Introduction
Prostate cancer (PCa) is one of the most frequently
diagnosed malignancies in men worldwide (1). The incidence of PCa in China has
increased (2,3), however, the mechanisms underlying the
development and progression of PCa remain to be fully elucidated.
Disease relapse and metastasis, as well as the development of
hormone-refractory disease, remain the leading causes of mortality.
Therefore, investigating the molecular mechanisms involved in the
progression of PCa is of major importance and may reveal novel
approaches for targeted PCa therapy.
With the advent of tiling-resolution genomic
microarrays and whole-genome and transcriptome sequencing
technologies, studies have revealed that at least 90% of the genome
is actively transcribed (4,5).
Several of these transcripts have emerged as critical regulators of
gene expression and determination of cell fate. A recently
identified group of transcripts, termed long noncoding RNAs
(lncRNAs), may contribute to a significant quantity of non-coding
RNA which makes up the human transcriptome (6,7).
lncRNAs are a class of noncoding RNAs >200 nucleotides in
length; they can be transcribed by RNA polymerase (Pol) II/Pol I,
and a number are transcribed by RNA Pol III. lncRNAs are involved
in several biological processes, including X chromosome
inactivation, nuclear structure, genomic imprinting and development
(8,9). lncRNA dysfunction has been associated
with cell fate determination and the pathogenesis of human disease,
including cancer (10).
Furthermore, several dysregulated lncRNAs are associated with the
carcinogenesis and growth of tumors, including breast cancer, colon
cancer, PCa, hepatocellular carcinoma and leukemia (11–15).
lncRNA-thrombospondin 4 (THBS4)-003 is a processed
transcript, which does not encode a protein product and has a
length of 558 bp, which is >200 bp and, thus fits well within
the definition for lncRNAs. lncRNA-THBS4-003 is located at
chromosome 5p14.1 and partially overlaps with the protein-coding
gene, THBS4 (16).
Mitogen-activated protein kinases (MAPKs) are a
family of kinases, which comprise one of the major signaling
pathways involved throughout the development of PCa. MAPKs can be
divided into three subfamilies: The extracellular-signal-regulated
kinases, the c-Jun N-terminal kinases, and p38 MAPK. It is well
known that p38 MAPK is capable of regulating several cellular
responses to cytokines and stress; however, previous data
demonstrated that p38 is also closely linked to the development of
different types of human cancer, through its ability to enhance
cancer cell migration and invasion (17). Matrix metalloproteinases (MMPs),
members of the zinc-dependent endoprotease family, are important
enzymes involved in degrading the dermal extracellular matrix
(ECM); in particular, MMP-9 is important in cancer cell invasion
and metastasis (18).
In the present study, THBS4 and lncRNA-THBS4-003
expression levels are analyzed in PCa tissue in comparison with
normal prostate tissue samples, and its potential biomedical
functions in vitro are investigated. The aim of the study is
to confirm whether lncRNA-THBS4-003 is a potential therapeutic
target due to its role in PCa migration and invasion.
Materials and methods
Ethical statement
The present study was approved by the Institutional
Review Board of the First Affiliated Hospital of Nanjing Medical
University (Nanjing, China). During recruitment, written informed
consent was obtained from all participants involved in the present
study.
Tissue collection
Primary PCa and adjacent non-tumor tissue samples
were collected from patients undergoing radical prostatectomy
between 2011 and 2013 at the Department of Urology, the First
Affiliated Hospital of Nanjing Medical University. Neither local
nor systemic treatment had been administered in these patients
prior to surgery. A total of 46 samples from 46 patients undergoing
radical prostatectomy were collected in this study. The size of
samples were 0.5cm3, at least containing 200 mg cells.
Following surgery, all the samples were immediately frozen and
stored in liquid nitrogen until further analysis. Only samples
containing >70% tumor cells were used for the extraction of
total RNA. All experiments were approved by the Research Ethics
Committee of Nanjing Medical University,. Detailed information on
each tissue donor is provided in Table
I.
 | Table IPatient characteristics. |
Table I
Patient characteristics.
| Characteristic | Number of PCa samples
(n=46) |
|---|
| Age (years) | |
| Median (range) | |
| T stage | |
| T1 | 6 |
| T2 | 22 |
| T3 | 10 |
| T4 | 8 |
| N stage | |
| N0 | 38 |
| N1 | 8 |
| M stage | |
| M0 | 46 |
| M1 | 0 |
| Gleason score | |
| <7 | 13 |
| 7 | 20 |
| >7 | 13 |
Bioinformatics microarray data
analysis
The microarray contained 8,277 lncRNA probes, which
were designed by Arraystar Human LncRNA Expression Microarray
(version 4.0; Arraystar, Inc., Rockville, MA, USA), based on the
RefSeq (http://www.ncbi.nlm.nih.gov/refseq/), UCSC Known genes
(http://genome.ucsc.edu/), and Ensembl (http://ensemblgenomes.org/)databases and associated
literature (19,20); 32,207 protein-coding transcripts
were used for microarray assays in three PCa tissue samples and
their matched non-tumor samples. Differentially expressed lncRNAs
and mRNAs, found to be statistically significant [P<0.05;
fold-change (FC)>2] between the two groups were identified by
comparing the normalized expression levels in the tumor and
non-tumor samples using a paired t-test. Hierarchical clustering
was then performed to analyze the differential lncRNA and mRNA
expression patterns.
Cell lines and cell culture
The DU145 and PC-3 human PCa cell lines were
purchased from the Institute of Biochemistry and Cell Biology of
the Chinese Academy of Sciences (Shanghai, China). The DU145 and
PC-3 cells (60-70% confluence) were cultured in F-12K (Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with
10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 mg/ml
streptomycin (Gibco; Thermo Fisher Scientific, Inc.) at 37°C with
5% CO2.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis
For the analyses of mRNA, total RNA was extracted
from 75 mg tissue samples and cultured cells using 1 ml TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocol, and centrifugation at 12,000 × g (15
min at 4°C). For RT-qPCR, 1 µg total RNA was reverse
transcribed into cDNA in a final volume of 20 µl, using
random primers and a High Capacity RNA-to-cDNA kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. The RT reaction was performed at 37°C for
60 min, followed by 95°C for 5 min. The expression levels of
lncRNA-THBS4-003 and THBS4 in the PCa cell lines and tissues were
then measured using qPCR, according to the standard protocol of the
SYBR Select Master mix (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The sequences of the PCR primers used were as
follows: lncRNA-THBS4-003, forward 5′-ATGAAGGCTCTGAGTTGGTG-3′ and
reverse 5′-CTTGGAAGTCCTCAGGGATG-3′; THBS4, forward
5′-GTTGCAGAACCTGGCATTCAG-3′ and reverse
5′-CCCTGGACCTGTCTTAGACTTCA-3′; and β-actin, forward
5′-ACTGGAACGGTGAAGGTGAC-3′ and reverse 5′-AGAGAAGTGGGGTGGCTTTT-3′.
Primers were synthesized by Invitrogen (Thermo Fisher Scientific,
Inc.). The mRNA detection reaction was performed under the
following conditions: 50°C for 2 min, 95°C for 2 min, 40 cycles at
95°C for 15 sec, and 60°C for 1 min. The expression levels of
lncRNA-THBS4-003 and THBS4 were normalized to that of β-actin and
calculated using the ΔΔCq method (21). The reactions were performed and
analyzed using an ABI StepOne plus system (Applied Biosystems;
Thermo Fisher Scientific, Inc.). All reactions were run in
triplicate.
Small interfering (si)RNA
transfection
Briefly, 1×105 cells were seeded into
six-well plates and cultured in complete growth media at 37°C until
the cell density reached 60–70%, prior to siRNA transfection using
Lipofectamine 2000 (Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocol. The cells were harvested after 48 h at
37°C for RT-qPCR and Western blot analyses. The sequences of the
siRNAs used in the present study were as follows:
5′-GGCAACAGCUACAGUACAATT-3′ and 5′-UUGUACUGUAGCUGUUGCCTT-3′ for
si-lncRNA-THBS4-003, 5′-GGCAGUUCUUGGGUCAAAUTT-3′ and
5′-AUUUGACCCAAGAACUGCCTT-3′ for si-THBS4, and
5′-UUCUCCGAACGUGUCACGUTT-3′ and 5′-ACGUGACACGUUCGGAGAATT-3′ for the
control siRNA (Invitrogen; Thermo Fisher Scientific, Inc.).
Cell migration and invasion assays
For the migration assays, 3×104 cells in
serum-free medium were placed in the upper chamber of a Transwell
(pore size, 8 mm; BD Biosciences, San Jose, CA, USA). For the
invasion assays, cells with 200 µl serum-free medium were
placed in upper chambers coated with Matrigel (BD Biosciences),
according to the manufacturer's protocol. Medium containing 10% FBS
was immediately added to the lower chamber as a chemoattractant.
Following incubation for 24 h at 37°C, the cells, which had not
migrated through the pores of the Transwell inserts were manually
removed using a cotton swab, and those cells on the lower surface
of the membrane were fixed in 95% ethanol and stained with 0.1%
crystal violet (Beyotime Institute of Biotechnology, Haimen,
China). The numbers of cells in five randomly-selected fields were
determined for each chamber, and the average value was calculated.
Each experiment was performed in triplicate. Matrigel invasion
assays were performed, as described previously (22).
Protein isolation and Western blot
analysis
The tissue samples and cells were washed twice in
ice-cold phosphate-buffered saline and then lysed in buffer
containing 20 mM Tris-HCl (pH 7.4; Bio-Rad Laboratories, Inc.,
Hercules, CA, USA), 137 mM NaCl, 10% glycerol, 1% Triton X-100, 2
mM EDTA, 25 mM β-glycerophosphate, 2 mM sodium pyrophosphate and
0.5 mM dithiothreitol (all purchased from Sigma-Aldrich, St. Louis,
MO, USA) with protease inhibitors at 4°C for 30 min. Subsequently,
cellular debris was removed by centrifugation of the lysate at
12,000 × g for 10 min at 4°C. The supernatants (50 µl) were
mixed with equal volumes of 2X sodium dodecyl sulfate (SDS) sample
buffer (Sigma-Aldrich) and heated to 100°C for 10 min. An equal
volume of sample was fractionated by SDS-PAGE on a 10% acrylamide
gel and transferred onto polyvinylidene difluoride membranes using
a Bio-Rad transfer system (version 4.62) (both purchased from
Bio-Rad Laboratories, Inc.), according to the manufacturer's
protocol. Following blocking of the non-specific binding sites with
5% nonfat milk in Tris-buffered saline with Tween 20, containing 50
mM Tris, 0.15 M NaCl and 0.1% Tween 20 (pH 7.6) for 1 h, the
membranes were probed with THBS4 rabbit polyclonal antibody
(1:2,000 dilution; cat. no. ab121094; Abcam, Cambridge, MA, USA),
p38 rabbit polyclonal antibody (1:1,000 dilution; cat. no. ab7952;
Abcam), rabbit polyclonal anti-MMP-9 antibody (1:1,000 dilution;
cat. no. ab38898; Abcam), or β-actin rabbit antibody (1:1,000
dilution; cat. no. 8457; Cell Signaling Technology, Inc., Danvers,
MA, USA) at 4°C overnight, followed by incubation with horseradish
peroxidase-conjugated goat anti-rabbit secondary antibodies
(1:1,000 dilution; cat. no. 7074; Cell Signaling Technology, Inc.)
for 1 h at 25°C. Immunoreactive bands were visualized using
enhanced chemiluminescence reagents (Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. Densitometric
analysis of the immunoblots was performed using QuantityOne
software (version 4.62; Bio-Rad Laboratories, Inc.). Protein levels
were determined by normalization to β-actin, and the mean ±
standard deviation was calculated from three individual
experiments.
Statistical analysis
The results are presented as the mean ± standard
error of the mean. Differences between groups were assessed for
significance using Student's t-test. P<0.05 was
considered to indicate a statistically significant difference. All
statistical analyses were performed using SPSS 11.0 for Windows
(SPSS Inc, Chicago, IL, USA).
Results
Expression of THBS4 is increased in
resected PCa tissue samples
In the present study, a microarray containing 8,277
lncRNA probes and 32,207 mRNA probes was used to identify
dysregulated mRNAs in three patients with PCa. Among these mRNAs,
354 were significantly upregulated and 350 were significantly
downregulated (P<0.05; FC>2; Fig. 1A). The most significantly
upregulated mRNA was THBS4 (P<0.05; FC>2).
THBS4 was selected to confirm the differential
expression levels in 46 paired PCa and adjacent non-tumor tissue
samples using RT-qPCR. The expression of THBS4 was significantly
higher in the PCa tissues, compared with that in the adjacent
non-tumor tissues (Fig. 1B).
Similarly, the results of the Western blot analyses
revealed that the protein expression level ofTHBS was higher in the
PCa tumor tissues, compared with the adjacent non-tumor tissues
collected from the same patients (Fig.
1C).
Expression of lncRNA-THBS4-003 is
increased in resected PCa tissue samples
The expression of lncRNA-THBS4-003 was analyzed in
46 primary PCa and adjacent non-tumor tissue samples using RT-qPCR.
The expression of lncRNA-THBS4-003 was significantly higher in the
tumor tissues, compared with non-tumor tissues (Fig. 2A).
In addition, patients with Gleason scores >7
exhibited higher expression levels of lncRNA-THBS4-003, compared
with the patients with lower Gleason scores (Fig. 2B).
Statistical analyses of the expression levels of
THBS4 and lncRNA-THBS4-003 revealed a Pearson's correlation
coefficient of 0.641 (P<0.0001), indicating a positive
correlation between the expression levels of THBS4 and
lncRNA-THBS4-003 in PCa (Fig.
2C).
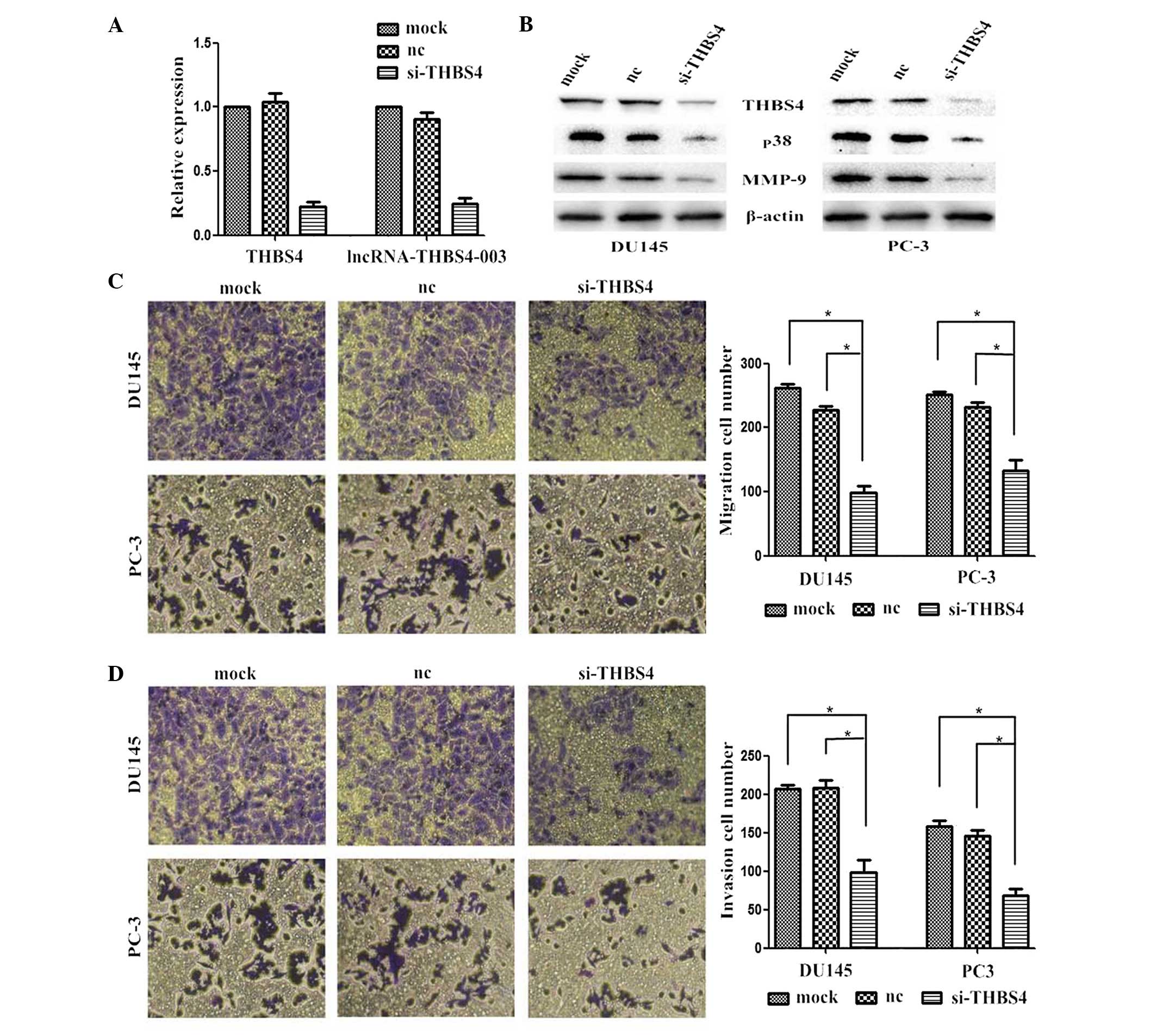
Effects of THBS4 knockdown on PCa cell
lines
In the present study, siRNA-mediated knockdown of
THBS4 was performed in the PCa cells to investigate the effects of
THBS4 on the migration and invasion of the PCa cells. The level of
silencing achieved was also analyzed by performing RT-qPCR and
Western blotting. The mRNA expression levels of THBS4 and
lncRNA-THBS4-003 were successfully reduced in the cells transfected
with si-THBS4 (Fig. 3A).
Suppressing the expression of THBS4 also reduced the expression
levels of p38 and MMP-9 (Fig. 3B).
Furthermore, the numbers of migrated and invaded cells transfected
with si-THBS4 were significantly lower, compared with the cells
transfected with the control (Fig. 3C
and D).
Knockdown of lncRNA-THBS4-003 inhibits
PCa cell migration and invasion in vitro
As shown in Fig.
4A, the expression levels of lncRNA-THBS4-003 in the PC-3 cells
transfected with siRNA were examined. lncRNA-THBS4-003 knockdown
significantly suppressed the migratory and invasive abilities of
the PCa cells (Fig. 4B and C).
Reciprocal regulation of lncRNA-THBS4-003
and THBS4 contribute to PCa cell line migration and invasion, and
regulate levels of MMP-9 through the MAPK signaling pathway
Knocking down the expression of THBS4 successfully
reduced the expression of lncRNA-THBS4-003 in the cells transfected
with si-THBS4 (Fig. 3A).
Statistical analyses of the expression levels of THBS4 and
lncRNA-THBS4-003 found a Pearson's correlation coefficient of 0.641
(P<0.0001; Fig. 2C). At 72 h
post-transfection, the protein levels of THBS4, p38 and MMP-9 were
significantly decreased in the cells transfected with
si-lncRNA-THBS4-003, compared with the cells transfected with the
control siRNA (Fig. 5).
Discussion
The present study is the first, to the best our out
knowledge, to report lncRNA-THBS4-003 as a potentially useful
biomarker for disease progression in patients with PCa.
In the present study, a microarray containing 8,277
lncRNA probes and 32,207 mRNA probes was used to identify
dysregulated mRNAs in three patients with PCa. A total of 354 mRNAs
were found to be significantly upregulated and 350 were
downregulated (P<0.05; FC>2). The most significantly
upregulated mRNA was THBS4 (P<0.05; FC>2). lncRNA-THBS4-003
is located at chromosome 5p14.1 and partially overlaps the
protein-coding gene, THBS4. Western blot and RT-qPCR analyses
revealed that lncRNA-THBS4-003 and the expression levels of THBS4
were higher in tumor tissues, compared with the adjacent non-tumor
tissues collected from the same patients. Statistical analyses of
THBS4 and lncRNA-THBS4-003 demonstrated a Pearson's correlation
coefficient of 0.641 (P<0.0001). The expression level of
lncRNA-THBS4-003 was significantly higher, compared with the
adjacent non-tumor tissues. Patients with Gleason scores >7
exhibited higher expression levels of lncRNA-THBS4-003, compared
with those with lower scores. The aberrant expression and function
of lncRNA-THBS4-003 in PCa remain to be elucidated. Using cell
migration and invasion assays to evaluate migratory and invasive
responses, lncRNA-THBS4-003 knockdown was found to significantly
decrease the migratory and invasive abilities of the PCa cells
in vitro, and to inhibit the expression levels of THBS4, p38
and MMP-9.
The thrombospondins (THBSs) are a family of five
extracellular calcium-binding proteins (THBS1, THBS2, THBS3, THBS4
and THBS5/COMP), which are important in diverse processes through
their interactions with the ECM. THBS4 is involved in several
critical processes, including cellular proliferation, attachment,
adhesion and migration, cytoskeletal organization, cell-to-cell
interactions, and the promotion of neurite outgrowth (23–26).
The role of THBS4 in cancer is well understood, and increasing
evidence suggests that THBS4 is involved in colorectal, gastric and
prostate carcinomas. THBS4 is reported to be expressed at high
levels by cancer-associated fibroblasts as a constituent of
desmoplastic stroma in prostate and gastric cancer (20,27–30).
Previous reports have shown that THBS4-induced
activation of p38-MAPK regulates vascular inflammation and
atherogenesis (20,31,32).
It is known that p38 MAPK is capable of regulating several cellular
responses to cytokines and stress; however, studies have
demonstrated that p38 is also closely associated with the
development of different types of human cancer through its ability
to elevate cancer cell migration and invasion in response to
various stimuli, including inflammatory factors (31). Evidence has shown that p38 MAPK
signals are involved in downregulating the expression of MMP-9,
which has been linked to tumor migration and invasion (32).
The present study demonstrated significant
downregulation in the protein levels of p38 and MMP-9 following the
suppression of lncRNA-THBS4-003 or THBS4. The numbers of migrated
and invaded cells transfected with si-lncRNA-THBS4-003 or si-THBS4
were also significantly lower, compared with the cells transfected
with control siRNA.
The present study revealed that the forced knockdown
of lncRNA-THBS4-003 or THBS4 decreased the in vitro
migratory and invasive abilities of PCa cells through the MAPK
signaling pathway. THBS4 is an adhesive glycoprotein that mediates
cell-to-cell and cell-to-matrix interactions and is involved in the
regulation of vascular inflammation. THBS4 and lncRNA-THBS4-003 can
promote angiogenesis in prostate tissue (33). In the present study, p38 and MMP-9
were decreased following knockdown of THBS4 and lncRNA-THBS4-003 in
prostate cancer cell lines. MMP-9 can promote angiogenesis, tumor
migration and invasion in prostate cancer. In conclusion, the
present study demonstrates that THBS4 and lncRNA-THBS4-003 serve a
significant role in PCa proliferation and migration via the MMP-9
and p38 MAPK signaling pathway.
Acknowledgments
This study was supported by the Priority Academic
Program Development of Jiangsu Higher Education Institutions (grant
no. JX10231801), the Program for Development of Innovative Research
Team at the First Affiliated Hospital of Nanjing Medical
University, the Provincial Initiative Program for Excellency
Disciplines of Jiangsu Province (grant no. BL2012027), the National
Natural Science Foundation of China (grant nos. 81201998 and
81372757) and the Natural Science Foundation of Jiangsu Province
(grant no. BK20141495).
References
|
1
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J,
Murray T and Thun MJ: Cancer statistics, 2008. CA: Cancer J Clin.
58:71–96. 2008.
|
|
2
|
Zeigler-Johnson CM, Rennert H, Mittal RD,
Jalloh M, Sachdeva R, Malkowicz SB, Mandhani A, Mittal B, Gueye SM
and Rebbeck TR: Evaluation of prostate cancer characteristics in
four populations worldwide. Can J Urol. 15:4056–4064.
2008.PubMed/NCBI
|
|
3
|
Peyromaure EM, Mao K, Sun Y, Xia S, Jiang
N, Zhang S, Wang G, Liu Z and Debré B: A comparative study of
prostate cancer detection and management in China and in France.
Can J Urol. 16:4472–4477. 2009.PubMed/NCBI
|
|
4
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
ENCODE Project Consortium; Birney E,
Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH,
Weng Z, Snyder M, Dermitzakis ET, et al: Identification and
analysis of functional elements in 1% of the human genome by the
ENCODE pilot project. Nature. 447:799–816. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang L, Duff MO, Graveley BR, Carmichael
GG and Chen LL: Genomewide characterization of non-polyadenylated
RNAs. Genome Biol. 12:R162011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kapranov P, St Laurent G, Raz T, Ozsolak
F, Reynolds CP, Sorensen PH, Reaman G, Milos P, Arceci RJ, et al:
The majority of total nuclear-encoded non-ribosomal RNA in a human
cell is 'dark matter' un-annotated RNA. BMC Biol. 8:1492010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Martin L and Chang HY: Uncovering the role
of genomic 'dark matter' in human disease. J Clin Invest.
122:1589–1595. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fenoglio C, Ridolfi E, Galimberti D and
Scarpini E: An emerging role for long non-coding RNA dysregulation
in neurological disorders. Int J Mol Sci. 14:20427–20442. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen
X, Zhang Q, Yan G and Cui Q: LncRNADisease: A database for
long-non-coding RNA-associated diseases. Nucleic Acids Res.
41(Database issue): D983–D986. 2013. View Article : Google Scholar :
|
|
11
|
Rinn JL, Kertesz M, Wang JK, Squazzo SL,
Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E and
Chang HY: Functional demarcation of active and silent chromatin
domains in human HOX loci by noncoding RNAs. Cell. 129:1311–1323.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pibouin L, Villaudy J, Ferbus D, Muleris
M, Prospéri MT, Remvikos Y and Goubin G: Cloning of the mRNA of
overexpression in colon carcinoma-1: A sequence overexpressed in a
subset of colon carcinomas. Cancer Genet Cytogenet. 133:55–60.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fu X, Ravindranath L, Tran N, Petrovics G
and Srivastava S: Regulation of apoptosis by a prostate-specific
and prostate cancer-associated noncoding gene, PCGEM1. DNA Cell
Biol. 25:135–141. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Calin GA, Liu CG, Ferracin M, Hyslop T,
Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, et
al: Ultraconserved regions encoding ncRNAs are altered in human
leukemias and carcinomas. Cancer Cell. 12:215–229. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lin R, Maeda S, Liu C, Karin M and
Edgington TS: A large noncoding RNA is a marker for murine
hepatocellular carcinomas and a spectrum of human carcinomas.
Oncogene. 26:851–858. 2007. View Article : Google Scholar
|
|
16
|
Brody MJ, Schips TG, Vanhoutte D,
Kanisicak O, Karch J, Maliken BD, Blair NS, Sargent MA, Prasad V
and Malkentin JD: Dissection of thrombospondin-4 domains involved
in intracellular adaptive endoplasmic reticulum stress-responsive
signaling. Mol Cell Biol. 36:2–12. 2015.PubMed/NCBI
|
|
17
|
da Silva HB, Amaral EP, Nolasco EL, de
Victo NC, Atique R, Jank CC, Anschau V, Zerbini LF and Correa RG:
Dissecting major signaling pathways throughout the development of
prostate cancer. Prostate cancer. 2013:9206122013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Birkedal-Hansen H, Moore WG, Bodden MK,
Windsor LJ, Birkedal-Hansen B, DeCarlo A and Engler JA: Matrix
metallo-proteinases: A review. Crit Rev Oral Biol Med. 4:197–250.
1993.
|
|
19
|
Subramanian A and Schilling TF:
Thrombospondin-4 controls matrix assembly during development and
repair of myotendinous junctions. eLife. 3:e023722014. View Article : Google Scholar :
|
|
20
|
Frolova EG, Pluskota E, Krukovets I, Burke
T, Drumm C, Smith JD, Blech L, Febbraio M, Bornstein P, Plow EF and
Stenina OI: Thrombospondin-4 regulates vascular inflammation and
atherogenesis. Circ Res. 107:1313–1325. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCt method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
22
|
Ding ZB, Shi YH, Zhou J, Shi GM, Ke AW,
Qiu SJ, Wang XY, Dai Z, Xu Y and Fan J: Liver-intestine cadherin
predicts micro-vascular invasion and poor prognosis of hepatitis B
virus-positive hepatocellular carcinoma. Cancer. 115:4753–4765.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Narouz-Ott L, Maurer P, Nitsche DP, Smyth
N and Paulsson M: Thrombospondin-4 binds specifically to both
collagenous and non-collagenous extracellular matrix proteins via
its C-terminal domains. J Biol Chem. 275:37110–37117. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Adams JC: Functions of the conserved
thrombospondin carboxy-terminal cassette in cell-extracellular
matrix interactions and signaling. Int J Biochem Cell Biol.
36:1102–1114. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Arber S and Caroni P: Thrombospondin-4, an
extracellular matrix protein expressed in the developing and adult
nervous system promotes neurite outgrowth. J Cell Biol.
131:1083–1094. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Stenina OI, Desai SY, Krukovets I, Kight
K, Janigro D, Topol EJ and Plow EF: Thrombospondin-4 and its
variants: Expression and differential effects on endothelial cells.
Circulation. 108:1514–1519. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Greco SA, Chia J, Inglis KJ, Cozzi SJ,
Ramsnes I, Buttenshaw RL, Spring KJ, Boyle GM, Worthley DL, Leggett
BA and Whitehall VL: Thrombospondin-4 is a putative
tumour-suppressor gene in colorectal cancer that exhibits
age-related methylation. BMC cancer. 10:4942010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Förster S, Gretschel S, Jöns T, Yashiro M
and Kemmner W: THBS4, a novel stromal molecule of diffuse-type
gastric adenocarcinomas, identified by transcriptome-wide
expression profiling. Mod Pathol. 24:1390–1403. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dakhova O, Ozen M, Creighton CJ, Li R,
Ayala G, Rowley D and Ittmann M: Global gene expression analysis of
reactive stroma in prostate cancer. Clin Cancer Res. 15:3979–3989.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gruel N, Lucchesi C, Raynal V, Rodrigues
MJ, Pierron G, Goudefroye R, Cottu P, Reyal F, Sastre-Garau X,
Fourquet A, et al: Lobular invasive carcinoma of the breast is a
molecular entity distinct from luminal invasive ductal carcinoma.
Eur J Cancer. 46:2399–2407. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang Q, Lan F, Wang X, Yu Y, Ouyang X,
Zheng F, Han J, Lin Y, Xie Y, Xie F, et al: IL-1β-induced
activation of p38 promotes metastasis in gastric adenocarcinoma via
upregulation of AP-1/c-fos, MMP2 and MMP9. Mol Cancer. 13:182014.
View Article : Google Scholar
|
|
32
|
Chien ST, Lin SS, Wang CK, Lee YB, Chen
KS, Fong Y and Shih YW: Acacetin inhibits the invasion and
migration of human non-small cell lung cancer A549 cells by
suppressing the p38α MAPK signaling pathway. Mol Cell Biochem.
350:135–148. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Frolova EG, Pluskota E, Krukovets I, Burke
T, Drumm C, Smith JD, Blech L, Febbraio M, Bornstein P, Plow EF and
Stenina OI: Thrombospondin-4 regulates vascular inflammation and
atherogenesis. Circ Res. 107:1313–1325. 2010. View Article : Google Scholar : PubMed/NCBI
|