Introduction
Esophageal cancer is the seventh most common type of
cancer and sixth leading cause of cancer-associated mortality
worldwide (1). There are 2 major
histological types of esophageal cancer, squamous cell carcinoma
(SCC) and adenocarcinoma. SSC is the most prevalent esophageal
cancer worldwide, particularly in developing countries (2). However, the incidence of esophageal
adenocarcinoma (EAC) has dramatically increased in the past 40
years (2). From 1975 to 2004, the
incidence of EAC among Caucasian American males increased by
>460% and over the same time period, the incidence among
Caucasian American females increased by 335% (3). Barrett's esophagus (BE) is a
meta-plastic lesion of the distal esophagus characterized by the
replacement of the normal stratified squamous epithelium by a
metaplastic, columnar-lined epithelium. Untreated BE can develop
dysplasia and progress to adenocarcinoma. Thus, BE is regarded as
the precursor to EAC. Patients with BE have a 30- to 60-fold
increased risk of developing EAC compared with the general
population (4–7).
MicroRNAs (miRNAs) are a class of small, non-coding
regulatory RNAs of ~22 nucleotides in length, that are important in
various aspects of biology (8).
Typically, miRNAs negatively regulate gene expression by binding to
the 3′-untranslated region (UTR) of target mRNAs, leading to mRNA
degradation and/or translational repression (9). Dysregulation of miRNAs is observed in
various types of human cancer, including esophageal cancer
(10,11). A large number of miRNAs have been
identified to act as oncogenes or tumor suppressor genes,
contributing to cancer development and progression (12). Compelling evidence suggests that
miRNAs may be novel molecular biomarkers for cancer detection and
targeted therapies (13).
DNA methylation is a crucial epigenetic mechanism
associated with the dysregulation of miRNAs in cancer (14). Several tumor-suppressing miRNAs
have been identified to be epigenetically silenced in esophageal
cancer. miR-375 expression is downregulated by hypermethylation of
its promoter in esophageal cancer compared with adjacent
non-tumorous tissues (15).
Additionally, miR-34a methylation is associated with its
downregulation in esophageal cancer (16). miR-193b functions as a tumor
suppressor in multiple malignancies, including melanoma (17), prostate (18), and breast (19) cancer. Downregulation of miR-193b
via DNA methylation has been previously observed in prostate cancer
(18). Despite these findings, the
importance of miR-193b expression and regulation in the
pathogenesis of esophageal cancer remains unclear.
Therefore, the present study aimed to investigate
the expression and epigenetic regulation of miR-193b in esophageal
cancer and patients with BE.
Materials and methods
Tissue samples
A total of 22 patients with esophageal cancer (16
males and 6 females) and 14 with BE (4 males and 10 females) were
enrolled in the current study. The patients were treated at the
Second Affiliated Hospital, Chongqing Medical University
(Chongqing, China). The mean age ± standard deviation of patients
with esophageal cancer and BE was 65±9 and 45±10 years,
respectively. Definitive histological diagnosis of esophageal
cancer or BE was performed for each patient. Surgical biopsies of
esophageal lesions and adjacent normal tissues (>5 cm from the
tumor margin) were obtained from all the patients with informed
consent. One part of the tissue samples was immediately snap-frozen
in liquid nitrogen and stored at −80°C prior to DNA or RNA
extraction. The remaining samples were fixed in 4% formaldehyde
(Sigma-Aldrich, St. Louis, MO, USA) overnight, embedded in
paraffin, sectioned and then stained with hematoxylin and eosin
(Sigma-Aldrich). The study was approved by the Ethics Committee of
Chongqing Medical University (approval number, CMU-2012-024).
Cell culture and demethylation
treatment
Human BE cell lines (B-T, B-T9 and B-T10),
esophageal cancer (EC) cell lines (EC109, TE-10 and SEG-1), and
normal esophageal squamous epithelial cells were obtained from the
American Type Culture Collection (Manassas, VA, USA). The cells
were cultured in RPMI 1640 medium (Invitrogen; Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS;
Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), 100
U/ml penicillin and 100 µg/ml streptomycin (Invitrogen;
Thermo Fisher Scientific, Inc.) in a humidified environment at 37°C
with a 5% CO2 atmosphere. For demethylation studies,
cells in 6-well plates (5×106 cells/well) were
serum-starved for 8 h and subsequently exposed to 5-azacytidine (10
µM; Sigma-Aldrich) in the presence of 10% FBS for a further
72 h. Cellular DNA and RNA were isolated and subjected to gene
expression and methylation analysis, as described below.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis of miR-193b
expression
Total RNA was isolated from the BC and EC cells
using the miRNA Isolation kit (Takara Biotechnology Co., Ltd.,
Dalian, China) and treaed with DNase I (Takara Biotechnology Co.,
Ltd.). The level of mature human miR-193b
(5′-AACUGGCCCUCAAAGUCCCGCU-3′) was measured using the TaqMan
MicroRNA Assay (Applied Biosystems; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) according to the manufacturer's protocol.
Briefly, RT was performed with an miRNA-specific stem-loop primer
(cat. no A25576; Applied Biosystems; Thermo Fisher Scientific,
Inc.) and the reaction was performed at 16°C for 30 min, followed
by 42°C for 30 min and 85°C for 5 min. qPCR was performed using 0.5
µg of cDNA and a TaqMan MicroRNA Assay kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.) using a 7900HT
Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The cycling conditions were as follows: 95°C for 2 min,
followed by 40 cycles of 95°C for 10 s and 60°C for 30 s. The
primer sequences were as follows: miR-193b, forward
5′-CTGACTCAGCTCGTTTGTGATG-3′ and reverse
5′-AGGTAAACTGGCCCTCAAAGT-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′
and reverse, 5′-AACGCTTCACGAATTTGCGT-3′. All reactions were
performed in triplicate. The relative miR-193b levels were
normalized to the level of U6 small nuclear RNA, calculated using
the comparative quantification cycle (ΔΔCq) method (20).
DNA methylation analysis
For determination of the methylation status of the
promoter region of miR-193b gene, genomic DNA was extracted from
the cells using the Genomic DNA Purification kit (Takara
Biotechnology Co., Ltd.) and treated with sodium bisulfite using
the EZ DNA Methylation-Gold kit (Zymo Research Corporation, Irvine,
CA, USA). Methylation-specific PCR (MSP) was performed with the Taq
Hot Start Version (Takara Biotechnology Co., Ltd.). The primers
specific for methylated and unmethylated sequences are as follows:
Methylation-specific sense primer, 5′-TTTTAGGTTTGTTTGTTGGGC-3′;
unmethylation-specific sense primer, 5′-GTTTTTAGGTTTGTTTGTTGGGT-3′;
and antisense primer, 5′-TCAAAAAATAAATCCCCATTCAC-3′. An
enzymatically methylated DNA was included as a positive control,
and unmethylated lymphocyte DNA as a negative control. PCR products
were separated on 2% agarose gel and stained with ethidium bromide
(Takara Biotechnology Co., Ltd.).
miR-193b methylation status was also measured by
bisulfite pyrosequencing, as described previously (21). In brief, genomic DNA was extracted
and subjected to bisulfite conversion. PCR amplification of the
region located ~2,000 bp upstream from the transcription site of
miR-193b was performed using the following primers: F
5′-TTTATTTAGCTGGAGATGGGGTG-3′; and R 5′-ACCACAGCCTCCAAAAGCCTC-3′.
The region analyzed by bisulfite pyrosequencing included 19 CpG
sites from the miR-193b promoter. PCR products were purified and
pyrosequencing was performed using a PyroMark Gold reagent kit
(Qiagen GmbH, Hilden, Germany) according to the manufacturer's
protocol.
Immunohistochemistry
Paraffin sections (4-µm thick) were
deparaffinized with xylene (Sigma-Aldrich), rehydrated in a graded
ethanol series and heated for 5 min at 100°C in the presence of 10
mM sodium citrate (pH 6.0; Sigma-Aldrich) to retrieve antigen.
Following blocking with normal goat serum for 30 min, sections were
incubated with a polyclonal rabbit anti-human Kirsten rat sarcoma
viral oncogene homolog (K-ras) antibody (1:1,000; ab84573; Abcam,
Cambridge, UK) overnight at 4°C. Subsequently, section were washed
with phosphate-buffered saline (pH 7.4; Thermo Fisher Scientific,
Inc.) and incubated with a biotin-labeled goat anti-mouse antibody
(ab6788; Abcam) and horseradish peroxidase-conjugated streptavidin
(Sigma-Aldrich) for 1 h at 37°C. Diaminobenzidine (Sigma-Aldrich)
was used as the peroxidase substrate to visualize the positive
staining. The nuclei were counterstained using hematoxylin
(Sigma-Aldrich). Slides were mounted and observed under a light
microscope (Leica DM750; Leica Microsystems GmbH, Wetzlar,
Germany). The primary antibody was omitted for negative controls
and human colon cancer tissue with strong K-Ras expression was used
as a positive control. The slides were examined independently by
two pathologists who were blinded to the clinical and pathological
data. Immunohistochemical results were quantified based on the
extent and intensity of staining. An index value expressed in
arbitrary units was calculated to grade the extent and intensity of
staining.
Statistical analysis
Statistical significance was evaluated by Student's
t-test or one-way analysis of variance followed by the Bonferroni
method for multiple comparisons between pairs. Comparison of
methylation rates in different tissues were performed using the
Fisher's exact test. P<0.05 was considered to indicate a
statistically significant difference.
Results
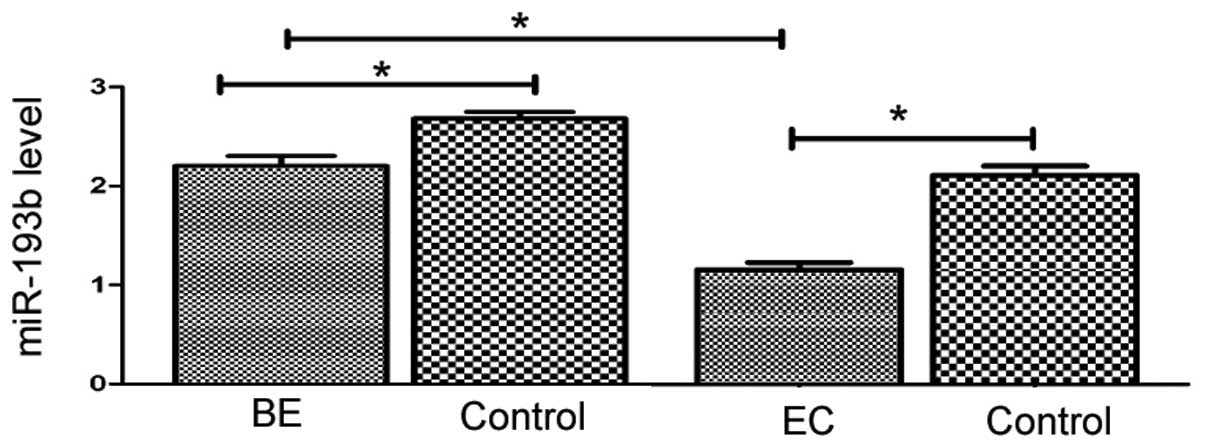
miR-193b expression is downregulated in
human BE and esophageal cancer tissues
The level of miR-193b was examined in human BE
tissues, esophageal cancer tissues, and their adjacent,
histologically normal tissues. The results demonstrated that
miR-193b expression was significantly downregulated in BE and
esophageal cancer tissues compared with the corresponding normal
tissues (P=0.002 and P=0.001, respectively; Fig. 1). Additionally, the expression
level of miR-193b in esophageal cancer tissues was significantly
reduced compared with the BE tissues (P= 0.002; Fig. 1).
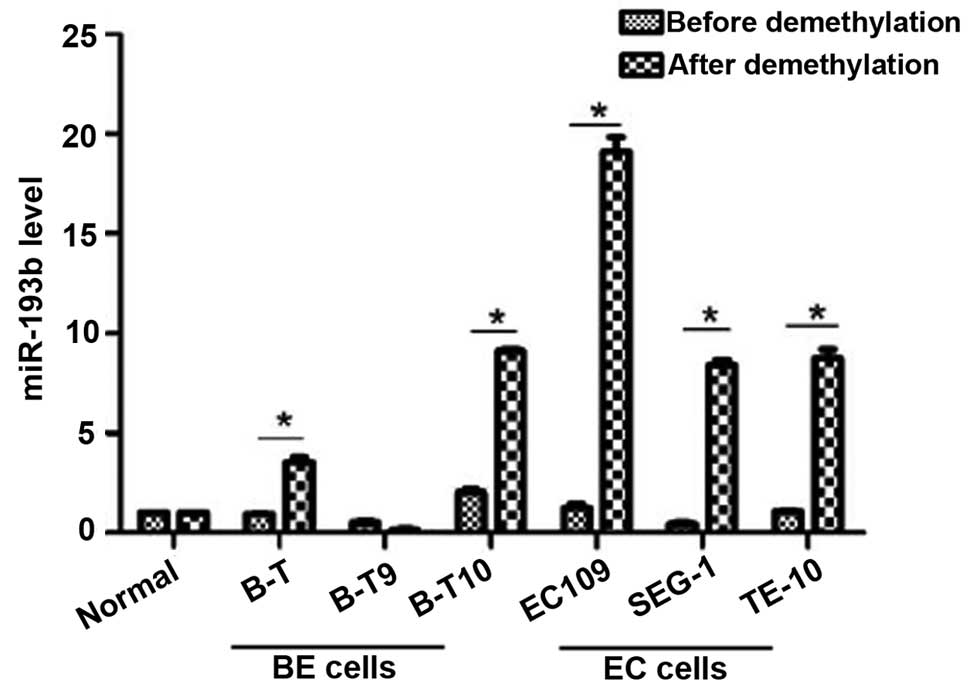
Upregulation of miR-193b by
hypomethylating agent 5-azacytidine
miR-193b expression levels in a number of BE and
esophageal cancer cell lines were analyzed by RT-qPCR following
demethylation by 5-azacytidine treatment. As demonstrated in
Fig. 2, demethylation treatment
resulted in a significant upregulation of miR-193b in Barrett's
esophagus (B-T and B-T10) and esophageal cancer (EC109, SEG-1 and
TE-10) cells, with greater increases observed in the esophageal
cancer cell lines. By contrast, the level of miR-193b in B-T9
Barrett's esophagus cells and normal esophageal squamous epithelial
cells were not observed to be significantly different following
exposure to 5-azacytidine (Fig.
2).
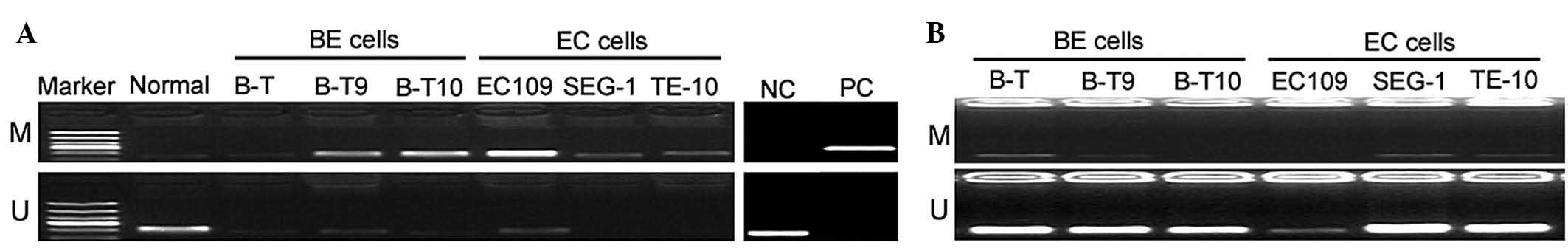
miR-193b promoter hypermethylation in
human BE and esophageal cancer cells
miR-193b methylation in human BE and esophageal
cancer cells was evaluated by MSP analysis. As presented in
Fig. 3A, miR-193b was
hypermethylated in the BE and esophageal cancer cell lines.
However, normal esophageal squamous epithelial cells exhibited low
methylation of the miR-193b promoter region. To confirm the
methylation status, cells were treated with 5-azacytidine prior to
methylation analysis. The results revealed weak or no methylation
of miR-193b in the 5-azacytidine-treated BE and esophageal cancer
cells (Fig. 3B).
miR-193b methylation in human BE and
esophageal cancer tissues
Table I presents
the methylation rates of miR-193b in human BE and esophageal cancer
tissues. miR-193b hyper-methylation was detected in 37.9±2.3% of BE
tissues and in 45.6±14.4% of esophageal cancer tissues. The
methylation rates in BE and esophageal cancer tissues were
significantly increased compared with the corresponding adjacent
normal esophageal tissues (P=0.038 and P=0.005, respectively).
 | Table IMethylation rates of microRNA-193b in
human Barrett's esophagus and esophageal cancer tissues. |
Table I
Methylation rates of microRNA-193b in
human Barrett's esophagus and esophageal cancer tissues.
| Sample | n | Rate (%) | P-value |
|---|
| Barrett's
esophagus | 14 | 37.9±2.3 | |
| Adjacent
tissuea | 14 | 28.7±2.9 | 0.038 |
| Esophageal
cancer | 22 | 45.6±14.4 | |
| Adjacent
tissueb | 22 | 17.5±2.9 | 0.005 |
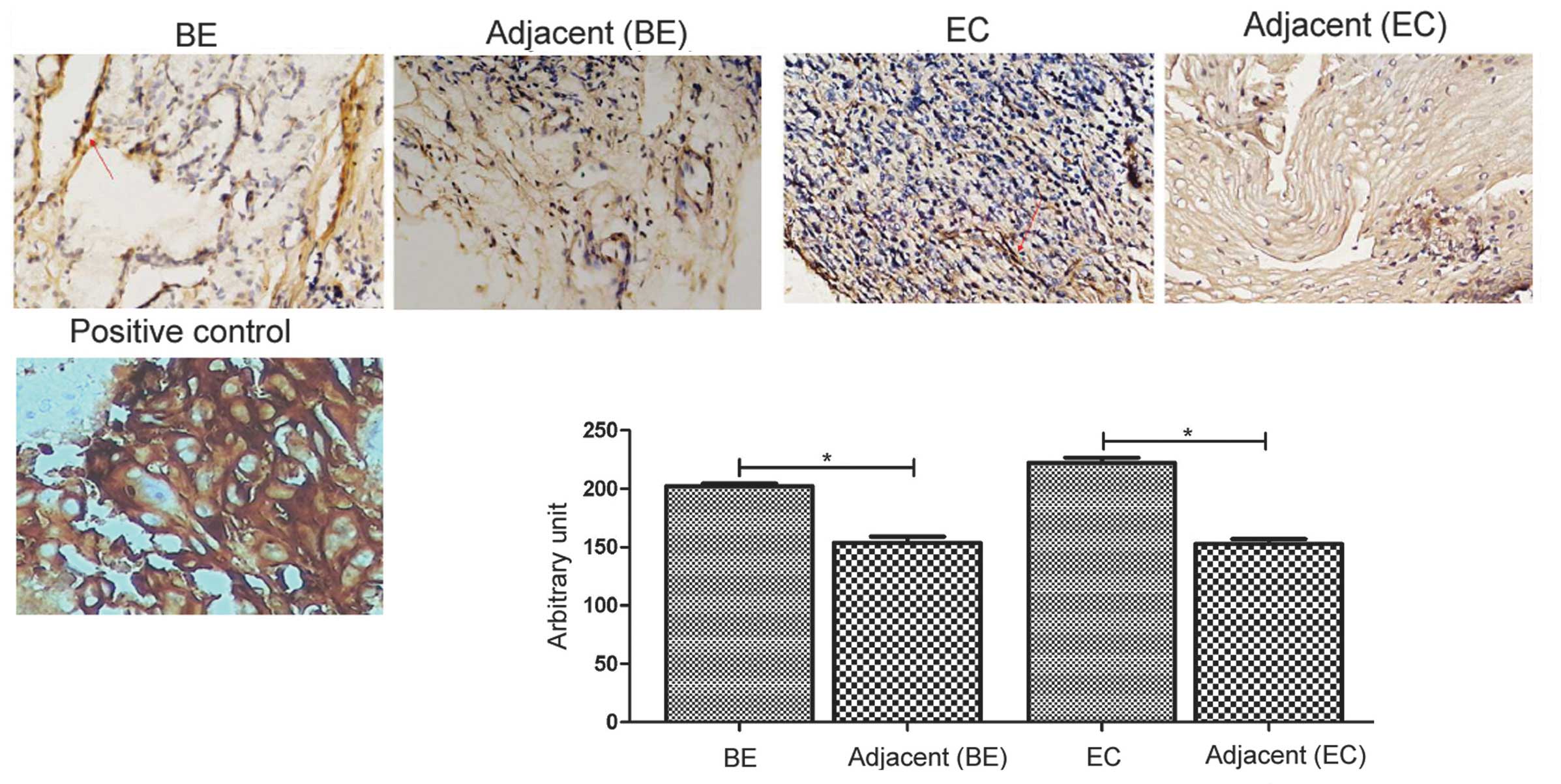
K-Ras protein expression in human BE and
esophageal cancer tissues
K-Ras was previously identified as a direct target
of miR-193b in epidermal squamous cell carcinoma (22). The current study investigated the
expression of K-Ras protein in esophageal cancer.
Immunohistochemistry demonstrated that human BE and esophageal
cancer tissues exhibited stronger K-Ras protein levels than the
corresponding adjacent normal tissues (P=0.005 and P=0.001,
respectively; Fig. 4).
Discussion
Dysregulation of miRNAs is an important mechanism of
tumorigenesis (10), and various
miRNAs have been identified to be aberrantly expressed in
esophageal cancer (23). The data
of the present study demonstrated that miR-193b was weakly
expressed in esophageal cancer tissues compared with adjacent
normal tissues. Additionally, the expression level of miR-193b in
esophageal cancer was significantly decreased compared with the
level in BE tissues. In agreement with the findings of the current
study, previous investigations have reported the downregulation of
miR-193b in several other malignancies (19,24,25).
For example, Gao et al (24) reported that miR-193b acts as a
tumor suppressor gene, and is down-regulated in acute myeloid
leukemia (AML). Using miRNA microarray technology, Wu et al
(25) profiled the miRNA
expression in endometrioid adenocarcinoma and observed that
miR-193b was downregulated in adenocarcinoma tissues compared with
adjacent non-tumorous endometrium. BE is a premalignant lesion that
predisposes patients to EAC (4).
The downregulation of miR-193b may contribute to the progression
from BE to EAC.
Epigenetic modification of promoter regions,
particularly DNA methylation, is frequently associated with the
downregulation of genes (26).
Epigenetic regulation of miRNA expression commonly occurs in cancer
(14). Rauhala et al
(18) reported that miR-193b DNA
is methylated in prostate cancer cells at a CpG island ~1 kb
upstream of the miRNA locus. Methylation-mediated silencing of
miR-193b has also been described in dedifferentiated liposarcoma
cells (27).
The current study demonstrated that miR-193b was
hypermethylated in esophageal cancer tissues and cells at its
promoter region. Aberrant methylation of the miR-193b promoter
occurred less frequently in BE tissues. By contrast, normal
esophageal squamous epithelial cells exhibited low methylation of
miR-193b compared with the BE and cancer cells. To confirm the
methylation status of miR-193b, the present study used the
hypomethylating agent 5-azacytidine to treat esophageal cancer and
BE cells, and examined the expression levels of miR-193b. Treatment
with 5-azacytidine resulted in a significant increase of miR-193b
expression compared with untreated cells. To the best of our
knowledge, these results provide the first evidence for DNA
methylation-mediated downregulation of miR-193b in BE and
esophageal cancer tissues.
The reduced expression levels of miR-193b in BE and
esophageal cancer tissues suggest that it negatively regulates the
pathogenesis of esophageal cancer. Indeed, miR-193b has been
previously demonstrated to act as a tumor suppressor gene in
several types of human cancer, including melanoma (17), AML (24), pancreatic cancer (28) and hepatocellular carcinoma
(29). However, miR-193b has been
demonstrated to enhance tumor progression in head and neck squamous
cell carcinoma (HNSCC) cells (30). Overexpression of miR-193b occurs in
HNSCC cells, and knockdown of its expression has been demonstrated
to result in reduced cell proliferation, migration, invasion and
tumorigenesis (30). Table II summarizes the biological roles
of miR-193b in human cancer. Additional direct evidence is required
to confirm the exact biological functions of miR-193b in esophageal
cancer.
 | Table IIBiological roles of microRNA-193b in
human cancer. |
Table II
Biological roles of microRNA-193b in
human cancer.
| Author, year | Type of cancer | MicroRNA-193b
function | Target | Refs. |
|---|
| Chen et al,
2010; Kaukoniemi et al, 2015 | Prostate cancer,
melanoma | Growth
suppression | cyclin D1 | (17,31) |
| Li et al,
2009; Mitra et al, 2015 | Breast cancer,
ovarian cancer | Inhibition of cell
invasion | uPA | (19,32) |
| Xu et al,
2010 | Hepatocellular
carcinoma | Inhibition of
proliferation, migration and invasion | cyclin D1,
ETS1 | (29) |
| Mets et al,
2015 | T-ALL | Tumor
suppression | MYB | (33) |
| Li et al,
2014; Jin et al, 2015 | Pancreatic
cancer | Inhibition of
growth and metastasis | STMN1, uPA,
K-Ras | (28,34) |
| Lenarduzzi et
al, 2013 | HNSCC | Acceleration of
tumor progression | Neurofibromin
1 | (30) |
| Zhong et al,
2014 | Glioma | Promotion of cell
proliferation | Smad3 | (35) |
A single miRNA can modulate hundreds of target genes
(36), and several targets of
miR-193b have been identified. Zhong et al (35) reported that miR-193b is capable of
accelerating human glioma cell proliferation via targeting Smad
family member 3. Neurofibromin 1 (NF1) is also a target of
miR-193b, and downregulation of NF1 promotes the HNSCC
aggressiveness induced by miR-193b (30). Gastaldi et al (22) identified K-Ras as a direct target
of miR-193b. Activation of the K-Ras oncogene has been implicated
in tumorigenesis (37). The data
of the present study demonstrated that human BE and esophageal
cancer tissues exhibited increased expression of K-Ras protein
compared with adjacent normal tissues. The upregulation of K-Ras in
BE and esophageal cancer tissues may be the result of the
epigenetic silencing of miR-193b.
A number of limitations of the present study should
be noted. The importance of miR-193b in the development and
progression of EC remain unclear. Additionally, it remains to be
determined whether the action of miR-193b in esophageal cancer is
mediated by targeting K-Ras. No information is available on the
action of miR-193b in xenograft in vivo models.
In conclusion, to the best of our knowledge, this is
the first report investigating epigenetic silencing of miR-193b via
DNA methylation in esophageal cancer and BE tissues. Downregulation
of miR-193b and upregulation of K-Ras may contribute to the
pathogenesis of esophageal cancer. The clinical and biological
relevance of miR-193b is unclear and requires further
exploration.
Acknowledgments
The current work was supported by the National
Natural Science Foundation of China (no. 81101827) and the Key
Program of Bureau of Health of Chongqing of China (no.
20111055).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Napier KJ, Scheerer M and Misra S:
Esophageal cancer: A review of epidemiology, pathogenesis, staging
workup and treatment modalities. World J Gastrointest Oncol.
6:112–120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brown LM, Devesa SS and Chow WH: Incidence
of adenocarcinoma of the esophagus among white americans by sex,
stage, and age. J Natl Cancer Inst. 100:1184–1187. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shaheen NJ and Richter JE: Barrett's
oesophagus. Lancet. 373:850–861. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hvid-Jensen F, Pedersen L, Drewes AM,
Sørensen HT and Funch-Jensen P: Incidence of adenocarcinoma among
patients with Barrett's esophagus. N Engl J Med. 365:1375–1383.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cameron AJ and Lomboy CT: Barrett's
esophagus: Age, prevalence, and extent of columnar epithelium.
Gastroenterology. 103:1241–1245. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wild CP and Hardie LJ: Reflux, Barrett's
oesophagus and adenocarcinoma: Burning questions. Nat Rev Cancer.
3:676–684. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ke XS, Liu CM, Liu DP and Liang CC:
MicroRNAs: Key participants in gene regulatory networks. Curr Opin
Chem Biol. 7:516–523. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nilsen TW: Mechanisms of microRNA-mediated
gene regulation in animal cells. Trends Genet. 23:243–249. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Melo SA and Esteller M: Dysregulation of
microRNAs in cancer: Playing with fire. FEBS Lett. 585:2087–2099.
2011. View Article : Google Scholar
|
|
11
|
Fassan M, Baffa R, Kiss A, Zaninotto G and
Rugge M: MicroRNA dysregulation in esophageal neoplasia: The
biological rationale for novel therapeutic options. Curr Pharm Des.
19:1236–1241. 2013.
|
|
12
|
Zhang B, Pan X, Cobb GP and Anderson TA:
MicroRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar
|
|
13
|
Hayes J, Peruzzi PP and Lawler S:
MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol
Med. 20:460–469. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Suzuki H, Maruyama R, Yamamoto E and Kai
M: DNA methylation and microRNA dysregulation in cancer. Mol Oncol.
6:567–578. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li X, Lin R and Li J: Epigenetic silencing
of microRNA-375 regulates PDK1 expression in esophageal cancer. Dig
Dis Sci. 56:2849–2856. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cui X, Zhao Z, Liu D, Guo T, Li S, Hu J,
Liu C, Yang L, Cao Y, Jiang J, et al: Inactivation of miR-34a by
aberrant CpG methylation in Kazakh patients with esophageal
carcinoma. J Exp Clin Cancer Res. 33:202014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen J, Feilotter HE, Paré GC, Zhang X,
Pemberton JG, Garady C, Lai D, Yang X and Tron VA: MicroRNA-193b
represses cell proliferation and regulates cyclin D1 in melanoma.
Am J Pathol. 176:2520–2529. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rauhala HE, Jalava SE, Isotalo J, Bracken
H, Lehmusvaara S, Tammela TL, Oja H and Visakorpi T: miR-193b is an
epigenetically regulated putative tumor suppressor in prostate
cancer. Int J Cancer. 127:1363–1372. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li XF, Yan PJ and Shao ZM: Downregulation
of miR-193b contributes to enhance urokinase-type plasminogen
activator (uPA) expression and tumor progression and invasion in
human breast cancer. Oncogene. 28:3937–3934. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−ΔΔC(t)) method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
21
|
Huang YW, Kuo CT, Chen JH, Goodfellow PJ,
Huang TH, Rader JS and Uyar DS: Hypermethylation of miR-203 in
endometrial carcinomas. Gynecol Oncol. 133:340–345. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gastaldi C, Bertero T, Xu N,
Bourget-Ponzio I, Lebrigand K, Fourre S, Popa A, Cardot-Leccia N,
Meneguzzi G, Sonkoly E, et al: miR-193b/365a cluster controls
progression of epidermal squamous cell carcinoma. Carcinogenesis.
35:1110–1120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zang W, Wang Y, Du Y, Xuan X, Wang T, Li
M, Ma Y, Li P, Chen X, Dong Z and Zhao G: Differential expression
profiling of microRNAs and their potential involvement in
esophageal squamous cell carcinoma. Tumour Biol. 35:3295–3304.
2014. View Article : Google Scholar
|
|
24
|
Gao XN, Lin J, Gao L, Li YH, Wang LL and
Yu L: MicroRNA-193b regulates c-Kit proto-oncogene and represses
cell proliferation in acute myeloid leukemia. Leuk Res.
35:1226–1232. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu W, Lin Z, Zhuang Z and Liang X:
Expression profile of mammalian microRNAs in endometrioid
adenocarcinoma. Eur J Cancer Prev. 18:50–55. 2009. View Article : Google Scholar
|
|
26
|
Issa JP: CpG island methylator phenotype
in cancer. Nat Rev Cancer. 4:988–993. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Taylor BS, DeCarolis PL, Angeles CV,
Brenet F, Schultz N, Antonescu CR, Scandura JM, Sander C, Viale AJ,
Socci ND and Singer S: Frequent alterations and epigenetic
silencing of differentiation pathway genes in structurally
rearranged liposarcomas. Cancer Discov. 1:587–597. 2011. View Article : Google Scholar
|
|
28
|
Li J, Kong F, Wu K, Song K, He J and Sun
W: miR-193b directly targets STMN1 and uPA genes and suppresses
tumor growth and metastasis in pancreatic cancer. Mol Med Rep.
10:2613–2620. 2014.PubMed/NCBI
|
|
29
|
Xu C, Liu S, Fu H, Li S, Tie Y, Zhu J,
Xing R, Jin Y, Sun Z and Zheng X: MicroRNA-193b regulates
proliferation, migration and invasion in human hepatocellular
carcinoma cells. Eur J Cancer. 46:2828–2836. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lenarduzzi M, Hui AB, Alajez NM, Shi W,
Williams J, Yue S, O'Sullivan B and Liu FF: MicroRNA-193b enhances
tumor progression via down regulation of neurofibromin 1. PLoS One.
8:e537652013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kaukoniemi KM, Rauhala HE, Scaravilli M,
Latonen L, Annala M, Vessella RL, Nykter M, Tammela TL and
Visakorpi T: Epigenetically altered miR-193b targets cyclin D1 in
prostate cancer. Cancer Med. 4:1417–1425. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mitra AK, Chiang CY, Tiwari P, Tomar S,
Watters KM, Peter ME and Lengyel E: Microenvironment-induced
downregulation of miR-193b drives ovarian cancer metastasis.
Oncogene. 34:5923–5932. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mets E, Van der Meulen J, Van Peer G,
Boice M, Mestdagh P, Van de Walle I, Lammens T, Goossens S, De
Moerloose B, Benoit Y, et al: MicroRNA-193b-3p acts as a tumor
suppressor by targeting the MYB oncogene in T-cell acute
lymphoblastic leukemia. Leukemia. 29:798–806. 2015. View Article : Google Scholar
|
|
34
|
Jin X, Sun Y, Yang H, Li J, Yu S, Chang X,
Lu Z and Chen J: Deregulation of the MiR-193b-KRAS Axis Contributes
to Impaired Cell Growth in Pancreatic Cancer. PLoS One.
10:e01255152015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhong Q, Wang T, Lu P, Zhang R, Zou J and
Yuan S: miR-193b promotes cell proliferation by targeting Smad3 in
human glioma. J Neurosci Res. 92:619–626. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Satoh J: Molecular network of microRNA
targets in Alzheimer's disease brains. Exp Neurol. 235:436–446.
2012. View Article : Google Scholar
|
|
37
|
Brink M, de Goeij AF, Weijenberg MP,
Roemen GM, Lentjes MH, Pachen MM, Smits KM, de Bruïne AP, Goldbohm
RA and van den Brandt PA: K-ras oncogene mutations in sporadic
colorectal cancer in The Netherlands Cohort Study. Carcinogenesis.
24:703–710. 2003. View Article : Google Scholar : PubMed/NCBI
|