Introduction
Glioblastoma is a highly malignant brain tumor,
characterized by poor prognosis and high recurrence rates. Despite
therapeutic strategies, including surgery, radiotherapy and
chemotherapy, the prognosis remains poor and the median survival of
patients is only 14.6 months (1,2).
Thus, developments of efficient therapeutic strategies and novel
therapeutic targets are urgently required.
MicroRNAs (miRNAs) are a group of endogenous
non-coding RNAs (length, 22 nt) that target mRNAs for cleavage or
translational repression. MiRNAs induce target gene silencing via
completely or partially complementing with the 3′-untranslated
region (3′-UTR) of specific mRNAs and this has been demonstrated to
be involved in various cellular processes (3–6).
Considered to be important gene regulators, miRNAs are emerging as
novel biomarkers for cancer. Previous studies have associated the
dysregulation of miRNAs with the initiation and development of
glioma, and a number of miRNAs are closely associated with glioma
staging and patient survival, and so have been suggested to be
novel diagnostic or prognostic markers (7–9).
Improved understanding of miRNA-mediated effects in glioma cells
may aid the future development of novel strategies in glioma
diagnosis and treatment.
MicroRNA-34a (miR-34a) is a highly conserved miRNA
observed in various species. In humans, there are three homologs,
miR34a, miR-34b and miR-34c. MiR-34a has been demonstrated to exert
a tumor suppressive effect in breast (10) and lung cancer, involving cell cycle
arrest, senescence and apoptosis (11,12).
A previous study by Gao et al (13) has demonstrated that miR-34a was
downregulated in glioma samples compared with normal brain tissue
samples, and that it is also associated with glioma grade and
prognosis. However, the possible roles and underlying mechanisms of
miR-34a in human glioma cells remains to be elucidated. The present
study aimed to demonstrate that miR-34a inhibits cell proliferation
and induces cell apoptosis of U87 glioma cells by targeting B-cell
lymphoma 2 (Bcl-2).
Materials and methods
Acquisition of tissue specimens
The protocol of the present study and acquisition of
tissue specimens was approved by the Biomedical Research Ethics
Committee of The Affiliated Hospital of North Sichuan Medical
College (Nanchong, China). A total of 20 glioma tissue specimens
were obtained from patients who received surgical treatment at the
Department of Neurosurgery at The Affiliated Hospital of North
Sichuan Medical College between June 2011 and December 2013.
Cells
U87 cells were obtained from the Institute of
Biochemistry and Cell Biology of the Chinese Academy of Sciences
(Shanghai, China) and cultured in Dulbecco's modified Eagle's
medium (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10%
fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc., Waltham,
MA, USA), 100 U/ml penicillin G and 100 µg/ml streptomycin
(Sigma-Aldrich). Cells were incubated in a humidified atmosphere
with 5% CO2 at 37°C.
Cell proliferation assay
To investigate cell proliferation, U87 cells were
diluted into single cell suspensions and seeded in 96-well plates
(1×104 cells/well). Following transfection with miR-34a
mimics or scramble mimics (miR-Ctrl) (Guangzhou RiboBio Co., Ltd.,
Guangzhou, China), 10 µl CCK-8 was added to each well at 24,
48 and 72 h, avoiding the production of air bubbles. The plates
were incubated at 37°C for 1–4 h and the absorbance was measured at
450 nm using an Epoch microplate spectrophotometer (BioTek
Instruments, Inc., Winooski, VT, USA). Experiments were
independently repeated three times.
Cell apoptosis analysis
Apoptosis was measured using the FITC Annexin V
Apoptosis Detection kit (BD Biosciences, Franklin Lakes, NJ, USA)
according to the manufacturer's protocol. U87 cells transfected
with miR-34a mimics or miR-Ctrl were washed and resuspended in 1X
binding buffer at a concentration of 1×106 cells/ml.
Cells (1×105) were incubated with 5 µl of Annexin
V-fluorescein isothiocyanate (FITC) and 5 µl of propidium
iodide (PI) for 15 min at room temperature in the dark. Following
staining, cells were diluted with 400 µl binding buffer and
directly detected by BD FACSCalibur™ (BD Biosciences) within 1 h.
The results were analyzed using FlowJo 7.6 software (Tree Star,
Inc. Ashland, OR, USA). All the cells positively stained for
Annexin V-FITC were considered to be apoptotic cells and
experiments were independently repeated three times.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
For quantitative detection of miR-34a and Bcl-2,
total RNA was extracted from cells or tissues using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocols. RNA samples were reverse transcribed to
cDNA using a PrimeScript RT Reagent Kit with gDNA Eraser (Takara
Biotechnology Co., Ltd., Dalian, China) with miR-34a specific
primers (5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACAACCA-3′;
Guangzhou RiboBio Co., Ltd.). The DNase reaction was performed by
incubation with gDNA Eraser at 42°C for 2 min, then RT was
performed at 37°C for 15 min and 85°C for 10 min. qPCR was
performed using SYBR Premix Ex Taq™ (Takara Biotechnology Co.,
Ltd.) in an ABI 7500 system (Applied Biosystems; Thermo Fisher
Scientific, Inc.). Primers used were as follows: Sense,
5′-GCGGCGGACCGTCACAGAATC-3′ for miR-34a; sense,
5′-GCATGGGTCCCCCGACGTTG-3′ and antisense,
5′-GCTCCGGCCAGAGGCCTCAA-3′ for Bcl-2; and sense,
5′-CGTGAAAAGACCCAGATCA-3′ and antisense, 5′-CACAGCCTGGATGGCTACGT-3′
for β-actin. The primers were obtained from Sangon Biotech Co.,
Ltd. (Shanghai, China). The mRNA expression of Bcl-2 was normalized
to β-actin mRNA and miR-34a was normalized to U6. The relative
expression of miR-34a and Bcl-2 was quantified with the
2−ΔΔCq method (14) and
experiments were independently repeated three times.
Caspase activity assays
The activities of caspase-3, -8 and -9 in U87 cells
were measured using caspase-3, -8 and -9 colorimetric assay kits
(R&D Systems, Inc., Minneapolis, MN, USA) following the
manufacturer's protocols. Briefly, U87 cells (1×104
cells/well) were seeded in 96-well plates and incubated for 24 h at
37°C. Following transfection with miR-34a mimics or scramble mimics
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), cells were washed with ice-cold
phosphate-buffered saline (PBS), and the activities of caspase-3,
-8 and -9 were determined using the kits according to the
manufacturer's protocols. The colorimetric substrates
DEVD-p-nitroaniline (pNA) for caspase-3, IETD-pNA for caspase-8 or
Ac-LEHD-pNA for caspase-9 were added to the cell lysates and
incubated for 1 h at 37°C. The chromophore pNA was released as a
result of cleavage of the substrates by caspase activity and
quantified spectrophotometrically at a wavelength of 405 nm using
the Epoch microplate spectrophotometer. Experiments were
independently repeated three times.
Plasmids construction and luciferase
assays
The Bcl-2 3′-UTR containing the miR-34a binding site
was cloned into a modified pGL3 vector (Promega Corporation,
Madison, WI, USA) immediately downstream of the luciferase gene.
Mutations in the 3′-UTR of the Bcl-2 gene with miR-34a target sites
deleted were generated with the QuikChange Site-Directed
Mutagenesis kit (Agilent Technologies, Inc., Santa Clara, CA<
USA). Approximately 1×105 U87 cells/well were seeded
into 24-well plates for 24 h prior to transfection. Cells were
co-transfected with 50 ng pGL3 firefly luciferase reporter, 10 ng
pRL-TK luciferase reporter and 50 nM miR-34a mimics or scramble
mimics using Lipofectamine® 2000. Cell lysates were
prepared using Passive Lysis Buffer (Promega Corporation) 48 h
after transfection, and luciferase activity was measured using the
Dual-Luciferase® Reporter assay system (Promega
Corporation). Dual-Glo® Luciferase assay reagent was
added to the plate (75 µl/well). Following incubation at
20-25°C for 15 min, firefly luciferase luminescence was measured by
an automatic multi-mode microplate reader (Infinite 200 PRO, Tecan
Group, Ltd., Männedorf, Switzerland). Subsequently,
Dual-Glo® Stop&Glo® reagent was added to
the plate (75µl/well) and incubated at 20-25°C for 15 min.
Renilla luminescence in the same wells was detected using the Epoch
microplate spectrophotometer. The ratio of firefly/Renilla
luminescence was calculated for each well. The sample well ratio
was normalized to the ratio from a control wells. The full length
Bcl-2 gene open reading frame (ORF) was amplified using PCR.
Following purification, the inserts and vector were digested by
EcoRI and XhoI (New England BioLabs, Inc., Ipswich,
MA, USA) at 37°C for 2 h, followed by electrophoresis and
extraction. Ligation and transformation was used to clone the Bcl-2
ORF into pCDNA3.1 (Invitrogen; Thermo Fisher Scientific, Inc.) to
generate the pCDNA3.1-Bcl-2 construct. Successful cloning was
confirmed by DNA sequencing.
Western blot analysis
For analysis by western blotting, cells were
harvested in ice-cold PBS 48 h after transfection. The cells were
lysed on ice in cold lysis buffer (Cell Signaling Technology, Inc.,
Danvers, MA, USA) supplemented with protease inhibitors
(Sigma-Aldrich). Protein concentrations were detected with the
Bradford assay (Bio-Rad Laboratories, Inc., Hercules, CA, USA) and
equal quantities of protein (50 µg) were analyzed by 10%
sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Gels
were electroblotted onto polyvinylidene difluoride membranes (EMD
Millipore, Billerica, MA, USA). Following blocking with 5% nonfat
dry milk at room temperature for 2 h, membranes were incubated with
polyclonal rabbit anti-Bcl-2 (cat. no. 2872) or monoclonal rabbit
anti-GAPDH (cat. no. 2118) antibodies (dilution, 1:1,000; Cell
Signaling Technology, Inc.) at 4°C overnight. The membranes were
then incubated with horseradish peroxidase-conjugated goat
anti-rabbit secondary antibodies (dilution, 1:1,000; cat. no. 7074;
Cell Signaling Technology, Inc.) and the protein was visualized
using SuperSignal West Femto Substrate enhanced chemiluminescence
kit (Pierce; Thermo Fisher Scientific, Inc.). A Bio-Rad ChemiDoc
imaging system (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was
used to visualize the proteins. GAPDH was used as a reference gene
to normalize protein expression levels. Experiments were
independently repeated three times.
Statistical analysis
All data were expressed as the mean ± the standard
error of the mean from at least three independent experiments.
Pearson's correlation coefficient was used to analyze the
correlation between groups. Data of experiments with more than two
groups were analyzed using analysis of variance and Tukey's test
Results were analyzed using SPSS 16.0 software (SPSS, Inc.,
Chicago, IL, USA) and GraphPad Prism 5.0 (GraphPad Software, Inc.,
La Jolla, CA, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
Transfection of miR-34a inhibited the
proliferation of U87 cells
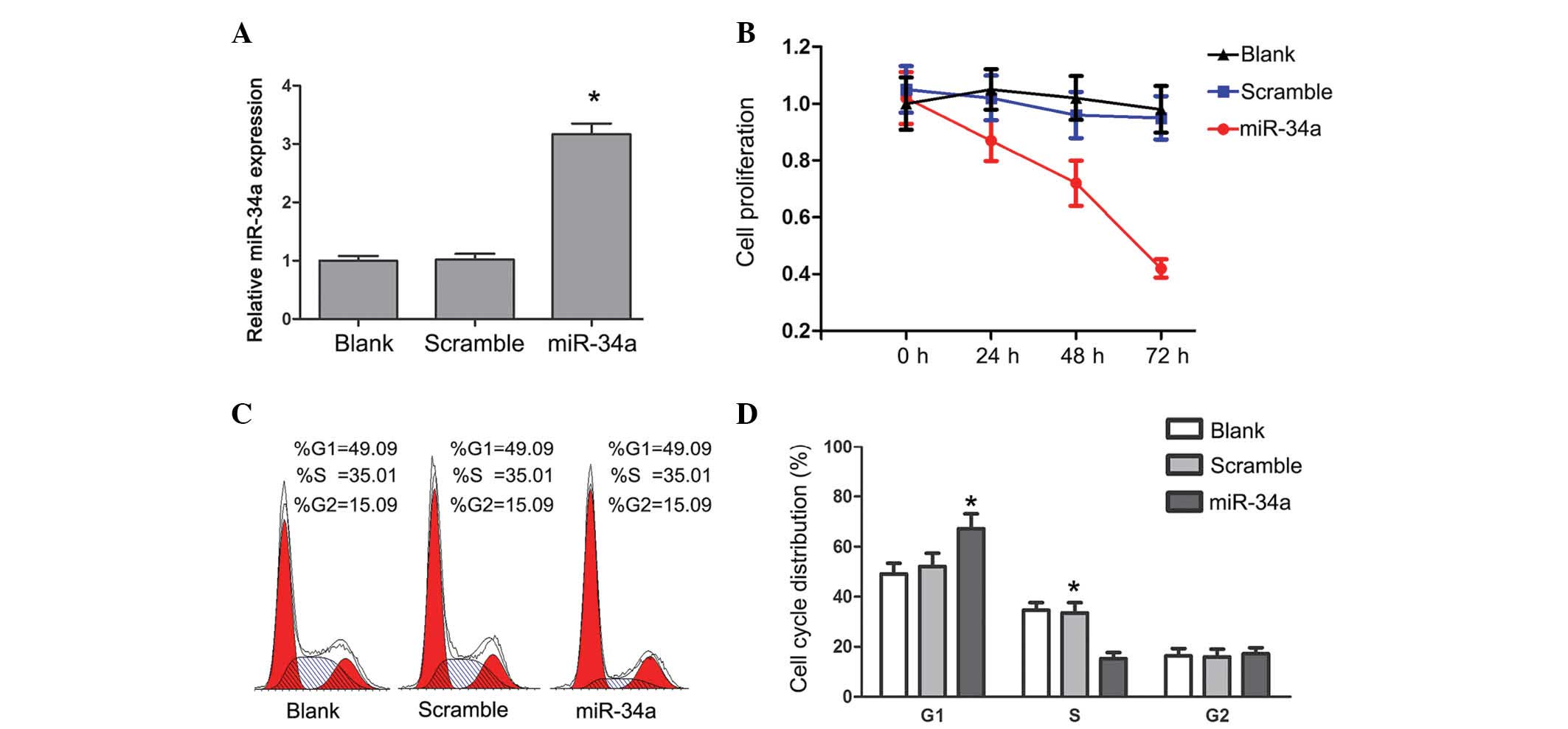
To investigate the association between miR-34a and
glioma cell growth, miR-34a or scramble mimics were transfected
into U87 cells. The miR-Ctrl is a scrambled sequence without the
ability to target any human gene. The level of miR-34a was 3-4 fold
higher in U87 cells following transfection with miR-34a mimics
compared with the control group (Fig.
1A; P<0.05). Proliferation of U87 cells was measured using
the CCK-8 assay. Overexpression of miR-34a resulted in a marked
decrease of proliferation in U87 cells (Fig. 1B). The effect of miR-34a on cell
cycle progression was analyzed by PI staining. The percentage of
cells in the S phase was decreased, and the percentage of cells in
G1 phase was increased (P<0.05) in U87 cells with
miR-34a transfection (Fig. 1C and
D), but not in cells transfected with the scramble mimics. The
expression level of miR-34a in glioma cells inhibited cell cycle
progression and proliferation.
Caspase-dependent apoptosis was induced
by miR-34a in U87 cells
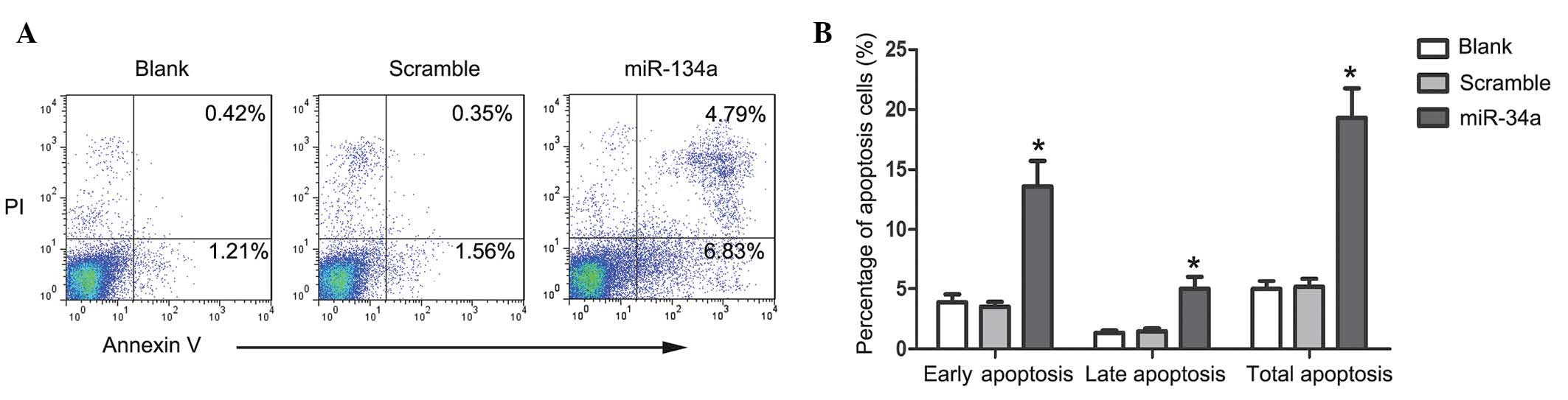
To investigate whether upregulation of miR-34a
influenced apoptosis of U87 cells, PI/Annexin V staining was
performed to detect apoptosis cells. Compared with scramble
mimic-transfected cells, the proportion of Annexin-V stained cells
was markedly increased 48 h after the transfection of miR-34a
(Fig. 2A). The percentage of early
apoptotic, late apoptotic and total apoptotic cells in the
miR-34a-transfected U87 cells was demonstrated to be increased
compared with the control group (Fig.
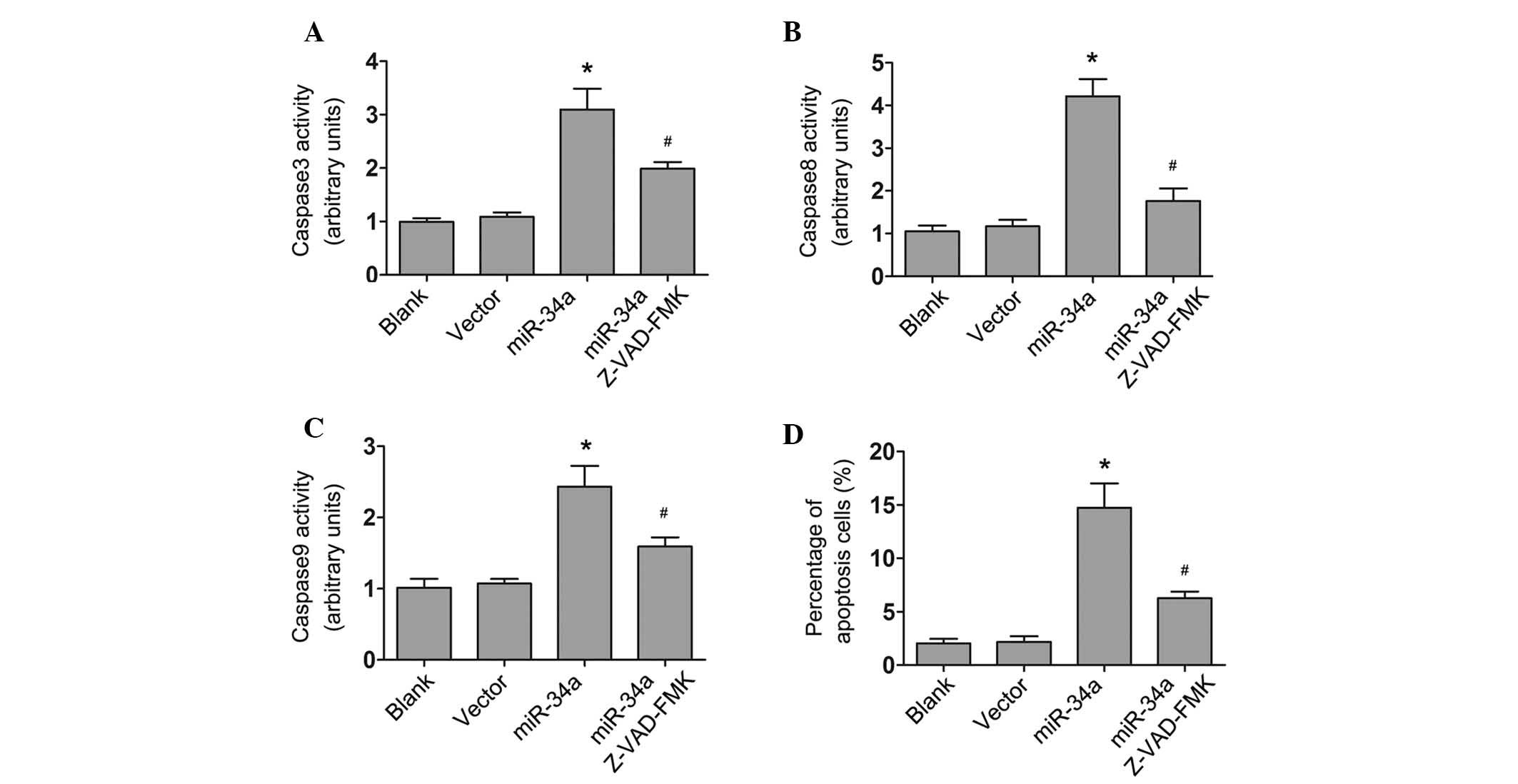
2B; all P<0.05). The activities of caspase-3, -8 and -9
significantly increased in miR-34a highly expressing cells
(P<0.05), but not in scramble mimic-transfected cells, which
suggests that miR-34a induced caspase-dependent apoptosis in U87
cells. Furthermore, U87 cells transfected with miR-34a or scramble
mimics were treated with an irreversible general caspase inhibitor,
z-VAD-FMK (Abcam, Cambridge, MA, US). The results demonstrated that
z-VAD-FMK (10µM) partially reversed the effect of miR-34a,
inhibiting the activity of caspase-3, -8 and -9 (P<0.05;
Fig. 3), indicating that miR-34a
induced caspase-dependent cell apoptosis.
Bcl-2 is a direct target of miR-34a in
glioma cells
Bcl-2 is a critical factor in apoptosis and has an
important role in the development and progression of cancer.
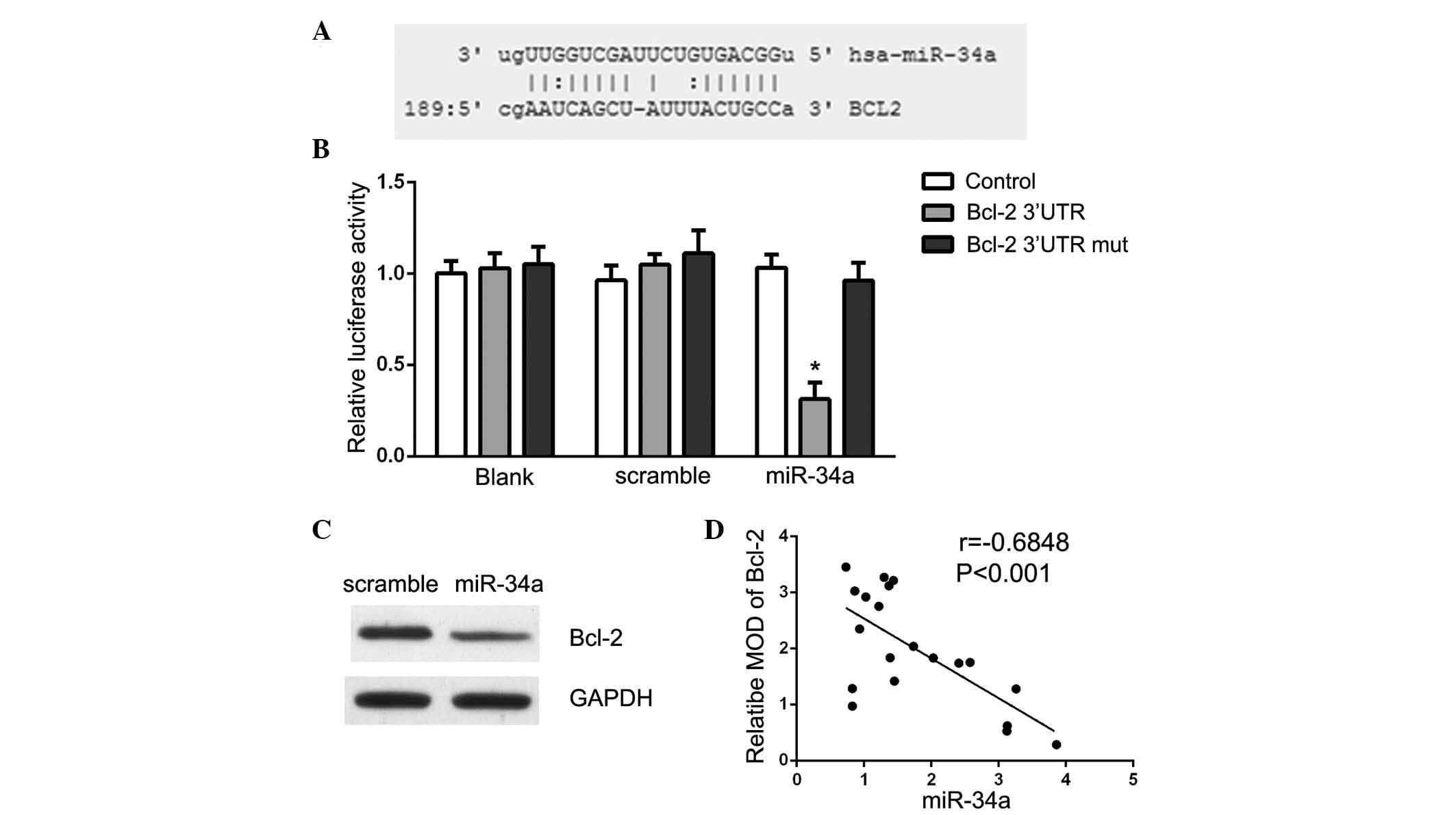
Targetscan (www.targetscan.org) and MicroRNA
(www.microrna.org) were used to search for
potential miR-34a targets. The analysis demonstrated that Bcl-2 is
a theoretical target gene of miR-34a in human cells (Fig. 4A). Luciferase reporter assays were
conducted to validate the prediction and Bcl-2 3′-UTR vectors and
miR-34a or scramble mimics were transfected into U87 cells. As
presented in Fig. 4B, a
significant decrease in luciferase activity upon miR-34a
transfection was observed, suggesting that miR-34a directly
suppressed Bcl-2 in U87 cells (P<0.05). The present study also
demonstrated that Bcl-2 protein expression was specifically
downregulated in miR-34a mimic-transfected cells (Fig. 4C). These data suggest that Bcl-2 is
a direct target of miR-34a in glioma. Furthermore, it was observed
that miR-34a levels were inversely correlated with the expression
of Bcl-2 in 20 glioma tissue samples (r=−0.684, P<0.001;
Fig. 4D).
miR-34a suppressed cell proliferation and
induced apoptosis partially by targeting Bcl-2
Data from the present study suggested that Bcl-2 was
a target gene of miR-34a in HepG2 cells, however, the interaction
between miR-34a and Bcl-2 in the regulation of cell proliferation
and apoptosis in glioma cells required further elucidation. To
investigate whether miR-34a mediated growth suppression in U87
cells by targeting Bcl-2, a novel construct containing the full ORF
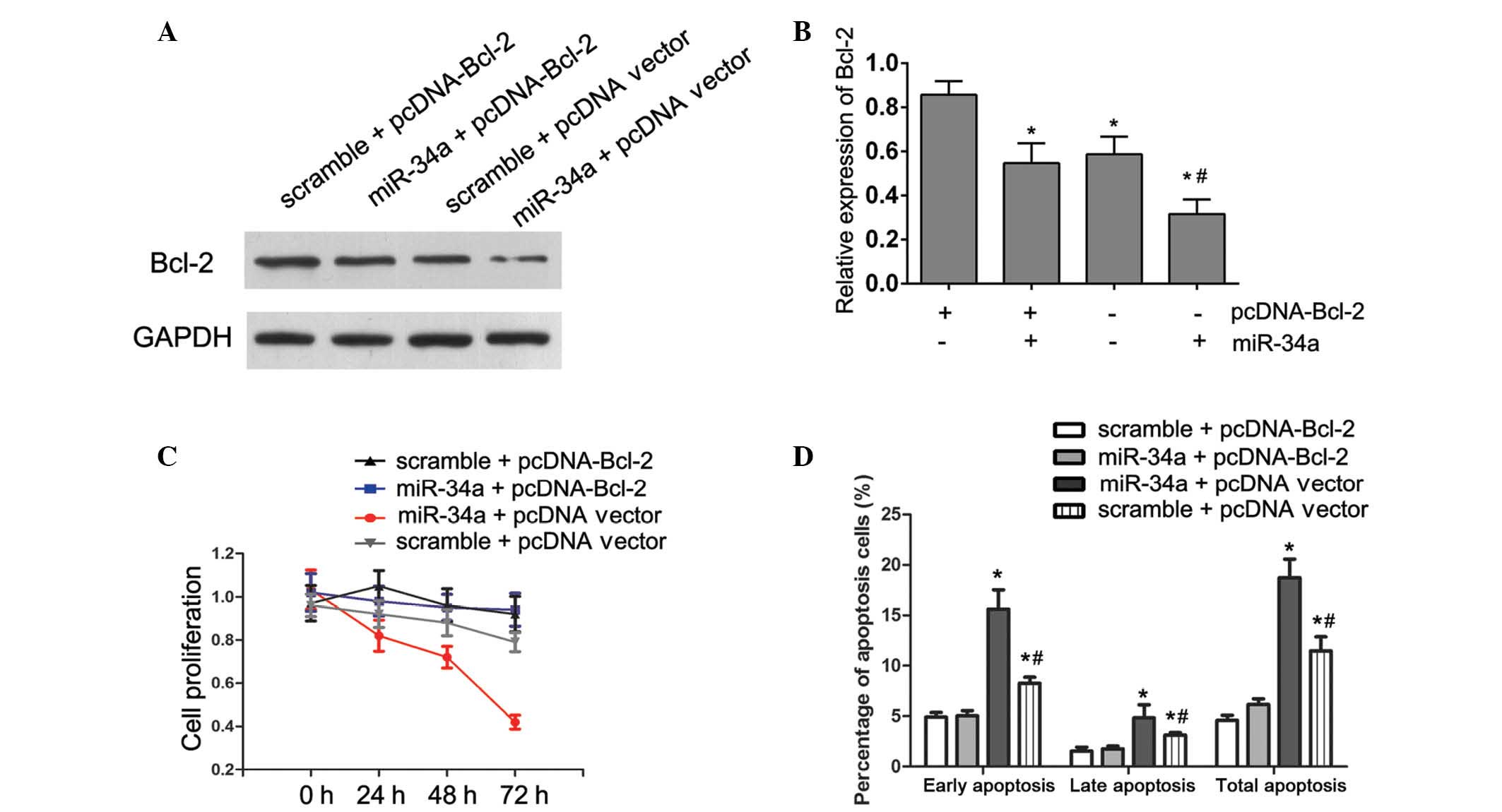
of Bcl-2 gene (pcDNA-Bcl-2). Following transfection of Bcl-2
construct into U87 cells that had been treated with miR-34a mimic
for 24 h, the expression of Bcl-2 was rescued (P=0.014; Fig. 5A and B). Consistent with the
restored expression of Bcl-2, increased cell proliferation
(Fig. 5C), accompanied by
decreased cell apoptosis (Fig. 5D)
was also observed in U87 cells transfected with pcDNA-Bcl-2
constructs following the treatment with miR-34a mimics
(P<0.001). These results demonstrated that the suppression of
cell growth by miR-34a was mediated via targeting Bcl-2.
Discussion
The results from the present study support the
hypothesis that miR-34a functions as a tumor suppressor in glioma
cells. Bcl-2 was identified as a direct target of miR-34a in U87
cells. Restored expression of Bcl-2 in U87 cells partially reversed
miR-34a-mediated apoptosis and inhibition of cell
proliferation.
MiRNAs are a group of small regulatory RNAs that are
involved in posttranscriptional gene expression regulation in a
sequence-specific manner. MiRNAs form ~1% of the genome and each of
which has hundreds of different conserved or non-conserved targets,
thus, they are key in various cellular processes, including
invasion, proliferation and apoptosis. Increased understanding the
physiological and disease-associated mechanisms of miRNAs may aid
development of novel strategies for diagnosis and treatment of
various diseases. Dysregulated miRNAs have been identified by
microarray and associated with glioma carcinogenesis via altering
expression of oncogenes or tumor suppressors to influence cell
proliferation, apoptosis, and tumor cell migration and invasion.
Among these miRNAs, miR-34a was reported to be underexpressed in
glioma (15), however, its exact
function and underlying mechanisms required further elucidation.
The present study demonstrated that restored expression of miR-34a
in the U87 human glioma cell line inhibited cell proliferation,
arrested cell cycle progression and induced cell apoptosis.
Furthermore, inhibition of caspase activity by Z-VAD-FMK suppressed
the apoptosis induced by miR-34a, indicating that miR-34a inhibited
the proliferation of U87 cells via the induction of
caspase-dependent apoptosis.
It is well understood that the intrinsic apoptosis
signaling pathway ultimately activates caspase-3, which may be
regulated by the Bcl-2 superfamily members, including
anti-apoptotic members, Bcl-2 and Bcl-xL, and pro-apoptotic
members, Bax, Bak and Bid (16,17).
It has been demonstrated that expression of Bcl-2 may be
downregulated by miR-34a in numerous types of cells, including Min6
cells (18), PC12 cells (19) and human hepatocellular carcinoma
cells (20). In the present study,
the putative binding site of miR-34a in Bcl-2 3′-UTR was observed
by a biological prediction program (21). Luciferase reporter assays indicated
that transfection of miR-34a resulted in a marked reduction of
luciferase activity by the luciferase expression constructs
carrying the target Bcl-2 3′-UTR fragment, but had no effect on
Bcl-2 3′-UTR mutated fragment. In addition, the protein expression
of Bcl-2 was significantly decreased following treatment with
miR-34a mimics demonstrating that miR-34a directly targets Bcl-2
mRNA in U87 cells. Furthermore, an inverse correlation was observed
between the miR-34a level and Bcl-2 expression in human glioma
tissue specimens, which was consistent with the results of Yang
et al (20) in
hepatocellular carcinoma. Using further rescue assays, the present
study demonstrated that miR-34a induced cell cycle arrest and
apoptosis was partially blocked by overexpression of Bcl-2. These
results imply that miR-34a-induced apoptosis was partially mediated
by silencing of Bcl-2.
In addition to Bcl-2, various genes have been
reported to participate in miR-34a-mediated tumor suppression,
including fos-related antigen 1, platelet derived growth factor
receptor and MET proto-oncogene, tyrosine receptor, were identified
to be targets of miR-34a involved in tumor migration and invasion
(10,22). MiR-34a also modulated the
sensitivity of doxorubicin-resistant MCF-7 cells to doxorubicin by
directly inhibiting Notch 1 (23).
Delivery of miR-34a to prostate cancer mouse models resulted in to
a marked decrease in tumor growth via inhibition of the c-Myc
oncogene (24). Li et al
(25) demonstrated that miR-34a
serves as a tumor suppressor in A172 human glioma cells,
predominantly by decreasing NADPH oxidase 2 expression. The
involvement of these genes in miR-34a-mediated tumor suppression in
glioma remains to be investigated.
In conclusion, the results from the present study
demonstrated that miR-34a was a tumor suppressor in glioma cells,
suppressing cell proliferation and inducing cell apoptosis via
targeting of Bcl-2. These findings aid understanding of the
underlying molecular mechanisms of glioma carcinogenesis and
suggest a potential use of miR-34a in future therapeutic strategies
to treat glioma.
Acknowledgments
The current study was supported by Youth Innovation
Project Plan of Sichuan Province Medical Scientific Research (grant
no. Q15079).
References
|
1
|
Koekkoek JA, Kerkhof M, Dirven L, Heimans
JJ, Reijneveld JC and Taphoorn MJ: Seizure outcome after
radiotherapy and chemotherapy in low-grade glioma patients: A
systematic review. Neuro Oncol. 17:924–934. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chiarelli PA, Kievit FM, Zhang M and
Ellenbogen RG: Bionanotechnology and the future of glioma. Surg
Neurol Int. 6(Suppl 1): S45–S58. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kawahara H, Imai T and Okano H: MicroRNAs
in neural stem cells and neurogenesis. Front Neurosci. 6:302012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Di Y, Lei Y, Yu F, Changfeng F, Song W and
Xuming M: MicroRNAs expression and function in cerebral ischemia
reperfusion injury. J Mol Neurosci. 53:242–250. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang Z, Chang H, Li Y, Zhang T, Zou J,
Zheng X and Wu J: MicroRNAs: potential regulators involved in human
anencephaly. Int J Biochem Cell Biol. 42:367–374. 2010. View Article : Google Scholar
|
|
6
|
Barger JF and Nana-Sinkam SP: MicroRNA as
tools and therapeutics in lung cancer. Respir Med. 109:803–812.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
An L, Liu Y, Wu A and Guan Y: microRNA-124
inhibits migration and invasion by down-regulating ROCK1 in glioma.
PloS one. 8:e694782013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li M, Li J, Liu L, Li W, Yang Y and Yuan
J: MicroRNA in human glioma. Cancers. 5:1306–1331. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Palumbo S, Miracco C, Pirtoli L and
Comincini S: Emerging roles of microRNA in modulating cell-death
processes in malignant glioma. J Cell Physiol. 229:277–286. 2014.
View Article : Google Scholar
|
|
10
|
Wu J, Wu G, Lv L, Ren YF, Zhang XJ, Xue
YF, Li G, Lu X, Sun Z and Tang KF: MicroRNA-34a inhibits migration
and invasion of colon cancer cells via targeting to Fra-1.
Carcinogenesis. 33:519–528. 2012. View Article : Google Scholar
|
|
11
|
Kato M, Paranjape T, Müller RU, Nallur S,
Gillespie E, Keane K, Esquela-Kerscher A, Weidhaas JB and Slack FJ:
The mir-34 microRNA is required for the DNA damage response in vivo
in C. elegans and in vitro in human breast cancer cells. Oncogene.
28:2419–2424. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Stahlhut C and Slack FJ: Combinatorial
action of MicroRNAs let-7 and miR-34 effectively synergizes with
Erlotinib to suppress non-small cell lung cancer cell
proliferation. Cell Cycle. 14:2171–2180. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gao H, Zhao H and Xiang W: Expression
level of human miR-34a correlates with glioma grade and prognosis.
J Neurooncol. 113:221–228. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schefe JH, Lehmann KE, Buschmann IR, Unger
T and Funke-Kaiser H: Quantitative real-time RT-PCR data analysis:
Current concepts and the novel “gene expression's CT difference”
formula. J Mol Med Berl. 84:901–910. 2006. View Article : Google Scholar
|
|
15
|
Guessous F, Zhang Y, Kofman A, Catania A,
Li Y, Schiff D, Purow B and Abounader R: microRNA-34a is tumor
suppressive in brain tumors and glioma stem cells. Cell Cycle.
9:1031–1036. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li Y, Qi H, Li X, Hou X, Lu X and Xiao X:
A novel dithiocarbamate derivative induces cell apoptosis through
p53-dependent intrinsic pathway and suppresses the expression of
the E6 oncogene of human papillomavirus 18 in HeLa cells.
Apoptosis. 20:787–795. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hockenbery DM, Oltvai ZN, Yin XM, Milliman
CL and Korsmeyer SJ: Bcl-2 functions in an antioxidant pathway to
prevent apoptosis. Cell. 75:241–251. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lin X, Guan H, Huang Z, Liu J, Li H, Wei
G, Cao X and Li Y: Downregulation of Bcl-2 expression by miR-34a
mediates palmitate-induced Min6 cells apoptosis. J Diabetes Res.
2014:2586952014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang M, Lou D, Chang X and Zhou Z: Micro
RNA alteration after paraquat induced PC12 cells damage and
regulatory mechanism of bcl-2. Zhonghua Lao Dong Wei Sheng Zhi Ye
Bing Za Zhi. 32:32–37. 2014.In Chinese. PubMed/NCBI
|
|
20
|
Yang F, Li QJ, Gong ZB, Zhou L, You N,
Wang S, Li XL, Li JJ, An JZ, Wang DS, et al: MicroRNA-34a targets
Bcl-2 and sensitizes human hepatocellular carcinoma cells to
sorafenib treatment. Technol Cancer Res Treat. 13:77–86. 2014.
|
|
21
|
Betel D, Wilson M, Gabow A, Marks DS and
Sander C: The http://microRNA.orgurisimplemicroRNA.org resource:
Targets and expression. Nucleic Acids Res. 36(Database issue):
D149–D153. 2008. View Article : Google Scholar
|
|
22
|
Peng Y, Guo JJ, Liu YM and Wu XL:
MicroRNA-34A inhibits the growth, invasion and metastasis of
gastric cancer by targeting PDGFR and MET expression. Biosci Rep.
34:e001122014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li XJ, Ji MH, Zhong SL, Zha QB, Xu JJ,
Zhao JH and Tang JH: MicroRNA-34a modulates chemosensitivity of
breast cancer cells to adriamycin by targeting Notch1. Arch Med
Res. 43:514–521. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yamamura S, Saini S, Majid S, Hirata H,
Ueno K, Deng G and Dahiya R: MicroRNA-34a modulates c-Myc
transcriptional complexes to suppress malignancy in human prostate
cancer cells. PloS One. 7:e297222012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li SZ, Hu YY, Zhao J, Zhao YB, Sun JD,
Yang YF, Ji CC, Liu ZB, Cao WD, Qu Y, et al: MicroRNA-34a induces
apoptosis in the human glioma cell line, A172, through enhanced ROS
production and NOX2 expression. Biochem Biophys Res Commun.
444:6–12. 2014. View Article : Google Scholar : PubMed/NCBI
|