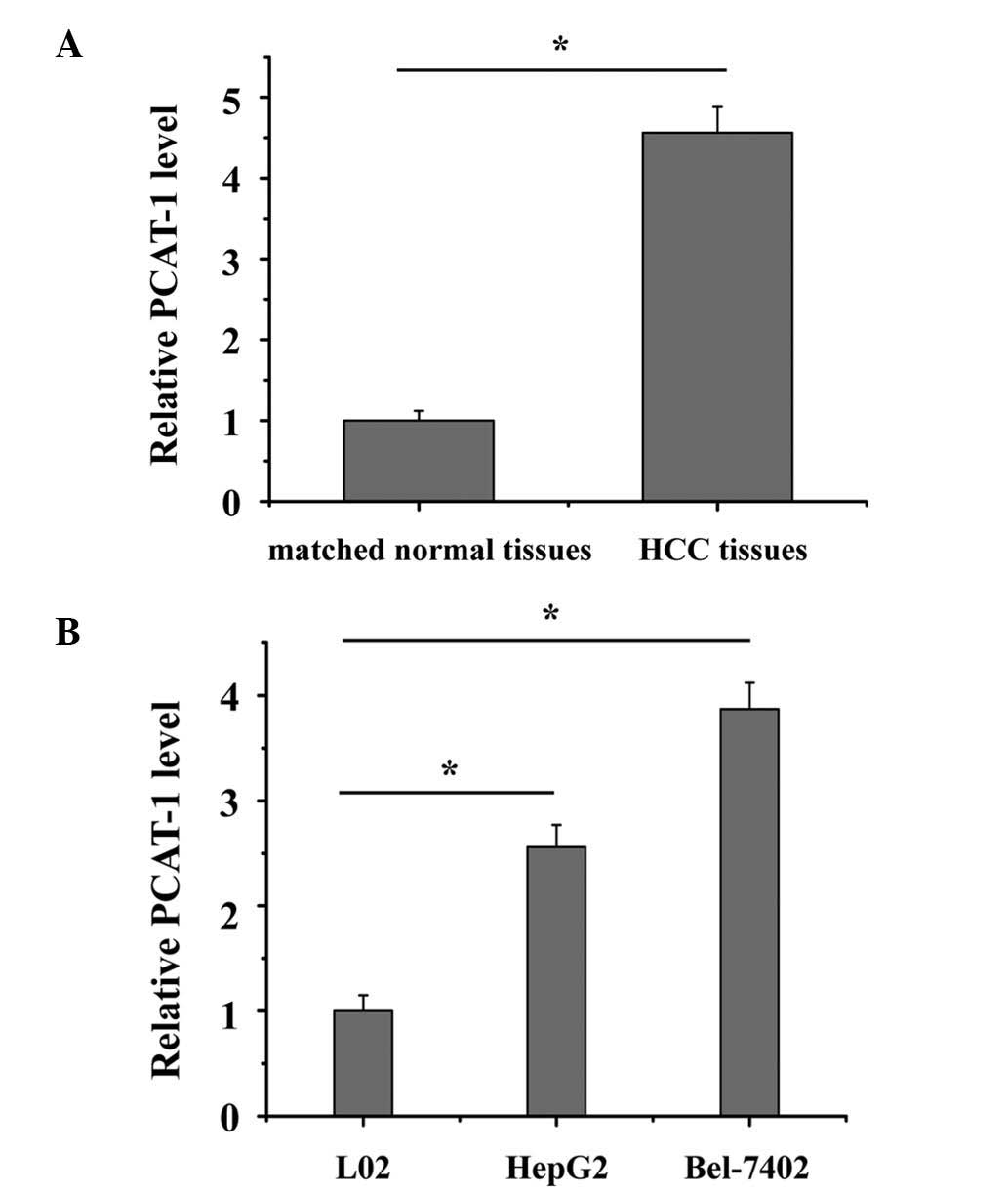
The relative expression level of PCAT-1 was
determined using RT-qPCR in a total of 82 patient HCC samples. As
presented in Fig. 1A, PCAT-1 was
significantly upregulated in HCC tissue compared with matched
normal tissue (P<0.05). The expression of PCAT-1 was also
significantly increased in HepG2 and Bel-7402 cells compared with
the L02 normal liver epithelial cell line (Fig. 1B; P<0.05). These results
indicated that PCAT-1 may exert an oncogenic effect in HCC. Each
experiment was performed in triplicate and repeated three
independent times.
Silencing and overexpression of PCAT-1
was effectively induced by use of shRNA and overexpression vectors
in HCC cell lines
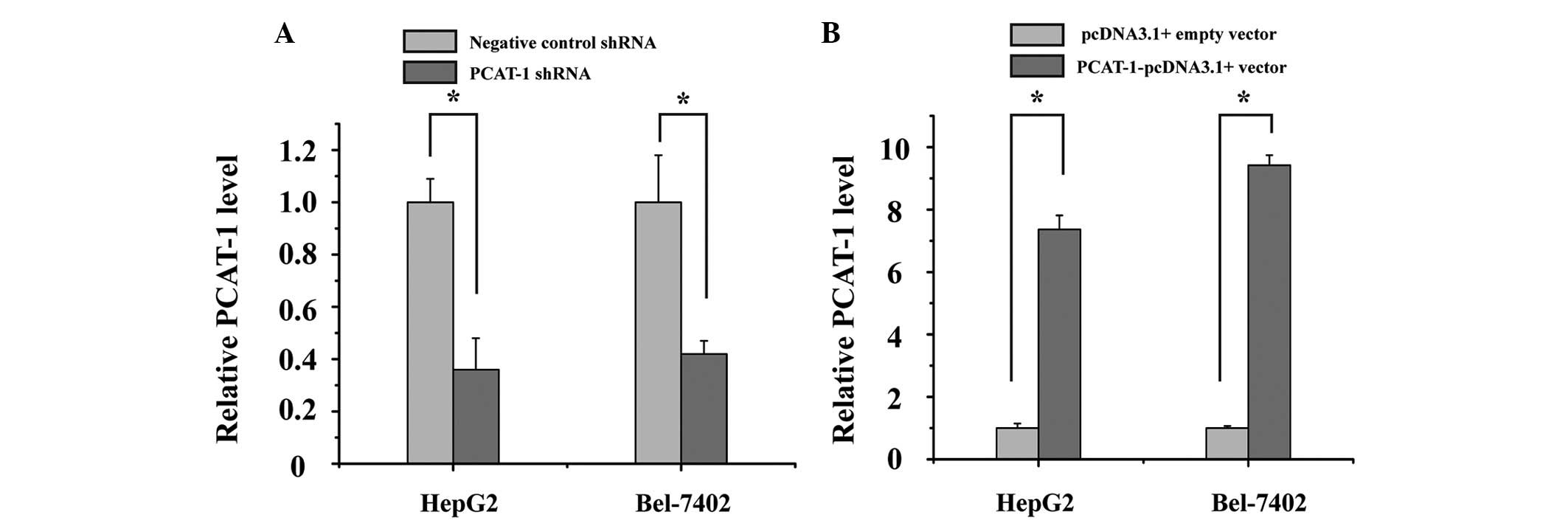
HepG2 and Bel-7402 cells were cultured and
transfected with PCAT-1 shRNA or negative control shRNA, and
PCAT-1-pcDNA3.1 + vector or pcDNA3.1 + empty vector. At 48 h after
transfection, the relative expression level of PCAT-1 was analyzed
by qPCR. The results demonstrated that the relative expression
level of PCAT-1 in HepG2 and Bel-7402 cells was significantly
decreased by the PCAT-1 shRNA (Fig.
2A; P<0.05), while PCAT-1 was increased by the
PCAT-1-pcDNA3.1+vector (Fig. 2B;
P<0.05).
PCAT-1 increases proliferation in HCC
cells
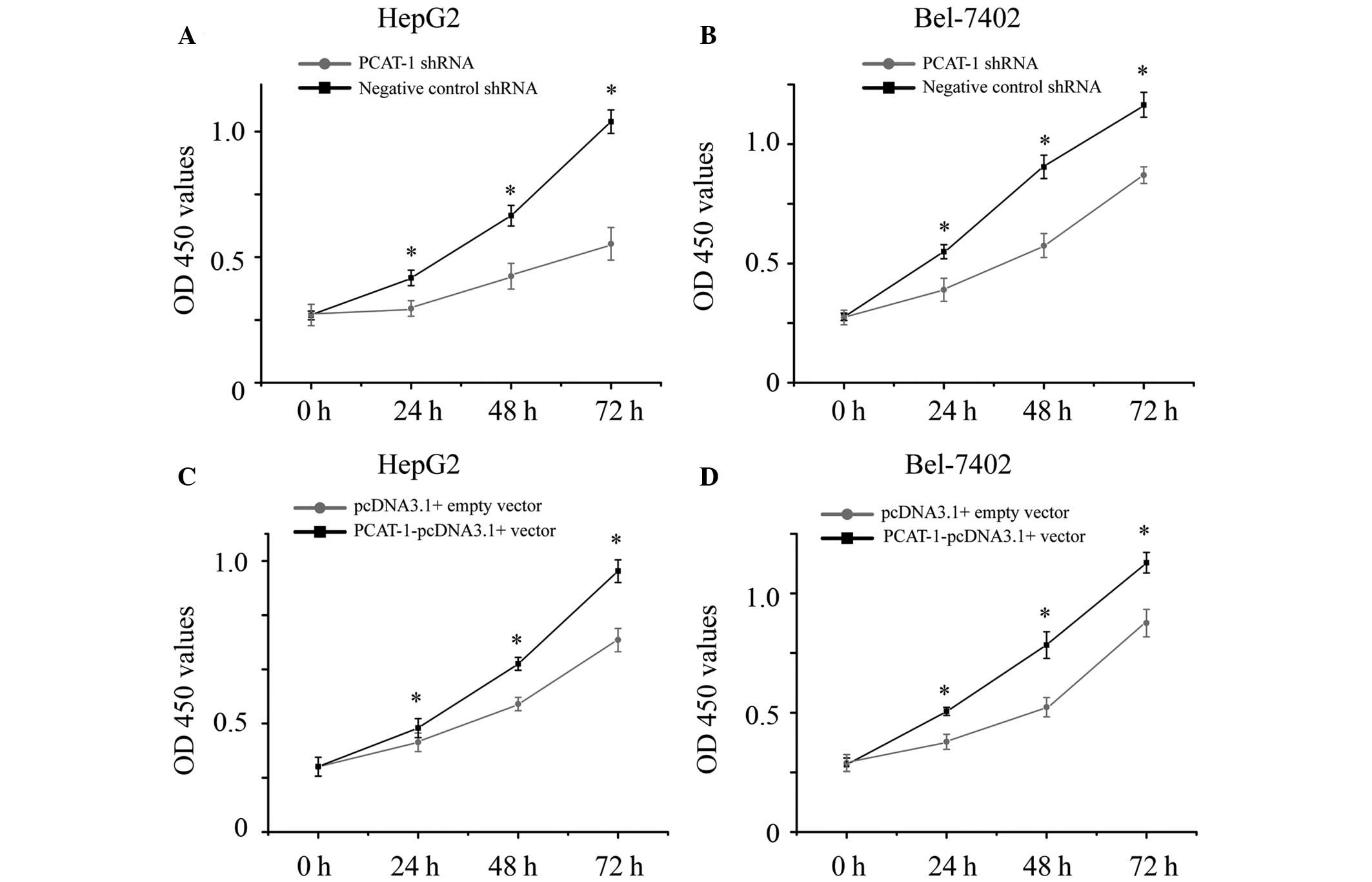
The present study also aimed to determine whether
PCAT-1 promoted cell proliferation in HCC. HepG2 and Bel-7402 cells
were transfected with PCAT-1 shRNA or negative control shRNA, and
PCAT-1-pcDNA3.1 + vector or pcDNA3.1 + empty vector. The cell
proliferation of the HCC cells were determined by CCK-8 assay.
Transfection of PCAT-1 shRNA resulted in cell growth arrest in the
HCC cells (Fig. 3A and B;
P<0.05), while transfection of PCAT-1-pcDNA3.1 + vector resulted
in promoting cell proliferation (Fig.
3C and D; P<0.05). These results demonstrated that PCAT-1
increases cell proliferation in HCC.
PCAT-1 exerts a positive effect on
migration in HCC cells
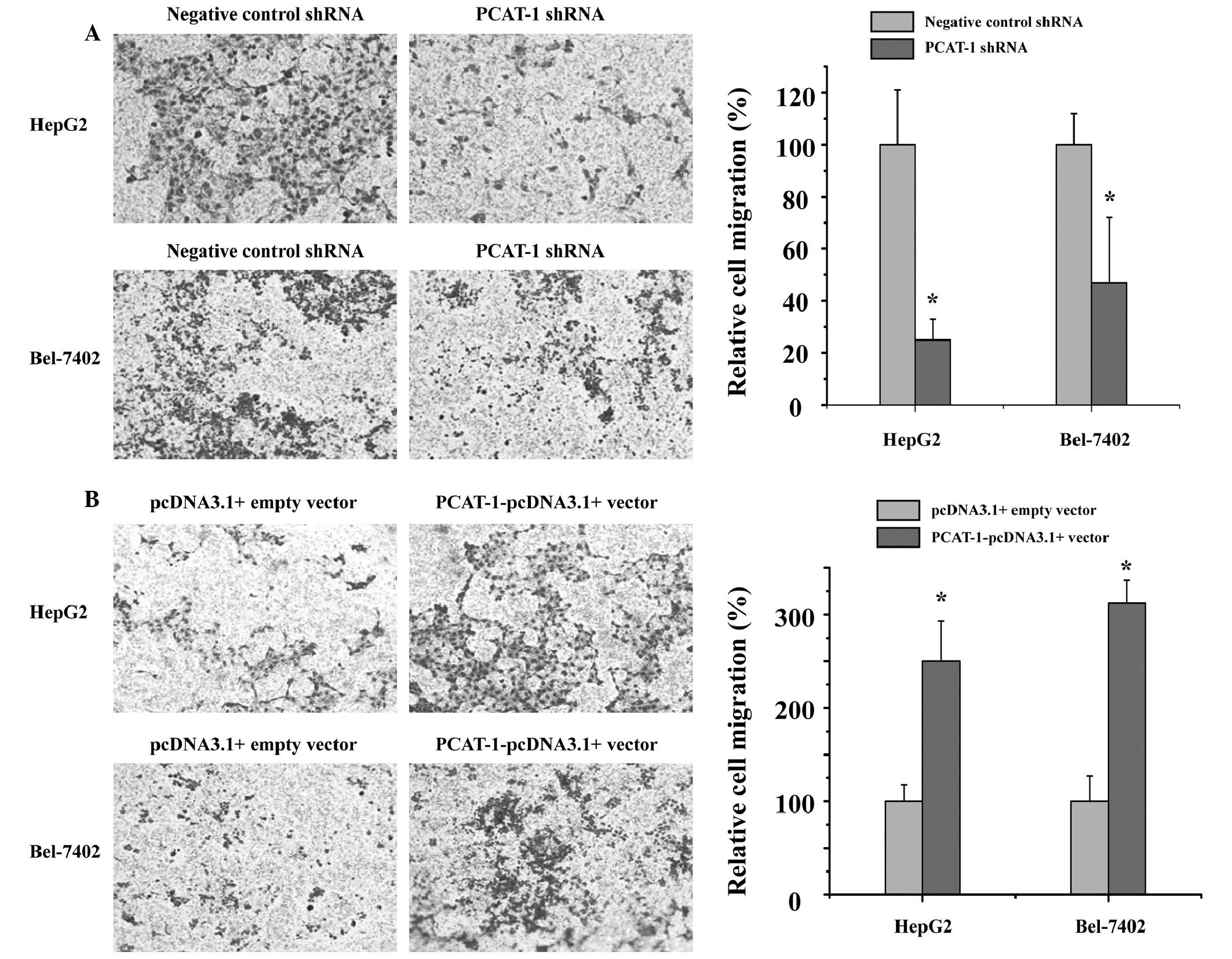
To investigate the potential role of PCAT-1 in HCC
metastasis, the present study detected whether PCAT-1 regulated the
capacity of HCC cells to migrate. The cell migration assay
demonstrated that cell migration was significantly inhibited in the
groups transfected with PCAT-1 shRNA compared with those in the
negative control shRNA group (Fig.
4A; P<0.05). As presented in Fig. 4B, overexpression of PCAT-1
significantly promotes HCC cell migration (P<0.05). These data
indicated that PCAT-1 has a positive effect on HCC cell
migration.
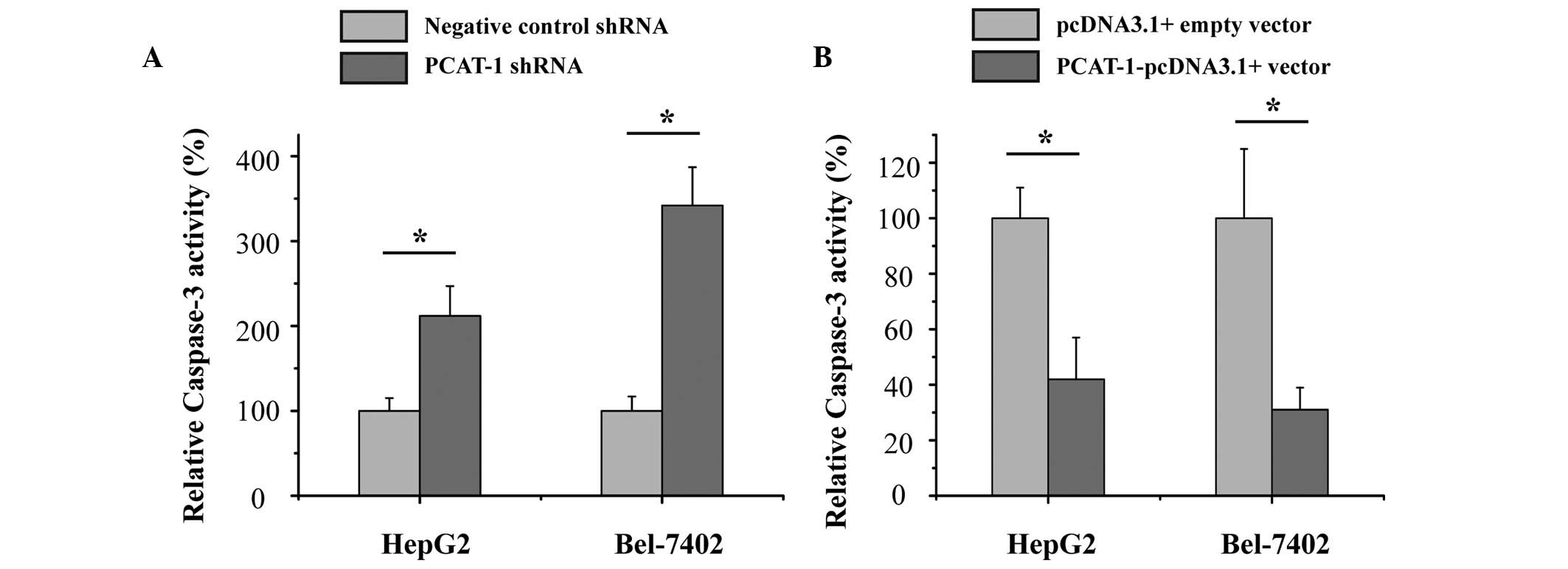
Upregulation of PCAT-1 inhibited
apoptosis in HCC cells
The current study also aimed to determine whether
PCAT-1 inhibited cell apoptosis in HCC. HepG2 and Bel-7402 HCC
cells were transfected with PCAT-1 shRNA or negative control shRNA,
and PCAT-1-pcDNA3.1 + vector or pcDNA3.1 + empty vector. After
transfection for 48 h, the relative activity of caspase-3 was
determined by ELISA assay. As presented in Fig. 5, the relative activity of caspase-3
was significantly increased by the PCAT-1 shRNA (P<0.05), while
the relative activity of caspase-3 was significantly decreased by
the PCAT-1-pcDNA3.1 + vector (P<0.05). These results suggest the
upregulation of PCAT-1 inhibited HCC apoptosis and suggests that
PCAT-1 exerts an oncogenic effect in hepatocellular carcinoma.
Discussion
LncRNAs represent an emerging group in
hepatocellular carcinoma, they may exert effects on tumor
proliferation and migration, and apoptosis (22–24).
PCAT-1, a newly-identified lncRNA, is located in the chromosome
8q24 gene desert and contributes to cell proliferation in prostate
cancer (18). Previous research
has demonstrated that PCAT-1 may suppress the breast cancer 2,
early onset tumor suppressor gene and produce a functional
deficiency in homologous recombination in sporadic cancers
(25). PCAT-1 promotes cell
proliferation in prostate cancer via stabilization of the Myc
proto-oncogene protein (26). A
recent study demonstrated that silencing PCAT-1 may induce cell
growth arrest and apoptosis in human bladder cancer (27).
The present study investigated the role of long
non-coding RNA PCAT-1 in HCC. It was observed that the expression
levels of PCAT-1 were upregulated in HCC tissue samples compared
with matched normal tissue. Furthermore, PCAT-1 expression levels
were also increased in the HepG2 and Bel-7402 HCC cell lines
compared with the L02 normal liver epithelial cell line. These
results suggest that PCAT-1 may exert an oncogenic effect in HCC
development.
To further elucidate the biological functions of
PCAT-1 in HCC, cell proliferation, migration and apoptosis were
detected in cells following PCAT-1 silencing and overexpression in
HepG2 and Bel-7402 HCC cell lines. Arrest of cell proliferation,
inhibition of migration and increased apoptosis were observed in
PCAT-1 shRNA-transfected HCC cells. Furthermore, acceleration of
cell proliferation, promotion of migration and decreased apoptosis
were observed in PCAT-1-pcDNA3.1 + vector-transfected HCC cells.
These results suggest that PCAT-1 has an oncogenic role in the
progression and development of HCC.
In conclusion, PCAT-1 exerts an oncogenic effect in
HCC. Further investigation is required to clarify the underlying
molecular mechanisms of PCAT-1 as a candidate biomarker for HCC
therapeutic strategies.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tang ZY, Ye SL, Liu YK, Qin LX, Sun HC, Ye
QH, Wang L, Zhou J, Qiu SJ, Li Y, et al: A decade's studies on
metastasis of hepatocellular carcinoma. J Cancer Res Clin Oncol.
130:187–196. 2004. View Article : Google Scholar
|
|
4
|
Yang Y, Nagano H, Ota H, Morimoto O,
Nakamura M, Wada H, Noda T, Damdinsuren B, Marubashi S, Miyamoto A,
et al: Patterns and clinicopathologic features of extrahepatic
recurrence of hepatocellular carcinoma after curative resection.
Surgery. 141:196–202. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Luk JM, Burchard J, Zhang C, Liu AM, Wong
KF, Shek FH, Lee NP, Fan ST, Poon RT, Ivanovska I, et al: DLK1-DIO3
genomic imprinted microRNA cluster at 14q32.2 defines a stemlike
subtype of hepatocellular carcinoma associated with poor survival.
J Biol Chem. 286:30706–30713. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Clark T, Maximin S, Meier J, Pokharel S
and Bhargava P: Hepatocellular Carcinoma: Review of epidemiology,
screening, imaging diagnosis, response assessment and treatment.
Curr Probl Diagn Radiol. 44:479–486. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang B and Zhang R: Regulatory non-coding
RNAs: Revolutionizing the RNA world. Mol Biol Rep. 41:3915–3923.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Du Y, Wang L, Wu H, Zhang Y, Wang K and Wu
D: MicroRNA-141 inhibits migration of gastric cancer by targeting
zinc finger E-box-binding homeobox 2. Mol Med Rep. 12:3416–3422.
2015.PubMed/NCBI
|
|
9
|
Wang T, Liu Y, Yuan W, Zhang L, Zhang Y,
Wang Z, Zhou X, Zhou H, Chu T, Hao Y, et al: Identification of
microRNAome in rat bladder reveals miR-1949 as a potential inducer
of bladder cancer following spinal cord injury. Mol Med Rep.
12:2849–2857. 2015.PubMed/NCBI
|
|
10
|
Li L and Ma HQ: MicroRNA216a inhibits the
growth and metastasis of oral squamous cell carcinoma by targeting
eukaryotic translation initiation factor 4B. Mol Med Rep.
12:3156–3162. 2015.PubMed/NCBI
|
|
11
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee JT and Bartolomei MS: X-inactivation,
imprinting and long noncoding RNAs in health and disease. Cell.
152:1308–1323. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu J, Shen L, Yao J, Li Y, Wang Y, Chen H
and Geng P: Forkhead box C1 promoter upstream transcript, a novel
long non-coding RNA, regulates proliferation and migration in
basal-like breast cancer. Mol Med Rep. 11:3155–3159. 2015.
|
|
15
|
Wang Y, Yao J, Meng H, Yu Z, Wang Z, Yuan
X, Chen H and Wang A: A novel long non-coding RNA,
hypoxia-inducible factor-2α promoter upstream transcript, functions
as an inhibitor of osteosarcoma stem cells in vitro. Mol Med Rep.
11:2534–2540. 2015.
|
|
16
|
Wang Y, Xu G, Chen W, Pan Q, Huang K, Pan
J, Zhang W and Chen J: Detection of long-chain non-encoding RNA
differential expression in non-small cell lung cancer by microarray
analysis and preliminary verification. Mol Med Rep. 11:1925–1932.
2015.
|
|
17
|
Shi X, Sun M, Liu H, Yao Y and Song Y:
Long non-coding RNAs: A new frontier in the study of human
diseases. Cancer Lett. 339:159–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Prensner JR, Iyer MK, Balbin OA,
Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso
CS, Kominsky HD, et al: Transcriptome sequencing across a prostate
cancer cohort identifies PCAT-1, an unannotated lincRNA implicated
in disease progression. Nat Biotechnol. 29:742–749. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ge X, Chen Y, Liao X, Liu D, Li F, Ruan H
and Jia W: Overexpression of long noncoding RNA PCAT-1 is a novel
biomarker of poor prognosis in patients with colorectal cancer. Med
Oncol. 30:5882013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shi WH, Wu QQ, Li SQ, Yang TX, Liu ZH,
Tong YS, Tuo L, Wang S and Cao XF: Upregulation of the long
noncoding RNA PCAT-1 correlates with advanced clinical stage and
poor prognosis in esophageal squamous carcinoma. Tumour Biol.
36:2501–2507. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang X, Xie X, Xiao YF, Xie R, Hu CJ, Tang
B, Li BS and Yang SM: The emergence of long non-coding RNAs in the
tumorigenesis of hepatocellular carcinoma. Cancer Lett.
360:119–124. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sun J, Bie B, Zhang S, Yang J and Li Z:
Long non-coding RNAs: Critical players in hepatocellular carcinoma.
Int J Mol Sci. 15:20434–20448. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang JL, Zheng L, Hu YW and Wang Q:
Characteristics of long non-coding RNA and its relation to
hepatocellular carcinoma. Carcinogenesis. 35:507–514. 2014.
View Article : Google Scholar
|
|
25
|
Prensner JR, Chen W, Iyer MK, Cao Q, Ma T,
Han S, Sahu A, Malik R, Wilder-Romans K, Navone N, et al: PCAT-1, a
long noncoding RNA, regulates BRCA2 and controls homologous
recombination in cancer. Cancer Res. 74:1651–1660. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Prensner JR, Chen W, Han S, Iyer MK, Cao
Q, Kothari V, Evans JR, Knudsen KE, Paulsen MT, Ljungman M, et al:
The long non-coding RNA PCAT-1 promotes prostate cancer cell
proliferation through cMyc. Neoplasia. 16:900–908. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu L, Liu Y, Zhuang C, Xu W, Fu X, Lv Z,
Wu H, Mou L, Zhao G, Cai Z and Huang W: Inducing cell growth arrest
and apoptosis by silencing long non-coding RNA PCAT-1 in human
bladder cancer. Tumour Biol. 36:7685–7689. 2015. View Article : Google Scholar : PubMed/NCBI
|