Introduction
Hirschsprung's disease (HSCR; also termed congenital
aganglionic megacolon) represents the predominant genetic cause of
functional intestinal obstruction, with an incidence of 1/5,000
live births, although there are differences in incidence among
ethnic groups and there is a 4:1 male:female gender bias (1). HSCR is characterized by the abnormal
development of the enteric nervous system (ENS), with aganglionosis
in a distal segment of the bowel leading to intestinal dysfunction.
HSCR typically occurs in infants and young children and the
patients lose their normal bowel reflexes and fecal discharge
barriers. Patients suffer from constipation, abdominal distension
and vomiting, which affect growth and development. HSCR is a
complex and heterogeneous disease; thus, there may be other unknown
factors contributing to ENS development and HSCR pathogenesis
(2). At present, the etiology of
HSCR is thought to be multigenetic and multifactorial in origin
(3). Although gaps in our
understanding of HSCR remain, advances in genetic testing have led
to an improved understanding of the disease (4).
Congenital anorectal malformations (ARMs) occur
commonly in humans, with a reported incidence of 1:1,000–5,000
newborns (5,6). The vast majority of children with
ARMs are born with the anus in the wrong position. Furthermore,
some patients have acute intestinal obstruction and difficultly in
defecating after birth. A minority of children with ARMs show no or
only mild symptoms (5,6). ARMs is thought to occur in the
developing embryo as a result of genetic factors; however, the
underlying pathogenesis of ARMs has yet to be fully elucidated.
Numerous technical advances have been made in the surgical
treatment of ARMs; however, patients with intermediate-type and
high-type ARMs often continue to have postoperative anal
dysfunctions.
Numerous signaling molecules have been shown to be
involved in the development of HSCR in the embryo, including RET,
endothelin receptor type B, endothelin 3, endothelin-converting
enzyme 1, SRY (sex determining regionY)-box 10 (SOX10), glial
cell-derived neurotrophic factor, neurturin, paired-like homeobox
2b (PHOX2B), transcription factor 4 (TCF4) and zinc finger E-box
binding homeobox 2 (ZEB2 or ZFHX1B) (7–11).
Those suggested to be involved in ARM include sonic hedgehog (SHH),
bone morphogenetic protein 4, HOX, fibroblast growth factor 10,
Wnt, Notch and Hedgehog (HH) (12–18).
Embryonic development is regulated by a number of complex signaling
pathways. A previous study indicated that the HH signaling pathway
exerts a key role in the initiation, patterning and morphogenesis
of the anus (13); and a
subsequent study reported the involvement of the HH signaling
pathway in the axial patterning of the gastrointestinal tract
(19).
The role of inductive interactions in animal
development has long been recognized; however, only recently have
the signaling molecules mediating these interactions been
identified (20). Prominent among
these are the Hedgehog (Hh) family of proteins, which are a group
of closely associated secreted proteins encoded by a gene family
originally identified through mutations of HH, a segment polarity
gene in Drosophila (20).
The HH proteins are evolutionarily conserved signaling molecules
that control the normal growth and patterning of diverse animals,
including Drosophila and humans (20). As with other signaling molecules,
HH is expressed in the notochord, neural tube, brain, polarizing
activities of the limbs of the development zone and the intestine
(20). Genetic analyses have
revealed that the majority of segment polarity genes are involved
in the transduction of the HH signal or the signal encoded by the
gene wingless (WNT), a member of the WNT family of signaling
proteins (21). Therefore, the
investigation of segment polarity genes has permitted the
delineation of these two signaling pathways, and efforts are under
way to characterize in molecular detail the precise biochemical
mechanisms underlying HH signal transduction (21). Similar to other signals regulating
embryonic development, members of the Hh family, including SHH,
Indian hedgehog homolog (IHH) and Desert hedgehog homolog (DHH),
are involved in a wide variety of processes, ranging from the
control of left-right asymmetry of the body to the specification of
individual types of cell within the neural tube and brain (21). In addition, as with other embryonic
signals, the aberrant activity of the HH signaling pathway has been
linked to abnormalities associated with specific diseases.
A previous study suggested that certain single
nucleotide polymorphisms (SNPs) in the HH gene may be responsible
for an underlying genetic predisposition to particular diseases
(22). Therefore, the present
study examined a larger cohort and extended the evaluation of the
genetic association between SNPs of the HH gene with HSCR and ARM.
Screening of the HH gene, in particular within the rs61730970,
rs200798148 and rs146535482 loci, was performed to identify genetic
polymorphisms associated with HSCR and ARM.
Materials and methods
Patients and controls
The present study was approved by the Ethics
Committee of the China Medical University (Shenyang, China; ethical
no. 2013PS07K). Blood samples (5 ml) from 200 HSCR (139 male and 61
female) and 100 ARM (67 male and 33 female) patients were collected
at the Departments of Pediatric Surgery of the Shengjing Hospital
of China Medical University and the Jinzhou Women and Children's
Hospital (Liaoning, China) between January 2008 and December 2013.
For the purposes of the study, patients with familial constipation
or other congenital gastrointestinal malformations were excluded.
Patients in the case groups of HSCR and ARM were aged between 0.5
and 3.5 years (average age, 1.5±0.3 years). The control group was
matched to the HSCR and ARM groups with respect to the ratio of
boys:girls and age (age range, 0.5 to 2 years).
Extraction of DNA
Blood samples (5 ml) were collected from the HSCR,
ARM and control group patients, and 200 μl venous blood
samples were treated with 15 g/l EDTA (Beijing bioco Laibo
Technology, Co., Ltd., Beijing, China) as an anticoagulant. Genomic
DNA was extracted using the Blood Genome DNA Extraction kit (cat.
no. 9450; Takara Biotechnology Co., Ltd., Dalian, China), according
to the manufacturer's protocol. The absorbance values at 260 and
280 nm (A260/A280) were
measured by spectrophotometry (NanoVue™ Plus Spectrophotometer; GE
Healthcare Life Sciences, Chalfont, UK), and an absorbance between
1.6 and 2 was considered to meet the experimental requirements. The
integrity and purity of DNA were determined by 1.5% agarose gel
electrophoresis. DNA templates were stored in a freezer at -70°C
until further use.
Polymerase chain reaction (PCR) and DNA
sequence analysis
The rs61730970, rs200798148 and rs146535482 loci
from the HH gene were selected for the SNP analysis, based on their
minor allele frequencies. Specific primers for the HH gene were
designed using the DNASTAR primer design program, and synthesized
by Liuhe Huada Gene Technology, Co., Ltd. (Beijing, China). The
primer sequences used for PCR were as follows: rs61730970-1, 5′-GGG
TTA GCA AGC ACT TCA-3′ and rs61730970-2, 5′-AAG CCA ACC TTT ATT CCA
CT-3′ (product size, 341 bp); rs200798148-1, 5′-TCC CTT ATC TCC TTC
ATC T-3′ and rs200798148-2, 5′-AAG CCA ACC TTT ATT CCA C-3′
(product size, 265 bp); rs146535482-1, 5′-CAC CTC AGC CTC ACA AAG
T-3′ and rs146535482-2, 5′-TTC CAT CTG GTC CAA GTA G-3′ (product
size, 303 bp). The primers exhibited no homology with other genes,
as determined from a Basic Local Alignment Search Tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi)
analysis. For PCR, a 50 μl reaction system included 10X PCR
buffer solution, 1 mmol/l MgCl2, 0.1 mmol/l
deoxynucleotides, 10 pmol/l each of the upstream and downstream
primers, 100 ng DNA template and 1 U Taq DNA polymerase
(Takara Biotechnology Co., Ltd.). The PCR conditions were as
follows: 95°C predegeneration for 3 min, then 95°C denatur- ation
for 30 sec, followed by annealing at 53–58°C for 30 sec and
elongation at 72°C for 1 min in 30 cycles, and finally, the
reaction was terminated by incubation at 72°C for 7 min. The PCR
products were electrophoresed on a 1.5% agarose gel containing the
DL 2000 DNA Marker, stained with ethidium bromide (both Beijing
bioco Laibo Technology, Co., Ltd.), and visualized using an
automatic gel documentation system (AzureSpot Analysis Software
System; Azure Biosytems, Dublin, CA, USA). Subsequently, the PCR
products were purified using 75% isopropanol (Beijing Qixiang
Hongye Trading, Co., Ltd., Beijiing, China) and sequenced using an
automatic sequencer (BGISEQ-1000; BGI Shenzhen, Shenzhen,
China).
Statistical analysis
Statistical analyses were conducted using SPSS
software, version 13.0 (SPSS, Inc., Chicago, IL, USA).
χ2 tests were performed to determine whether each
polymorphism was in Hardy-Weinberg equilibrium within the normal
control and patient groups. The unconditional logistic regression
model was used to analyze the association between the risk of each
SNP and HSCR and ARM. Multiplicative interactions have previously
been considered to be suitable for the detection of possible
gene-environment interactions (23,24).
The χ2 test was also used to determine the significance
of the association of the allele frequencies between the normal
control group and the patient groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
PCR amplification of HH gene
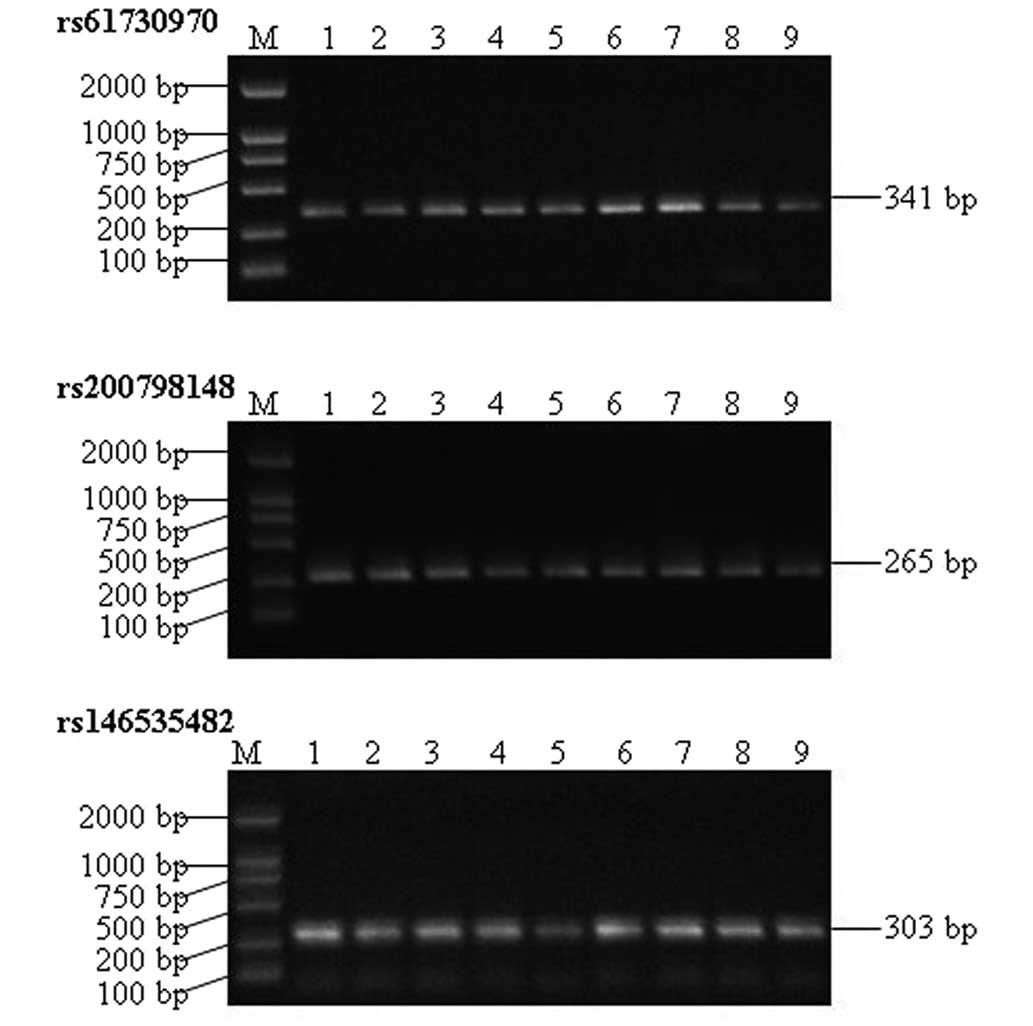
PCR amplification of the HH gene was successfully
performed. The amplified segments of rs61730970 (601 bp),
rs200798148 (401 bp) and rs146535482 (503 bp) were of lengths 341,
265 and 303 bp, respectively, which were in accordance with the
theoretical lengths. The amplification products were highly
expressed, without any apparent non-specific bands (Fig. 1).
Distribution of rs61730970, rs200798148
and rs146535482 allele and genotype frequencies in the HSCR, ARM
and control groups
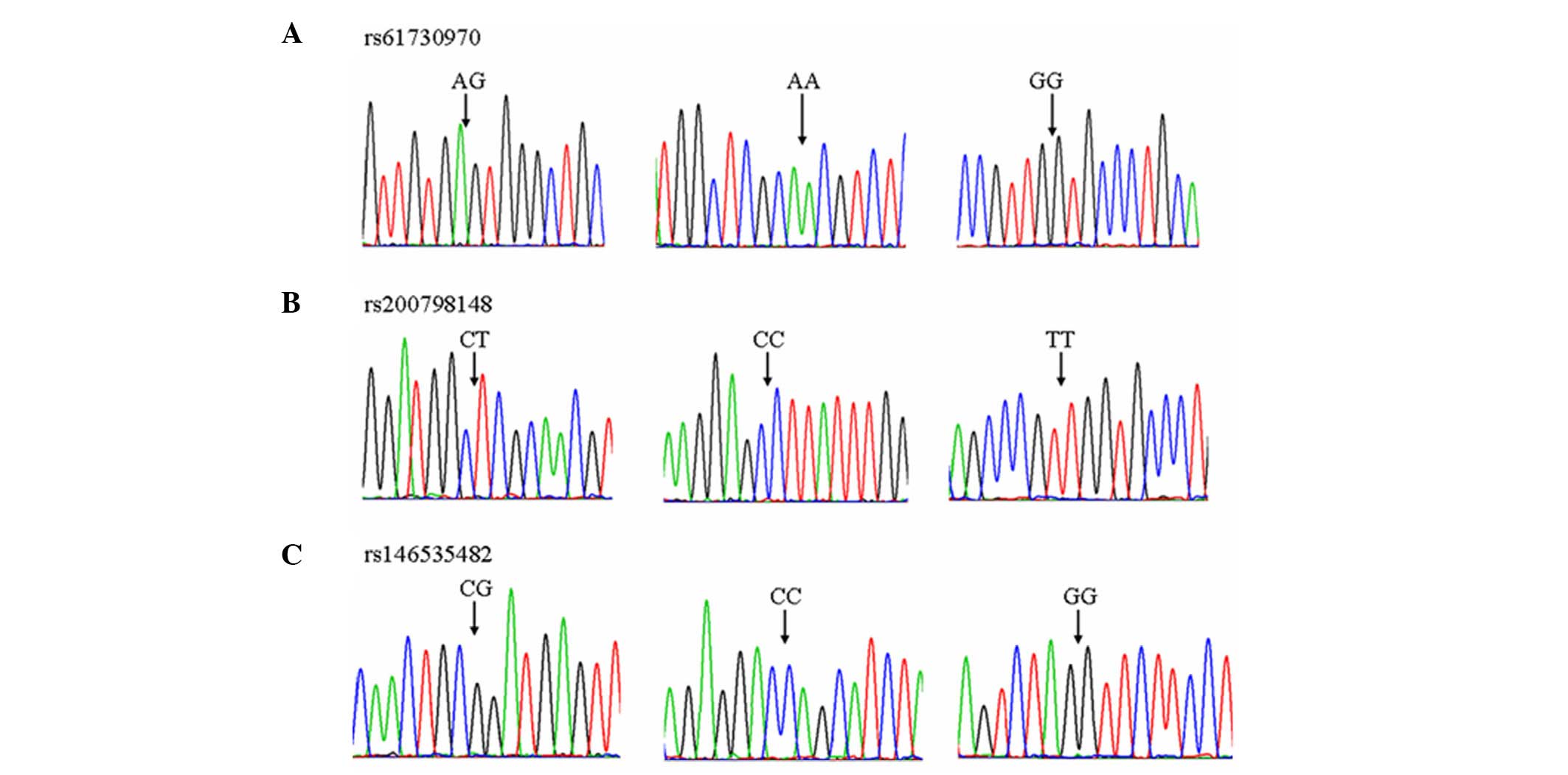
Genotype distributions in the three SNPs were in
accordance with the Hardy-Weinberg equilibrium (P>0.05; Fig. 2). rs61730970, rs200798148 and
rs146535482 of the HH gene genotype were revealed to be
particularly associated with a greater risk of HSCR and ARM
(Tables I and II).
 | Table IAllele and genotype frequency
distribution in patients with Hirschsprung's disease, and
controls. |
Table I
Allele and genotype frequency
distribution in patients with Hirschsprung's disease, and
controls.
| Polymorphism | Type | HSCR | Controls |
X2 | P-value | OR (95% CI) |
|---|
| rs61730970 | AA | 107 | 76 | | | 1.00 (ref.) |
| AG | 74 | 98 | 8.465 | 0.004 | 0.536
(0.352–0.817) |
| GG | 19 | 26 | 0.009 | 0.923 | 1.033
(0.532–2.007) |
| A | 288 | 250 | | | |
| G | 112 | 150 | 8.195 | 0.004 | 1.543
(1.146–2.078 |
| rs200798148 | CC | 102 | 73 | | | 1.00 (ref.) |
| CT | 77 | 99 | 7.419 | 0.006 | 0.557
(0.365–0.850) |
| TT | 21 | 28 | 0.012 | 0.911 | 1.037
(0.547–1.966) |
| C | 281 | 245 | | | |
| T | 119 | 155 | 7.194 | 0.007 | 1.494
(1.113–2.004) |
| rs146535482 | CC | 101 | 72 | | | 1.00 (ref.) |
| CG | 78 | 96 | 6.381 | 0.012 | 0.579
(0.379–0.886) |
| GG | 21 | 32 | 0.448 | 0.504 | 1.238
(0.662–2.316) |
| C | 280 | 240 | | | |
| G | 120 | 160 | 8.791 | 0.003 | 1.556
(1.161–2.085) |
 | Table IIAllele and genotype frequency
distribution in patients with anorectal malformations, and
controls. |
Table II
Allele and genotype frequency
distribution in patients with anorectal malformations, and
controls.
| Polymorphism | Type | ARM | Controls |
X2 | P-value | OR (95% CI) |
|---|
| rs61730970 | AA | 53 | 36 | | | 1.00 (ref.) |
| AG | 35 | 52 | 5.872 | 0.115 | 0.475
(0.260–0.871) |
| GG | 12 | 14 | 0.202 | 0.653 | 0.817
(0.337–1.976) |
| A | 141 | 122 | | | |
| G | 59 | 78 | 4.008 | 0.045 | 1.528
(1.008–2.316) |
| rs200798148 | CC | 59 | 37 | | | 1.00 (ref.) |
| CT | 32 | 51 | 9.343 | 0.002 | 0.393
(0.215–0.720) |
| TT | 9 | 12 | 0.130 | 0.718 | 0.837
(0.317–2.209) |
| C | 150 | 125 | | | |
| T | 50 | 75 | 7.273 | 0.007 | 1.800
(1.172–2.765) |
| rs146535482 | CC | 55 | 36 | | | 1.00 (ref.) |
| CG | 33 | 46 | 5.902 | 0.015 | 0.470
(0.254–0.867) |
| GG | 12 | 18 | 0.028 | 0.867 | 1.076
(0.475–2.534) |
| C | 143 | 118 | | | |
| G | 57 | 82 | 6.891 | 0.009 | 1.743
(1.149–2.645) |
Regarding the genotype level in rs61730970, HSCR was
negatively correlated with GG homozygosity, and positively
correlated with AG heterozygosity and AA homozygosity (P=0.008),
which revealed that the risk of HSCR was significantly increased
among patients with the AG or AA genotype. In the SNP of rs61730970
of the HSCR group, the allele frequencies revealed a trend for a
significant association with the G allele (P=0.004). Regarding the
genotype level in rs200798148, HSCR was negatively correlated with
TT homozygosity and positively correlated with CT heterozygosity
and CC homozygosity (P=0.014), which revealed that the risk of HSCR
was significantly increased among patients with the CT or CC
genotype. In the SNP of rs200798148 of the HSCR group, the allele
frequencies revealed a trend for a significant association with the
T allele (P=0.007).
Regarding the geno-type level in rs146535482, HSCR
was negatively correlated with GG homozygosity, and positively
correlated with CG heterozygosity and CC homozygosity (P=0.011),
revealing that the risk of HSCR was significantly increased among
patients with the CG or CC genotype. In the SNP of rs146535482 of
the HSCR group, the allele frequencies revealed a trend for a
significant association with the G allele (P=0.003). The differ-
ences in the genotypes and allele distributions were statistically
significant among various clinical types in the HSCR variants
rs61730970, rs200798148 and rs146535482 (Table III).
 | Table IIIAllele and genotype frequency
distribution in HSCR. |
Table III
Allele and genotype frequency
distribution in HSCR.
| Polymorphism | Group | Case (n) | Genotype frequency
(%) | Allele
frequency |
|---|
| | | AA | AG | GG | A | G |
| rs61730970 | HSCR | 200 | 107 (53.50) | 74 (37.00) | 19 (9.50) | 288 (72.00) | 112 (28.00) |
| Controls | 200 | 98 (49.00) | 76 (38.00) | 26 (13.00) | 250 (62.50) | 150 (37.50) |
| | |
X2=9.689 P=0.008 |
X2=8.195 | P=0.004 |
| | | CC | CT | TT | C | T |
| rs200798148 | HSCR | 200 | 102 (51.00) | 77 (38.50) | 21 (10.50) | 281 (70.25) | 119 (29.75 |
| Controls | 200 | 73 (36.50) | 99 (49.50) | 28 (14.00) | 245 (61.25) | 155 (38.75) |
| | |
X2=8.556 P=0.014 |
X2=7.194 | P=0.007 |
| | | CC | CG | GG | C | G |
| rs146535482 | HSCR | 200 | 101 (50.50) | 78 (39.00) | 21 (10.50) | 280 (70.00) | 120 (30.00 |
| Controls | 200 | 72 (36.00) | 96 (48.00) | 32 (16.00) | 240 (60.00) | 160 (40.00) |
| | |
X2=9.006 P=0.011 |
X2=8.791 | P=0.003 |
For the genotype level in rs61730970, ARM was
negatively correlated with GG homozygosity and positively
correlated with AG heterozygosity and AA homozygosity (P=0.049),
revealing that the risk of ARM was significantly increased among
patients with the AG or AA genotype. In the SNP of rs61730970 of
the ARM group, the allele frequencies revealed a trend for a
significant association with the G allele (P=0.045). For the
genotype level in rs200798148, ARM was negatively correlated with
TT homozygosity and positively correlated with CT heterozygosity
and CC homozygosity (P=0.007), which revealed that the risk of ARM
was significantly increased among patients with the CT or CC
genotype. In the SNP of rs200798148 of the ARM group, the allele
frequencies revealed a trend for a significant association with the
T allele (P=0.007). For the genotype level in rs146535482, ARM was
negatively correlated with GG homozygosity and positively
correlated with CG heterozygosity and CC homozygosity (P=0.026),
which revealed that the risk of ARM was significantly increased
among patients with the CG or CC genotype. In the SNP of
rs146535482 of the ARM group, the allele frequencies revealed a
trend for a significant association with the G allele (P=0.009).
The differences in the genotypes and allele distributions were
statistically significant among the various clinical types in the
ARM variants rs61730970, rs200798148 and rs146535482 (Table IV).
 | Table IVAllele and genotype frequency
distribution in ARM. |
Table IV
Allele and genotype frequency
distribution in ARM.
| Polymorphism | Group | Case (n) | Genotype frequency
(%) | Allele
frequency |
|---|
| | | AA | AG | GG | A | G |
| rs61730970 | ARM | 100 | 53 (26.50) | 35 (17.50) | 12 (6.00) | 141 (70.50) | 59 (29.50) |
| Controls | 100 | 36 (18.00) | 50 (25.00) | 14 (7.00) | 122 (61.00) | 78 (39.00) |
| | |
X2=6.048 P=0.049 |
X2=4.008 | P=0.045 |
| | | CC | CT | TT | C | T |
| rs200798148 | ARM | 100 | 59 (29.50) | 32 (16.00) | 9 (4.50) | 150 (75.00) | 50 (25.00) |
| Controls | 100 | 37 (18.50) | 51 (25.50 | 12 (6.00) | 125 (62.50) | 75 (37.50) |
| | |
X2=9.820 P=0.007 |
X2=7.273 | P=0.007 |
| | | CC | CG | GG | C | G |
| rs146535482 | ARM | 100 | 55 (27.50) | 33 (16.50) | 12 (6.00 | 143 (71.50) | 57 (28.50) |
| Controls | 100 | 36 (18.00) | 46 (23.00) | 18 (9.00) | 118 (59.00) | 82 (41.00) |
| | |
X2=7.306 P=0.026 |
X2=6.891 | P=0.009 |
Sequence variants of rs61730970,
rs200798148 and rs146535482 in the HSCR and ARM groups
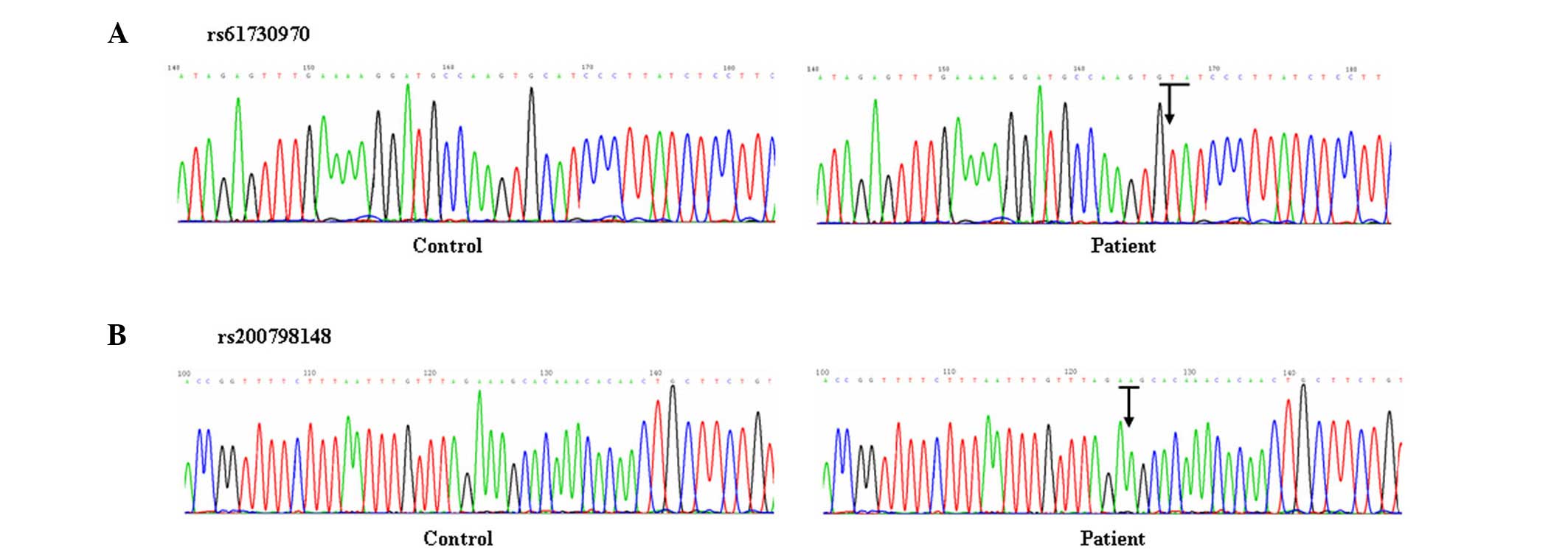
In the HSCR group, a loss of heterozygosity and SNPs
were detected in the rs61730970 and rs200798148 sequences by
sequencing. For rs61730970, a substitution was detected at the
166th codon of GCA →GTA (alanine → valine) in 17
patients; for rs200798148, a loss of heterozygosity was detected in
21 patients, and the sequence lost one 'A' at its 125th
codon (Fig. 3). Sequencing of
rs146535482 did not identify any abnormalities associated with
HSCR.
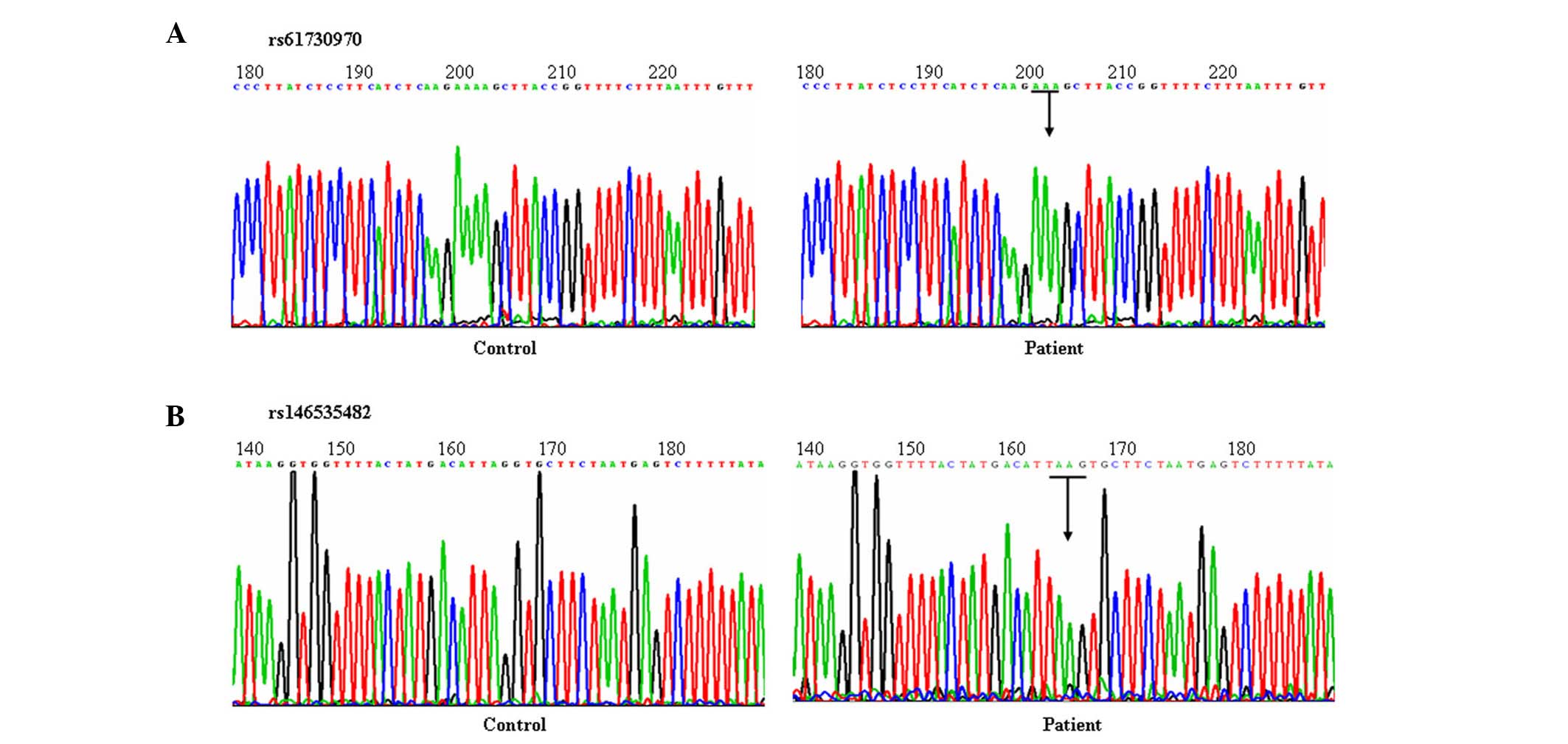
In the ARM group, a loss of heterozygosity and SNPs
were detected in the rs61730970 and rs146535482 sequences. For
rs61730970, a loss of heterozygosity was detected in 19 patients
and one 'A' was lost at the at the 203rd codon; for
rs146535482, a substitution was detected at the 166th
codon of AGG → AAG (arginine → lysine) in 33 patients (Fig. 4). Sequencing of rs200798148 did not
identify any abnormalities associated with ARM.
Discussion
HSCR is a common congenital malformation of the
gastro- intestinal tract. It is thought to be due to stagnation of
the digestive tract into nerve cells during embryonic development.
In addition, it has been suggested that environmental and genetic
factors may have an important role in the occurrence of HSCR
(1,3).
The vast majority of ARM children do not have an
anus in the normal anal position, some patients have acute
intestinal obstruction after birth and some have difficultly in
defecating. A small minority of ARM children show no or only mild
symptoms (5,6). Despite its clinical relevance, the
development of ARM remains poorly understood in the context of
development of the hindgut (25).
The genetics of ARM are extremely complex and numerous genes have
previously been associated with the pathogenesis, including BMPs,
HOX, SHH, and so forth (14,15,20).
However, at present, no common consensus exists regarding the
pathogenesis of ARM.
HSCR and ARM are complex diseases, and at least 17
genes have been associated with the pathogenesis of HSCR and ARM.
HH family proteins are secreted signaling molecules that control
animal development and the balancing of tissues (26,27).
They are produced as precursors that are activated by autocatalytic
processing to generate sterol modification, in particular
palmitoylation, activity in the N-terminal signaling domain
(28–30). Lipid modifications influence HH
signaling, although its precise function has yet to be fully
elucidated (31). The HH pathway
can be activated by any one of the three homologs of the
Drosophila proteins, namely SHH, DHH or IHH, of which SHH is
the best characterized signaling protein involved in early
patterning and cell-fate specification in various systems (32). In all mammals investigated, SHH,
IHH and DHH are expressed in the developing gut tube. SHH and IHH
are expressed in the epithelium, whereas DHH is specifically
expressed in Schwann cells, peripheral nerves and endothelial cells
(21). In the mouse, during gut
organogenesis, SHH and IHH are co-expressed in gut primitive
endoderm epithelium at the early somite stage (8.5 days
post-coitum), prior to the enteric neural crest cells reaching the
stomach, which usually occurs at embryonic day 10.5 (E10.5). At the
latter stages, from early stomach development (E10) to the
initiation of epithelial cytodifferentiation at E15.5, SHH and IHH
are persistently, although differentially, expressed in the
developing guts (21).
The precursor protein of HH protein has a molecular
weight of ~45 kDa. The C-terminal portion of the HH precursor has
zymogen activity; HH is cleaved into a C-terminal peptide with a
molecular weight of ~25 kDa of no known function, and an N-terminal
fragment (HH-N), which constitutes the biologically active portion
of HH (33). During
autoprocessing, a cholesterol group is attached to the C-terminus
of HH-N, which is expressed in the form designated as HH-Np
(34). It is considered that
cholesterol helps to maintain the HH-Np cell membrane, and also
limits the scope of HH activity. However, in mice that were
modified to express a form of Shh lacking the cholesterol
modification (N-SHH), short-range HH signaling was maintained,
while long-range signaling was defective, resulting in the loss of
digits and proper patterning in the developing limb, thereby
suggesting differential requirements for the cholesterol HH
signaling pathway (35). HH
proteins are further modified by palmitoylation at a highly
conserved N-terminal cysteine residue (36). Thus far, the function of the HH
protein in the signaling pathway is relatively unexplored in other
mammals.
The aim of the present study was to determine the
role of the ENS on the development of the HH cascade in HSCR and
ARM. Particular loci in the HH gene (rs61730970, rs200798148 and
rs146535482) exhibited SNPs and loss of heterozygosity in a number
of patients with HSCR and ARM, and these variations occurred much
less frequently in the normal control patients. Heterozygous change
was observed for rs61730970 in 17 patients, whereas a loss of
rs200798148 correlated with the loss of heterozygosity in 21
patients of the HSCR group. By contrast, with rs61730970, a loss of
heterozygosity was observed in 19 patients, while the heterozygous
change with rs146535482 was observed with 33 patients in the ARM
group. Furthermore, the presence of a heterozygous form of the
rs61730970 codon (alanine at position 166 → valine) in the HSCR
group, and a heterozygous form of the rs146535482 codon (arginine
at position 166 → lysine) in the ARM group, may represent the key
mutation.
The present study also aimed to examine the risk and
potential association of HH gene polymorphisms in Chinese patients
with HSCR and ARM. The association between specific genotypes and
HSCR and ARM was assessed using logistic regression analysis, or an
examination of the 95% confidence interval. The risk of HSCR and
ARM increased as presumptive high-risk genotypes increased for the
combined genotypes of the HH heterozygosity genotype. Essentially,
the results suggested that polymorphisms in the HH gene, in
particular at the rs61730970, rs200798148 and rs146535482 loci,
were associated with susceptibility to HSCR and ARM. In terms of
the genotype and allele distribution of rs61730970, rs200798148,
and rs146535482, various changes were shown to be statistically
significant, which may affect the clinical performance. In
addition, the sequence analysis performed in the present study
revealed that the HH gene may influence the risk of common
developmental abnormalities.
In conclusion, the present study demonstrated an
association between genetic polymorphisms at the HH gene and HSCR
and ARM. An increased risk of HSCR and ARM was most evident for
patients who possessed the heterozygosity-associated alleles,
indicating that the effect was present in the Chinese population
examined in the present study. These results suggested that
heterozygosity variations in the HH gene have an important role in
the development of HSCR and ARM, and the present study may provide
novel insights into the pathogenesis of HSCR and ARM.
Acknowledgments
This study was supported by grants from the National
Natural Science Foundation of China (grant nos. 81270436 and
81170334) and the Shenyang Science and Technology Plan Project
(grant no. F13-316-1-01).
References
|
1
|
Amiel J, Sproat-Emison E, Garcia-Barcelo
M, Lantieri F, Burzynski G, Borrego S, Pelet A, Arnold S, Miao X,
Griseri P, et al: Hirschsprung disease, associated syndromes and
genetics: A review. J Med Genet. 45:1–14. 2008. View Article : Google Scholar
|
|
2
|
Saeed A, Barreto L, Neogii SG, Loos A,
McFarlane I and Aslam A: Identification of novel genes in
Hirschsprung disease pathway using whole genome expression study. J
Pediatr Surg. 47:303–307. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Passarge E and Bruder E: Genetic bases of
Hirschsprung's disease. Pathologe. 28:113–118. 2007.In German.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
El-Nachef W and Grikscheit T: Enteric
nervous system cell replacement therapy for Hirschsprung disease:
Beyond tissue-engineered intestine. Eur J Pediatr Surg. 24:214–218.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rintala RJ: Congenital anorectal
malformations: Anything new? J Pediatr Gastroenterol Nutr. 48(Suppl
2): S79–S82. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bán G, Németh P and Túri S: Neurovesical
dysfunction in children with anorectal malformations. Orv Hetil.
147:1645–1649. 2006.In Hungarian.
|
|
7
|
Sánchez-Mejías A, Fernández RM,
López-Alonso M, Antiñolo G and Borrego S: New roles of EDNRB and
EDN3 in the pathogenesis of Hirschsprung disease. Genet Med.
12:39–43. 2010. View Article : Google Scholar
|
|
8
|
Pan ZW, Lou J, Luo C, Yu L and Li JC:
Association analysis of the SOX10 polymorphism with Hirschsprung
disease in the Han Chinese population. J Pediatr Surg.
46:1930–1934. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fernandez RM, Ruiz-Ferrer M, Lopez-Alonso
M, Antiñolo G and Borrego S: Polymorphisms in the genes encoding
the 4 RET ligands, GDNF, NTN, ARTN, PSPN, and susceptibility to
Hirschsprung disease. J Pediatr Surg. 43:2042–2047. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kwon MJ, Lee GH, Lee MK, Kim JY, Yoo HS,
Ki CS, Chang YS, Kim JW and Park WS: PHOX2B mutations in patients
with Ondine-Hirschsprung disease and a review of the literature.
Eur J Pediatr. 170:1267–1271. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gregory-Evans CY, Vieira H, Dalton R,
Adams GG, Salt A and Gregory-Evans K: Ocular coloboma and high
myopia with Hirschsprung disease associated with a novel ZFHX1B
missense mutation and trisomy 21. Am J Med Genet A. 131:86–90.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Seifert AW, Harfe BD and Cohn MJ: Cell
lineage analysis demonstrates an endodermal origin of the distal
urethra and perineum. Dev Biol. 318:143–152. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
van den Brink GR: Hedgehog signaling in
development and homeostasis of the gastrointestinal tract. Physiol
Rev. 87:1343–1375. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Torihashi S, Hattori T, Hasegawa H,
Kurahashi M, Ogaeri T and Fujimoto T: The expression and crucial
roles of BMP signaling in development of smooth muscle progenitor
cells in the mouse embryonic gut. Differentiation. 77:277–289.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zacchetti G, Duboule D and Zakany J: Hox
gene function in vertebrate gut morphogenesis: The case of the
cecum. Development. 134:3967–3973. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fairbanks TJ, De Langhe S, Sala FG,
Warburton D, Anderson KD, Bellusci S and Burns RC: Fibroblast
growth factor 10 (Fgf10) invalidation results in anorectal
malformation in mice. J Pediatr Surg. 39:360–365; discussion
360–365. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tai CC, Sala FG, Ford HR, Wang KS, Li C,
Minoo P, Grikscheit TC and Bellusci S: Wnt5a knock-out mouse as a
new model of anorectal malformation. J Surg Res. 156:278–282. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ngan ES, Garcia-Barceló MM, Yip BH, Poon
HC, Lau ST, Kwok CK, Sat E, Sham MH, Wong KK, Wainwright BJ, et al:
Hedgehog/Notch-induced premature gliogenesis represents a new
disease mechanism for Hirschsprung disease in mice and humans. J
Clin Invest. 121:3467–3478. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lees C, Howie S, Sartor RB and Satsangi J:
The hedgehog signalling pathway in the gastrointestinal tract:
Implications for development, homeostasis, and disease.
Gastroenterology. 129:1696–1710. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Odent S, Atti-Bitach T, Blayau M, Mathieu
M, Aug J, Delezo de AL, Gall JY, Le Marec B, Munnich A, David V and
Vekemans M: Expression of the Sonic hedgehog (SHH) gene during
early human development and phenotypic expression of new mutations
causing holoprosencephaly. Hum Mol Genet. 8:1683–1689. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chan LH, Wang W, Yeung W, Deng Y, Yuan P
and Mak KK: Hedgehog signaling induces osteosarcoma development
through Yap1 and H19 overexpression. Oncogene. 33:4857–4866. 2014.
View Article : Google Scholar
|
|
22
|
Casey JP, Magalhaes T, Conroy JM, Regan R,
Shah N, Anney R, Shields DC, Abrahams BS, Almeida J, Bacchelli E,
et al: A novel approach of homozygous haplotype sharing identifies
candidate genes in autism spectrum disorder. Hum Genet.
131:565–579. 2012. View Article : Google Scholar :
|
|
23
|
Lu J, Yang L, Zhao H, Liu B, Li Y, Wu H,
Li Q, Zeng B, Wang Y, Ji W and Zhou Y: The polymorphism and
haplotypes of PIN1 gene are associated with the risk of lung cancer
in Southern and Eastern Chinese populations. Hum Mutat.
32:1299–1308. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang L, Li Y, Cheng M, Huang D, Zheng J,
Liu B, Ling X, Li Q, Zhang X, Ji W, et al: A functional
polymorphismatmi- croRNA-629-binding site in the 3′-untranslated
region of NBS1 gene confers an increased risk of lung cancer in
Southern and Eastern Chinese population. Carcinogenesis.
33:338–347. 2012. View Article : Google Scholar
|
|
25
|
Moore SW: Associations of anorectal
malformations and related syndromes. Pediatr Surg Int. 29:665–676.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ingham PW, Nakano Y and Seger C:
Mechanisms and functions of Hedgehog signalling across the metazoa.
Nat Rev Genet. 12:393–406. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
McMahon AP, Ingham PW and Tabin CJ:
Developmental roles and clinical significance of hedgehog
signaling. Curr Top Dev Biol. 53:1–114. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Porter JA, Ekker SC, Park WJ, von Kessler
DP, Young KE, Chen CH, Ma Y, Woods AS, Cotter RJ, Koonin EV and
Beachy PA: Hedgehog patterning activity: Role of a lipophilic
modification mediated by the carboxy-terminal autoprocessing
domain. Cell. 86:21–34. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Porter JA, Young KE and Beachy PA:
Cholesterol modification of hedgehog signaling proteins in animal
development. Science. 274:255–259. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pepinsky RB, Zeng C, Wen D, Rayhorn P,
Baker DP, Williams KP, Bixler SA, Ambrose CM, Garber EA, Miatkowski
K, et al: Identification of a palmitic acid-modified form of human
Sonic hedgehog. J Biol Chem. 273:14037–14045. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mann RK and Beachy PA: Novel lipid
modifications of secreted protein signals. Annu Rev Biochem.
73:891–923. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pettigrew CA, Asp E and Emerson CP Jr: A
new role for Hedgehogs in juxtacrine signaling. Mech Dev.
131:137–149. 2014. View Article : Google Scholar :
|
|
33
|
Jiang SQ and Paulus H: A high-throughput,
homogeneous, fluorescence polarization assay for inhibitors of
hedgehog protein autoprocessing. J Biomol Screen. 15:1082–1087.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tokhunts R, Singh S, Chu T, D'Angelo G,
Baubet V, Goetz JA, Huang Z, Yuan Z, Ascano M, Zavros Y, et al: The
full-length unprocessed hedgehog protein is an active signaling
molecule. J Biol Chem. 285:2562–2568. 2010. View Article : Google Scholar :
|
|
35
|
Palm W, Swierczynska MM, Kumari V,
Ehrhart-Bornstein M, Bornstein SR and Eaton S: Secretion and
signaling activities of lipoprotein-associated hedgehog and
non-sterol-modified hedgehog in flies and mammals. PLoS Biol.
11:e10015052013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lee JD, Kraus P, Gaiano N, Nery S, Kohtz
J, Fishell G, Loomis CA and Treisman JE: An acylatable residue of
Hedgehog is differentially required in Drosophila and mouse limb
development. Dev Biol. 233:122–136. 2001. View Article : Google Scholar : PubMed/NCBI
|