Introduction
Thiazolidinediones (TZDs) are therapeutic agents
commonly used to treat patients with type 2 diabetes, they have
been demonstrated to affect bone metabolism (1). It has been reported that long-term
usage of TZDs in diabetic patients induced bone loss and a higher
risk of fracture (2,3).
Bone homeostasis depends on the balance between bone
formation by osteoblasts and bone resorption by osteoclasts
(4). Bone loss and osteoporosis
occur when bone resorption exceeds bone formation, and
osteopetrosis may occur when bone formation predominates (5). The molecular osteoprotegerin (OPG) /
receptor activator of nuclear factor-κB ligand (RANKL) / RANK axis
is important in the regulation of homeostasis (6). OPG and RANKL are secreted by
osteoblasts, RANKL mediates osteoclastogenesis by binding RANK
expressed by osteoclasts, whereas OPG acts as a decoy receptor of
RANKL to prevent osteoclastogenesis (6). Thus, osteoclastogenesis depends on
the ratio of OPG to RANKL secreted by osteoblasts (6,7).
TZDs act via peroxisome proliferator-activated
receptor γ (PPARγ), which is a member of the nuclear receptor
super-family of transcription factors (8), and the predominant factor involved in
adipose metabolism. Activation of PPARγ results in adipogenic
differentiation in various kinds of progenitor cells (9,10). A
number of studies have demonstrated that TZDs inhibit
osteoblastogenesis directly, but its effect on osteoclastogenesis
remains to be determined. Few studies have investigated the effect
of TZDs on the OPG/RANKL/RANK system and the paracrine regulation
of osteoclastogenesis.
The present study investigated the direct effect of
pioglitazone (PIO), a TZD, on osteoblastogenesis and
osteoclastogenesis, and the paracrine mechanisms by which PIO
affects osteoclastogenesis were also investigated by performing a
co-culture system of a pre-osteoblastic cell line with bone marrow
mononuclear cells.
Materials and methods
Reagents
Recombinant murine macrophage colony-stimulating
factor (M-CSF) and RANKL were purchased from R&D Systems, Inc.
(Minneapolis, MN, USA). The ALP Staining kit and TRAP Staining kit
were obtained from Sigma-Aldrich (St. Louis, MO, USA). Anti-RANKL
(#4816; rabbit polyclonal; 1:1,000) antibody was obtained from Cell
Signaling Technology, Inc. (Danvers, MA, USA) and the anti-OPG
(BA1475-1; rabbit polyclonal; 1:100–400) antibody, OPG and RANKL
ELISA kits were obtained from Wuhan Boster Biological Technology,
Co., Ltd. (Wuhan, China). PIO was obtained from Santa Cruz
Biotechnology, Inc. (Dallas, TX, USA).
Cell culture
The MC3T3-E1 murine pre-osteoblastic cell line was
purchased from the American Type Culture Collection (Manassas, VA,
USA). Cells were cultured in α-modified essential medium (α-MEM;
Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) supplemented
with 100 U/ml penicillin, 100 μg/ml streptomycin and 10%
fetal calf serum (FCS; Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) at 37°C in a humidified atmosphere of 5%
CO2. The media was changed every 3 days.
Isolation of monocytes
Bone marrow cells were isolated from the femora of
C57BL/6 mice (male; age, 6–8 weeks; weight, 18–22 g; Beijing HFK
Bioscience Co., Ltd., Beijing, China). The mice were housed
separately in a temperature- and humidity-controlled (20–26°C and
40–70%, respectively) environment, with a 12/12 h light/dark cycle
and free access to food and water. The mice were sacrificed by
CO2 inhalation and cervical dislocation. The cells were
cultured in α-MEM supplemented with 100 U/ml penicillin, 100
μg/ml streptomycin and 15% FCS at 37°C in a humidified
atmosphere of 5% CO2. After 24 h of culture, the
non-adherent BMMCs were collected. All procedures were approved by
the Animal Care and Use Committee, Tongji Medical College, Huazhong
University of Science and Technology (Wuhan, China).
Osteoblast differentiation
For differentiation studies, MC3T3-E1 cells were
cultured in 6-well plates (Corning Incorporated, Corning, NY, USA)
at a density of 5×106 cells/well. After 24 h, the
culture medium was replaced with conditioned medium [α-MEM
supplemented with 10% FCS, 10 mM β-glycerophosphate
(Sigma-Aldrich), 50 μg/ml L-ascorbic acid (Sigma-Aldrich)
and 100 nM dexamethasone (Wuhan Boster Biological Technology, Co.,
Ltd.)] with dimethyl sulfoxide (DMSO; as control; Santa Cruz
Biotechnology, Inc.), PIO 0.1 μM or PIO 1 μM for 14
days. The media were changed every 3 days. For the 24 h prior to
harvesting, the medium of each well was changed to 2 ml conditioned
medium without FCS, and the medium was collected for ELISA
analysis. The harvested cells were used to conduct ALP staining and
activity measurement, reverse transcription-quantitative polymerase
chain reaction (RT-qPCR) and western blotting.
Osteoclast differentiation
BMMCs were cultured in 96-well plates (Corning
Incorporated) at a density of 5,000 cells/well in conditioned
medium containing 50 ng/ml M-CSF and 50 ng/ml RANKL and were
treated with DMSO, 0.1 μM PIO or 1 μM PIO. After 4
days, the cells were harvested and TRAP staining and activity
measurement, and western blotting were conducted.
OPG and RANKL ELISA
OPG and RANKL levels in the medium of osteoblast
differentiation were analyzed using a commercially available ELISA
kit according to the manufacturer's protocols. Each sample was
assessed 3 times.
ALP activity and TRAP activity
measurement
For measurement of ALP activity, the harvested cells
were washed with phosphate-buffered saline (PBS; Gibco; Thermo
Fisher Scientific, Inc.) twice prior to the addition of lysis
buffer [1.5 M Tris-HCl (pH 9.2), 0.1 M ZnCl2, 0.5 M
MgCl2-6H2O, and Triton X-100; Wuhan Boster
Biological Technology, Co., Ltd.], and the cells in each well were
sonicated for 10 sec. The sonicated samples were added to a
substrate solution containing 4-nitrophenyl phosphate
(Sigma-Aldrich), 1.5 M alkaline buffer solution (Sigma-Aldrich) and
H2O and incubated for 20 min at 37°C. Subsequently, 2 N
NaOH was added to the samples to stop the reaction. Absorbance of
the samples was measured at a wavelength of 405 nm using a
microplate reader (Multiskan™ FC; Thermo Fisher Scientific,
Inc.).
For measurement of TRAP activity, the harvested
cells were fixed with 10% formalin (Sigma-Aldrich) for 10 min and
95% ethanol for 1 min, and then 100 μl of citrate buffer (50
mM; pH 4.6; Sigma-Aldrich) containing 10 mM sodium tartrate
(Sigma-Aldrich) and 5 mM p-nitrophenylphosphate (Sigma-Aldrich) was
added to the wells containing fixed cells in the plates. Following
incubation for 1 h, enzyme reaction mixtures in the wells were
transferred to new plates containing an equal volume of 0.1 N NaOH.
Absorbance was measured at a wavelength of 405 nm using a
Multiskan™ FC. Each experiment was performed in triplicate.
Co-culture of BMMCs with MC3T3-E1
cells
Using a 6-well Transwell plate (Corning
Incorporated), MC3T3-E1 cells were cultured in the lower layer at a
density of 5×106 cells/well. After 24 h, the α-MEM
medium was changed to α-MEM medium containing DMSO, 0.1 μM
PIO or 1 μM PIO, and BMMCs were cultured in the upper layer
of the Transwell plate at the density of 5,000 cells/well. The
media was changed every 3 days. After 7 days, the cells in the
upper layer were harvested to conduct TRAP staining.
RNA isolation and RT-qPCR analysis
Total RNA was isolated using TRIzol (Invitrogen;
Thermo Fisher Scientific, Inc.). DNase I, RNase-free (#EN0521) and
the RevertAid First Strand cDNA Synthesis kit (all from Thermo
Fisher Scientific, Inc.) were used for reverse transcription. The
reverse trancription reaction was conducted with total RNA, Oligo
(dT)18 primers and nuclease-free water made up to a total volume of
12 μl. This mixture was then incubated at 65°C for 5 min,
chilled on ice, spun down and placed back on ice. The 5× reaction
buffer (4 μl), RiboLock RNase Inhibitor (1 μl), 10 mM
dNTP Mix (2 μl), RevertAid M-MuLV RT (200 U/μl; 1
μl) were then added to a total volume of 20 μl, the
mixture was mixed gently and centrifuged prior to incubation for 60
min at 42°C. For qPCR, Thermo Fisher Scientific, Inc. Maxima SYBR
Green qPCR Master Mix (2×) Thermal cycling was conducted, which
used a three-step cycling protocol: Pre-treatment at 50°C for 2
min, 1 cycle of 95°C for 10 min; 1 cycle of denaturation at 95°C
for 15 sec; 40 cycles of annealing at 60°C for 30 sec and extension
at 72°C for 30 sec. The forward and reverse primers were obtained
from Invitrogen (Thermo Fisher Scientific, Inc.) and are presented
in Table I. Data are presented as
the mean ± standard deviation for at least three independent
experiments. Gene expression analysis was performed using RT-qPCR
(iCycler iQ5 System; Bio-Rad Laboratories, Inc., Hercules, CA, USA)
and normalized to 18S RNA.
 | Table IList of primers. |
Table I
List of primers.
| Gene | Primer | Product size | Accession no. |
|---|
| 18S | F:
TTCGAACGTCTGCCCTATCAA | 50 | M35283.1 |
| R:
ATGGTAGGCACGGGGACTA | | |
| PPARγ | F:
GGAAGACCACTCGCATTCCTT | 121 | NM_001127330 |
| R:
GTAATCAGCAACCATTGGGTCA | | |
| RUNX2 | F:
GACTGTGGTTACCGTCATGGC | 84 | NM_001146038 |
| R:
ACTTGGTTTTTCATAACAGCGGA | | |
| ALP | F:
GCCTTACCAACTCTTTTGTGCC | 61 | NM_007431 |
| R:
GCTTGCTGTCGCCAGTAAC | | |
| OCN | F:
CTGACCTCACAGATCCCAAGC | 187 | NM_031368 |
| R:
TGGTCTGATAGCTCGTCACAAG | | |
| RANK | F:
CCAGGAGAGGCATTATGAGCA | 94 | AF019046.1 |
| R:
ACTGTCGGAGGTAGGAGTGC | | |
| Cathepsin K | F:
GAAGAAGACTCACCAGAAGCAG | 136 | NM_007802 |
| R:
CTGTATTCCCCGTTGTGTAGC | | |
| TRAP | F:
CACTCCCACCCTGAGATTTGT | 118 | NM_001102405 |
| R:
CATCGTCTGCACGGTTCTG | | |
ALP and TRAP staining
Harvested cells were rinsed three times with PBS and
fixed for 15 min in 4% paraformaldehyde (Wuhan Boster Biological
Technology, Co., Ltd.) at 4°C. The cells were stained with ALP and
TRAP using kits according to the manufacturer's protocols. The
staining was analyzed using an OsteoMeasure system (Osteometrics,
Inc., Atlanta, GA, USA) connected to a Axioskop microscope (Zeiss,
Oberkochen, Germany).
Western blotting
Total cell lysates were obtained by lysing cells in
radioimmunoprecipitation assay buffer (Wuhan Boster Biological
Technology, Co., Ltd.) containing 50 mM Tris-HCl, 150 mM NaCl, 1%
NP-40, 0.1% SDS, 0.5% sodium deoxycholate, 2 mM sodium fluoride, 1
mM EDTA, 1 mM EGTA and a protease inhibitor cocktail. Protein
concentration was determined using a Pierce BCA protein assay kit
(Thermo Fisher Scientific, Inc.). The proteins (20 μg) were
separated by 10% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis, transferred to nitrocellulose membranes, blocked
with bovine serum albumin (Wuhan Boster Biological Technology, Co.,
Ltd.) in Tris-buffered saline with 0.1% Tween-20 (TBST) for 1 h at
room temperature, and incubated with primary antibodies (anti-OPG
and anti-RANKL) overnight at 4°C. The membrane was washed three
times with TBST and incubated with the horseradish
peroxidase-conjugated secondary antibodies (BA1039; goat
anti-rabbit monoclonal; 1:1,000; Wuhan Boster Biological
Technology, Co., Ltd.) for 1 h at room temperature, then washed
three further times with TBST. Protein detection was conducted with
a Pierce enhanced chemiluminescence detection system (Thermo Fisher
Scientific, Inc.). Densitometry was conducted using Quantity One
software (Bio-Rad Laboratories, Inc.).
Statistical analysis
All data were presented as the mean ± standard
deviation. Statistical analysis was performed using one-way
analysis of variance with SPSS software, version 12.0 (SPSS, Inc.,
Chicago, IL, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
PIO inhibits the differentiation of
MC3T3-E1 cells into osteoblasts
To investigate the effect of PIO on osteoblast
differentiation, MC3T3-E1 cells were cultured in osteoblastic
differentiation media with or without PIO. ALP staining and ALP
activity served as biomarkers of osteoblastic differentiation
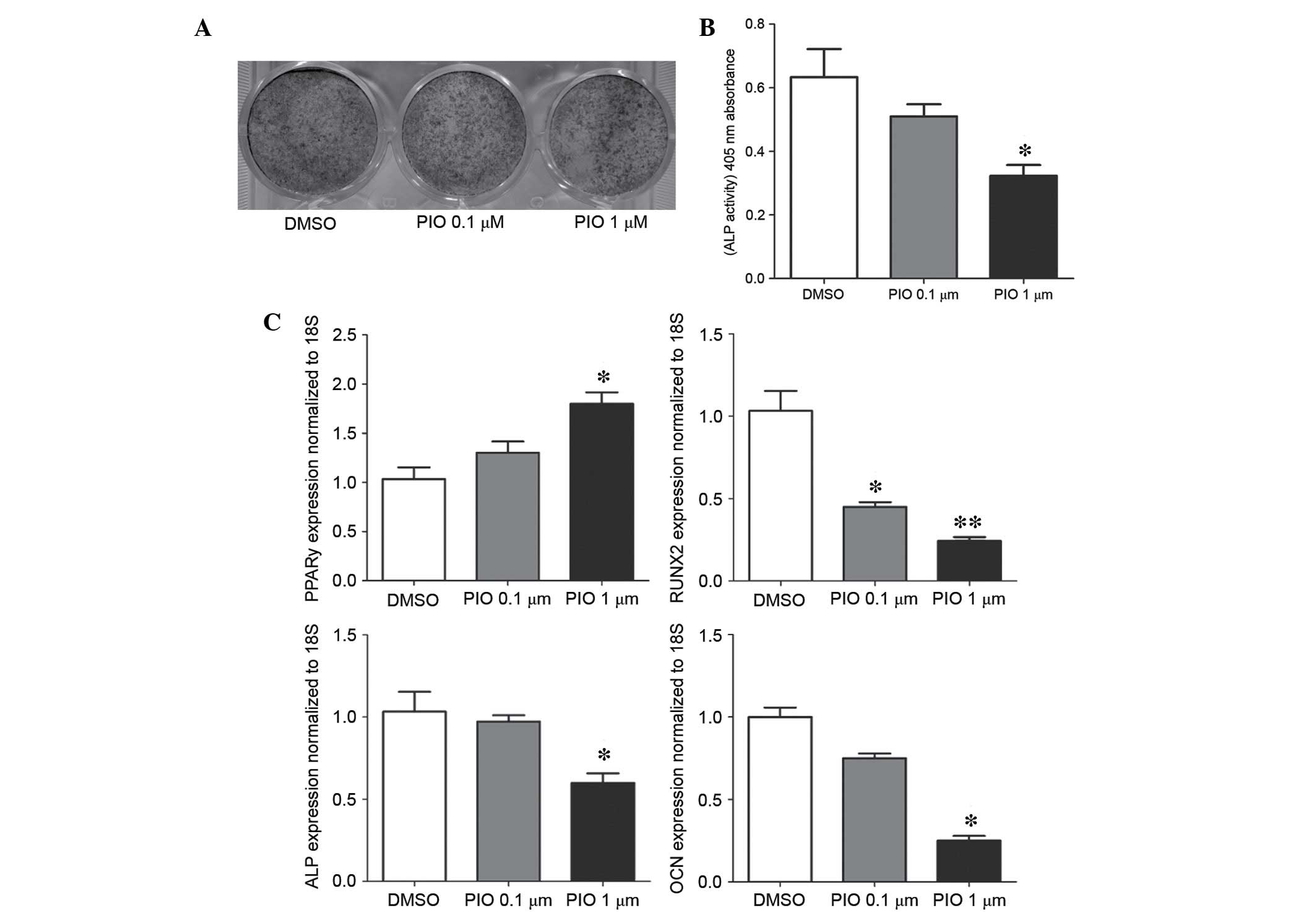
(Fig. 1). As presented in Fig. 1A and B, 1 μM PIO
significantly reduced the number of ALP positive osteoblasts and
ALP activity when compared with the control group (P<0.05). In
addition, expression levels of osteoblastic genes, Runt-related
transcription factor 2 (RUNX2) (P<0.01), ALP (P<0.05) and
osteocalcin (OCN; P<0.05) were significantly decreased following
treatment with 1 μM PIO (Fig.
1C). By contrast, PPARγ was significantly increased (P<0.05;
Fig. 1C). No significant
alterations were observed following 0.1 μM PIO treatment,
apart from in RUNX2 expression.
PIO promotes the differentiation of BMMCs
into osteoclasts
The effect of pioglitazone on osteoclast
differentiation was investigated in BMMCs by culturing the cells in
the presence of RANKL (50 ng/ml) and M-CSF (50 ng/ml) with or
without PIO, and detecting the number of TRAP-positive cells and
TRAP activity, which indicate the degree of osteoclast
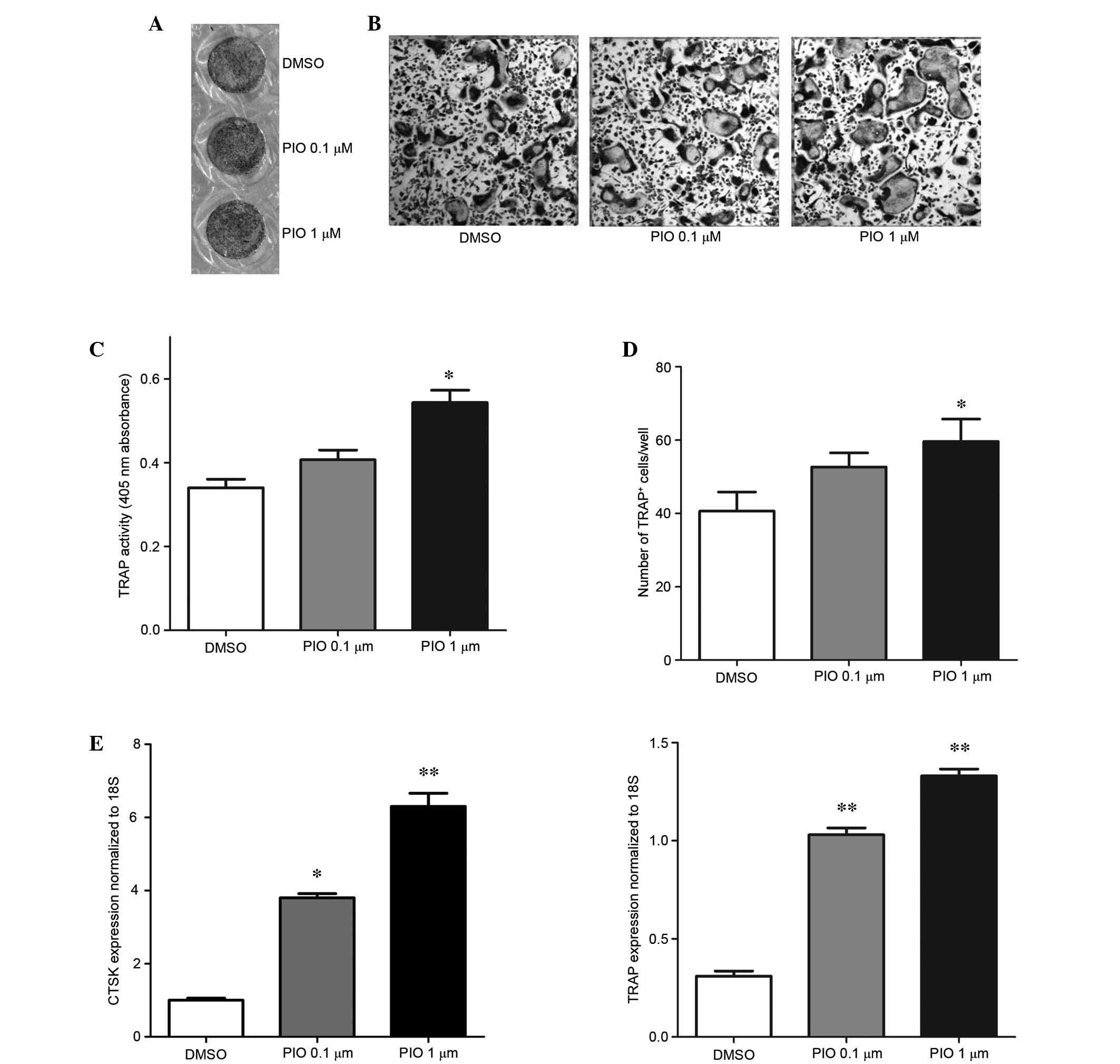
differentiation (Fig. 2). As
presented in Fig. 2A,
RANKL-mediated osteoclast differentiation was promoted by 1
μM PIO. The number of TRAP-positive cells (Fig. 2C and D) and TRAP activity (Fig. 2B) were significantly increased
following treatment with 1 μM PIO. In addition, the gene
expression levels of TRAP and cathepsin K, which indicate the
function of osteoclasts, were significantly increased (P<0.05;
Fig. 2E). Treatment with 0.1
μM PIO did not significantly affect the osteoclast
differentiation of BMMCs.
PIO decreases the OPG/RANKL ratio in
osteoblasts
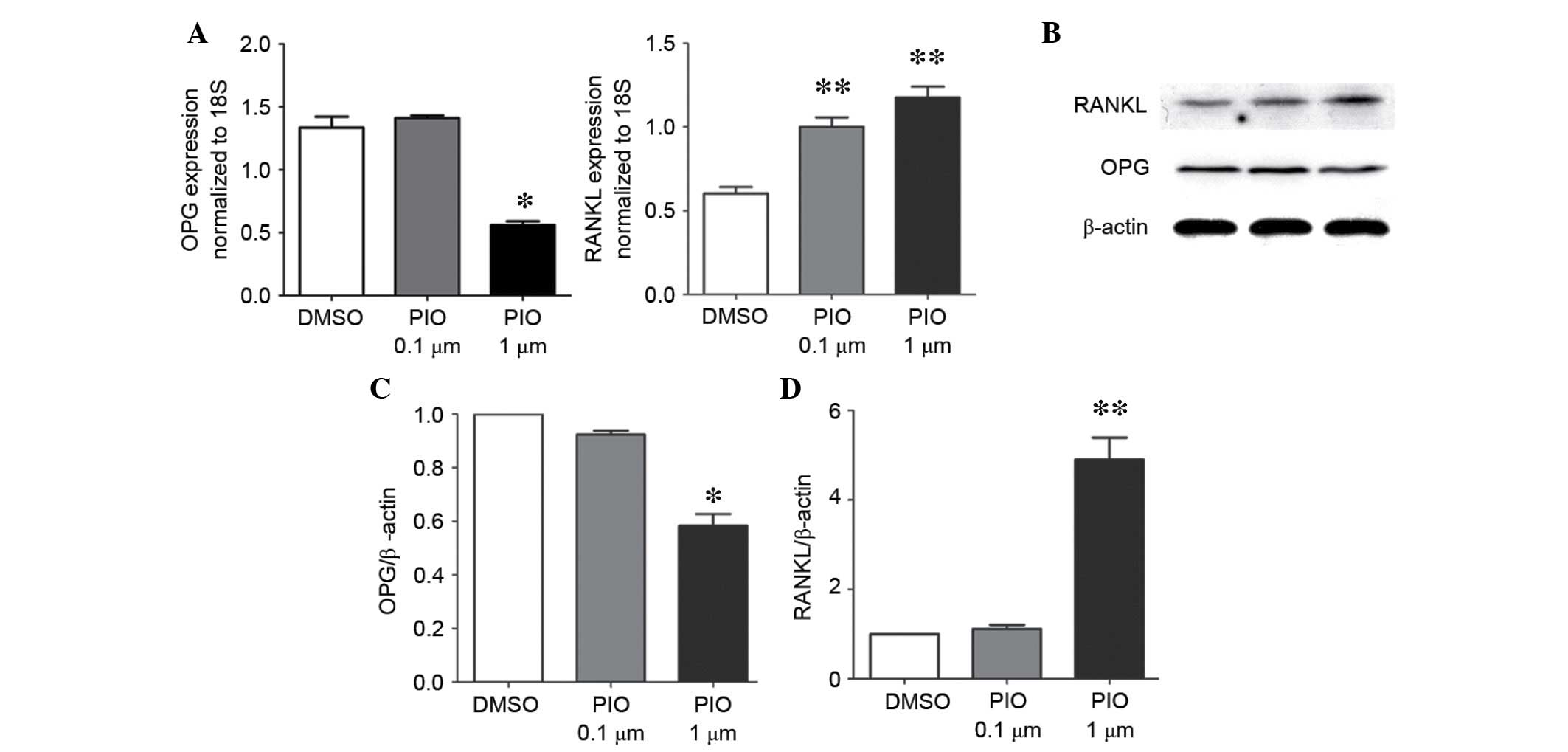
The MC3T3-E1 cells were treated with PIO or DMSO for
7 days, and the mRNA and protein expression levels of OPG and RANKL
were analyzed using RT-qPCR and western blotting, respectively. As
presented in Fig. 3, the 1
μM PIO group exhibited a significant decrease of the ratio
of OPG/RANKL mRNA expression levels (P<0.05) due to decreased
OPG and increased RANKL expression levels compared with the control
group. Furthermore, the protein expression levels of OPG, RANKL and
ratio of OPG to RANKL demonstrated the same trend of change as
observed for the mRNA expression levels. The cells treated with 0.1
μM PIO demonstrated no significant difference compared with
that of the control group.
PIO promotes osteoclastogenesis by
modulating the osteoblast and osteoclast cross-talk
The OPG and RANKL expression levels in the
osteoblast culture medium of MC3T3-E1 cells were detected by ELISA.
Following PIO treatment, OPG expression levels decreased and RANKL
expression levels increased, compared with DMSO-treated controls
(Table II). Few TRAP positive
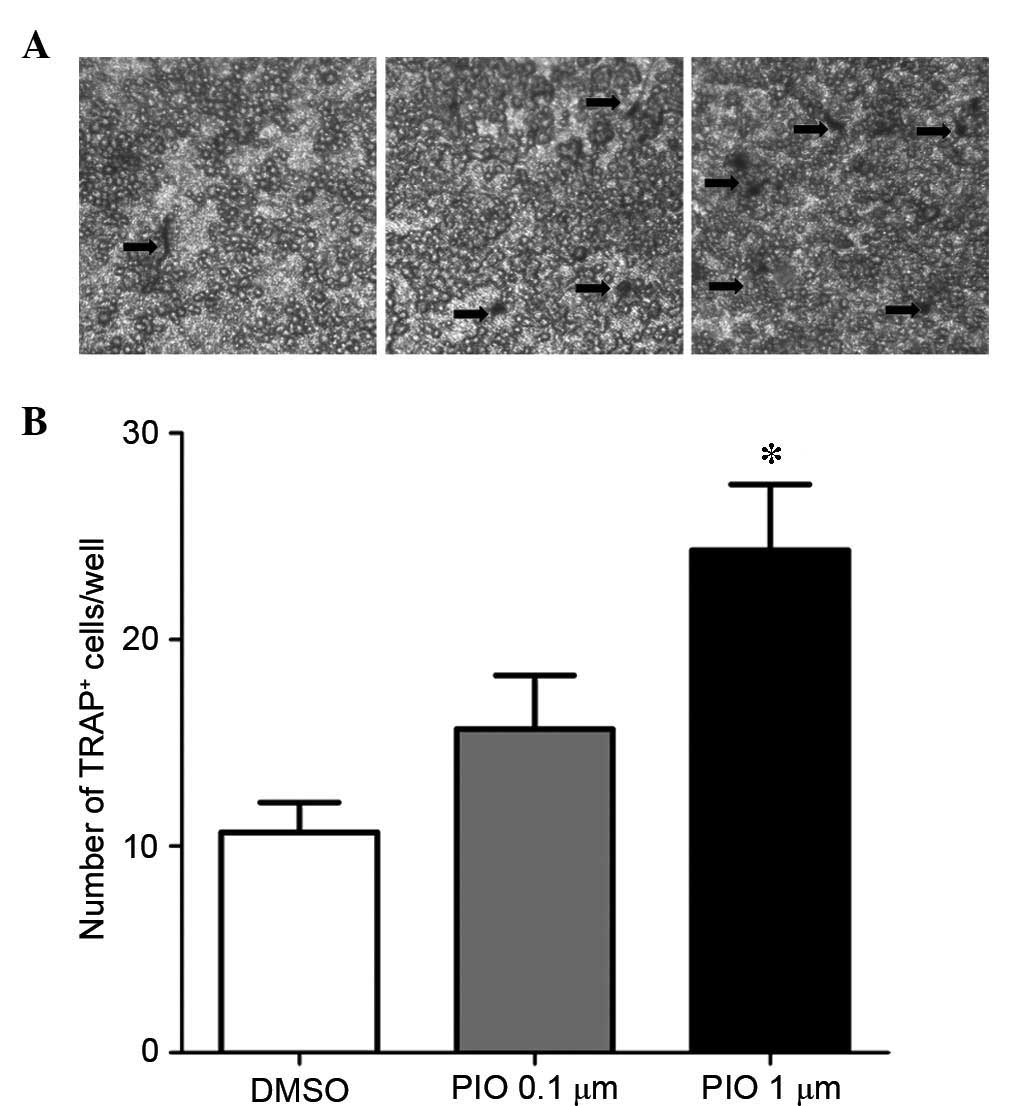
cells were observed in BMMCs co-cultured with MC3T3-E1 cells
without PIO treatment after 7 days (Fig. 4A). Following addition of PIO, an
increased number of TRAP positive cells were observed (Fig. 4). Furthermore, the number of
TRAP-positive cells among BMMCs in the PIO-treated co-culture
system was significantly increased compared with BMMCs untreated
with PIO (P<0.05; Fig. 4B).
Notably, in these experiments, 0.1 μM PIO demonstrated the
same effects as 1 μM pioglitazone, however, the difference
was not statistically significant (Fig. 4A and B).
 | Table IIThe OPG and RANKL levels in
osteoblast culture medium (detected by ELISA). |
Table II
The OPG and RANKL levels in
osteoblast culture medium (detected by ELISA).
| Gene | DMSO (n=3) | PIO 0.1 μM
(n=3) | PIO 1 μM
(n=3) |
|---|
| OPG (pg/ml) | 476.5±26.8 | 431.6±26.18 | 326.12±34.4a |
| RANKL (pg/ml) | 792.5±43.1 |
815.5±33.2 | 1265.1±79.2b |
Discussion
Pioglitazone inhibited osteoblast
differentiation and promoted osteoclast formation
As one of thiazolidinediones that are commonly
prescribed for patients with diabetes, PIO is a PPARγ agonist,
which may result in bone loss and an increased fracture rate in
diabetic patients (2). The present
study demonstrated that PIO inhibited osteoblast differentiation
and promoted osteoclast formation directly, and enhanced
osteoclastogenesis via the OPG/RANKL/RANK system, which is involved
in paracrine regulation of osteoclastogenesis.
PIO negatively regulates osteoblast
differentiation
Consistently with previous studies, the present
study demonstrated that PIO inhibited osteoblastogenesis.
Osteoblasts and adipocytes are commonly derived from mesenchymal
stem cells (MSCs), and their differentiation is reliant on
different stimulating signals. Activation of core binding
factor-α1/RUNX2 results in osteoblast formation from MSCs, however,
activation of PPARγ results in adipocyte formation (11,12).
Competition exists between the differentiation of the two cell
types, previous studies have shown that TDZs promote
adipocytogenesis at expense of osteoblastogenesis (13–16).
Furthermore, silencing PPARγ using synthetic small interfering RNA
inhibited adipocyte differentiation and induced osteoblastic
differentiation (17). However,
the underlying mechanisms remain to be elucidated. In the present
study, PIO was observed to upregulate PPARγ and downregulate RUNX2
in parallel with inhibition of osteoblastogenesis. Results from the
current study support the findings of Jeon et al (18) that activation of PPARγ interfered
with the transactivation ability of RUNX2 and suppressed its
expression, thus resulting in the inhibition of OCN (an
osteoblast-specific protein) expression. Furthermore, apoptosis of
osteoblasts has been proven to be accelerated by TZDs. By contrast,
Bruedigam et al (19)
demonstrated that rosiglitazone promoted osteoblastogenesis and
produced a large quantity of reactive oxygen species and increased
apoptosis, which resulted in attenuation of osteoblastogenesis.
PIO increases osteoclastogenesis
Osteoclasts are derived from bone marrow
hematopoietic stem cells, which may be stimulated to differentiate
by RANKL signaling and PPARγ activation (20). TZDs have been reported to affect
osteoclast differentiation, however, results from various previous
studies are conflicting. Chan et al (21) demonstrated that ciglitazone, a TZD,
suppressed multinucleated osteoclast formation in a dose-dependent
manner. Furthermore, Cho et al (15) demonstrated that rosiglitazone
attenuated osteoclast formation and bone resorption by preventing
RANK and enhancing PPARγ2 expression in osteoclasts. By contrast,
Wan et al (22)
demonstrated that activation of PPARγ stimulated osteoclastogenesis
and bone resorption, and deletion of PPARγ prevented osteoclast
formation and resulted in osteopetrosis. Wu et al (23) also demonstrated that rosiglitazone
markedly increased the differentiation of mice bone marrow cells
into osteoclasts. In the present study, administration of exogenous
RANKL and M-CSF, which are required for osteoclast differentiation,
demonstrated that PIO exerts a direct effect on promoting
differentiation of BMMCs into osteoclasts. Furthermore, contrary to
results from Cho et al (15), the RANK expression levels in the
cells remained unchanged, which indicated that this effect of PIO
may involve PPARγ but not the RANKL-RANK response. Although c-Fos
induction, TNF receptor-associated factor 6 and downstream
extracellular signal-regulated kinase signaling, PPARγ coactivator
1β and estrogen-related receptor α have been reported to be
associated with enhancement of PPARγ-mediated osteoclastogenesis
(22,24), the underlying mechanisms remain to
be elucidated.
PIO decreases the OPG/RANKL ratio in
osteoblasts
The molecular OPG/RANKL/RANK axis participates in
regulating bone metabolism, provides paracrine regulation for
osteoclastogenesis (25). RANKL,
expressed and secreted by osteoblasts, binds the extracellular RANK
domain of pre-osteoclasts, and results in expression of specific
genes involved in osteoclast differentiation and bone resorption
(26). Furthermore, OPG, secreted
by osteoblasts, acts as a soluble receptor antagonist for RANKL, to
prevent it binding to RANK and decreases osteoclastogenesis
(6). Thus, the OPG/RANKL ratio and
the expression of RANK in pre-osteoclasts are key in
osteoclastogenesis (6,7). However, studies concerning the effect
of TZDs on this paracrine regulation are rare.
In the present study, it was observed that the
OPG/RANKL ratio decreased, due to a decreased OPG expression level
and an elevated RANKL expression level, in osteoblasts in response
to treatment with PIO. Furthermore, the expression levels of RANK
remained unchanged following PIO treatment. Thus, the results of
the present study suggest, in addition to the direct effect, PIO
may positively regulate osteoclastogenesis via influencing the
OPG/RANKL/RANK axis in the co-culture system mimicking the in
vivo bone marrow microenvironment. Consistent with these
findings, Lazarenko et al (27) demonstrated that PPARγ activation in
osteoblasts promotes osteoclast differentiation by inducing the
expression of RANKL. Previous in vitro and in vivo
studies have demonstrated that TZD treatment lowers OPG levels
(28–30). Contrary to results in the present
study, Cho et al (15)
demonstrated that rosiglitazone inhibited the RANK protein
expression in monocytes induced by RANKL. This difference may be
attributed to the higher dose of TZD used. In future experiments a
larger range of doses of TZDs should be evaluated.
In conclusion, the present study demonstrates that
PIO suppresses osteoblastogenesis and enhances osteoclastogenesis
directly. It also decreases the OPG/RANKL ratio in osteoblasts, to
promote osteoclast formation via a paracrine mechanism.
Acknowledgments
The present study was supported by the National
Natural Sciences Research Program of China (grant no.
81070691).
References
|
1
|
Derosa G: Efficacy and tolerability of
pioglitazone in patients with type 2 diabetes mellitus: Comparison
with other oral antihyperglycaemic agents. Drugs. 70:1945–1961.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
McDonough AK, Rosenthal RS, Cao X and Saag
KG: The effect of thiazolidinediones on BMD and osteoporosis. Nat
Clin Pract Endocrinol Metab. 4:507–513. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Montagnani A and Gonnelli S: Antidiabetic
therapy effects on bone metabolism and fracture risk. Diabetes Obes
Metab. 15:784–791. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Raggatt LJ and Partridge NC: Cellular and
molecular mechanisms of bone remodeling. J Biol Chem.
285:25103–25108. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lazner F, Gowen M, Pavasovic D and Kola I:
Osteopetrosis and osteoporosis: Two sides of the same coin. Hum Mol
Genet. 8:1839–1846. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Khosla S: Minireview: The OPG/RANKL/RANK
system. Endocrinology. 142:5050–5055. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Eriksen EF: Cellular mechanisms of bone
remodeling. Rev Endocr Metab Disord. 11:219–227. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lehmann JM, Moore LB, Smith-Oliver TA,
Wilkison WO, Willson TM and Kliewer SA: An antidiabetic
thiazolidinedione is a high affinity ligand for peroxisome
proliferator-activated receptor gamma (PPAR gamma). J Biol Chem.
270:12953–12956. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tontonoz P, Hu E and Spiegelman BM:
Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a
lipid-activated transcription factor. Cell. 79:1147–1156. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
MacDougald OA and Lane MD: Transcriptional
regulation of gene expression during adipocyte differentiation.
Annu Rev Biochem. 64:345–373. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gimble JM, Robinson CE, Wu X, Kelly KA,
Rodriguez BR, Kliewer SA, Lehmann JM and Morris DC: Peroxisome
proliferator-activated receptor-gamma activation by
thiazolidinediones induces adipogenesis in bone marrow stromal
cells. Mol Pharmacol. 50:1087–1094. 1996.PubMed/NCBI
|
|
12
|
Mabilleau G, Chappard D and Baslé MF:
Cellular and molecular effects of thiazolidinediones on bone cells:
A review. Int J Biochem Mol Biol. 2:240–246. 2011.PubMed/NCBI
|
|
13
|
Wang L, Li L, Gao H and Li Y: Effect of
pioglitazone on transdifferentiation of preosteoblasts from rat
bone mesenchymal stem cells into adipocytes. J Huazhong Univ Sci
Technolog Med Sci. 32:530–533. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu L, Aronson J and Lecka-Czernik B:
Rosiglitazone disrupts endosteal bone formation during distraction
osteogenesis by local adipocytic infiltration. Bone. 52:247–258.
2013. View Article : Google Scholar
|
|
15
|
Cho ES, Kim MK, Son YO, Lee KS, Park SM
and Lee JC: The effects of rosiglitazone on osteoblastic
differentiation, osteoclast formation and bone resorption. Mol
Cells. 33:173–181. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Viccica G, Francucci CM and Marcocci C:
The role of PPARγ for the osteoblastic differentiation. J
Endocrinol Invest. 33(Suppl 7): S9–S12. 2010.
|
|
17
|
Yamashita A, Takada T, Nemoto K, Yamamoto
G and Torii R: Transient suppression of PPARgamma directed ES cells
into an osteoblastic lineage. FEBS Lett. 580:4121–4125. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jeon MJ, Kim JA, Kwon SH, Kim SW, Park KS,
Park SW, Kim SY and Shin CS: Activation of peroxisome
proliferator-activated receptor-gamma inhibits the Runx2-mediated
transcription of osteocalcin in osteoblasts. J Biol Chem.
278:23270–23277. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bruedigam C, Eijken M, Koedam M, van de
Peppel J, Drabek K, Chiba H and van Leeuwen JP: A new concept
underlying stem cell lineage skewing that explains the detrimental
effects of thiazolidinediones on bone. Stem Cells. 28:916–927.
2010.PubMed/NCBI
|
|
20
|
Teitelbaum SL and Ross FP: Genetic
regulation of osteoclast development and function. Nat Rev Genet.
4:638–649. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chan BY, Gartland A, Wilson PJ, Buckley
KA, Dillon JP, Fraser WD and Gallagher JA: PPAR agonists modulate
human osteoclast formation and activity in vitro. Bone. 40:149–159.
2007. View Article : Google Scholar
|
|
22
|
Wan Y, Chong LW and Evans RM: PPAR-gamma
regulates osteoclastogenesis in mice. Nat Med. 13:1496–1503. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu H, Li L, Ma Y, Chen Y, Zhao J, Lu Y and
Shen P: Regulation of selective PPARγ modulators in the
differentiation of osteoclasts. J Cell Biochem. 114:1969–1977.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wei W, Wang X, Yang M, Smith LC, Dechow
PC, Sonoda J, Evans RM and Wan Y: PGC1beta mediates PPARgamma
activation of osteoclastogenesis and rosiglitazone-induced bone
loss. Cell Metab. 11:503–516. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hofbauer LC, Khosla S, Dunstan CR, Lacey
DL, Boyle WJ and Riggs BL: The roles of osteoprotegerin and
osteoprotegerin ligand in the paracrine regulation of bone
resorption. J Bone Miner Res. 15:2–12. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Muruganandan S, Roman AA and Sinal CJ:
Adipocyte differentiation of bone marrow-derived mesenchymal stem
cells: Cross talk with the osteoblastogenic program. Cell Mol Life
Sci. 66:236–253. 2009. View Article : Google Scholar
|
|
27
|
Lazarenko OP, Rzonca SO, Hogue WR, Swain
FL, Suva LJ and Lecka-Czernik B: Rosiglitazone induces decreases in
bone mass and strength that are reminiscent of aged bone.
Endocrinology. 148:2669–2680. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Park JS, Cho MH, Nam JS, Yoo JS, Ahn CW,
Cha BS, Kim KR and Lee HC: Effect of pioglitazone on serum
concentrations of osteoprotegerin in patients with type 2 diabetes
mellitus. Eur J Endocrinol. 164:69–74. 2011. View Article : Google Scholar
|
|
29
|
Sultan A, Avignon A, Galtier F, Piot C,
Mariano-Goulart D, Dupuy AM and Cristol JP: Osteoprotegerin,
thiazolidinediones treatment and silent myocardial ischemia in type
2 diabetic patients. Diabetes Care. 31:593–595. 2008. View Article : Google Scholar
|
|
30
|
Krause U, Harris S, Green A, Ylostalo J,
Zeitouni S, Lee N and Gregory CA: Pharmaceutical modulation of
canonical Wnt signaling in multipotent stromal cells for improved
osteoinductive therapy. Proc Natl Acad Sci USA. 107:4147–4152.
2010. View Article : Google Scholar : PubMed/NCBI
|