Introduction
Sepsis is a systemic inflammatory response mediated
by various innate immune cells, including neutrophiles, monocytes
and macrophages, upon severe infection (1). Normally, the moderate production of
pro-inflammatory cytokines, including tumor necrosis factor
(TNF)-α, interleukin (IL)-1, IL-6 and IL-8 assist in confining
infection and tissue damage; with the eradication of infectious
agents, the inflammatory response can recover to homeostasis.
However, excessive and prolonged production of inflammatory
cytokines can lead to an overwhelming inflammatory response, which
is referred to as sepsis (2). The
mortality rates of severe sepsis can reach as high as 70% and the
number of cases of sepsis continues to increase due to the
continued increase in the number of immunocompromised patients
(3,4). However, the molecular mechanism of
sepsis remains to be fully elucidated. Studies have revealed that
several mechanisms may contribute to the occurrence of sepsis,
including the continued activation of neutrophils and
macrophages/monocytes, upregulation of lymphocyte costimulatory
molecules (5,6), rapid lymphocyte apoptosis and delayed
neutrophil apoptosis, and excessive necrosis of cells and tissues
(7,8). Of these, the overwhelming production
of TNF-α is considered to be important in the occurrence and
development of sepsis (9).
Sirtuin 1 (sirT1), an NAD+-dependent
deacetylase, has been well-established as a major component in the
regulation of cellular stress responses, including apoptosis,
autophagic DNA damage repair and metabolic disorders through
histone and non-histone deacetylation. It is not surprising that
SirT1 has a number of roles in multiple tissues through its effects
on diverse physiological processes (10,11).
Previous studies have further revealed the role of SirT1 in sepsis.
For example, nuclear SirT1 has been found to guide RelB to promote
mitochondrial biogenesis, which alters bioenergetics during sepsis
adaptation (12). Resveratrol, an
activator of SirT1, has been revealed to protect against
sepsis-induced liver injury through promoting the SirT1-mediated
nucleocytoplasmic translocation of high mobility group box 1
(HMGB1) (13). Other previous
studies have examined the association between SirT1 and
inflammatory cytokines in sepsis. For example, acute hyperglycemia
in sepsis is considered to repress the transcription and
translation of SirT1, and then promote the transcription and
translation of TNF-α and IL-1β (14). Studies have shown that SirT1
inhibits acute lung inflammation during sepsis by repressing the
inflammasome activation pathway, including the activation of
nuclear factor (NF)-κB, signal transducer and activator of
transcription 3 and extracellular signal-regulated kinase (ERK)1/2
(15). In particular, the NF-κB
transcription factor, RelA/p65, is considered to be the primary
target of SirT1 in regulating the transcription of TNF-α through
deacetylating RelA/p65 in innate cells (16,17).
It is well-known that SirT1 is also involved in
chromatin compaction and gene silencing through deacetylating
H4K16, H3K9 and H1K26 (18,19).
Studies have also shown that SirT1 deacetylates H4K16 and promotes
termination of the NF-κB-dependent transcription of TNF-α during
initial lipopolysaccharide (LPS) stimulation in normal THP-1 cells
(20). However, the exact role of
SirT1 in epigenetic modifications of inflammatory gene promoters in
sepsis remains to be fully elucidated. In the present study,
epigenetic modifications by SirT1 and resveratrol were assayed in a
cell model of sepsis, namely LPS-mediated tolerance in THP-1
promonocytes (21), to reveal
whether SirT1 activation can repress the transcription of TNF-α in
the sepsis model, and offer potential as a promising candidate for
sepsis therapy.
Materials and methods
Cell culture model of sepsis and
resveratrol treatment
THP-1 cells, obtained from the American Type Culture
Collection (Manassas, VA, USA) were maintained in Gibco RPMI-1640
medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10 U/ml penicillin G, 10 µg/ml streptomycin, 2 mM
L-glutamine and 10% FBS (HyClone; GE Healthcare Life Sciences,
Logan, UT, USA) at 37°C and 5% CO2 in a humidified incubator. The
sepsis phenotype (LPS-tolerant) was mimicked by stimulation of the
THP-1 cells with LPS (0111:B4; 1 µg/ml) overnight (21). The normal and LPS-tolerant group
THP-1 cells (1×106 cells/sample) were washed once with
RPMI-1640, re-suspended in FBS supplemented RPMI-1640 medium at
1×106 cells/ml, and stimulated by 1 µg/ml LPS
(Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) for 3 h at
37°C and 5% CO2. The normal+resveratrol group and
tolerant+resveratrol group were treated with resveratrol (10
µmol/l; Sigma-Aldrich; Merck Millipore) (22) for 30 min at 37°C and 5%
CO2 prior to the LPS stimulation described above.
Chromatin immunoprecipitation (ChIP)
assay
To assess the binding of SirT1, H3K9me2 and H4K16ace
around the NF-κB binding site of the TNF-α proximal promoter in the
SirT1-treated tolerant THP-1 cells, ChIP assays (Upstate
Biotechnology, Inc., Lake Placid, NY, USA) were performed with the
following modifications. Proteins from 5×106 cells in
each sample were cross-linked to DNA using 1% formaldehyde for 10
min at room temperature. Each sample with sheared chromatin was
divided into two sample groups, providing an ‘input’ sample, which
was not incubated with any antibodies. The other sample was
incubated with antibodies specific for SirT1 (cat. no. sc-135792;
1:500; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA),
H3K9me2 (cat. no. 6814-25; 1:600; BioVision, Inc., Milpitas, CA,
USA), H4K16ace (cat. no. sc-8662; 1:500; Santa Cruz Biotechnology,
Inc.), and IgG (1:800; cat. no. sc-2027; Santa Cruz Biotechnology,
Inc.) for the negative control at 4°C overnight.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
The ChIP DNA for each treatment group were analyzed
quantitatively through the amplification of a sequence in the human
TNF-α proximal promoter region containing the κB3 site at −98 bp,
relative to the transcription start site (23). The primers were as follows:
Forward, 5′TACCGCTTCCTCCAGATGAG-3′ and reverse,
5′-TGCTGGCTGGGTGTGCCAA-3′. The probe was:
5′-6-FAMTTGGTGGAGAAACC-TAMRA-3′ (Sangon Biotech Co., Ltd.,
Shanghai, China). The PCR reaction (total 20 µl) contained 2 µl
DNA, 10 µl 2x TaqMan Universal Master Mix, 300 nM each primer and
100 nM dNTPs. The PCR procedure was as follows: 2 min at 50°C, 10
min at 95°C, followed by 40 cycles with 15 sec at 95°C and 15 sec
at 60°C, using an ABI 7500 fast detection system (Thermo Fisher
Scientific, Inc.). Data were normalized to the input DNA and are
presented as the fold change, relative to DNA from the untreated
cells.
To measure the mRNA expression of TNF-α, total RNA
was isolated from cells using a Qiagen RNA mini kit (Qiagen, Inc.,
Valencia, CA, USA) according to the manufacturer's protocol. The
RNA (1 µg) was reverse transcribed into cDNA in a 20 µl volume
containing 5 mM MgCl2, 1 mM dNTP, 2.5 µM Oligo d (T),
2.5 U/µl MuLV Reverse Transcriptase (Thermo Fisher Scientific,
Inc.). The qPCR was performed using 3 µl cDNA TNF-α and GAPDH
predesigned TaqMan primer/probe sets (Thermo Fisher Scientific,
Inc.) under the conditions described above. Data were normalized to
GAPDH mRNA, which was analyzed using GAPDH pre-designed TaqMan
primer/probe kits (Thermo Fisher Scientific, Inc.) and are
presented as the fold change, relative to mRNA from the untreated
cells. Sample data were normalized to GAPDH mRNA values and are
presented as the fold changes, relative to mRNA from the untreated
cells (23).
PCR analysis
A standard PCR reaction (total 25 µl) was composed
of 2 µl ChIPed DNA or 2 µl Input DNA, 1 µM of each primer (as
above), 2 mM MgCl2, 0.2 M dNTPs and 0.03 U/µl AmpliTaq
Gold DNA polymerase (Thermo Fisher Scientific, Inc.) was performed
to confirm the results of the RT-qPCR analysis. The PCR conditions
were as follows: 1 cycle at 94°C for 5 min, 30 cycles at 94°C, 58°C
and 72°C for 30 sec each, and a final cycle at 72°C for 5 min.
Equal volumes of PCR product was visualized using 1.5% agarose gels
and images were captured using Quantity One Imager version 4.6.2
(Bio-Rad Laboratories, Inc., Hercules, CA, USA).
SirT1 histone deacetylase (HDAC)
activity assay
SirT1 activity was assayed according to a previous
study (24). Briefly, the THP-1
cells were homogenized using a Diagenode Bioruptor sonicator (Tosho
Denki Co., Ltd., Tokyo, Japan). Total protein was extracted from
1.5×106 cells/sample (21). Briefly, cells were collected and
washed twice with PBS, resuspended in 100 µl RIPA buffer (cell
lysis buffer containing 0.45% NaCl, 0.5% deoxycholate, 0.5% Triton
X-100, 0.05% sodium dodecyl sulfate, 0.005 M Tris) and incubated on
ice for 20 min. Next, the cells were vortexed for 10 sec and
centrifuged at 12,000 × g for 5 min, supernatant was
retained. Protein quantity was assayed with Bradford method protein
assay kit (Amresco, LLC, Solon, OH, USA). Each sample (containing
30 µg/10 µl total protein) was used for the measurement of SirT1
activity. The activity of HDAC was determined using an SirT1
Fluorimetric Drug Discovery kit (Enzo Life Sciences, Inc.,
Farmingdale, NY, USA) according to the manufacturer's protocol. The
THP-1 cell protein extracts were incubated in assay buffer with
β-nicotinamide adenine dinucleotide (NAD+) substrate at
37°C for 45 min. The fluorescence density was determined using a
multimode detector (DTx880; Beckman Coulter, Brea, CA). The SirT1
activity was determined, relative to that in the untreated control
cells.
Western blot analysis
Total nuclear protein was assayed using western blot
analysis. The methods for protein extraction are the same as
described previously (21).
Whole-cell protein (20 µg) or nuclear protein (30 µg) was separated
by SDS-PAGE and transferred to PVDF membranes. SirT1 antibodies
(cat no. sc-135791; 1:800; Santa Cruz Biotechnology, Inc.) were
used to visualize and quantify protein levels following incubation
at 37°C for 30 min using Image Quant software 4.6.2 (GE Healthcare
Life Sciences). IgG (cat. no. sc-69917; 1:1,000) was used as
negative control.
Results
Resveratrol treatment suppresses the
mRNA transcription of TNF-α in LPS-tolerant THP-1 cells
To assess whether resveratrol treatment repressed
the mRNA transcription of TNF-α in a cell model of sepsis, RT-qPCR
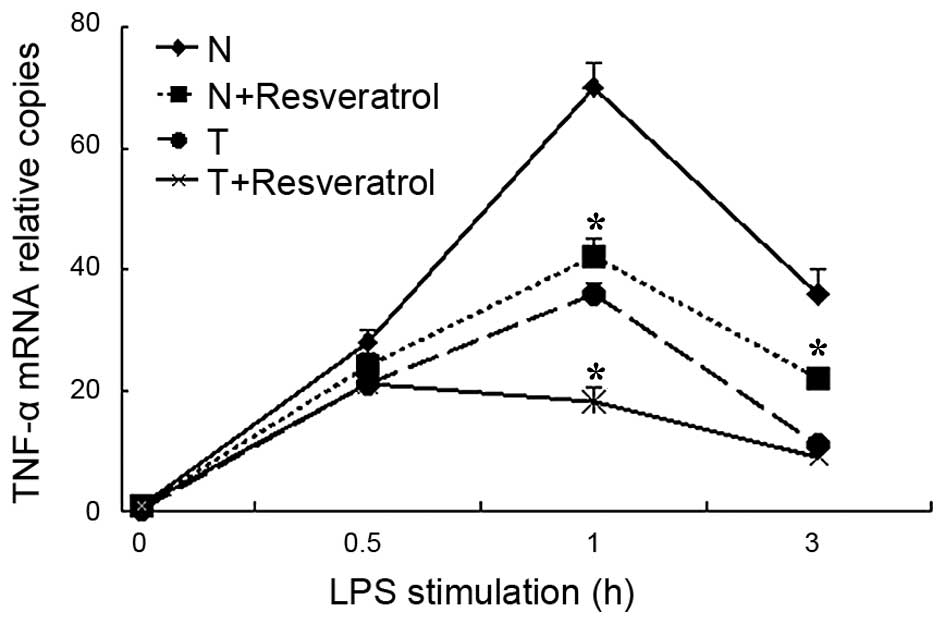
analysis was used to assess the mRNA levels of TNF-α. As indicated
in Fig. 1, the mRNA levels of
TNF-α rapidly and markedly increased in the normal cells stimulated
with LPS, and then reduced over the 4 h period. By contrast, in the
tolerant cells, the peak increase in the mRNA transcription of
TNF-α was relatively lower following LPS re-stimulation, compared
with the normal cells, which indicated that the sepsis model using
THP-1 cells had been successfully established. Following
resveratrol treatment for 30 mins, the mRNA levels of TNF-α
decreased significantly in the tolerant+resveratrol group, compared
with the tolerant group, particularly at the 1 h time point. In the
normal cells, resveratrol treatment also suppressed the mRNA
transcription of TNF-α, as indicated in the normal+resveratrol
group, compared with the normal group.
Protein levels of SirT1 are not
altered significantly with resveratrol treatment in tolerant
cells
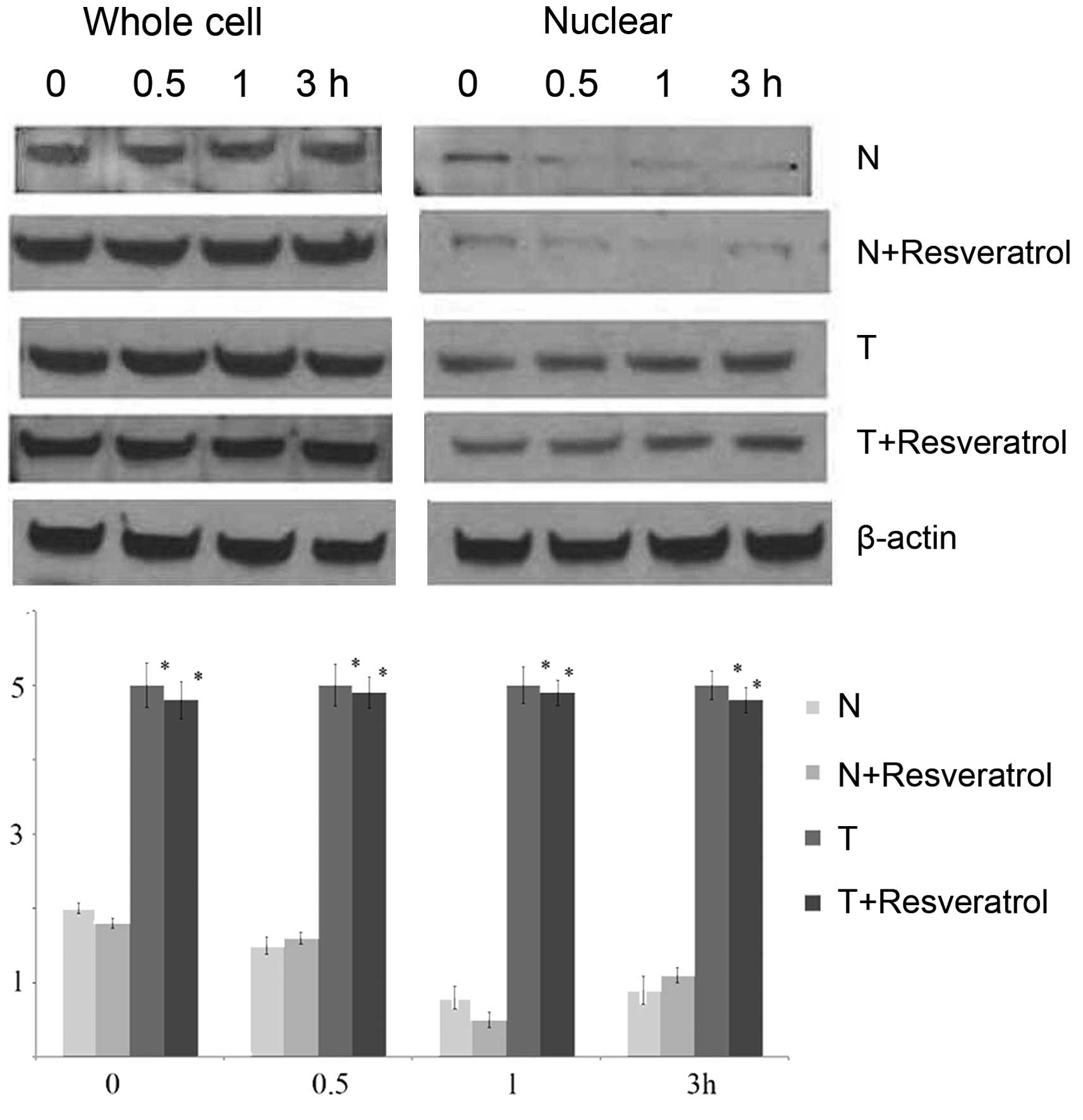
To analyze whether resveratrol treatment affected
the protein transcription and translation of SirT1, western blot
analysis was used to measure the protein levels of SirT1 in the
THP-1 cells of each group 0, 0.5, 1 and 3 h following stimulation
by LPS. As shown in Fig. 2,
nuclear SirT1 protein decreased and then recovered partially in the
normal and normal+resveratrol groups following LPS stimulation.
However, the nuclear protein levels of SirT1 remained substantially
higher over the 4 h assessment period in the tolerant group and
tolerant+resveratrol group. In addition, the nuclear protein levels
of SirT1 in the tolerant group and tolerant+resveratrol groups were
significantly higher, compared with those in the normal and
normal+resveratrol groups. Of note, the whole cell protein levels
of SirT1 in the normal+resveratrol group, tolerant group and
tolerant+resveratrol group were higher, compared with that in the
normal group. However, it was clear that resveratrol treatment had
no effect on the protein levels of SirT1 in the tolerant cells.
Resveratrol treatment promotes SirT1
activity and the binding of SirT1 to the TNF-α promoter in tolerant
cells
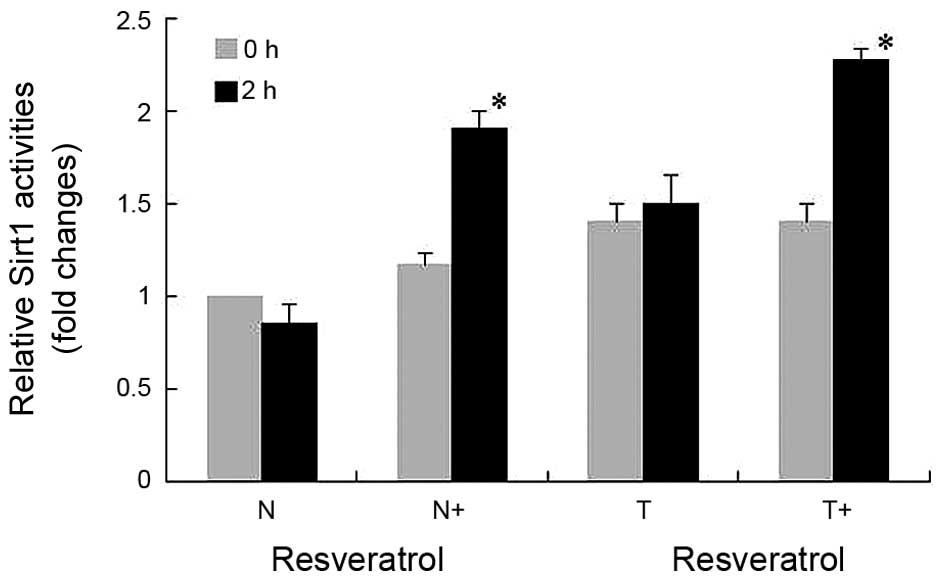
To investigate the effect of resveratrol on SirT1
activity, THP-1 cells in each group were collected at 0 and 2
h-post LPS stimulation, and activities of SirT1 HDAC were analyzed
(Fig. 3). Although the activities
of SirT1 in the normal cells (N group) were reduced 2 h-post LPS
stimulation, the decreases were not statistically significant. No
significant alterations were found in the activities of SirT1 in
the normal cells or tolerant cells at 2 h-post LPS stimulation.
However, the activity of SirT1 in the tolerant cells was higher,
compared with that in the normal cells. Following resveratrol
treatment, the activities of SirT1 were increased significantly in
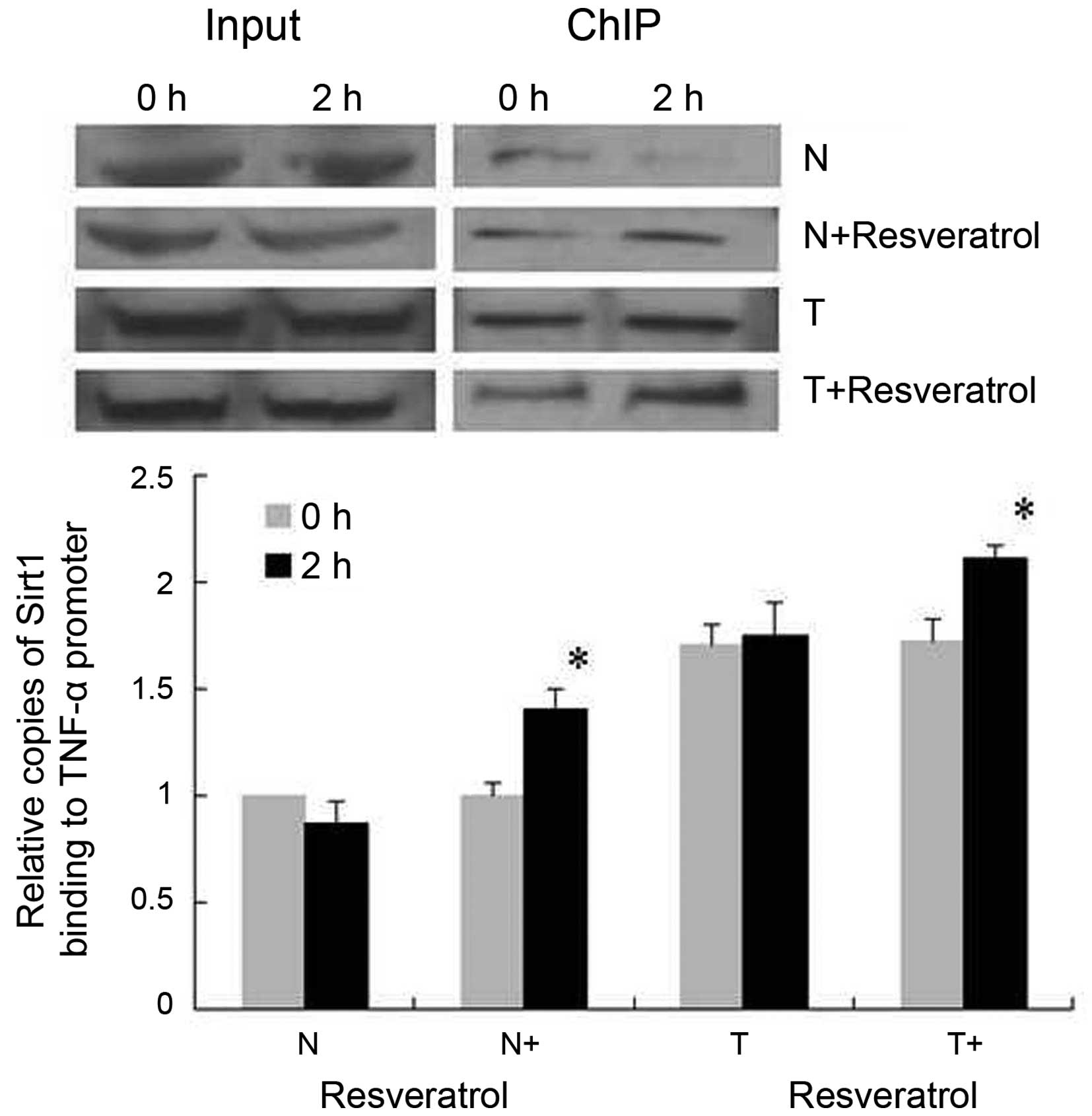
the normal cells and tolerant cells. To further assess the binding
of SirT1 to the TNF-α promoter κB3 site at the core promoter, the
functionally important NF-κB site for the activation of TNF-α
transcription (23), ChIP assays
were used. It was shown that, in the normal group, binding of SirT1
to the TNF-α promoter was decreased 2 h post-LPS stimulation
(Fig. 4), whereas binding of SirT1
was increased significantly in the normal+resveratrol group. In the
tolerant group, the binding of SirT1 remained consistently higher,
compared with that in the normal cells under LPS stimulation, and
resveratrol promoted the binding of SirT1 to the TNF-α promoter, as
indicated in the tolerant+resveratrol group.
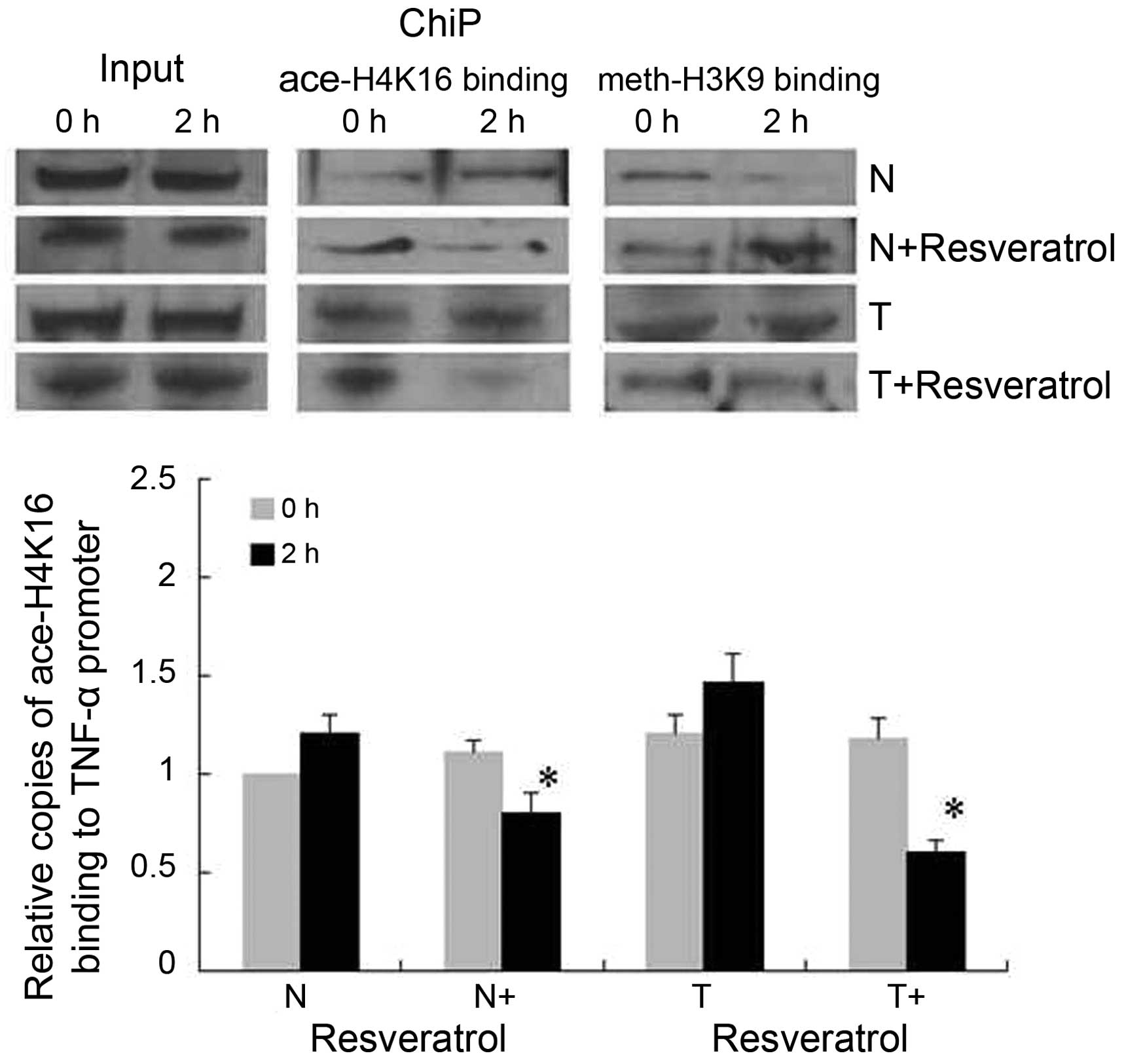
Binding of H4K16ace to the TNF-α
promoter decreases with resveratrol treatment in tolerant
cells
The epigenetic regulation in the TNF-α promoter was
assessed using ChIP assays to analyze the binding of H4K16ace and
H3K9me2 to the κB3 site (Fig. 5).
In the normal group, H4K16ace was increased and H3K9me2 was
decreased at 2 h post-LPS stimulation. In the normal+resveratrol
group, H4K16ace binding decreased with an accompanied increase in
H3K9me2 binding at 2 h post-LPS stimulation. In the tolerant cells,
prominent H4K16ace and H3K9me2 binding to the TNF-α promoter was
sustained constantly regardless of the time course of LPS
stimulation, whereas H4K16ace binding decreased significantly with
resveratrol treatment (tolerant+resveratrol group) without effects
on H3K9me2 binding.
Discussion
In the present study, the SirT1 activator,
resveratrol, successfully repressed the transcription of TNF-α
through deacetylating H4K16 at the TNF-α promoter, which indicated
that resveratrol may be a promising therapeutic candidate for
sepsis. Human SirT1 consists of 747 amino acids, divided into four
major regions: N-terminal domain (residues 1–182), allosteric site
(residues 183–243), catalytic core (residues 244–498) and
C-terminal domain (residues 499–747) (25). SirT1 catalyzes the protein
deacetylation reaction in its catalytic core, which consists of two
subdomains for NAD+ and substrate binding (26). Adjacent to the N-terminal in the
catalytic core is the compacted allosteric domain, to which SirT1
activators, including resveratrol, bind and positively regulate
sirtuin activity (27).
Resveratrol has been shown to activate SirT1 and suppress the
overexpression of pro-inflammatory molecules in a dose-dependent
manner in a mouse model of sepsis induced by LPS (28). The results of the present further
supported that resveratrol decreased the transcription of TNF-α
under LPS stimulation in tolerant cells and in normal cells.
A previous study indicated that the protein levels
of SirT1 decrease transiently, followed by a substantial increase
between 8 and 24 h of LPS stimulation, in THP-1 cells, which is
attributed to increased SirT1 protein synthesis in tolerant cells
(20). The present study also
found higher protein levels of SirT1 in tolerant cells, compared
with normal cells. However, rather than analyzing alterations in
whole cell SirT1 protein, the present study assessed the levels of
nuclear SirT1 protein in each group. In normal THP-1 cells, the
level of nuclear SirT1 protein decreased initially, and then
recovered partially under LPS stimulation, which was similar to the
observations in whole cell SirT1 protein reported previously
(20). It is possible that nuclear
SirT1 repressed the transcription of inflammatory genes in basal
conditions by repressing TNF-α promoter transcription. Under LPS
stimulation, nuclear SirT1 decreased, possibly through transferring
to the cytosol, indicated by the fact that no significant
alterations were found in total SirT1 protein in the present study.
In accordance with this, the transcription of TNF-α was
increased.
In addition to activating SirT1, resveratrol has
been revealed to increase the mRNA and protein levels of SirT1 in
Wistar rats, although the mechanism remains to be elucidated
(29). In the present study,
elevated protein levels of SirT1 were observed in normal cells
treated with resveratrol, compared with untreated cells. However,
resveratrol appeared to have no effect on the protein level of
SirT1 in tolerant cells. These results suggested that, in the
tolerant cells, the transcription and translation of SirT1 may have
peaked and responded minimally to resveratrol treatment.
Accordingly, the nuclear protein level of SirT1 in the LPS-tolerant
cells was sustained at high levels, and binding of SirT1 to the
TNF-α promoter was more marked, compared with that in normal cells
in the present study. This indicated that SirT1 constantly
repressed the transcription of TNF-α in tolerant cells, which
showed a high mRNA level of TNF-α, but were hyporesponsive to LPS
re-stimulation (30).
Studies have revealed that SirT1 limits inflammation
through several non-histone proteins. It has been reported that
SirT1 deacetylates H4K16 and promotes silencing of the
transcription of TNF-α on initial LPS stimulation in normal THP-1
cells (20). The present study
demonstrated that SirT1 was involved in epigenetic modifications in
the TNF-α promoter of cells in sepsis. The binding and activity of
SirT1 at the TNF-α promoter were increased in tolerant cells,
compared with normal cells. Resveratrol treatment further promoted
the activity and binding of SirT1 to the TNF-α promoter in the
tolerant cells. As expected, H3K9 methylation decreased with LPS
stimulation in normal cells due to chromatin relaxation and gene
transcription (31). Resveratrol
appeared to repress the transcription of TNF-α through promoting
H3K9 methylation in normal cells. However, resveratrol had no
effect on H3K9 methylation in tolerant cells, which already had
higher levels of H3K9 methylation at the TNF-α promoter, compared
with normal cells. Of note, the contradictory finding of the
coexistence of a high level of H4K16 acetylation and H3K9
methylation in the TNF-α promoter in tolerant cells demonstrated
that pro- and anti-inflammatory activities were active at the same
time. These results indicated that resveratrol further promoted the
activity of SirT1 in LPS-tolerant THP-1 cells and repressed the
transcription TNF-α through the deacetylation of H4K16 without
affecting the methylation of H3K9. Taken together, the results of
the present study indicated that resveratrol, as an activator of
SirT1, offers potential as an infective subsidiary treatment to
alleviate inflammation in patients with sepsis.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (grant no. 81201250) and the National
Key Technology Support Program (grant no. 2012BAI11B05).
References
|
1
|
Stearns-Kurosawa DJ, Osuchowski MF,
Valentine C, Kurosawa S and Remick DG: The pathogenesis of sepsis.
Annu Rev Pathol. 6:19–48. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cai B, Deitch EA and Ulloa L: Novel
insights for systemic inflammation in sepsis and hemorrhage.
Mediators Inflamm. 2010:6424622010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Russell JA: Management of sepsis. N Engl J
Med. 355:1699–1713. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Matsuda A, Jacob A, Wu R, Aziz M, Yang WL,
Matsutani T, Suzuki H, Furukawa K, Uchida E and Wang P: Novel
therapeutic targets for sepsis: Regulation of exaggerated
inflammatory responses. J Nippon Med Sch. 79:4–18. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nolan A, Weiden M, Kelly A, Hoshino Y,
Hoshino S, Mehta N and Gold JA: CD40 and CD80/86 act
synergistically to regulate inflammation and mortality in
polymicrobial sepsis. Am J Respir Crit Care Med. 177:301–308. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Flohé SB, Agrawal H, Schmitz D, Gertz M,
Flohé S and Schade FU: Dendritic cells during polymicrobial sepsis
rapidly mature but fail to initiate a protective Th1-type immune
response. J Leukoc Biol. 79:473–481. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Roger PM, Hyvernat H, Ticchioni M, Kumar
G, Dellamonica J and Bernardin G: The early phase of human sepsis
is characterized by a combination of apoptosis and proliferation of
T cells. J Crit Care. 27:384–393. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Paunel-Görgülü A, Kirichevska T, Lögters
T, Windolf J and Flohé S: Molecular mechanisms underlying delayed
apoptosis in neutrophils from multiple trauma patients with and
without sepsis. Mol Med. 18:325–335. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Russell JA, Boyd J, Nakada T, Thair S and
Walley KR: Molecular mechanisms of sepsis. Contrib Microbiol.
17:48–85. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Simmons GE Jr, Pruitt WM and Pruitt K:
Diverse roles of SIRT1 in cancer biology and lipid metabolism. Int
J Mol Sci. 16:950–965. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Giblin W, Skinner ME and Lombard DB:
Sirtuins: Guardians of mammalian healthspan. Trends Genet.
30:271–286. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu TF, Vachharajani V, Millet P,
Bharadwaj MS, Molina AJ and McCall CE: Sequential actions of
SIRT1-RELB-SIRT3 coordinate nuclear-mitochondrial communication
during immunometabolic adaptation to acute inflammation and sepsis.
J Biol Chem. 290:396–408. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu W, Lu Y, Yao J, Li Z, Chen Z, Wang G,
Jing H, Zhang X, Li M, Peng J and Tian X: Novel role of
resveratrol: Suppression of high-mobility group protein box 1
nucleocytoplasmic translocation by the upregulation of sirtuin 1 in
sepsis-induced liver injury. Shock. 42:440–447. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jia Y, Zheng Z, Wang Y, Zhou Q, Cai W, Jia
W, Yang L, Dong M, Zhu X, Su L and Hu D: SIRT1 is a regulator in
high glucose-induced inflammatory response in RAW264.7 cells. PLoS
One. 10:e01208492015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gao R, Ma Z, Hu Y, Chen J, Shetty S and Fu
J: SirT1 restrains lung inflammasome activation in a murine model
of sepsis. Am J Physiol Lung Cell Mol Physiol. 308:L847–L853. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schug TT, Xu Q, Gao H, Peres-da-Silva A,
Draper DW, Fessler MB, Purushotham A and Li X: Myeloid deletion of
SIRT1 induces inflammatory signaling in response to environmental
stress. Mol Cell Biol. 30:4712–4721. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang Z, Kahn BB, Shi H and Xue BZ:
Macrophage alpha1 AMP-activated protein kinase (alpha1AMPK)
antagonizes fatty acid-induced inflammation through SIRT1. J Biol
Chem. 285:19051–19059. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Imai S, Armstrong CM, Kaeberlein M and
Guarente L: Transcriptional silencing and longevity protein Sir2 is
an NAD-dependent histone deacetylase. Nature. 403:795–800. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vaquero A, Scher M, Lee D,
Erdjument-Bromage H, Tempst P and Reinberg D: Human SirT1 interacts
with histone H1 and promotes formation of facultative
heterochromatin. Mol Cell. 16:93–105. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu TF, Yoza BK, El Gazzar M, Vachharajani
VT and McCall CE: NAD+-dependent SIRT1 deacetylase
participates in epigenetic reprogramming during endotoxin
tolerance. J Biol Chem. 286:9856–9864. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen X, Yoza BK, El Gazzar M, Hu JY,
Cousart SL and McCall CE: RelB sustains IkappaBalpha expression
during endotoxin tolerance. Clin Vaccine Immunol. 16:104–110. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li J, Qu X, Ricardo SD, Bertram JF and
Nikolic-Paterson DJ: Resveratrol inhibits renal fibrosis in the
obstructed kidney: Potential role in deacetylation of Smad3. Am J
Pathol. 177:1065–1071. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
El Gazzar M, Yoza BK, Chen X, Hu J,
Hawkins GA and McCall CE: G9a and HP1 couple histone and DNA
methylation to TNFalpha transcription silencing during endotoxin
tolerance. J Biol Chem. 283:32198–32208. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hou J, Chong ZZ, Shang YC and Maiese K:
Early apoptotic vascular signaling is determined by SirT1 through
nuclear shuttling, forkhead trafficking, bad, and mitochondrial
caspase activation. Curr Neurovasc Res. 7:95–112. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Autiero I, Costantini S and Colonna G:
Human Sirt-1: Molecular modeling and structure-function
relationships of an unordered protein. PLoS One. 4:e73502008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sharma A, Gautam V, Costantini S, Paladino
A and Colonna G: Interactomic and pharmacological insights on human
Sirt-1. Front Pharmacol. 3:402012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kim EJ, Kho JH, Kang MR and Um SJ: Active
regulator of SIRT1 cooperates with SIRT1 and facilitates
suppression of p53 activity. Mol Cell. 28:277–290. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li T, Zhang J, Feng J, Li Q, Wu L, Ye Q,
Sun J, Lin Y, Zhang M, Huang R, et al: Resveratrol reduces acute
lung injury in a LPS-induced sepsis mouse model via activation of
SirT1. Mol Med Rep. 7:1889–1895. 2013.PubMed/NCBI
|
|
29
|
Franco JG, de Moura EG, Koury JC, Trotta
PA, Cordeiro A, Souza LL, Almeida NA, Lima Nda S, Pazos-Moura CC,
Lisboa PC and Passos MC: Resveratrol reduces lipid peroxidation and
increases sirtuin 1 expression in adult animals programmed by
neonatal protein restriction. J Endocrinol. 207:319–328. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen X, El Gazzar M, Yoza BK and McCall
CE: The NF-kappaB factor RelB and histone H3 lysine
methyltransferase G9a directly interact to generate epigenetic
silencing in endotoxin tolerance. J Biol Chem. 284:27857–27865.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
El Gazzar M, Yoza BK, Chen X, Garcia BA,
Young NL and McCall CE: Chromatin-specific remodeling by HMGB1 and
linker histone H1 silences proinflammatory genes during endotoxin
tolerance. Mol Cell Biol. 29:1959–1971. 2009. View Article : Google Scholar : PubMed/NCBI
|