Introduction
Nasopharyngeal carcinoma (NPC) is the most common
type of cancer originating in the nasopharynx, and has a higher
incidence in certain regions of East Asia and Africa than in other
parts of the world. NPC is caused by a combination of factors:
Viral, environmental influences and heredity (1). It has been shown that NPC is
sensitive to radiotherapy and chemotherapy, with a cure rate of
~70% (2,3). The viral influence is associated with
infection with Epstein-Barr virus (EBV) (4). EBV-encoded RNA signals are present in
all NPC cells, and early diagnosis of the disease is possible
through the detection of raised antibodies against EBV. However, a
number of genes are reported to contribute to the risk of NPC
according to studies regarding genetic linkage and association
(5,6). Therefore, separate efforts are
required to investigate the underlying molecular mechanisms of
carcinogenesis.
In recent years, a class of novel non-coding RNAs
termed microRNAs (miRNAs) have been identified in plants and
animals. MicroRNAs (miRNAs) are 18–26 nucleotides long and
post-transcriptionally regulate gene expression in multicellular
organisms by affecting the stability and translation of mRNAs. In
the process of tumor formation, the abnormal expression or the loss
of the dynamic balance between oncogenes and tumor suppressor genes
leads to tumorigenesis and cancer development. miRNAs, important
regulatory factors of gene expression, are also involved in tumor
formation and progression. Considerable evidence has demonstrated
critical functions of miRNAs in diverse biological processes, such
as proliferation (7–15), apoptosis (16–23),
angiogenesis (24–30), cell differentiation (31–33),
adhesion and metastasis (34) of
tumor cells. Therefore, downregulation of the expression of certain
miRNAs may result in the development of cancer. Previous studies
have confirmed the presence of cancer-specific miRNAs in numerous
types of cancers, such as breast cancer (35), lung cancer (36) and hepatocellular carcinoma
(37).
The incidence of NPC involves changes in the
expression of oncogenes and tumor suppressor genes. The present
study aimed to determine the effects of miR-23b on the phenotypes
of NPC cells as well as identify its target genes, in order to
investigate the molecular mechanisms underlying the involvement of
miR-23b in the initiation and progression of NPC.
Materials and methods
Patient and samples
Samples were obtained from 17 patients (3 men and 14
women) with NPC who underwent complete resection at the Huai'an
First People's Hospital from April 2010 to January 2014. NPC tissue
biopsies were obtained at the time of diagnosis prior to any
therapy, and the cancerous tissue sections were immediately frozen
at −80°C following removal from the patients. Regarding the primary
tumor stage, 5 patients had a T1–2 stage and 12 patients had a T3–4
stage according to the 2008 Chinese staging system (38). In terms of clinical stage, 7
patients were in stage I–II and 7 patients in stage III–IV
(information regarding the clinical stage of the remaining patients
was unavailable). The clinicopathological characteristics of the
patients with NPC are summarized in Table I. The present study was approved by
the ethics committee of Huai'an First People's Hospital (Huai'an,
China), and written-informed consent was obtained from all the
study participants.
 | Table IClinicopathological features of the
patients with nasopharyngeal carcinoma. |
Table I
Clinicopathological features of the
patients with nasopharyngeal carcinoma.
| Variable | No. of
patients | Percentage |
|---|
| Gender |
| Male | 14 | 82.36 |
| Female | 3 | 17.64 |
| Age |
| ≥46 | 14 | 82.36 |
| <46 | 3 | 17.64 |
| Primary tumor
stage |
| T1–2 | 5 | 29.41 |
| T3–4 | 12 | 70.59 |
| Clinical stage |
| I–II | 7 | 41,17 |
| III–IVa | 7 | 41,17 |
miRNA target prediction
TargetScan (http://www.targetscan.org/), PicTar (http://pictar.mdc-berlin.de/) and miRBase (http://www.mirbase.org/) were used to predict miRNA
targets.
Cell culture and transfection
CNE1 and CNE2z cells (1×106/ml; obtained
from the department of ENT, Huai'an First People's Hospital) were
cultured in RPMI-1640 (Gibco-BRL, Carlsbad, CA, USA) containing 10%
heat-inactivated fetal bovine serum (Gibco-BRL), 100 IU
penicillin/ml (Gibco-BRL) and 0.1 mg streptomycin/ml (Gibco-BRL) in
a humidified 5% (v/v) atmosphere of CO2 at 37°C. The
cells were transfected with Lipofectamine 2000 (Invitrogen Life
Technologies, Carlsbad, CA, USA), according to the manufacturer's
instructions. Briefly, the cells were seeded (1×106) to
90% confluence at transfection, and 4 μl of Lipofectamine
2000 were diluted in 250 μl Opti-Minimal Essential Medium
(MEM; Invitrogen Life Technologies). miR23b mimics, miR23b control,
anti-sense oligonucleotides (ASO) miR23b, and ASO control (2
μg of each; Guangzhou RiboBio Co., Ltd., Guangzhou, China)
were then diluted in 250 μl Opti-MEM, and the diluted DNA
was further diluted with Lipofectamine 2000 (1:1 ratio). The
solution was incubated for 5 min at room temperature, and the
DNA-lipid complex was added to the cells.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
To detect the relative level of transcription,
RT-qPCR was performed. Briefly, cDNA was generated through reverse
transcription using M-MLV reverse transcriptase (Promega
Corporation, Madison, WI, USA) with 2 μg large RNA extracted
from the cells. Total RNA was separated into large and small RNAs,
which were isolated using the mirVana miRNA Isolation kit
(Invitrogen Life Technologies). cDNA (1 μg) was used for the
amplification of E-cadherin and β-actin, which was used as an
endogenous control for the PCR reaction. PCR was performed under
the following conditions: 94°C for 4 min followed by 40 cycles of
94°C for 1 min, 56°C for 1 min, 72°C for 1 min, using an iQ5
Real-Time PCR detection system (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). The relative fold-change in the transcripts was
calculated with the 2−ΔΔCt method (39). The primers used were as follows:
E-cadherin forward, 5′-CAATCTCAAGCTCATGG-3′, and reverse,
5′-CCATTCGTTCAAGTAGTC-3′; β-actin forward,
5′-ATGCCAACACAGTGCTGTCTGG-3′, and reverse,
5′-TACTCCTGCTTGCTGATCCACAT-3′; miR-23b forward,
5′-CGCGGCCGCTAGTATTATGTT-3′, and reverse, 5′-CACATTTTAAAAAACATA-3′;
and U6 forward, 5′-TGCGGGTGCTCGCTTCGGCAGC-3′, and reverse,
5′-CCAGTGCAGGGTCCGAGGT-3′.
Western blotting
Cultured cells were lysed in
radioimmuno-precipitation assay buffer (containing 0.1% SDS, 1%
Triton X-100, 1 mM MgCl2 and 10 mM Tris-HCl; pH 7.4;
Invitrogen Life Technologies) at 4°C for 25 min. The lysates were
collected and cleared by centrifugation at 10,000 × g for 10 min,
and protein concentration was determined. Briefly, total cell
lysates (50 μg) were fractionated by 10% SDS-PAGE. Proteins
were electroblotted onto nitrocellulose membranes (Bio-Rad
Laboratories, Inc.). Nonspecific binding sites of membranes were
saturated with 5% skimmed milk in Tris-buffered saline with
Tween-20 solution (TBST; 100 mmol/l Tris-Cl, pH 7.5; 150 mmol/l
NaCl and 0.1% Tween-20) and incubated for 2 h with antibodies at
room temperature. The following antibodies were used: Monoclonal
mouse anti-human E-cadherin (1:100; cat. no. ab1416; Abcam,
Cambridge, UK) and monoclonal mouse anti-human GAPDH (1:1,000; cat.
no. ab8245; Abcam). After four washes with TBST, the filters were
incubated with polyclonal goat anti-mouse peroxidase-conjugated
secondary antibody (Sigma-Aldrich, Carlsbad, CA, USA) in 5% skimmed
milk in TBST solution for 1 h at room temperature. Reactions were
developed using enhanced chemiluminiscence (Perkin Elmer, Waltham,
MA, USA).
Cell proliferation assay
CNE1 cells were seeded in a 96-well plate at 8,000
cells per well the day prior to transfection. The cells were
transfected with 0.2 μg/well miR-23b, anti-miR-23b or
control vector (Gene Pharma, Shanghai, China). An MTT assay was
used to measure the number of viable, proliferating cells at 12, 24
and 48 h after transfection. The absorbance at 570 nm was measured
using a μQuant Universal Microplate spectrophotometer
(Bio-Tek Instruments Inc., Winooski, VT, USA).
Colony formation assay
After transfection, CNE1 cells were counted and
seeded in 6-well plates (in triplicate) at 50, 60 and 75 cells per
well. Fresh culture medium was provided every three days. Colonies
were counted only if they contained >50 cells, and the number of
colonies was counted from the 6th day after seeding and then the
cells were stained using crystal violet (Beyotime Institute of
Biotechnology, Shanghai, China). The rate of colony formation was
calculated with the equation: Colony formation rate = (number of
colonies/number of seeded cells) × 100.
Transwell assay
CNE1 cells (2×105) were transiently
transfected with or without anti-miR-23b. Posttransfection (48 h),
1.5×105 cells were resuspended in 300 μl serum
free medium and seeded to the transwell of uncoated polycarbonate
membranes with 8.0 μm pores (BD Biosciences, Franklin Lakes,
NJ, USA) with the bottom supplemented with 800 μl complete
medium (Gibco-BRL). After 20 h incubation, cells that had migrated
to the other side of the Transwell chamber were stained with 0.005%
crystal violet. Ten images were randomly captured using an IX71
microscope (Olympus Corporation, Tokyo, Japan) and the cells were
counted.
Enhanced green fluorescent protein (EGFP)
reporter assay
Cells were cotransfected with miR-23b mimics or
miR-23b control, together with a pcDNA3/EGFP-E-cadherin
3′-untranslated region (UTR) or a mutant UTR with a 4 base mutation
in the complementary reporter vector seed sequence (all Guangzhou
RiboBio Co., Ltd.). The pDsRed2-N1 red fluorescent protein (RFP)
expression vector (Clontech Laboratories, Inc., Mountain View, CA,
USA) was used as an internal control. A total of 48 h
post-transfection, the cells were lysed with
radioimmunoprecipitation assay lysis buffer (150 mM NaCl, 50 mM
Tris-HCl pH 8.0, 1% Triton X-100, 0.1% SDS; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) to isolate the proteins. EGFP
and RFP intensity was subsequently detected using an F-4500
fluorescence spectrophotometer (Hitachi, Ltd., Tokyo, Japan).
Statistical analysis
Data are expressed as the mean ± standard deviation.
P<0.05 was considered to indicate a statistically significant
difference. Students-Newman-Keuls test was used to determine
significance. The data were analyzed using SPSS 17.0 (SPSS Inc.,
Chicago, IL, USA).
Results
miR-23b expression level in human NPC and
the correlation with clinicopathological characteristics
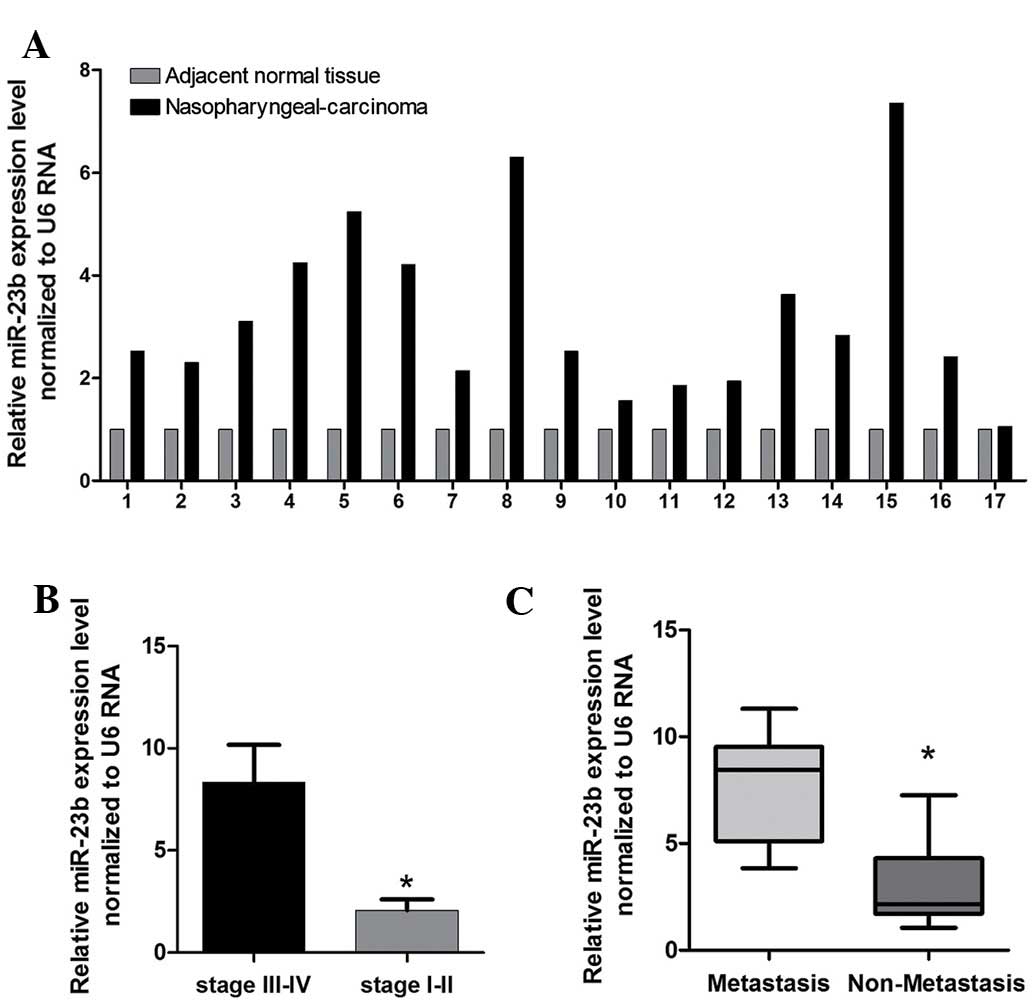
RT-qPCR was used to detect differential expression
of miR-23b in 17 paired samples of human NPC tissue and
corresponding adjacent normal tissues. The results showed that the
expression of miR-23b in NPC was significantly higher than that in
the corresponding adjacent normal tissues (Fig. 1A). Staging of NPC is based on
clinical and radiological examination. The majority of patients
present with stage III or IV disease. A higher level of expression
of miR-23b was associated with pathological tumor-node-metastasis
stage (Fig. 1B), and a higher
level of miR-23b was associated with pM stage (P<0.05,
metastasis vs. no metastssis) in patients with NPC (Fig. 1C). These data suggested that
changes in the expression of miR-23b could be involved in NPC
progression.
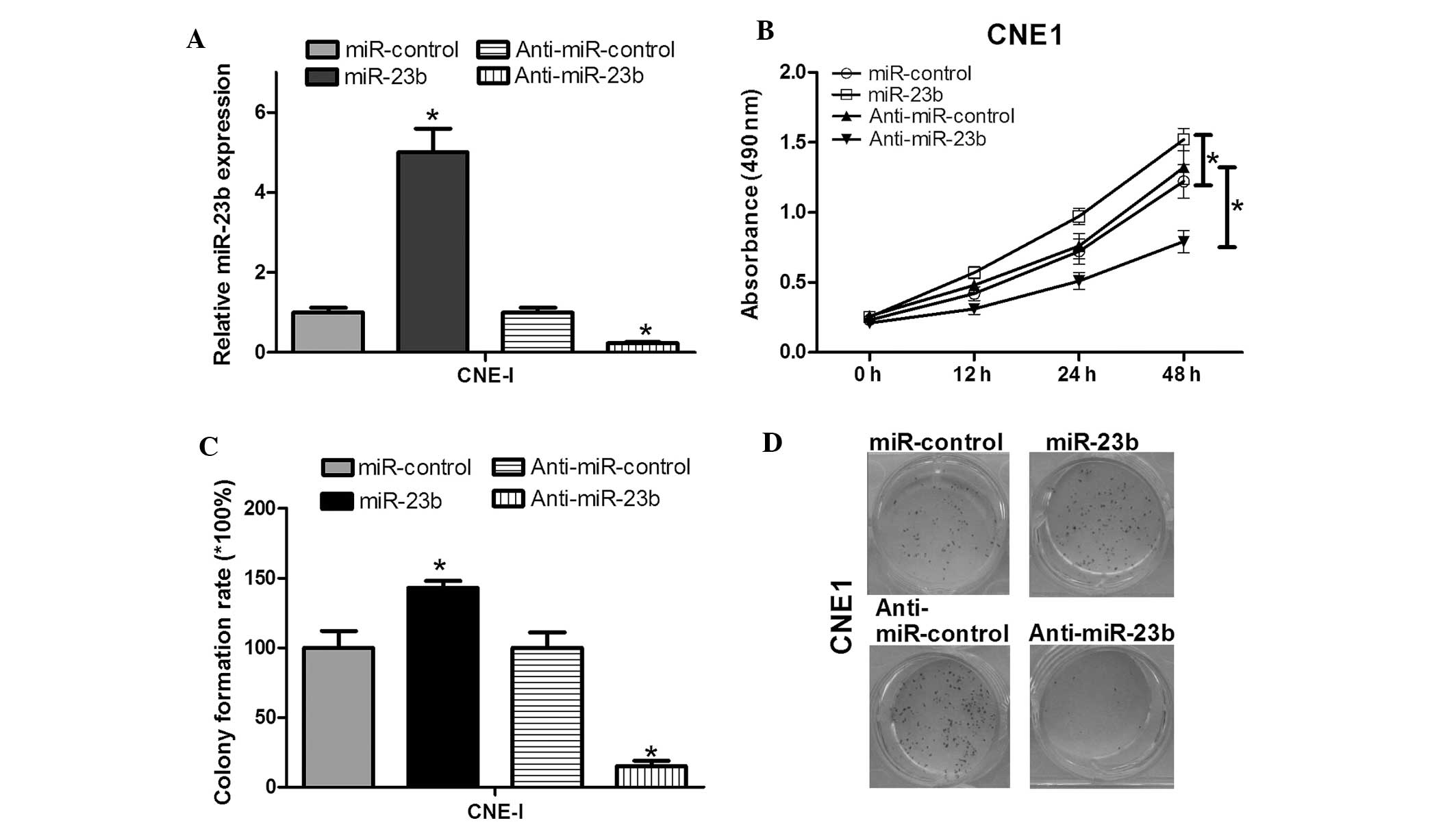
Overexpression of miR-23b enhances NPC
cell proliferation
In order to investigate the effects of miR-23b on
NPC cell proliferation, the miR-23b expression vector was
constructed. After transfection of CNE1 cells, the validity of
miR-23b and anti-miR-23b ectopic expression was determined by
RT-qPCR. The results revealed that the miR-23b expression level was
significantly higher in the miR-23b group than in the control group
(Fig. 2A). In addition,
anti-miR-23b could downregulate the miR-23b expression level. To
test the effects of miR-23b on NPC cell proliferation, cell growth
was investigated by an MTT assay and it was demonstrated that
miR-23b could enhance CNE1 cell growth (Fig. 2B). A colony formation assay was
conducted to further confirm the effects of miR-23b on cell
proliferation. The colony formation rate of CNE1 cells transfected
with anti-miR-23b was significantly lower than that of the control
group (Fig. 2C). In addition, the
colony formation rate of CNE1 cells transfected with miR-23b was
significantly higher than the control group (Fig. 2C). These two experiments showed
that miR-23b was involved in enhancing the cell growth and
proliferation of NPC cells. Downregulating miR-23b resulted in the
inhibition of cell viability and proliferation.
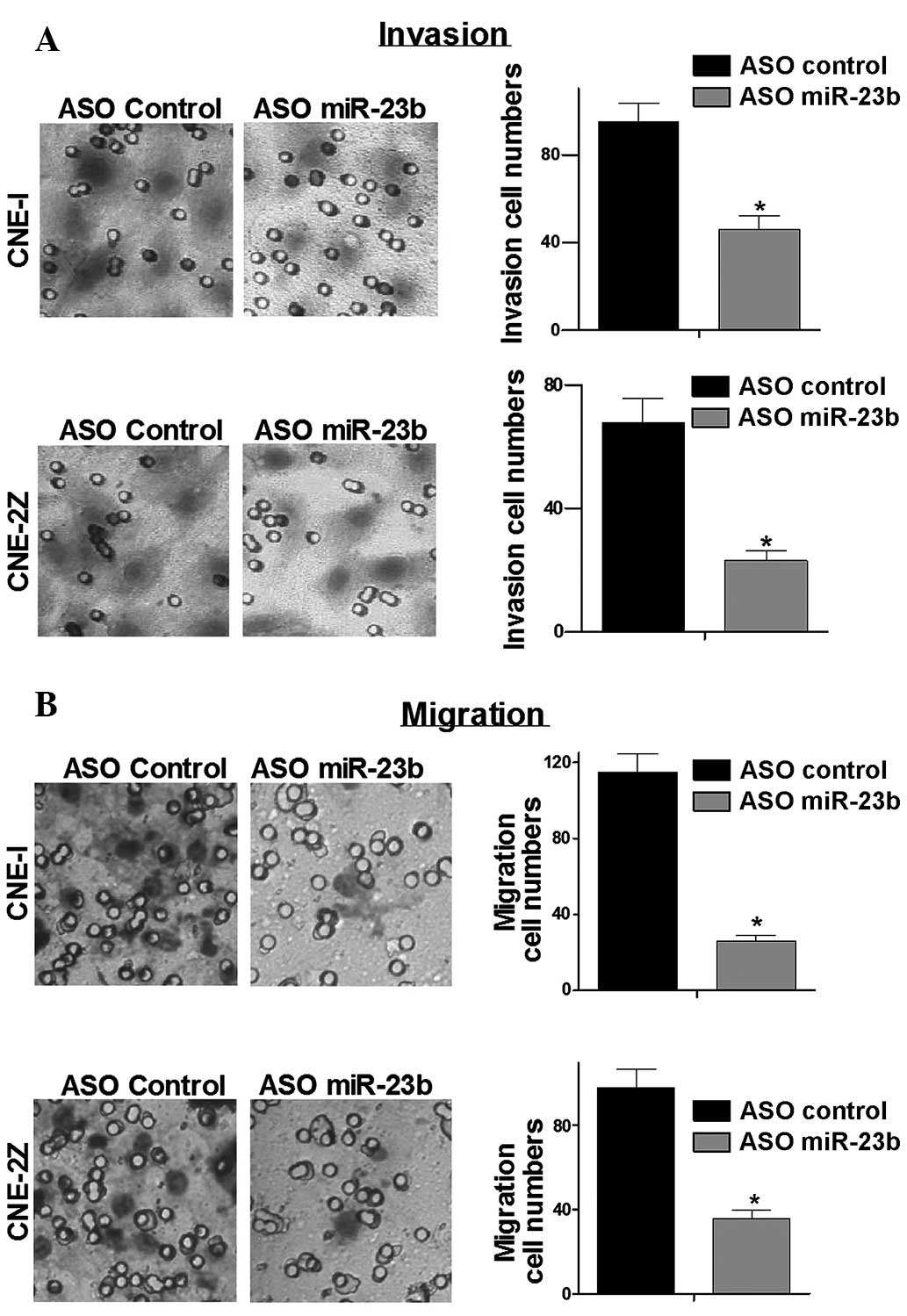
Low expression of miR-23b inhibits cell
migration and invasion of NPC cells
To test whether low expression of miR-23b affects
cancer cell migration and invasion, Transwell assays were
performed. Transwell assays demonstrated that low expression of
miR-23b significantly reduced the migration and invasion capacity
of NPC cells (Fig. 3). These data
demonstrated that low expression of miR-23b suppressed migration
and invasion of NPC cell lines.
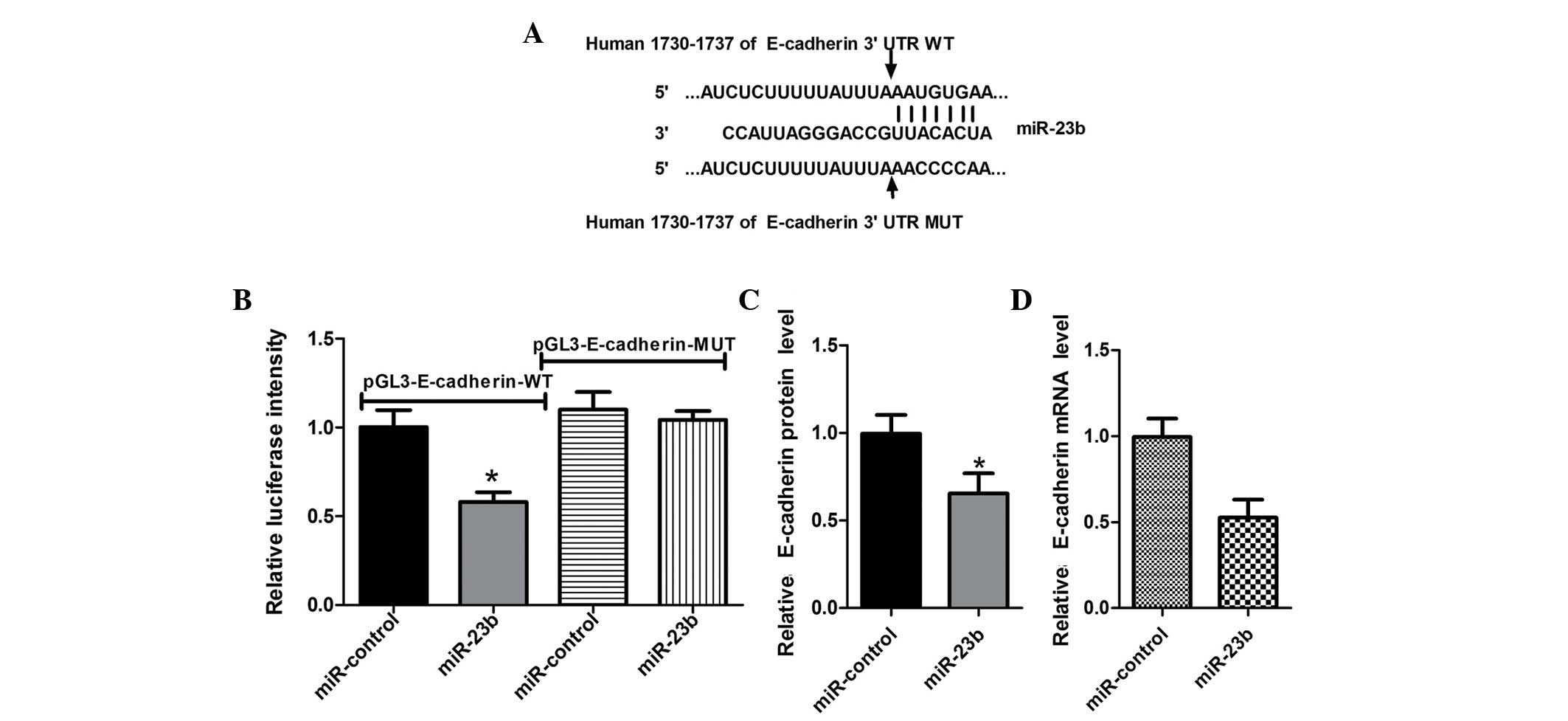
miR-23b directly targets the E-cadherin
3′-untranslated region (UTR) in NPC cells
To determine the target gene mediating the function
of miR-23b, bioinformatics methods were used to predict potential
target genes. It was identified that the 3′UTR of E-cadherin mRNA
contains miR-23b complementary binding sites (Fig. 4A). To validate that E-cadherin can
be directly targeted by miR-23b, an EGFP reporter assay was
performed using engineered EGFP reporter vectors that had either
the wild-type 3′UTR of E-cadherin or a mutant UTR with a 4-base
mutation in the complementary seed sequence (Fig. 4A). pDsRed2-NI was also
cotransfected for normalization. After CNE1 cells were
cotransfected with pGL3-E-cadherin-WT and miR-23b mimics or
miR-control, overexpression of miR-23b significantly repressed EGFP
expression, compared with the control group (Fig. 4B). By contrast, EGFP expression
levels by mutants of E-cadherin 3′UTR binding sites were not
influenced by overexpression of miR-23b (Fig. 4B), indicating that miR-23b could
bind to the specific sites of the E-cadherin mRNA 3′UTR and
negatively regulate the expression of the E-cadherin gene.
miR-23b exhibits a negative regulatory
role at the E-cadherin posttranscriptional level
miRNAs regulate target genes at the
post-transcriptional level by binding their target genes 3′UTR to
silence the gene function (40).
CNE1 cells were transfected with miR-23b in order to examine
whether miR-23b depresses endogenous E-cadherin expression through
translational repression, the expression of E-cadherin protein was
examined by western blotting. The results showed that
overexpression of miR-23b resulted in a decrease in the expression
level of E-cadherin protein (Fig.
4C), suggesting that miR-23b negatively regulates endogenous
E-cadherin protein expression through a translational repression
mechanism. Furthermore, a high expression level of miR-23b in CNE1
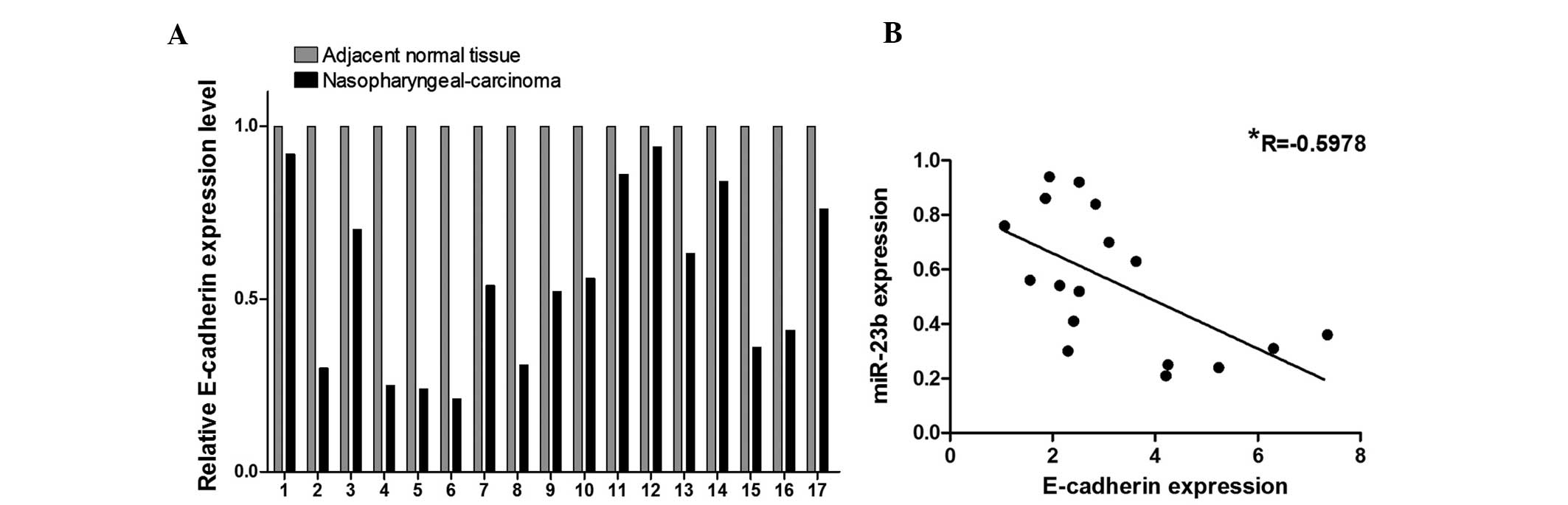
cells also decreased the endogenous E-cadherin mRNA level (Fig. 4D). In the 17 pairs of NPC tissues,
the expression level of E-cadherin in NPC tissues was identified to
be significantly lower than that in the matched adjacent normal
tissues (Fig. 5A). The expression
level of E-cadherin was negatively correlated with miR-23b
expression (Fig. 5B). These data
suggest that miR-23b negatively regulates the expression of
E-cadherin through mRNA cleavage at the post-transcriptional
level.
Discussion
Transformation of malignant tumors is regulated by
the synergy of multiple genes, including overexpression of
oncogenes and low expression or even loss of function of tumor
suppressor genes. Recent studies have demonstrated that the
regulation of oncogenes and tumor suppressor genes not only
occurred at the transcriptional level, but also at the
post-transcriptional level. miRNAs, as important regulatory
factors, are involved in the altered gene expression that occurs in
human carcinogenesis. In recent years, miRNA-mediated
post-transcriptional gene silencing and its relevance in tumor
formation have become the focus of attention in miRNA research.
Tumor cells and normal cells have significantly different in miRNA
expression profiles. The majority of miRNA genes are located in
chromosomal regions that frequently display amplification, deletion
or translocation in human cancer. In cancer, miRNA genes that
appear in high frequency are closely associated with variability of
the genome, suggesting that miRNAs may participate in cancer
development and progression. Detection of differential expression
of miRNAs in human NPC may determine the role of miRNAs in cancer
and function of their target genes, and provide a novel direction
for the diagnosis and treatment of human NPC.
The present study aimed to identify a novel miRNA
that regulates the expression of E-cadherin, and evaluate its
effects on cell phenotype using NPC cells. Initially, RT-qPCR was
conducted and demonstrated that miR-23b was significantly
upregulated in human NPC tissue, compared with the adjacent normal
tissue. The results suggested that miR-23b may be important in the
development of human NPC. Therefore, it was hypothesized that
miR-23b was a positive factor in carcinogenesis in human NPC cells
due to high expression levels of miR-23b in human NPC tissue. Cell
growth viability was determined using the MTT assay to detect the
correlation between miR-23b and the growth capacity of CNE1 NPC
cells. The cell growth viability of CNE1 cells transfected with
anti-miR-23b was significantly decreased when compared with the
control group (Fig. 2B). Moreover,
overexpression of miR-23b increased cell growth and viability when
compared with the control group (Fig.
2B). It was also demonstrated that low expression of miR-23b
significantly reduced the migration and invasion capacity of NPC
cells (Fig. 3A and B).
Secondly, bioinformatics analyses predicted an
miR-23b binding site on the E-cadherin transcript (Fig. 4A). Experimental evidence indicated
that E-cadherin was a target of miR-23b. The ability of miR-23b to
regulate E-cadherin expression was likely direct as it bound to the
3′UTR of E-cadherin mRNA complementarily to the miR-23b seed
region. The EGFP fluorescence intensity of pGL3-E-cadherin-WT was
specifically responsive to miR-23b overexpression (Fig. 4B). However, mutation of the miR-23b
binding site abolished the effect of miR-23b on the regulation of
EGFP fluorescence intensity (Fig.
4B). Conversely, endogenous E-cadherin protein expression was
decreased in CNE1 cells transfected with miR-23b, while it was
increased in CNE1 cells transfected with anti-miR-23b (Fig. 4C). In addition, it was observed
that the change in miR-23b expression effected the E-cadherin mRNA
level. These results suggested that miR-23b regulated E-cadherin
protein expression.
E-cadherin is a classical member of the cadherin
super-family. Cadherins comprise of a family of calcium-dependent
adhesion glycoproteins that mediate cell-cell binding to maintain
differentiated tissue structure and morphogenesis. Loss of
E-cadherin function or expression has been implicated in cancer
progression and metastasis (41).
Downregulation of E-cadherin decreases the strength of cellular
adhesion within a tissue, resulting in an increase in cellular
motility. This in turn may allow cancer cells to cross the basement
membrane and invade surrounding tissues.
In conclusion, it was demonstrated that miR-23b is
important in the regulation of E-cadherin gene expression. The
effect of miRNAs on NPC cell expression occurred at the mRNA and
transcriptional levels, and at least in part through targeting
E-cadherin. However, miR-23b may be capable of controlling
tumor-specific gene(s), consequently favoring cell growth and
migration. Therefore, this study suggests that targeting miR-23b
may provide a promising strategy for inhibiting tumor proliferation
and metastasis.
References
|
1
|
Zhang F and Zhang J: Clinical hereditary
characteristics in nasopharyngeal carcinoma through Ye-Liang's
family cluster. Chin Med J (Engl). 112:185–187. 1999.
|
|
2
|
Wei WI and Sham JS: Nasopharyngeal
carcinoma. Lancet. 365:2041–2054. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cao SM, Simons MJ and Qian CN: The
prevalence and prevention of nasopharyngeal carcinoma in China.
Chin J Cancer. 30:114–119. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lo KW, Chung GT and To KF: Deciphering the
molecular genetic basis of NPC through molecular, cytogenetic, and
epigenetic approaches. Semin Cancer Biol. 22:79–86. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Feng BJ, Huang W, Shugart YY, Lee MK,
Zhang F, Xia JC, Wang HY, Huang TB, Jian SW, Huang P, et al:
Genome-wide scan for familial nasopharyngeal carcinoma reveals
evidence of linkage to chromosome 4. Nat Genet. 31:395–399.
2002.PubMed/NCBI
|
|
6
|
Zhou G, Zhai Y, Cui Y, Zhang X, Dong X,
Yang H, He Y, Yao K, Zhang H, Zhi L, et al: MDM2 promoter SNP309 is
associated with risk of occurrence and advanced lymph node
metastasis of nasopharyngeal carcinoma in Chinese population. Clin
Cancer Res. 13:2627–2633. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M
and Croce CM: Human microRNA genes are frequently located at
fragile sites and genomic regions involved in cancers. Proc Natl
Acad Sci USA. 101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Takamizawa J, Konishi H, Yanagisawa K,
Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y,
et al: Reduced expression of the let-7 microRNAs in human lung
cancers in association with shortened postoperative survival.
Cancer Res. 64:3753–3756. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yanaihara N, Caplen N, Bowman E, Seike M,
Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et
al: Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Johnson SM, Grosshans H, Shingara J, Byrom
M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D and Slack
FJ: RAS is regulated by the let-7 microRNA family. Cell.
120:635–647. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee YS and Dutta A: The tumor suppressor
microRNA let-7 represses the HMGA2 oncogene. Genes Dev.
21:1025–1030. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Felli N, Fontana L, Pelosi E, Botta R,
Bonci D, Facchiano F, Liuzzi F, Lulli V, Morsilli O, Santoro S, et
al: MicroRNAs 221 and 222 inhibit normal erythropoiesis and
erythroleukemic cell growth via kit receptor down-modulation. Proc
Natl Acad Sci USA. 102:18081–18086. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Visone R, Russo L, Pallante P, De Martino
I, Ferraro A, Leone V, Borbone E, Petrocca F, Alder H, Croce CM and
Fusco A: MicroRNAs (miR)-221 and miR-222, both overexpressed in
human thyroid papillary carcinomas, regulate p27Kip1 protein levels
and cell cycle. Endocr Relat Cancer. 14:791–798. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chou CK, Chen RF, Chou FF, Chang HW, Chen
YJ, Lee YF, Yang KD, Cheng JT, Huang CC and Liu RT: MiR-146b is
highly expressed in adult papillary thyroid carcinomas with high
risk features including extrathyroidal invasion and the BRAF
(V600E) mutation. Thyroid. 20:489–494. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Voorhoeve PM, le Sage C, Schrier M, Gillis
AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A,
et al: A genetic screen implicates miRNA-372 and miRNA-373 as
oncogenes in testicular germ cell tumors. Cell. 124:1169–1181.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chan JA, Krichevsky AM and Kosik KS:
MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells.
Cancer Res. 65:6029–6033. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhu S, Si ML, Wu H and Mo YY: MicroRNA-21
targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol
Chem. 282:14328–14336. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Si ML, Zhu S, Wu H, Lu Z, Wu F and Mo YY:
MiR-21-mediated tumor growth. Oncogene. 26:2799–2803. 2007.
View Article : Google Scholar
|
|
19
|
Li J, Huang H, Sun L, Yang M, Pan C, Chen
W, Wu D, Lin Z, Zeng C, Yao Y, et al: MiR-21 indicates poor
prognosis in tongue squamous cell carcinomas as an apoptosis
inhibitor. Clin Cancer Res. 15:3998–4008. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cimmino A, Calin GA, Fabbri M, Iorio MV,
Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et
al: MiR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl
Acad Sci USA. 102:13944–13949. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun
S, Hong L, Liu J and Fan D: MiR-15b and miR-16 modulate multidrug
resistance by targeting BCL2 in human gastric cancer cells. Int J
Cancer. 123:372–379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
He L, Thomson JM, Hemann MT,
Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe
SW, Hannon GJ and Hammond SM: A microRNA polycistron as a potential
human oncogene. Nature. 435:828–833. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Matsubara H, Takeuchi T, Nishikawa E,
Yanagisawa K, Hayashita Y, Ebi H, Yamada H, Suzuki M, Nagino M,
Nimura Y, et al: Apoptosis induction by antisense oligonucleotides
against miR-17-5p and miR-20a in lung cancers overexpressing
miR-17-92. Oncogene. 26:6099–6105. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fish JE, Santoro MM, Morton SU, Yu S, Yeh
RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY and Srivastava D:
MiR-126 regulates angiogenic signaling and vascular integrity. Dev
Cell. 15:272–284. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang S, Aurora AB, Johnson BA, Qi X,
McAnally J, Hill JA, Richardson JA, Bassel-Duby R and Olson EN: The
endothelial-specific microRNA miR-126 governs vascular integrity
and angiogenesis. Dev Cell. 15:261–271. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kuhnert F, Mancuso MR, Hampton J,
Stankunas K, Asano T, Chen CZ and Kuo CJ: Attribution of vascular
phenotypes of the murine Egfl7 locus to the microRNA miR-126.
Development. 135:3989–3993. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dews M, Homayouni A, Yu D, Murphy D,
Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT and
Thomas-Tikhonenko A: Augmentation of tumor angiogenesis by a
Myc-activated microRNA cluster. Nat Genet. 38:1060–1065. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Würdinger T, Tannous BA, Saydam O, Skog J,
Grau S, Soutschek J, Weissleder R, Breakefield XO and Krichevsky
AM: MiR-296 regulates growth factor receptor overexpression in
angiogenic endothelial cells. Cancer Cell. 14:382–393. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Poliseno L, Tuccoli A, Mariani L,
Evangelista M, Citti L, Woods K, Mercatanti A, Hammond S and
Rainaldi G: MicroRNAs modulate the angiogenic properties of HUVECs.
Blood. 108:3068–3071. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kuehbacher A, Urbich C, Zeiher AM and
Dimmeler S: Role of dicer and drosha for endothelial microRNA
expression and angiogenesis. Circ Res. 101:59–68. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu N, Papagiannakopoulos T, Pan G, Thomson
JA and Kosik KS: MicroRNA-145 Regulates OCT4, SOX2 and KLF4 and
represses pluripotency in human embryonic stem cells. Cell.
137:647–658. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Di Leva G, Calin GA and Croce CM:
MicroRNAs: Fundamental facts and involvement in human diseases.
Birth Defects Res C Embryo Today. 78:180–189. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen CZ, Li L, Lodish HF and Bartel DP:
MicroRNAs modulate hematopoietic lineage differentiation. Science.
303:83–86. 2004. View Article : Google Scholar
|
|
34
|
Ma L, Young J, Prabhala H, Pan E, Mestdagh
P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S,
et al: MiR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin
and cancer metastasis. Nat Cell Biol. 12:247–256. 2010.PubMed/NCBI
|
|
35
|
Iorio MV, Ferracin M, Liu CG, Veronese A,
Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M,
et al: MicroRNA gene expression deregulation in human breast
cancer. Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yanaihara N, Caplen N, Bowman E, Seike M,
Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et
al: Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gramantieri L, Ferracin M, Fornari F,
Veronese A, Sabbioni S, Liu CG, Calin GA, Giovannini C, Ferrazzi E,
Grazi GL, et al: Cyclin G1 is a target of miR-122a, a microRNA
frequently down-regulated in human hepatocellular carcinoma. Cancer
Res. 67:6092–6099. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chinese Committee for Staging of
Nasopharyngeal Carcinoma: Report on revision of the Chinese 1992
staging system for nasopharyngeal carcinoma. J Radiat Oncol.
3:233–240. 2013.
|
|
39
|
Hou F, Wang L, Wang H, Gu J, Li M, Zhang
J, Ling X, Gao X and Luo C: Elevated gene expression of S100A12 is
correlated with the predominant clinical inflammatory factors in
patients with bacterial pneumonia. Mol Med Rep. 11:4345–4352.
2015.PubMed/NCBI
|
|
40
|
Gregory RI and Shiekhattar R: MicroRNA
biogenesis and cancer. Cancer Res. 65:3509–3512. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shirakawa T, Miyahara Y, Tanimura K,
Morita H, Kawakami F, Itoh T and Yamada H: Expression of
epithelial-mesenchymal transition-related factors in adherent
placenta. Int J Gynecol Pathol. 34:584–589. 2015. View Article : Google Scholar : PubMed/NCBI
|