Introduction
Alveolar bone loss is associated with osteoporosis
in postmenopausal women (1). It
takes place rapidly during the early postmenopausal years prior to
stabilization approximately six years after menopause, owing to a
decline in estrogen levels (2).
Estrogen (3), bisphosphonates
(4) and parathyroid hormone (PTH)
(5) have been employed to inhibit
alveolar bone loss in postmenopausal women; however, overt adverse
reactions can be caused by long-term treatment with these drugs
(6–9). Thus, it is necessary to develop
alternative therapeutic agents for preventing and treating alveolar
bone loss.
In a previous study, it was demonstrated that
Rhizoma Dioscoreae extract (RDE) has a protective effect on
alveolar bone loss in ovariectomized (OVX) rats via regulation of
Wnt and p38 mitogen-activated protein kinase (MAPK) signaling,
using gene microarray and bioinformatics (10). Although the protective effect of
RDE was partially explained, it was hypothesized that RDE could
achieve its effect by regulating other signaling pathways or
targets. Therefore, considering the important biological
significance of protein-protein interaction (PPI) networks in the
majority of molecular processes, our previous data (10) was analyzed with the predictive
analysis method based on the PPI network to explore the potential
pathways or targets regulated by RDE (11).
Materials and methods
Differentially expressed genes
As previously reported (10), our group demonstrated that RDE
exhibits a protective effect on alveolar bone loss in OVX rats via
regulation of Wnt and p38 MAPK signaling. A microarray analysis for
gene expression profiling in alveolar bone samples from the two
group rats was performed, and significant differentially expressed
genes between two groups (cutoff of 3) were recorded. Bone tissue
samples were obtained between the three molars and incisor of left
mandibles. In the present study, the significant differentially
expressed genes that were determined in our previous study were
used. Methods of animal treatment, specimen preparation and
microarray protocols are described in detail in our previous study
(10). The present study was
approved by the ethics committee of Institute of Basic Theory,
China Academy of Chinese Medical Sciences (Beijing, China).
Construction of the PPI network
PPI represents an essential outline for the analysis
of homeostasis and self-organization in living organisms. In cells
or organisms, PPIs are important cellular events that form the
basis for multiple signal transduction pathways or various
transcriptional regulatory networks, further affecting the majority
of molecular processes (12).
Considering the importance of PPIs, a PPI network was established
to explore the mechanisms underlying the effect of RDE on the
prevention of bone loss.
In this study, BisoGenet (13), a Cytoscape (14) plugin (version 2.8.2), was used for
building the PPI network. BisoGenet uses four databases to collect
the information on PPI networks involving relevant genes. The four
databases were BIND (Biomolecular Interaction Network Database;
www.bind.ca), BioGRID (General Repository for
Interaction Datasets; thebiogrid.org), DIP (Database of Interacting
Proteins; dip.doe-mbi.ucla.edu/dip/), IntAct (Database system and
analysis tools for protein interaction data; ebi.ac.uk/intact/),
and MINT (Molecular Interactions Database; http://www.ebi.ac.uk/intact/). A PPI network was
established based on the differentially expressed genes from
microarray data analysis.
Detection of molecular clusters in the
PPI network
Detecting molecular clusters in a PPI network is a
common method to identify key biological functions in complex
signaling pathways.
The database was integrated and the PPI network was
generated an analyzed using the MCODE plugin in Cytoscape to
analyze the PPI network features. MCODE can execute molecular
complex detection (MCODE) algorithm to detect molecule clusters in
the interactome network (15).
Molecule clusters with P<0.05, with at least four nodes and one
seed node were considered significant.
Functional annotation of molecular
clusters in the PPI network
Database for Annotation, Visualization, and
Integrated Discovery (DAVID) Bioinformatics Resources 6.7 (16), a versatile functional annotation
tool for discovering the biological significance of genes, was used
to obtain biological process information of gene ontology (GO) for
genes in the PPI Network's molecular clusters, with a threshold
EASE score set at 0.1. In the analysis, enriched GO terms were
considered to be statistically significant with P<0.05 and false
discovery rate (FDR) <0.1.
Pathway analysis of molecular clusters in
the PPI network
To perform a pathway analysis, the names of proteins
in clusters were uploaded into IPA (Ingenuity Systems, Redwood
City, CA, USA). Based on two parameters, the most significant
canonical pathways associated with the dataset were identified by
IPA: i) Ratio of the number of proteins mapped to a pathway divided
by the total number of proteins in the given canonical pathway and
(ii) a P-value showing a degree of association between the
canonical pathway and the dataset proteins.
Western blotting analysis
Western blotting was performed to validate protein
expression of key genes in clusters. The rat alveolar bone,
extracted as previously reported (10), of three groups (SHAM group, n=6;
OVX group, n=6; RDE group, n=6) were used in western blotting.
After alveolar bone was homogenized in liquid nitrogen, proteins
were extracted and dissolved in radioimmunoprecipitation assay
buffer (Thermo Fisher Scientific, Inc., Waltham, MA, USA)
containing phosphatase and protease inhibitors. The insoluble
constituents were removed by centrifugation at 5,000 × g for 5 min
at 4°C. Protein concentrations were determined with the
bicinchoninic acid assay reagent (Pierce, Rockford, IL, USA). Total
protein (80 μg) were resolved by electrophoresis on 12%
sodium dodecyl sulfate-polyacrylamide gels (Bio-Rad Laboratories,
Inc., Hercules, CA, USA) and transferred onto nitrocellulose
membranes (Hybond-ECL; GE Healthcare, Piscataway, NJ, USA).
Membranes were blocked with 5% non-fat dry milk for 1 h, incubated
overnight at 4°C with antibodies against the following: Rabbit
anti-rat polyclonal antibody against cyclin-dependent kinase 1
(CDK1; cat no. ab47594; 1:1,000 dilution) (Abcam, Cambridge, MA,
USA) and rabbit anti-rat polyclonal antibody against β-actin (cat
no. A2066; 1:25,000 dilution; Sigma-Aldrich, St. Louis, MO, USA).
Subsequently, membranes were washed and incubated for 1 h with
horseradish peroxidase-linked antibody (1:1,000 dilution, Cell
Signaling Technology, Inc., Beverly, MA, USA). Immunoreactive
proteins were detected using an enhanced chemiluminescence kit
(PerkinElmer, Waltham, MA, USA), and bands quantified with the
Quantity One software (version 4.0; Bio-Rad, Hercules, CA, USA) by
densitometry, with β-actin used as a loading control. Normalized
data are expressed as fold increases compared with SHAM or OVX
control.
Statistical analysis
The analyses were conducted using SPSS (version
13.0; SPSS Inc., Chicago, IL, USA). The difference between the
groups regarding the evaluated parameters was assessed by analysis
of variance followed by the least significant difference test. The
data of all groups passed the Kolmogorov-Smirnov test of normality.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Differentially expressed genes
In total, 380 differentially expressed genes
(≥3-fold) between the RDE and OVX groups were identified.
Specifically, 205 genes were upregulated, and 175 downregulated
(data not shown).
PPI network
Based on the differentially expressed genes (data
not shown), a biological network showing protein-protein
interactions was established. The PPI network was visualized using
Cytoscape. In the PPI network, the nodes represent proteins, while
the edges reflect the biological relationships between two given
nodes. A total of 631 nodes and 1,273 edges were obtained in the
PPI network (data not shown).
Molecular clusters in the PPI
network
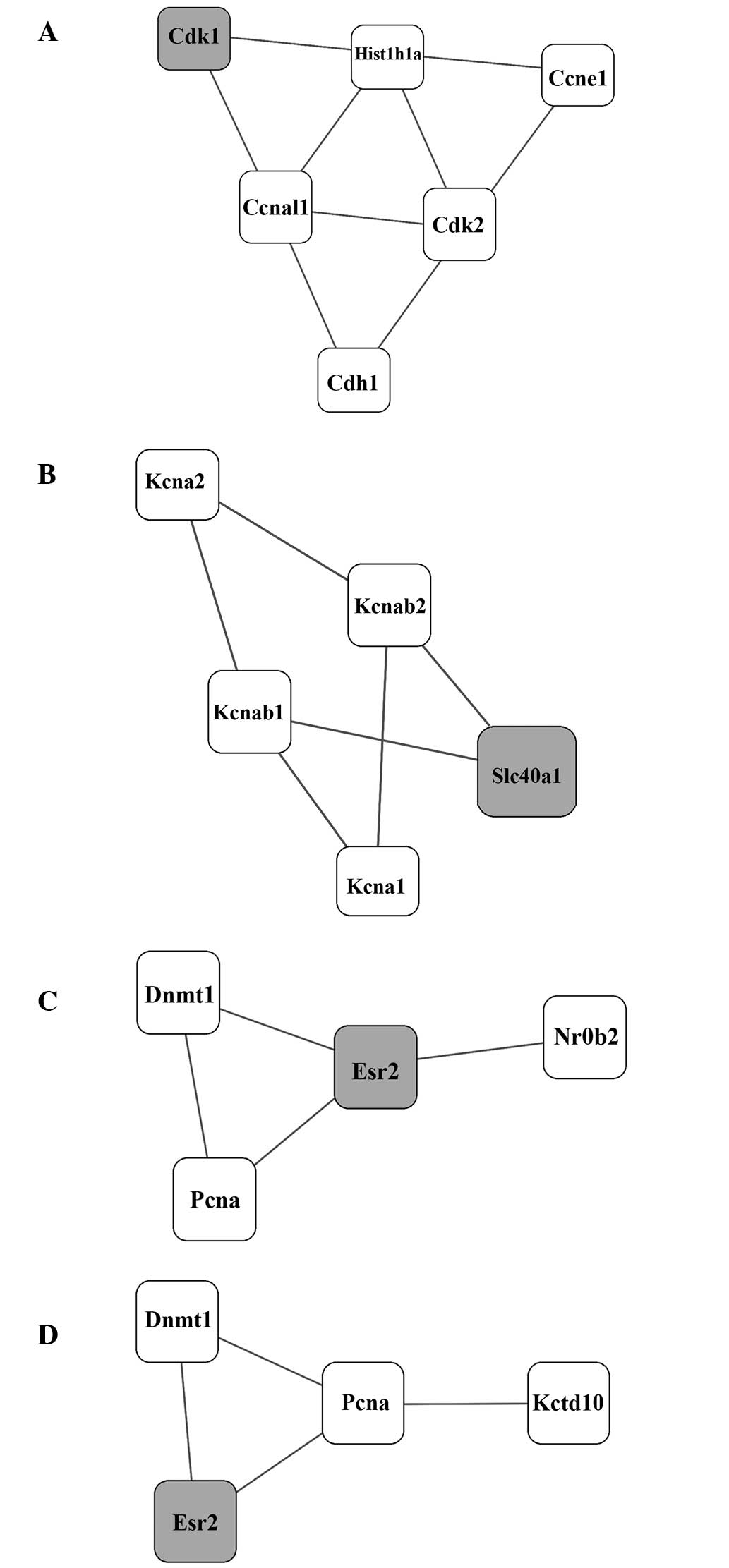
Using the MCODE algorithm, four molecular clusters
(A–D) in the PPI network were identified with smallest P-values,
and were considered the significant molecular clusters (Table I and Fig. 1A–D).
 | Table IMolecular clusters in the
protein-protein interaction network. |
Table I
Molecular clusters in the
protein-protein interaction network.
| Cluster | P-value | Members |
|---|
| Cluster A | 0.017641 | Cdk1 Hist1h1a Cdh1
Cdk2 Ccna1 Ccne1 |
| Cluster B | 0.025993 | Slc40a1 Kcnab2 Kcna2
Kcna1 Kcnab1 |
| Cluster C | 0.026880 | Dnmt1 Pcna Esr2
Nr0b2 |
| Cluster D | 0.049713 | Dnmt1 Pcna Esr2
Kctd10 |
Biological processes associated with
molecular clusters in the PPI network
The DAVID tool was used to retrieve biological
process information associated with the four molecular clusters. Of
the four clusters, only two (A and B) were functionally annotated
(Table II).
 | Table IIKey biological processes associated
with the clusters. |
Table II
Key biological processes associated
with the clusters.
| Cluster | Biological
process | Term | P-value | FDR |
|---|
| Cluster A | GOTERM_BP_FAT | Cell division |
8.70×10−4 |
1.0×10−2 |
| GOTERM_BP_FAT | Response to
drug |
4.20×10−3 |
5.0×10−2 |
| GOTERM_BP_FAT | Cell cycle |
8.20×10−3 |
9.5×10−2 |
| Cluster B | GOTERM_BP_FAT | Metal ion
transport |
1.20×10−6 |
8.5×10−4 |
| GOTERM_BP_FAT | Cation
transport |
2.70×10−6 |
1.9×10−3 |
| GOTERM_BP_FAT | Potassium ion
transport |
6.30×10−6 |
4.4×10−3 |
| GOTERM_BP_FAT | Ion transport |
9.20×10−6 |
6.4×10−3 |
| GOTERM_BP_FAT | Monovalent
inorganic cation transport |
5.40×10−5 |
3.7×10−2 |
Biological pathways associated with
molecular clusters in the PPI network
Table III
summarizes the top IPA canonical pathways (P<0.001) which are
associated with the molecular clusters in the PPI network and
include the seed genes. Of the four clusters, only Cluster A was
associated with biological pathways in the IPA pathway database.
Table III shows that these
pathways were predominantly associated with the cell cycle.
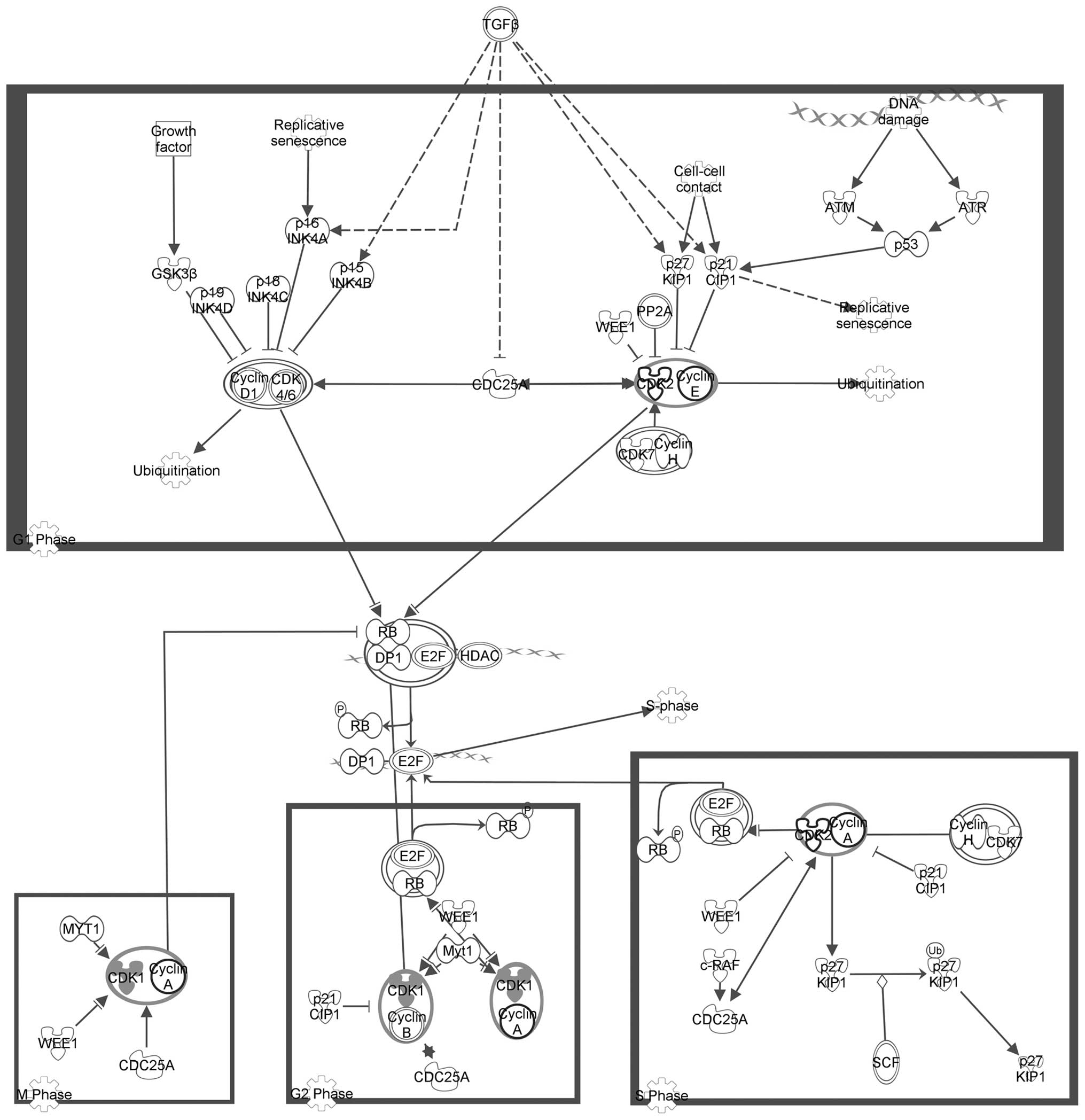
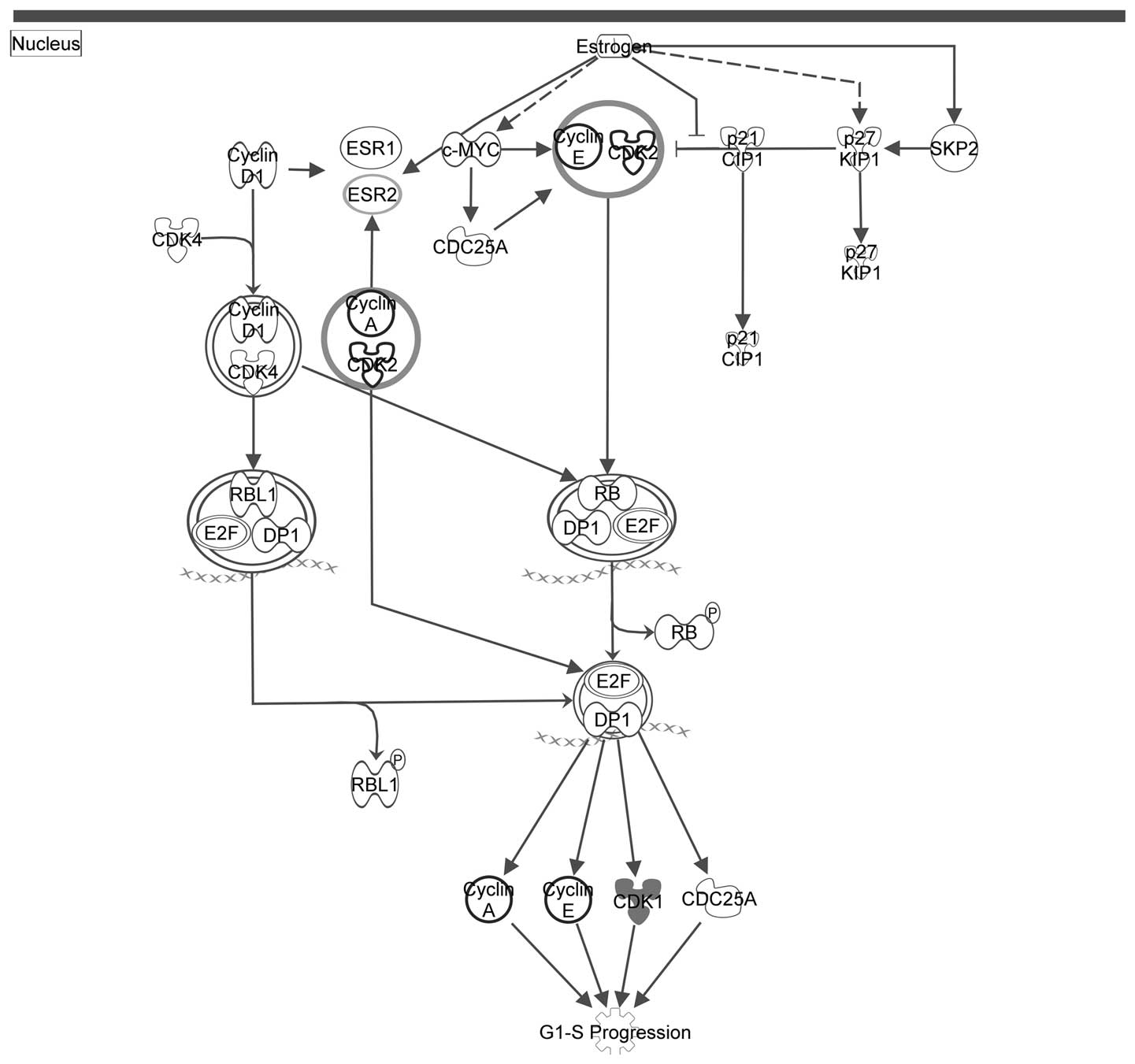
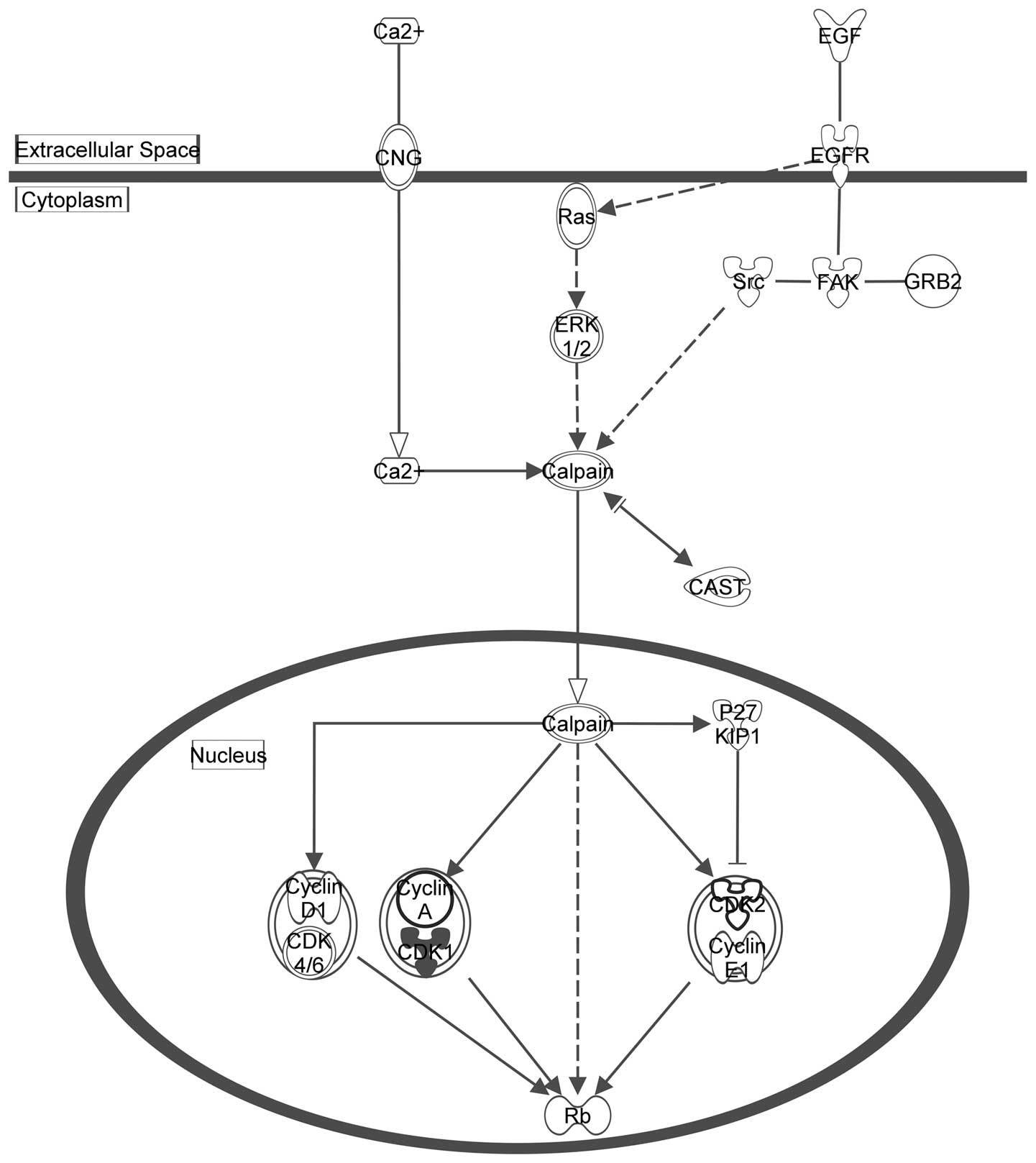
Figs. 2Figure 3–4 depict the three pathways involved in
the cell cycle (displayed in Table
III) with the seed genes are highlighted in gray.
 | Table IIIKey canonical pathways associated
with the clusters. |
Table III
Key canonical pathways associated
with the clusters.
| Cluster | Pathways | P-value |
|---|
| Cluster A | Estrogen-mediated
S-phase entry |
4.54×10−5 |
| Cyclins and cell
cycle regulation |
1.73×10−4 |
| Regulation of
cellular mechanics by calpain protease |
3.82×10−4 |
Protein expression by western
blotting
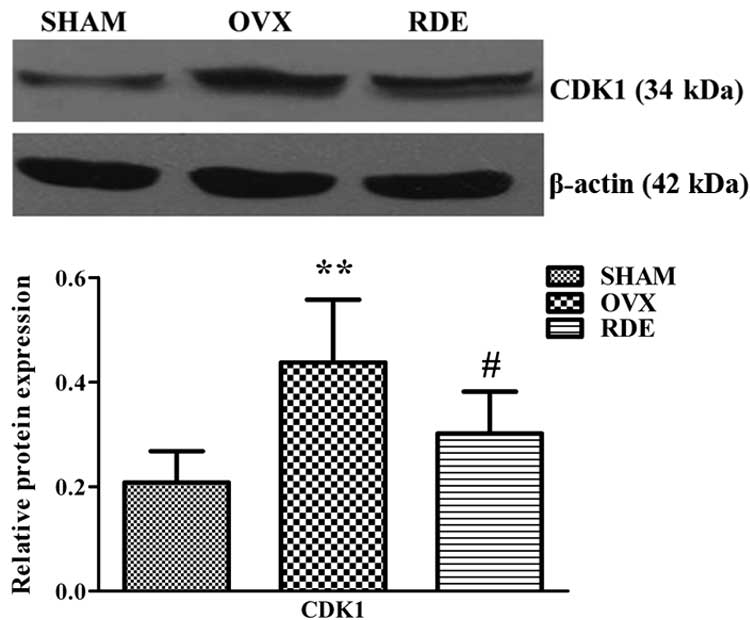
Protein expression of CDK1 was measured in alveolar
bone of rats from the SHAM, OVX and RDE groups. Notably, changes in
the level of CDK1 protein was in agreement with mRNA expression.
Indeed, CDK1 was downregulated in RDE-treated rats compared with
the OVX group (Fig. 5).
Discussion
In the present study, a huge PPI network was
established based on the differentially expressed genes found in
our previous study. To understand the complexity of this network,
one of the most accepted methods was used, analyzing modules or
molecular clusters. A molecular cluster is composed of a number of
interrelated molecules, has a stable structure, and reflects a
specific biological function. Table
II shows the biological processes of two significant molecular
clusters in the PPI network. Cluster A and B were shown to be
associated with the cell cycle and ion transport, respectively.
In addition to biological function analysis, pathway
prediction is another important method to extract potential
signaling pathway information from molecular networks, and
contributes to understanding the mechanisms of a given biological
process (17). Table III presents the key canonical
pathways of cluster A; the only significant molecular cluster in
the generated PPI network. As shown in Table III, cluster A was associated with
the cell cycle. As shown in Table
III, clusters A and C were associated with the cell cycle and
potassium ion channel, respectively.
Potassium channels are major intracellular signaling
pathways at the bone cell surface, through which cell signals are
transferred across to the nuclear material for subsequent cellular
activation (18). In osteoclasts,
potassium ion channels include an inward rectifier channel
regulated by G proteins, and a transient outward rectifier channel
regulated by cell-matrix interactions and extracellular cations
such as calcium and hydrogen (19). Solute carrier family 40 (SLC40A1)
is a cell membrane protein involved in iron export from the cells
to the blood (20). The
relationship between SLC40A1 and potassium channels remains
unknown, and it may be interesting to identify the role of SLC40A1
in regulating potassium channels.
As only cluster A was assigned both function and
pathway annotation, it was further assessed. The cell cycle is
modulated by multiple regulatory molecules, including cyclins and
cyclin-dependent kinases (CDKs). CDKs are a family of protein
kinases that are essential for cell cycle progression. They bind to
specific cyclins and are further activated (21). CDK1 is known to be a highly
conserved protein, and functions as a serine/threonine kinase. CDK1
binds to cyclins A and B, and is involved in modulating the S, G2
and M phases of the cell cycle (22).
The rat OVX model is the most commonly used and
extensively studied animal model of early postmenopausal
osteoporosis. Unlike late postmenopausal osteoporosis (23), early postmenopausal osteopenia,
including alveolar bone loss, is characterized by high bone
turnover that can reproducibly be induced in the OVX animal model
(24). In high-turnover bone loss,
the marked increase in the bone turnover rate is triggered by an
imbalance between bone resorption and formation, with resorption
exceeding formation (25). OVX may
raise the number of pre-osteoblasts in the S phase due in part to
the inhibition of osteoprogenitor cell proliferation in rodent
models (26,27). With regard to osteoclasts, studies
have indicated that RANKL induces RAW264.7 cell proliferation and
promotes cell cycle transition (28), whereas Herba Epimedii flavonoids
may suppress this process (29).
Thus, excessive bone formation and resorption in high-turnover bone
remodeling are associated with promoting cell cycle
progression.
The S phase is an important process common to all
cell types during which chromosomes are replicated; loss of normal
replication control is a hallmark of cancer (30). In breast cancer cells, estrogen and
its receptors can stimulate G1-S phase transition via cyclins or
CDKs (31,32); specifically, CDK2 and CDK1 are
essential in S phase control (33). In the present study, it was
demonstrated that RDE was able to inhibit the expression of CDK1 in
the alveolar bone of OVX rats. The retarding cell cycle progression
in S phase may be one of mechanisms by which RDE protects against
alveolar bone loss.
Calpains are non-lysosomal, calcium-dependent
cysteine proteases, which are highly conserved and found in cells
of organisms, ranging from mammals to Drosophila. They are pivotal
proteases in limited proteolysis of various structural proteins,
regulatory proteins and the tumor-suppressor protein retinoblastoma
(Rb). Enhanced calpain activity is also associated with cell cycle
progression. Calpain promotes transformed cells to go through G1,
enhances hyper-phosphorylation of the Rb protein and increases
protein expression levels of cyclin D, cyclin A, CDK2 and CDK1
(34). Calpains are involved in
bone remodeling, namely cytoskeletal remodeling, integrin-mediated
cell migration, cell differentiation and apoptosis (35–38).
We speculated that the retarding cell cycle progression in G1 phase
may be another mechanism by which RDE protects against alveolar
bone loss.
GO and pathway analysis data showed that the cluster
A was associated with cell cycle regulation and was the most
important in the PPI network. CDK1 in this cluster is a key
positive cell cycle regulator; notably, CDK1 was shown to be
downregulated at the gene level following RDE treatment in our
previous study (10). Here, the
change in CDK1 protein levels corroborated gene expression data
(Fig. 5). Based on the results
described above, it was inferred that RDE may prevent alveolar bone
loss by downregulating CDK1 and further inhibiting cell cycle
progression in alveolar bone tissues.
A few limitations of this study should be mentioned.
MCODE used in the present study is only one of the multiple
algorithms that can be used to determine molecular clusters in PPI
networks, the other molecular clusters or functional modules with
important biological significance may be identified with the use of
the other algorithms.
Using PPI network analysis, this study predicted
that delayed cell cycle progression in bone remodeling via CDK1
downregulation may be a mechanism behind the anti-osteopenic effect
of RDE on alveolar bone. Studies are warranted to further
investigate the mechanisms underlying the anti-osteopenic effect of
RDE.
Acknowledgments
This study was supported by the National Natural
Science Foundation of China (grant nos. 81102680 and 81473450), the
Fundamental Research Funds for the Central Public Welfare Research
Institutes (grant no. YZ-1409) and the Beijing Foundation for
Science and Technology Development of Traditional Chinese Medicine
(grant no. JJ2015-54).
References
|
1
|
Sultan N and Rao J: Association between
periodontal disease and bone mineral density in postmenopausal
women: A cross sectional study. Med Oral Patol Oral Cir Bucal.
16:e440–e447. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Streckfus CF, Johnson RB, Nick T, Tsao A
and Tucci M: Comparison of alveolar bone loss, alveolar bone
density and second metacarpal bone density, salivary and gingival
crevicular fluid interleukin-6 concentrations in healthy
premenopausal and postmenopausal women on estrogen therapy. J
Gerontol A Biol Sci Med Sci. 52:M343–M351. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Civitelli R, Pilgram TK, Dotson M,
Muckerman J, Lewandowski N, Armamento-Villareal R,
Yokoyama-Crothers N, Kardaris EE, Hauser J, Cohen S and Hildebolt
CF: Alveolar and postcranial bone density in postmenopausal women
receiving hormone/estrogen replacement therapy: A randomized,
double-blind, placebo-controlled trial. Arch Intern Med.
162:1409–1415. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Palomo L, Bissada NF and Liu J:
Periodontal assessment of postmenopausal women receiving
risedronate. Menopause. 12:685–690. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu J, Cao Z and Li C: Intermittent PTH
administration: A novel therapy method for periodontitis-associated
alveolar bone loss. Med Hypotheses. 72:294–296. 2009. View Article : Google Scholar
|
|
6
|
Strom BL, Schinnar R, Weber AL, Bunin G,
Berlin JA, Baumgarten M, DeMichele A, Rubin SC, Berlin M, Troxel AB
and Rebbeck TR: Case-control study of postmenopausal hormone
replacement therapy and endometrial cancer. Am J Epidemiol.
164:775–786. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Woo SB, Hellstein JW and Kalmar JR:
Narrative [corrected] review: Bisphosphonates and osteonecrosis of
the jaws. Ann Intern Med. 144:753–761. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rizzoli R, Reginster JY, Boonen S, Bréart
G, Diez-Perez A, Felsenberg D, Kaufman JM, Kanis JA and Cooper C:
Adverse reactions and drug-drug interactions in the management of
women with postmenopausal osteoporosis. Calcif Tissue Int.
89:91–104. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Clemett D and Spencer CM: Raloxifene: A
review of its use in postmenopausal osteoporosis. Drugs.
60:379–411. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang Z, Xiang L, Bai D, Wang W, Li Y, Pan
J, Liu H, Wang S, Xiao GG and Ju D: The protective effect of
Rhizoma Dioscoreae extract against alveolar bone loss in
ovariectomized rats via regulating Wnt and p38 MAPK signaling.
Nutrients. 6:5853–5870. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu Z, Zhao X and Chen L: Identifying
responsive functional modules from protein-protein interaction
network. Mol Cells. 27:271–277. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Real-Chicharro A, Ruiz-Mostazo I,
Navas-Delgado I, Kerzazi A, Chniber O, Sánchez-Jiménez F, Medina MA
and Aldana-Montes JF: Protopia: A protein-protein interaction tool.
BMC Bioinformatics. 10(Suppl 12): S172009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Martin A, Ochagavia ME, Rabasa LC, Miranda
J, Fernandez-de-Cossio J and Bringas R: BisoGenet: A new tool for
gene network building, visualization and analysis. BMC
Bioinformatics. 11:912010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gomez SM: Prediction of protein-protein
interaction networks. Curr Protoc Bioinformatics. Chapter 8: Unit
8.2. 2003, View Article : Google Scholar
|
|
18
|
McDonald F: Ion channels in osteoblasts: A
story of two intracellular organelles. Surgeon. 2:63–69. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Supanchart C and Kornak U: Ion channels
and transporters in osteoclasts. Arch Biochem Biophysics.
473:161–165. 2008. View Article : Google Scholar
|
|
20
|
Ward DM and Kaplan J: Ferroportin-mediated
iron transport: Expression and regulation. Biochim Biophys Acta.
1823:1426–1433. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li JM and Brooks G: Cell cycle regulatory
molecules (cyclins, cyclin-dependent kinases and cyclin-dependent
kinase inhibitors) and the cardiovascular system; potential targets
for therapy? Eur Heart J. 20:406–420. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dorée M and Hunt T: From Cdc2 to Cdk1:
When did the cell cycle kinase join its cyclin partner? J Cell Sci.
115:2461–2464. 2002.PubMed/NCBI
|
|
23
|
Tchetina EV, Maslova KA, Krylov MY and
Myakotkin VA: Association of bone loss with the upregulation of
survival-related genes and concomitant downregulation of Mammalian
target of rapamycin and osteoblast differentiation-related genes in
the peripheral blood of late postmenopausal osteoporotic women. J
Osteoporos. 2015:8026942015.PubMed/NCBI
|
|
24
|
Wronski TJ, Lowry PL, Walsh CC and
Ignaszewski LA: Skeletal alterations in ovariectomized rats. Calcif
Tissue Int. 37:324–328. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Garnero P, Sornay-Rendu E, Chapuy MC and
Delmas PD: Increased bone turnover in late postmenopausal women is
a major determinant of osteoporosis. J Bone Miner Res. 11:337–349.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Turner RT, Backup P, Sherman PJ, Hill E,
Evans GL and Spelsberg TC: Mechanism of action of estrogen on
intramembranous bone formation: Regulation of osteoblast
differentiation and activity. Endocrinology. 131:883–889.
1992.PubMed/NCBI
|
|
27
|
Orlić I, Borovecki F, Simić P and
Vukicević S: Gene expression profiling in bone tissue of
osteoporotic mice. Arh Hig Rada Toksikol. 58:3–11. 2007. View Article : Google Scholar
|
|
28
|
Li CH, Zhao JX, Sun L, Yao ZQ, Deng XL,
Liu R and Liu XY: AG490 inhibits NFATc1 expression and STAT3
activation during RANKL induced osteoclastogenesis. Biochem Biophys
Res Commun. 435:533–539. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang D, Zhang J, Fong C, Yao X and Yang
M: Herba epimedii flavonoids suppress osteoclastic differentiation
and bone resorption by inducing G2/M arrest and apoptosis.
Biochimie. 94:2514–2522. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wuarin J and Nurse P: Regulating S phase:
CDKs, licensing and proteolysis. Cell. 85:785–787. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Prall OW, Sarcevic B, Musgrove EA, Watts
CK and Sutherland RL: Estrogen-induced activation of Cdk4 and Cdk2
during G1-S phase progression is accompanied by increased cyclin D1
expression and decreased cyclin-dependent kinase inhibitor
association with cyclin E-Cdk2. J Biol Chem. 272:10882–10894. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Vondracek J, Kozubik A and Machala M:
Modulation of estrogen receptor-dependent reporter construct
activation and G0/G1-S-phase transition by polycyclic aromatic
hydrocarbons in human breast carcinoma MCF-7 cells. Toxicol Sci.
70:193–201. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hayashi S and Yamaguchi M:
Kinase-independent activity of Cdc2/cyclin A prevents the S phase
in the Drosophila cell cycle. Genes Cells. 4:111–122. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Suzuki K, Hata S, Kawabata Y and Sorimachi
H: Structure, activation, and biology of calpain. Diabetes.
53(Suppl 1): S12–S18. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hayashi M, Koshihara Y, Ishibashi H,
Yamamoto S, Tsubuki S, Saido TC, Kawashima S and Inomata M:
Involvement of calpain in osteoclastic bone resorption. J Biochem.
137:331–338. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Murray EJ, Tram KK, Spencer MJ, Tidball
JG, Murray SS and Lee DB: PTH-mediated osteoblast retraction:
Possible participation of the calpain pathway. Miner Electrolyte
Metab. 21:184–188. 1995.PubMed/NCBI
|
|
37
|
Shimada M, Greer PA, McMahon AP, Bouxsein
ML and Schipani E: In vivo targeted deletion of calpain small
subunit, Capn4, in cells of the osteoblast lineage impairs cell
proliferation, differentiation and bone formation. J Biol Chem.
283:21002–21010. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Murray SS, Grisanti MS, Bentley GV, Kahn
AJ, Urist MR and Murray EJ: The calpain-calpastatin system and
cellular proliferation and differentiation in rodent osteoblastic
cells. Exp Cell Res. 233:297–309. 1997. View Article : Google Scholar : PubMed/NCBI
|