Introduction
Hematological malignancies (HMs) are clonal
malignant disorders originating from cells of the bone marrow or
the lymphatic system (1). Despite
advances in HM treatment, including combination chemotherapy,
targeted therapy and bone marrow transplantation, the disease
relapses and the five-year survival rate of patients with HMs
remains low (2,3). Therefore, additional therapeutic
strategies are required. Previous studies on the involvement of
immunogenicity in cancer development have suggested cancer vaccines
as a promising and innovative alternative for the treatment of HMs
(4–6). The use of cancer vaccines is an
approach for active immunotherapy, in which a formulation of
specific tumor-associated antigen (TAA) is delivered, together with
immunomodulatory agents, to patients with cancer. This activates
the host's immune system to mount an immune response, predominantly
through cytotoxic T lymphocyte (CTL)-mediated cytolysis, against
TAA and eliminates cancer cells (7). Therefore, the key for developing a
successful cancer vaccination against solid or hematological types
of cancer is to identify specific TAAs, which are selectively
presented by tumor cells or dendritic cells (8).
Epidermal growth factor receptor pathway substrate 8
(EPS8) is a 97-kDa protein originally identified as a substrate for
kinase activity of epidermal growth factor (EGF) receptor, and
augments the mitogenic signaling downstream of EGF (9). The constitutive phosphorylation of
EPS8 has been demonstrated to promote the malignant transformation
in tumor cells (10). Previous
studies have shown that EPS8 is important in several types of solid
tumor, including colon, pancreatic, pituitary and cervical cancer
(11–14), where the amplification of EPS8 is
associated with tumor progression, acquired drug resistance and
poor prognosis (15). Aside from
solid tumor types, our previous study demonstrated a significant
correlation between elevated expression levels of EPS8 and poor
overall survival rates in patients with acute myeloid leukemia
(AML) (16). Considering the
biological significance of EPS8 in cancer, using the full-length
EPS8 protein as the cancer vaccine in a murine model of breast
carcinoma, He et al (17)
showed that the EPS8 vaccine induced a marked CTL response in
vitro and effectively prevented tumor growth in vivo
(17). These findings suggest that
EPS8 is an ideal target antigen for the development of antitumor
vaccines.
CTL epitopes are peptides presented by the human
leukocyte antigen (HLA) class I molecules and recognized by CTLs.
In the present study, the HLA-A*1101 allele was selected for
investigation as it is the most common HLA-I allele within the
Chinese population (18,19). The present study identified five
potential EPS8 peptides, which bind with high affinity to
HLA-A*1101. Among these, P380 showed the highest immunogenicity in
CTLs, indicating it as a promising immunotherapeutic target for the
treatment of HMs.
Materials and methods
Cell lines
The K562 human erythroleukemia cell line, the THP-1
human acute monocytic leukemia cell line and the SW480 colon cancer
cell line were obtained from the Type Culture Collection of the
Chinese Academy of Sciences (Shanghai, China) and were routinely
preserved in the Department of Hematology laboratory Zhujiang
Hospital, Southern Medical University (Guangzhou, China). The
HLA-A*1101+ K562 cell line (20) was obtained from the Hematology
Institute of Jinan University (Guangzhou, China). The
HLA-A*1101+ K562 cell line in which the expression of
EPS8 was absent was obtained from the transient transfection of
HLA-A*1101+ K562 cells with small interfering RNA
(siRNA) specifically targeting EPS8. All cell lines were cultured
in RPMI-1640 medium (Hyclone; GE Healthcare Life Sciences, Logan,
UT, USA) supplemented with 10% fetal calf serum, 100 IU/ml
penicillin and 100 µg/ml streptomycin (Biological Industries, Beit
Haemek, Israel) in a humidified 37°C incubator with 5% CO2.
siRNA transfection
siRNA targeting EPS8 and control siRNA (NC siRNA)
(21), which targeted no known
human genes, were synthesized by Guangzhou RiboBio Co., Ltd.
(Guangzhou, China). The sequences were as follows: si-h-EPS8101,
5′-GGUGGAUGUUAGAAGUCGA dTdT-3′, si-h-EPS8102,
5′-GGACACAAUUGAUUUCUUA dTdT-3′ and si-h-EPS8103,
5′-GAUCCACCUUAUACUCAUA dTdT-3′.
To knock down the expression of EPS8, the
HLA-A*1101+ K562 cells were seeded into 24-well plates
at a density of 1×105 cells/well and allowed to grow to
~80% confluence. Subsequently, 50 nM EPS8 siRNA or NC siRNA was
transiently transfected into the cells using
Lipofectamine® RNAiMAX transfection reagent at room
temperature (Life Technologies; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) according to the manufacturer's protocol. At 24,
48 and 72 h post-transfection, the cells were harvested and lysed
in RIPA buffer containing the proteinase inhibitor,
phenylmethanesulfonylfluoride, to extract total proteins.
Western blot analysis
The protein expression levels of EPS8 in the cell
lines were detected using western blot analysis. Briefly, the
protein concentrations were determined from the cell lysates using
a BCA protein assay (Keygen Biotech., Nanjing, China). A 30 µg
quantity of total protein from each sample was electrophoresed on a
10% SDS-polyacrylamide gel and transferred onto a PVDF membrane.
Following blocking in 5% skim milk for 1 h at room temperature, the
membrane was incubated overnight at 4°C in 5% skim milk containing
anti-EPS8 primary antibody (cat. no. ab12488; 1:1,000; Abcam,
Cambridge, UK) or anti-GAPDH antibody (cat. no. KC-5G5; 1:1,000;
Kangcheng Bio-Tech, Shanghai, China) as a loading control.
Following three washes with 0.5% TBS-Tween 20, the membrane was
incubated in 5% skim milk containing horseradish peroxidase
(HRP)-conjugated secondary antibody (cat. no. 4030–05; 1:2,000;
Southern Biotechnology Associates Inc., Birmingham, AL, USA). The
signal was detected using chemiluminescent HRP substrate and
blotted onto chemiluminescence-sensitive film (Merck Millipore,
Billerica, MA, USA).
HLA phenotyping
The HLA-A phenotypes of the cell lines were
determined using flow cytometry with phycoerythrin-conjugated
anti-human HLA-A, B and C antibodies, as described previously
(20,22).
Epitope prediction and peptide
synthesis
The EPS8-derived peptides containing
HLA-A*1101-binding motifs were predicted using two computer
algorithms: Bioinformatics and Molecular Analysis Section (BIMAS;
www.bimas.cit.nih.gov/molbio/hla_bind/) (23) and SYFPEITHI (www.syfpeithi.de/) (24). Peptides among the top 20 peptides
from the BIMAS prediction, and with a cut-off score of ≥20 from the
SYFPEITHI prediction met the two criteria for selection. The
predicted candidate peptides were synthesized using
fluorenylmethyloxycarbonyl chemistry (Zhongtai Biological
Technology, Hangzhou, China), with >95% purity, as determined
using reverse-phase high performance liquid chromatography, and
expected molecular weight, as confirmed using mass spectrometry.
Reverse-phase high-performance liquid chromatography involves the
separation of molecules on the basis of hydrophobicity. The
separation depends on the hydrophobic binding of the solute
molecule from the mobile phase to the immobilized hydrophobic
ligands attached to the stationary phase, i.e. the sorbent.
Mass spectrometry is an analytical technique that ionizes chemical
species and sorts the ions based on their mass to charge ratio. In
simpler terms, a mass spectrum measures the masses within a sample.
The lyophilized peptides were dissolved in DMSO at a concentration
of 2 mg/ml and stored at −20°C until further use.
Isolation of peripheral blood
mononuclear cells (PBMCs) and induction of peptide-specific
CTLs
The present study was approved by the Institutional
Ethics Committee of Southern Medical University (Guangzhou, China)
and informed consent was obtained from all healthy donors. A total
of 10 healthy donors (3 female and 7 male, average age 20.5 years)
were recruited between April and August 2015 in Zhujiang Hospital,
Southern Medical University. PBMCs were purified using standard
Ficoll-Hypaque density gradient centrifugation at 800 × g
for 30 min at room temperature (Dakewe Biotech Co., Ltd., Shenzhen,
China). HLA-A*1101 phenotypic analyses of the donors were performed
using a PCR-SBT tying kit (BGI Tech, Shenzhen, China). To generate
peptide-specific CTLs, the HLA-A*1101+ PBMCs were
incubated with 10 µmol/l candidate peptides (P380, P529, P70, P82,
P30) in RPMI-1640 medium at 37°C in a humidified incubator
containing 5% CO2. As the negative control, no peptide (P0) was
added to the PBMCs. Recombinant human interleukin 2 (rhIL-2) at a
concentration of 50 U/ml was added into the medium on the second
day. The same quantity of candidate peptides and rhIL-2 were added
to the PBMCs every 7 days. On day 3 following the third
stimulation, PBMCs were collected as effector cells.
Enzyme-linked immunospot (ELISPOT)
assay
The production of interferon-γ (IFN-γ) from the
peptide-specific effector cells was analyzed using an ELISPOT
assay. Briefly, the effector cells, prepared as described above,
were seeded into 96-well assay plates (Dakewe Biotech Co., Ltd.) at
a density of 1×105/100 µl/well in serum-containing
RPMI-1640 medium. For restimulation, 1×104
HLA-A*1101+ K562 cells, as antigen-presenting cells,
were irradiated (5 Gy) to prevent further proliferation, and were
added together with different peptides (P380, P70, P82, P30 and
P529) or medium alone (negative control). Following overnight
culture at 37°C and extensive washing using a washing buffer 5–7
times for 30–60 sec each time, to remove free peptides, the
production of IFN-γ was detected using biotinylated anti-IFN-γ
antibody (cat nos. DKW22-1000-048/DKW22-1000-096; 1:100; Dakewe
Biotech Co., Ltd.), followed by incubation at 37°C for 1 h with
streptavidin-HRP solution and 3-amino-9-ethyl carbazole substrate.
The numbers of IFN-γ-positive spot-forming cells (SFCs) were
counted by Dakota Biotechnology Co., Ltd. (Shenzhen, China). As the
positive control, the lymphocyte mitogen, phytohemagglutinin (PHA)
stimulation was used. As a background control, X–VIVO15 serum-free
medium (Lonza, Basel, Switzerland) was used. Each condition was
assessed in triplicate.
Cytotoxicity assay
The cytolytic activity of the effector cells was
assessed using flow cytometry. In brief, the target cells were
labeled with carboxyfluorescein succinimidyl ester (CFSE;
BioLegend, Inc., San Diego, CA, USA) according to the
manufacturer's protocol. The effector cells (1×106/ml)
were mixed with the target cells at different ‘effector to target’
(E/T) ratios (40:1, 20:1, 10:1) and incubated for 4 h in a 37°C
incubator with 5% CO2. The cell mixture was then washed twice with
PBS and stained with propidium iodide (PI; 2.5 µg/ml, BioLegend,
Inc.) at room temperature in the dark for 15 min. Each condition
was assessed in triplicate. Flow cytometric analysis was performed
on a Beckman Coulter ACL Elite (Beckman Coulter, Brea, CA, USA),
with the results reported as a percentage of CFSE and PI double
positive cells.
Statistical analysis
All data are presented as the mean ± standard
deviation from at least three independent experiments and were
analyzed using GraphPad Prism 5 software (GraphPad Software, Inc.,
La Jolla, CA, USA). The levels of differences were analyzed using
one-way analysis of variance for one independent variable or
factorial analysis of variance for more than one independent
variable, with LSD post-hoc analysis. P<0.05 was considered to
indicate a statistically significant difference.
Results
Phenotypic characterization of target
cell lines
To characterize the expression of EPS8 and HLA-A
phenotypes of the target cell lines used in the present study,
western blot analysis was performed to examine the levels of EPS8,
and flow cytometry was performed to detect the surface expression
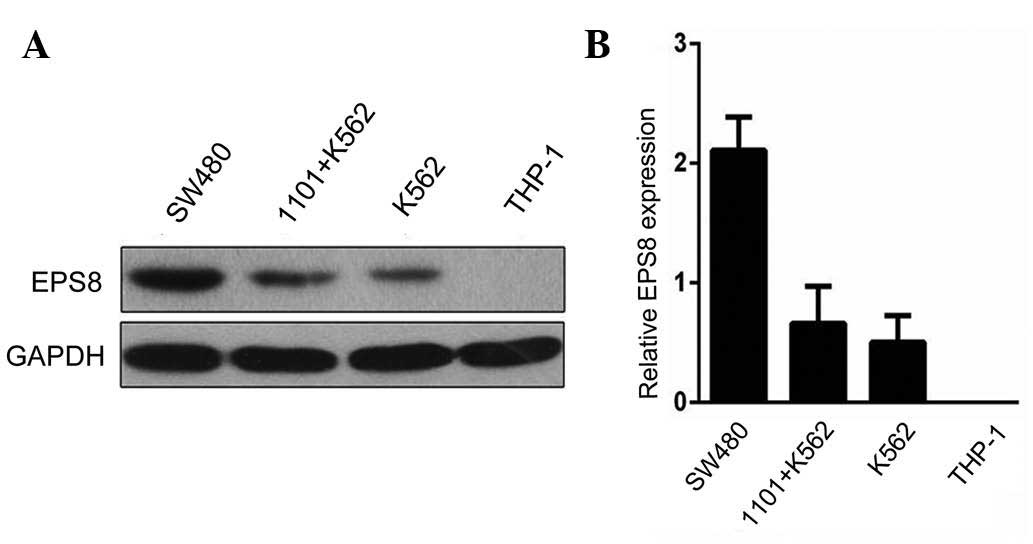
of HLA-A*1101. As shown in Fig. 1A and
B, the expression of EPS8 was highest in the SW480 human colon
cell line, a cancer cell line known to express EPS8 (13), followed by the K562 human
erythroleukemia cell line and the HLA-A*1101+K562 cell
line. No EPS8 was detected in the THzP-1 human acute monocytic
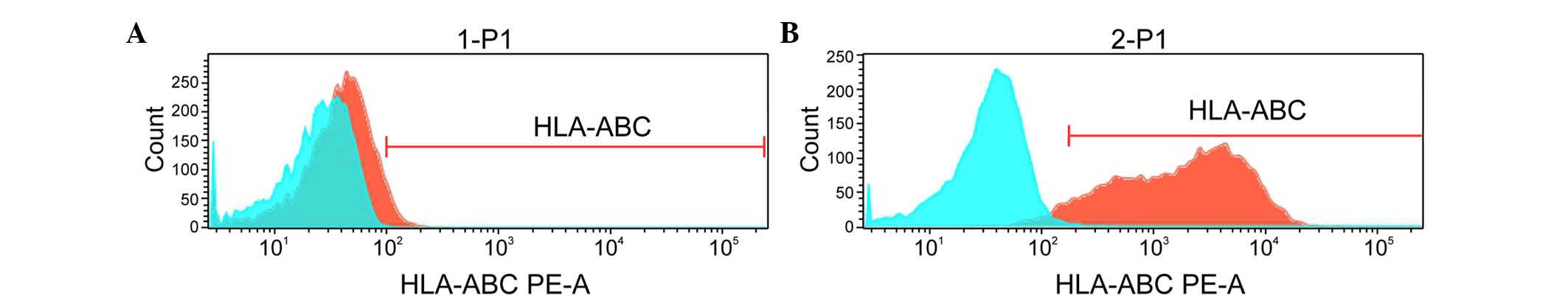
leukemia cell line. Using flow cytometry, it was found that the
HLA-A*1101+EPS8+ K562 and
HLA-A*1101+EPS8− K562 cells were positive for
the expression of HLA-A*1101, whereas the K562 and THP-1 cells were
negative (Fig. 2).
Generation of
HLA-A*1101+EPS8− K562 cells
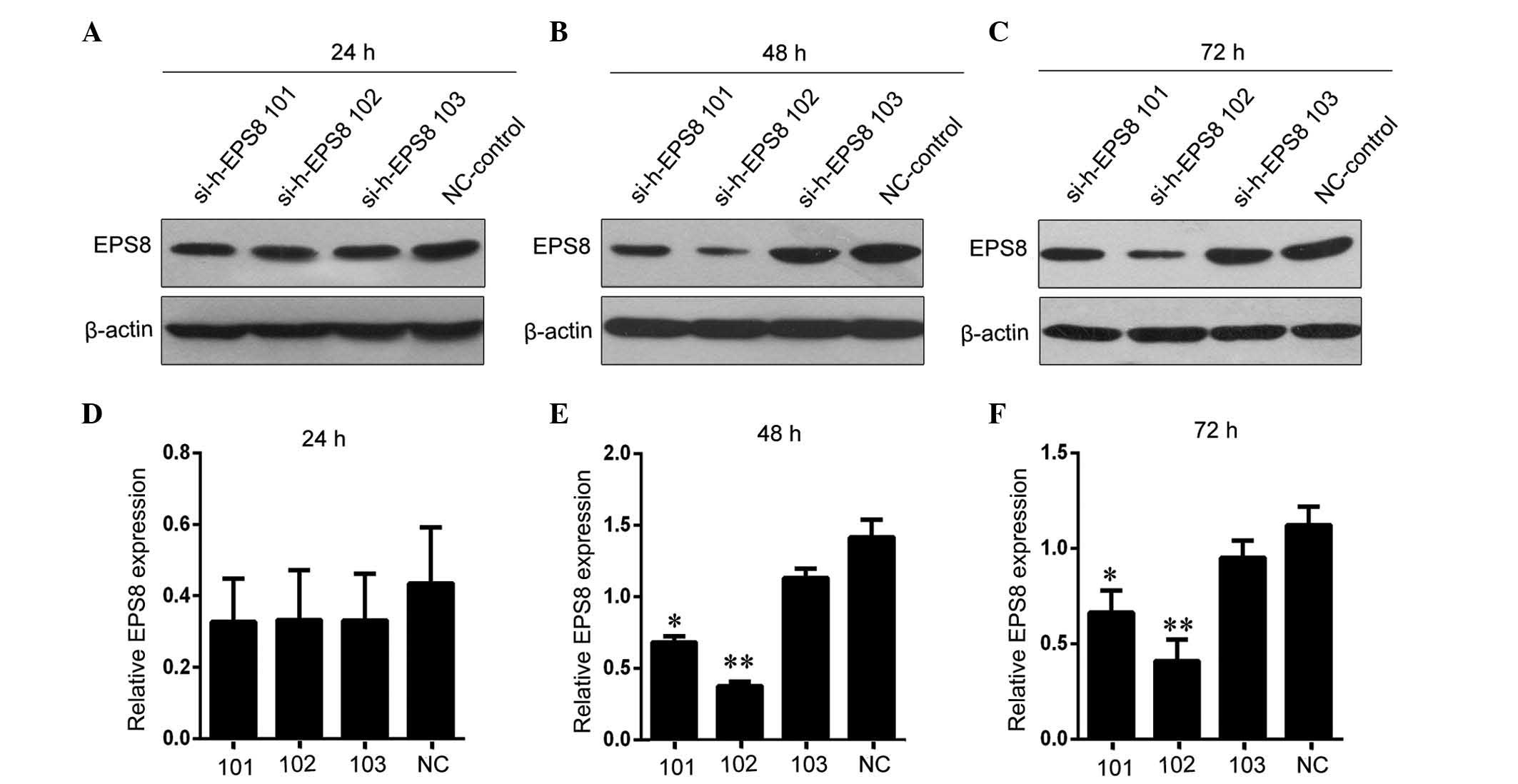
To generate HLA-A*1101+EPS8−
K562 target cells, EPS8-specific siRNA was transiently transfected
into HLA-A*1101+EPS8+ K562 cells. As shown in
Fig. 3, of the three EPS8 siRNA
sequences assessed, the si-h-EPS8 102 sequence had the most marked
effect on reducing the level of EPS8 at 48 h post-transfection,
compared with the levels in the cells transfected with control
siRNA. Therefore, this transfection condition was used for the
following experiments in the present study.
Prediction of HLA-A*1101-restricted
CTL epitopes of EPS8
To identify peptides on EPS8 with the highest
binding affinities to HLA-A*1101, the present study used two
algorithms, BIMAS and SYFPEITHI, to scan the complete amino acid
sequence of EPS8. A total of five nine-amino-acid peptides meeting
the selection criteria from the two algorithms were identified
(Table I). Based on the position
of the starting amino acid, in order from highest to lowest ranking
as predicted by the two algorithms, these five peptides were P380,
P529, P82, P30 and P70, respectively.
 | Table I.Characteristics of predicted EPS8 CTL
epitopes restricted to the HLA-A*1101 allele. |
Table I.
Characteristics of predicted EPS8 CTL
epitopes restricted to the HLA-A*1101 allele.
| Position | Length | Sequence | BIMAS rank | SYFEITHI score |
|---|
| 380 | 9 | SVLSPLLNK | 1 | 29 |
| 529 | 9 | KTQPKKYAK | 2 | 21 |
| 82 | 9 | ITVDDGIRK | 3 | 26 |
| 30 | 9 | QTDREHGSK | 4 | 20 |
| 70 | 9 | LTTFVLDRK | 5 | 20 |
Secretion of IFN-γ from in vitro
peptide-primed CTLs
In response to antigen presentation, CTLs secrete
cytokines, including IFN-γ, which was used as a functional
measurement of the peptide-stimulated CTLs to determine whether
EPS8-derived peptides are potent enough to elicit a functional CTL
response. PBMCs were isolated from HLA-A*1101+ healthy
donors, primed with each of the EPS8-derived peptides, and
re-exposed to EPS8 presented by
HLA-A*1101+EPS8+ K562 cells, following which
the production of INF-γ was examined using an ELISPOT assay, in
which the SFCs represented CTLs secreting IFN-γ. As a positive
control, the lymphocyte mitogen, PHA, was used, which is known to
stimulate the production of IFN-γ from PBMCs (25). As the negative controls, CTL with
no priming (P0-CTL) but re-exposed o EPS8 presented by
HLA-A*1101+EPS8+ K562 cells
(P0-CTL+HLA-A*1101+EPS8+ K562),
peptide-primed CTLs without re-exposure to EPS8 presented by
HLA-A*1101+EPS8+ K562 cells (P-CTL) or
peptide-free non-primed CTLs. For all five peptides examined, PHA
led to the highest number of SFCs, suggesting that these
peptide-primed lymphocytes were capable of non-specifically
responding to lymphocyte mitogens. The second highest number of
SFCs was observed in the peptide-primed CTLs with antigen presented
by HLA-A*1101+EPS8+ K562 cells, which was
significantly higher, compared with the number of SFCs from the
P0-CTL+HLA-A*1101+EPS8+ K562 group
(P<0.05), indicating that these peptides specifically augmented
the CTL responses following antigen presentation. As expected, the
P-CTLs and the P0-CTLs exposed to X–VIVO medium presented
with minimal, if any, IFN-γ-producing activity (Fig. 4A). When comparing between the five
peptides, variability was observed in their capacity to induce the
secretion of IFN-γ from CTLs following antigen presentation. P380
presented with significantly higher activity, compared to the other
four peptides (P<0.05; Fig.
4B), consistent with its ranking predicted by the BIMAS and
SYFPEITHI algorithms.
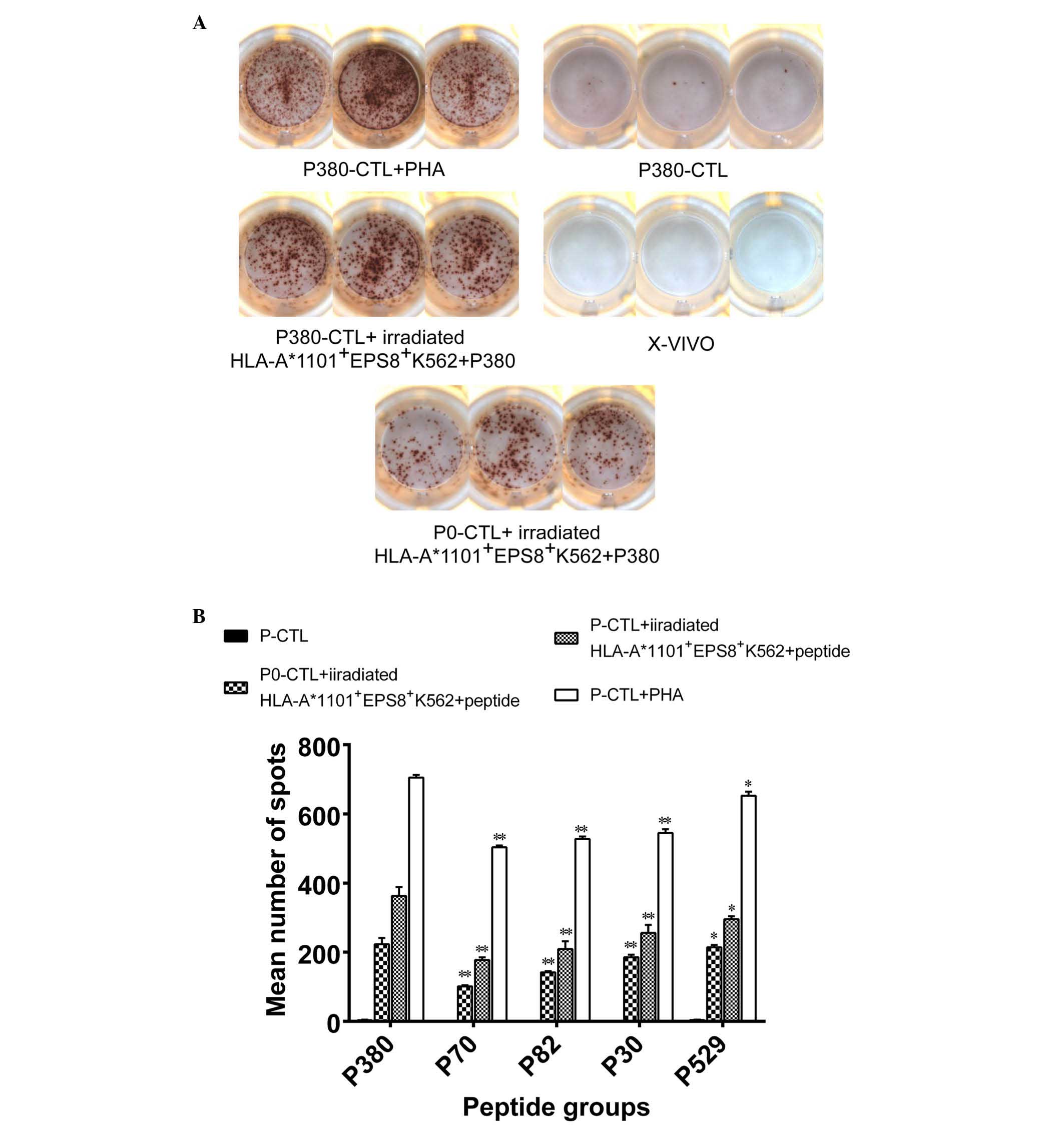 | Figure 4.Production of IFN-γ by PBMCs primed
with different peptides. Peptide-primed PBMCs were exposed to
irradiated EPS8-presenting HLA-A*1101+ K562 cells and
the production of IFN-γ was detected using an ELISPOT assay. PHA
treatment was used as a positive control; P0-CTLs or P-CTLs were
used as negative controls. (A) Representative ELISPOT images from
each condition is shown. (B) Quantification on the numbers of
spot-forming cells is shown. *P<0.05 and **P<0.001, compared
with the P380 group. IFN-γ interferon-γ; PBMCs, peripheral blood
mononuclear cells; EPS8, epidermal growth factor receptor pathway
substrate 8; HLA, human leukocyte antigen; P0, PBMCs with no
peptide priming; P-CTL, PBMCs not exposed to EPS8-presenting
HLA-A*1101+ K562 cells; ELISPOT, enzyme-linked
immunospot; X–VIVO, X–VIVO15 serum-free medium; CTL, control; P,
P380. |
Cytotoxicity of peptide-primed
CTLs
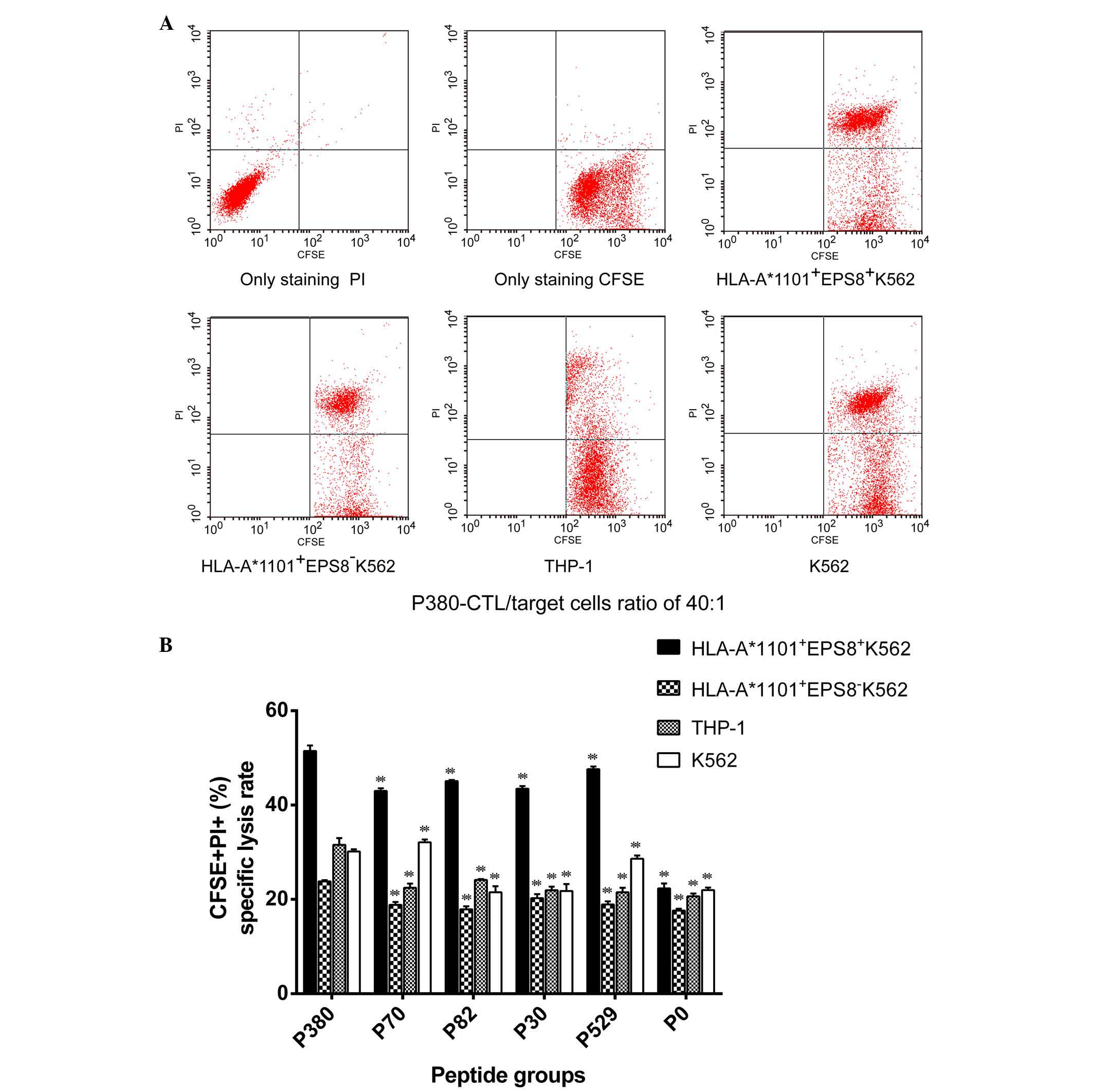
To examine the functional capability of
peptide-primed CTLs (effector cells) to cause antigen-bearing tumor
cell (target cell) death, the effector and target cells were mixed
at different E/T ratios, and the death of the CFSE-labeled target
cells was quantified using PI staining. Cytotoxicity was measured
as the percentage of CFSE+PI+ cells. For
P380, the percentages of cytotoxicity for the E/T ratios of 10:1,
20:1 and 40:1 were 21.57±11.37, 28.68±11.57 and 34.27±10.85%,
respectively (P<0.05 between every two groups), with the highest
cytotoxicity observed at the E/T ratio of 40:1. When examining the
cytotoxicity of the five peptides using this ratio, it was found
that all five peptides significantly augmented the cytotoxic
activity of the CTLs against HLA-A*1101+EPS8+
K562 cells, but not against the
HLA-A*1101+EPS8− K562, THP-1 or K562 cells,
compared with the P0-CTLs (Fig.
5A). These results suggested that CTLs primed with these
peptides show a high levels of specificity for MHC restriction and
tumor antigen presentation. When comparing between the five
peptides, P380, P529 and P82 induced similar magnitudes of toxicity
in the CTLs (P>0.05), which were significantly higher, compared
with those conferred by P70 and P30 (P<0.05; Fig. 5B).
Discussion
In the present study, systematic screening for novel
immunogenic HLA-A*1101-restricted CTL epitopes derived from EPS8
was performed, and their potential as candidate cancer vaccines
targeting HMs were characterized. A total of five nine-amino-acid
peptides were identified using two computer programs, BIMAS and
SYFPEITH, and these were P380, P529, P82, P30 and P70. Based on the
predicted scores, those with a higher score, indicating a higher
binding capacity to the HLA*A1101 molecule, were considered ideal
antigen peptides. However, as not all peptides binding to HLA
molecules are optimal T cell epitopes (26), it is essential to confirm the
activities of predicted peptides through in vitro and in
vivo experiments.
EPS8 complexes with multiple signaling molecules
include Ras/Rac, Src, Src homology 2 domain-containing, son of
sevenless homolog-1, E3b1 and focal adhesion kinase to regulate EGF
downstream phenotypes (27–32).
Biologically, EPS8 modulates various cellular behaviors, including
cell proliferation, cell motility/invasiveness, remodeling actin
cytoskeleton and membrane protrusion and vesicular trafficking
(27). The functional significance
of EPS8, together with its aberrant expression and correlation with
prognosis in several types of cancer, including colon cancer
(13), thyroid cancer (12) and breast cancer (33), suggest that it has potential as a
target for cancer therapy (34).
Our previous finding on the correlation between EPS8 and AML
(16) prompted the investigation
of the feasibility of EPS8 as an immunotherapeutic target for HMs.
In support of this hypothesis, our previous study identified eight
native peptides and generated four modified peptides from EPS8,
which show high-affinity binding to HLA-A2.1 molecules. The CTLs
primed by these peptides presented with augmented immune
activities, as demonstrated by the secretion of IFN-γ and capacity
to induce tumor cell death in a variety tissue types, in an
HLA-A2.1-restricted and Eps8-specific manner (35).
To the best of our knowledge, no
HLA-A*1101-restricted epitopes derived from EPS8 have been
identified previously. Therefore, the present study followed the
standard procedure already established (36) and started by predicting peptide
sequences, which bind to the HLA-A*1101 molecule with high
affinity. By combining two different predicting algorithms, five
sequences were obtained. Following in vitro synthesis of
these peptides to high purity, their immunogenic activity was
examined by stimulating PBMCs from healthy donors positive for the
HLA-A*1101 allele.
CTLs are a subtype of lymphocytes, which cause death
of target cells presenting with specific antigens in a major
histocompatibility complex (MHC) I-restricted manner (37). By labeling target tumor cells with
CFSE and staining them with PI, it is possible to quantify the
death of target cells induced by the peptide-primed CTLs. It was
found that, among the five peptides identified, P380, P529 and P80
primed CTLs with similar cytolytic activity against tumor cells
positive for HLA-A*1101 and EPS8, but not the tumor cells negative
for either or both of these two molecules, suggesting strict
specificity on MHC I molecule and tumor antigen. These three
peptides also showed higher binding affinity to the HLA-A*1101
molecule, as predicted by the two computer-based algorithms,
corroborating the effectiveness of these algorithms in predicting
MHC-binding motifs.
Another phenotypic assay commonly used for CTLs is
the production and secretion of antitumor cytokines, including
IFN-γ, in response to antigen presentation (38). In the present study, when the
production of INF-γ by CTLs was analyzed using an ELISPOT assay, it
was shown that all five peptides significantly activated the CTLs
to produce IFN-γ in response to antigen presentation, and P380
showed the highest immunogenic activity among the five
peptides.
As the present study was limited by the blood
samples, it was possible to obtain from the donors, it was not
possible sort the CD8+ CTLs from the PBMCs or use
professional antigen presenting cells, including dendritic cells,
for antigen presentation. These limitations may weaken the
phenotypes observed, including the number of SFCs observed in the
IFN-γ ELISPOT assay or the percentage of
CFSE+PI+ cells detected using flow cytometry.
However, the data obtained in the present study demonstrated, with
statistical significance, that the peptides identified were TAA
candidates. In addition, the present study did not identify a cell
line positive for HLA-A*1101, but completely negative for EPS8,
making it challenging to characterize the binding stability of
identified peptides. However, the functional assays of the
production of IFN-γ and cytolysis indirectly confirmed that these
peptides were able to effectively prime CTLs in the PBMCs. In the
present study, all functional characterizations on peptide-primed
CTLs were performed in vitro. The immunogenic
characteristics of these peptides may vary between in vitro
and in vivo conditions, therefore, it is important to
further assess their immunogenic activities using a suitable murine
model and with blood samples from patients with HMs. Although the
present study focused on HMs and used the K562 human
erythroleukemia cell line, the peptides identified may also target
solid tumors, which requires examination in future
investigations.
In conclusion, the present study identified five
nine-amino-acid HLA-A*1101-restricted, EPS8-derived CTL epitopes,
which started from amino acid positions 380, 529, 82, 30 and 70 on
the EPS protein. All five peptides presented with good binding
affinity to the HLA-A*1101 molecule. The CTLs primed with these
peptides produced increased IFN-γ in response to EPS8 presentation,
and specifically recognized and lysed target tumor cells expressing
ESP8 and HLA-A*1101. Therefore, these five peptides may become
promising TAAs for the development of immunotherapy targeting
EPS-expressing HMs and/or solid tumors.
Acknowledgements
The present study was supported by the Natural
Science Foundation of China (grant no. 81372249), Science and
Technology Planning Project of Guangdong Province, China (grant no.
2013B091500072), Project of Department of Education of Guangdong
Province, China (grant no. 2014GKXM029).
Glossary
Abbreviations
Abbreviations:
|
AML
|
acute myeloid leukemia
|
|
CFSE
|
carboxyfluorescein succinimidyl
ester
|
|
P380-CTL
|
CTLs induced by P380
|
|
CTLs
|
cytotoxic T lymphocytes
|
|
E/T
|
effector to target
|
|
ELISPOT
|
enzyme-linked immunosorbent spot
|
|
EGF
|
epidermal growth factor
|
|
EPS8
|
epidermal growth factor receptor
pathway substrate 8
|
|
HM
|
hematological malignancy
|
|
HLA
|
human leukocyte antigen
|
|
IFN-γ
|
interferon-γ
|
|
MW
|
molecular weight
|
|
PBMCs
|
peripheral blood mononuclear cells
|
|
PI
|
propidium iodide
|
|
rhIL-2
|
recombinant human interleukin 2
|
|
siRNA
|
small interfering RNA
|
|
SFC
|
spot forming cells
|
|
SD
|
standard deviation
|
|
TAAs
|
tumor-associated antigens
|
|
LAAs
|
leukemia-associated antigens
|
References
|
1
|
Vardiman JW, Thiele J, Arber DA, Brunning
RD, Borowitz MJ, Porwit A, Harris NL, Le Beau MM,
Hellström-Lindberg E, Tefferi A and Bloomfield CD: The 2008
revision of the World Health Organization (WHO) classification of
myeloid neoplasms and acute leukemia: Rationale and important
changes. Blood. 114:937–951. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fielding AK, Richards SM, Chopra R,
Lazarus HM, Litzow MR, Buck G, Durrant IJ, Luger SM, Marks DI,
Franklin IM, et al: Outcome of 609 adults after relapse of acute
lymphoblastic leukemia (ALL); an MRC UKALL12/ECOG 2993 study.
Blood. 109:944–950. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schmid C, Labopin M, Nagler A,
Niederwieser D, Castagna L, Tabrizi R, Stadler M, Kuball J,
Cornelissen J, Vorlicek J, et al: Treatment, risk factors, and
outcome of adults with relapsed AML after reduced intensity
conditioning for allogeneic stem cell transplantation. Blood.
119:1599–1606. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bocchia M, Defina M, Aprile L and
Sicuranza A: Peptide vaccines for hematological malignancies: A
missed promise? Int J Hematol. 99:107–116. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mohamed YS, Dunnion D, Teobald I, Walewska
R and Browning MJ: In vitro evaluation of human hybrid cell lines
generated by fusion of B-lymphoblastoid cells and ex vivo tumour
cells as candidate vaccines for haematological malignancies.
Vaccine. 30:6578–6587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shah GL, Shune L, Purtill D, Devlin S,
Lauer E, Lubin M, Bhatt V, McElrath C, Kernan NA, Scaradavou A, et
al: Vaccine responses in adult and pediatric cord blood
transplantation recipients treated for hematologic malignancies.
Biol Blood Marrow Transplant. 21:2160–2166. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Melero I, Gaudernack G, Gerritsen W, Huber
C, Parmiani G, Scholl S, Thatcher N, Wagstaff J, Zielinski C,
Faulkner I and Mellstedt H: Therapeutic vaccines for cancer: An
overview of clinical trials. Nat Rev Clin Oncol. 11:509–524. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mellman I, Coukos G and Dranoff G: Cancer
immunotherapy comes of age. Nature. 480:480–489. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fazioli F, Minichiello L, Matoska V,
Castagnino P, Miki T, Wong WT and Di Fiore PP: Eps8, a substrate
for the epidermal growth factor receptor kinase, enhances
EGF-dependent mitogenic signals. Embo J. 12:3799–3808.
1993.PubMed/NCBI
|
|
10
|
Matoskova B, Wong WT, Salcini AE, Pelicci
PG and Di Fiore PP: Constitutive phosphorylation of eps8 in tumor
cell lines: Relevance to malignant transformation. Mol Cell Biol.
15:3805–3812. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen YJ, Shen MR, Chen YJ, Maa MC and Leu
TH: Eps8 decreases chemosensitivity and affects survival of
cervical cancer patients. Mol Cancer Ther. 7:1376–1385. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Griffith OL, Melck A, Jones SJ and Wiseman
SM: Meta-analysis and meta-review of thyroid cancer gene expression
profiling studies identifies important diagnostic biomarkers. J
Clin Oncol. 24:5043–5051. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Maa MC, Lee JC, Chen YJ, Chen YJ, Lee YC,
Wang ST, Huang CC, Chow NH and Leu TH: Eps8 facilitates cellular
growth and motility of colon cancer cells by increasing the
expression and activity of focal adhesion kinase. J Biol Chem.
282:19399–19409. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang H, Patel V, Miyazaki H, Gutkind JS
and Yeudall WA: Role for EPS8 in squamous carcinogenesis.
Carcinogenesis. 30:165–174. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gorsic LK, Stark AL, Wheeler HE, Wong SS,
Im HK and Dolan ME: EPS8 inhibition increases cisplatin sensitivity
in lung cancer cells. PLoS One. 8:e822202013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang L, Cai SH, Xiong WY, He YJ, Deng L
and Li YH: Real-time quantitative polymerase chain reaction assay
for detecting the eps8 gene in acute myeloid leukemia. Clin Lab.
59:1261–1269. 2013.PubMed/NCBI
|
|
17
|
He YJ, Zhou J, Zhao TF, Hu LS, Gan JY,
Deng L and Li Y: Eps8 vaccine exerts prophylactic antitumor effects
in a murine model: A novel vaccine for breast carcinoma. Mol Med
Rep. 8:662–668. 2013.PubMed/NCBI
|
|
18
|
Lee TD, Zhao TM, Mickey R, Sun YP, Lee G,
Song CX, Cheng DZ, Zhou S, Ding SQ, Cheng DX, et al: The
polymorphism of HLA antigens in the Chinese. Tissue Antigens.
32:188–208. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lin M, Chu CC, Chang SL, Lee HL, Loo JH,
Akaza T, Juji T, Ohashi J and Tokunaga K: The origin of Minnan and
Hakka, the so-called ‘Taiwanese’, inferred by HLA study. Tissue
Antigens. 57:192–199. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zha XF, Zhou YB, Yang LJ, Chen SH, Li B,
Yan XJ and Li YQ: Establishment of stable subline of K562 cells
expressing human leucocyte antigen a1101. Zhongguo Shi Yan Xue Ye
Xue Za Zhi. 19:1112–1116. 2011.(In Chinese). PubMed/NCBI
|
|
21
|
Cattaneo MG, Cappellini E and Vicentini
LM: Silencing of Eps8 blocks migration and invasion in human
glioblastoma cell lines. Exp Cell Res. 318:1901–1912. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fu J, Li L and Bouvier M: Adenovirus
E3-19K proteins of different serotypes and subgroups have similar,
yet distinct, immunomodulatory functions toward major
histocompatibility class I molecules. J Biol Chem. 286:17631–17639.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Parker KC, Bednarek MA and Coligan JE:
Scheme for ranking potential HLA-A2 binding peptides based on
independent binding of individual peptide side-chains. J Immunol.
152:163–175. 1994.PubMed/NCBI
|
|
24
|
Rammensee H, Bachmann J, Emmerich NP,
Bachor OA and Stevanović S: SYFPEITHI: Database for MHC ligands and
peptide motifs. Immunogenetics. 50:213–219. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Deenadayalan A, Maddineni P and Raja A:
Comparison of whole blood and PBMC assays for T-cell functional
analysis. BMC Res Notes. 6:1202013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Doytchinova IA and Flower DR: Predicting
class I major histocompatibility complex (MHC) binders using
multivariate statistics: Comparison of discriminant analysis and
multiple linear regression. J Chem Inf Model. 47:234–238. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cunningham DL, Creese AJ, Auciello G,
Sweet SM, Tatar T, Rappoport JZ, Grant MM and Heath JK: Novel
binding partners and differentially regulated phosphorylation sites
clarify Eps8 as a multi-functional adaptor. PLoS One. 8:e615132013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Scita G, Nordstrom J, Carbone R, Tenca P,
Giardina G, Gutkind S, Bjarnegård M, Betsholtz C and Di Fiore PP:
EPS8 and E3B1 transduce signals from Ras to Rac. Nature.
401:290–293. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Scita G, Tenca P, Areces LB, Tocchetti A,
Frittoli E, Giardina G, Ponzanelli I, Sini P, Innocenti M and Di
Fiore PP: An effector region in Eps8 is responsible for the
activation of the Rac-specific GEF activity of Sos-1 and for the
proper localization of the Rac-based actin-polymerizing machine. J
Cell Biol. 154:1031–1044. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Schoenherr C, Serrels B, Proby C,
Cunningham DL, Findlay JE, Baillie GS, Heath JK and Frame MC: Eps8
controls Src- and FAK-dependent phenotypes in squamous carcinoma
cells. J Cell Sci. 127:5303–5316. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Funato Y, Terabayashi T, Suenaga N, Seiki
M, Takenawa T and Miki H: IRSp53/Eps8 complex is important for
positive regulation of Rac and cancer cell motility/invasiveness.
Cancer Res. 64:5237–5244. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Innocenti M, Frittoli E, Ponzanelli I,
Falck JR, Brachmann SM, Di Fiore PP and Scita G: Phosphoinositide
3-kinase activates Rac by entering in a complex with Eps8, Abi1 and
Sos-1. J Cell Biol. 160:17–23. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yao J, Weremowicz S, Feng B, Gentleman RC,
Marks JR, Gelman R, Brennan C and Polyak K: Combined cDNA array
comparative genomic hybridization and serial analysis of gene
expression analysis of breast tumor progression. Cancer Res.
66:4065–4078. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang TP, Chiou HL, Maa MC and Wang CJ:
Mithramycin inhibits human epithelial carcinoma cell proliferation
and migration involving downregulation of Eps8 expression. Chem
Biol Interact. 183:181–186. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li Y, Zhou W, Du J, Jiang C, Xie X, Xue T
and He Y: Generation of cytotoxic T lymphocytes specific for native
or modified peptides derived from the epidermal growth factor
receptor pathway substrate 8 antigen. Cancer Immunol Immunother.
64:259–269. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Okuyama R, Aruga A, Hatori T, Takeda K and
Yamamoto M: Immunological respons es to a multi-peptide vaccine
targeting cancer-testis antigens and VEGFRs in advanced pancreatic
cancer patients. Oncoimmunology. 2:e270102013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Williams MA and Bevan MJ: Effector and
memory CTL differentiation. Annu Rev Immunol. 25:171–192. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li R, Qian J, Zhang W, Fu W, Du J, Jiang
H, Zhang H, Zhang C, Xi H, Yi Q and Hou J: Human heat shock
protein-specific cytotoxic T lymphocytes display potent antitumour
immunity in multiple myeloma. Br J Haematol. 166:690–701. 2014.
View Article : Google Scholar : PubMed/NCBI
|