Introduction
Gastric cancer remains to be the third leading cause
of cancer mortality, worldwide; however, the incidence rates have
declined in Western Europe (1).
Helicobacter pylori (H. pylori) infection is regarded
as a primary factor in the development of atrophic gastritis, and
gastric cancer may subsequently occur (2,3).
Gastric cancer occurs via numerous stages, beginning with chronic
inflammation, atrophic alterations, intestinal metaplasia and
dysplasia (4). From the majority
of patients with an H. pylori infection, only a small
percentage of patients have the poorest clinical outcome, such as
diagnosis with gastric cancer. However, a specific factor which may
aid in determining the variation of the clinical outcome remains to
be identified, the specific outcome may be the result of an
interaction of bacterial virulence factors, host immune system and
the environment. Additionally, a previous study determined that
genetic variation affected the clinical course of H. pylori
infection (5).
A microRNA (miRNA) is an endogenous small
non-protein-coding RNA cleaved from a precursor of the 70–100 bp
hairpin form (6,7). miRNAs, which bind to the
3′-untranslated region (UTR) of target genes, regulate the mRNA
expression post-transcriptionally (6). Previous studies which examined
expression of miRNAs using clinical samples, determined that the
expression and function of miRNAs may be associated with cancer
pathogenesis (8–10) including gastric cancer (11). Mishra et al (12) determined that polymorphism of pri-,
pre- and mature miRNA may affect miRNA function and potentially
influence various gene expression levels and signaling pathways. It
has also been reported that the processing and target selection of
miRNAs may be influenced by variations in the coding region of
miRNA genes, including pri-, pre- and mature miRNAs (13). The authors of the present study
previously reported that miR-27a genetic variation may be
associated with gastric mucosal atrophy (14).
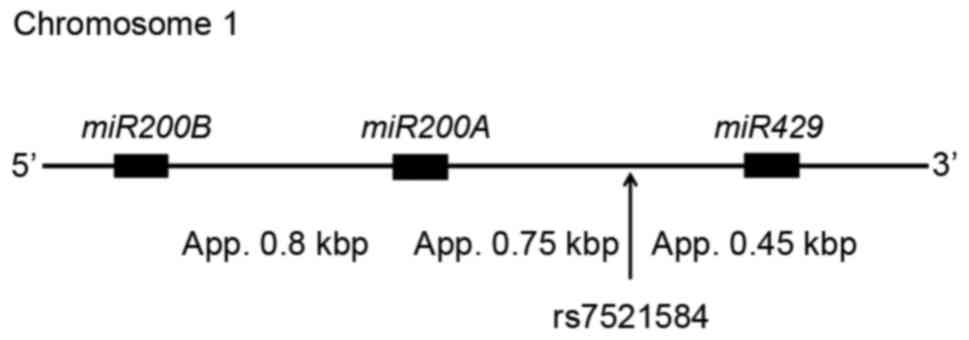
The present study, investigated the region of ~2 kbp
at chromosome 1, containing miRNA (miR)-200b, miR-200a and miR-429
(Fig. 1). From this region, 5
miRNAs are yielded and may regulate various gene expression levels.
It is possible that genetic variations in this region may influence
the clinical outcome of some diseases. The rs7521584 polymorphism
reported in National Center for Biotechnology Information single
nucleotide polymorphism (SNP) database was selected, and the
present study investigated the association of this polymorphism
with the development of gastric mucosal atrophy and metaplastic
change as a pre-malignant condition.
Materials and methods
Clinical samples and DNA
extraction
A total of 393 patients with no neoplastic lesions,
who had a medical examination in Fujita Health University Hospital
(Toyoake, Japan) or Kanazawa Medical University Hospital
(Uchinada-machi, Japan) from April 2005 to March 2014 were
recruited for the present study. All patients underwent upper
gastrointestinal endoscopy due to reported abdominal discomfort and
were diagnosed with gastritis or no abnormal appearance. Patients
with severe systemic diseases, malignancies in other organs and
those that had received nonsteroidal anti-inflammatory drugs,
antibiotics and H. pylori eradication treatment were
excluded.
All histological diagnoses were preformed using
biopsy specimens obtained from the antrum by the Division of
Pathology of each hospital. The severity of chronic gastritis was
classified according to the updated Sydney system (15) by a pathologist who had no access to
any clinical information. According to the severity of atrophic
gastritis, the subjects were divided into the following 2 groups:
The atrophic gastritis (AG) group (atrophy score ≥1 and metaplasia
score ≥1) and the no atrophy (NA) group.
Genomic DNA was isolated from the blood samples
using QIAamp DNA blood kit (Qiagen, Inc., Valencia, CA, USA)
according to the manufacturer's protocol. H. pylori
infection status was determined by serology, histological
examination and the urea breath test. A total of 251 patients were
diagnosed as H. pylori positive when at least one of the
diagnostic tests was positive. In addition, serum pepsinogen (PG)
I/II levels were quantified in 123/393 patients.
The Ethical Committee of Fujita Health University
and Kanazawa Medical University approved the protocol and written
informed consent was obtained from all patients.
SNP detection
In order to determine the genotype of patients, the
present study used polymerase chain reaction-single strand
conformation polymorphism (PCR-SSCP) as previously described
(14). Primer pairs were
synthesized and the sequences were as follows: rs7521584 forward,
5′-AACAGTGGCCTCTCTCACGTGGT-3′ and reverse,
5′-TTGCAGATGGAAAAGATGAAACAAT-3′. PCR was performed in a volume of
20 µl containing 0.1 µg DNA using Ex Taq DNA polymerase (Takara Bio
Inc., Otsu, Japan). The DNA was denatured at 95°C for 3 min,
followed by 35 cycles at 95°C for 30 sec, 54°C for 40 sec and 72°C
for 45 sec, with a final extension step at 72°C for 5 min.
Subsequently, 2 µl of the PCR products were denatured using 10 µl
formamide (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) at
90°C for 5 min. SSCP was performed at 18°C using a GenePhor DNA
separation system with GeneGel Excel 12.5/24 (GE Healthcare Life
Sciences, Chalfont, UK), subsequently the denatured single strand
DNA bands were detected using a DNA Silver Staining kit (GE
Healthcare Life Sciences).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA from whole blood samples of healthy
patients with 5GG, 5GT and 5TT genotypes was extracted using TRIzol
reagent (Thermo Fisher Scientific, Inc., Waltham, MA, USA)
according to the manufacturer's instructions. The Mir-X miRNA
qRT-PCR SYBR kit (Takara Bio, Inc.) was used to quantify miRNA
expression, according to the manufacturer's instructions. Total RNA
(0.8 µg) was reverse transcribed using the Mir-X™ miRNA
First-Strand Synthesis kit according the manufacturer's protocol.
The PCR reactions were performed using miRNA-specific primers
(miR-429, 5′-TAATACTGTCTGGTAAAACCGT-3′; miR-200a-3p,
5′-TAACACTGTCTGGTAACGATGT-3′ for miR-200a-3p). The thermocycling
conditions were as follows: initial denaturation at 95°C for 10
sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 20 sec.
U6 was used as the reference gene (5-GGG CAG GAA GAG GGC CTA T-3).
All data were analyzed using the 2−ΔΔCq method (16).
Statistical analysis
The Hardy-Weinberg equilibrium of the allele was
assessed using a χ2 test. The age data were expressed as
the mean ± standard deviation. Analyses were performed using Stata
(version 13; StataCorp LP, College Station, TX, USA). The mean ages
between the 2 groups were compared using Student's t-test. The
ratios of H. pylori infection status and gender were
compared using Fisher's exact test. Differences of genotype
frequencies were determined with a two-sided Fisher's exact test.
The odds ratio (OR) and 95% confidence intervals (CI) were
estimated by logistic regression following an adjustment for age,
gender and H. pylori infection status. Each of the Sydney
system scores between the 2 groups was compared using a
Mann-Whitney U test. The association between age and PG I/II ratio
was determined using one-way analysis of variance. P<0.05 was
considered to indicate a statistically significant difference.
Results
Characteristics of subjects and
genotype frequencies
Single strand DNA of rs7521584 was clearly separated
by SSCP (Fig. 2). A total of 393
patients participated in the present study. The patients were
classified into the AG (n=139) and NA groups (n=254), their
characteristics are summarized in Table I. The mean age of the AG group was
significantly greater compared with the NA group. The male/female
ratio and H. pylori positivity were also significantly
greater in the AG group compared with the NA group.
 | Table I.Characteristics of the subjects and
frequency of genotypes. |
Table I.
Characteristics of the subjects and
frequency of genotypes.
| Characteristic | Total | NA group | AG group | P-value |
|---|
| Number of
patients | 393 | 254 | 139 |
|
| Mean age ± standard
deviation | 60.0±13.3 | 58.0±13.7 | 63.6±11.8 |
<0.0001a |
| Male:female | 233:160 | 132:122 | 101:38 |
<0.0001a |
| H. pylori
positive rate rs7521584 G>T | 251/393 | 118/254 | 133/139 |
<0.0001a |
| GG | 139 | 88 | 51 |
|
| GT | 203 | 137 | 66 |
|
| TT | 51 | 29 | 22 |
|
| T allele
frequency | 38.8% | 38.4% | 39.6% | NS |
The distribution of genotypes in the patients
recruited in the present study were in Hardy-Weinberg equilibrium
(P=0.09). No significant difference between the genotype
distribution of AG and NA groups was identified (Table I).
Association between the rs7521584
polymorphism and atrophic gastritis
The present study used logistic regression analysis
following an adjustment for age, gender and H. pylori
infection status, to determine that the rs7521584 TT genotype was
significantly associated with the severity of atrophic gastritis
(OR, 2.41; 95% CI, 1.10–5.25; P=0.027; Table II). In the H. pylori
infected subjects, TT genotype was also significantly associated
with the atrophic gastritis (OR, 3.31; 95% CI, 1.35–8.12; P=0.0089;
Table II). In addition, in the
subjects younger than 60 years of age, this genotype was positively
associated with atrophic gastritis (OR, 3.15; 95% CI, 1.03–9.61;
P=0.044; Table II).
 | Table II.Association between rs7521584 and
gastric mucosal atrophy. |
Table II.
Association between rs7521584 and
gastric mucosal atrophy.
| Characteristic | No. of
subjects | GG | GT | TT | OR (95%
CI)a | P-value |
|---|
| rs7521584
G>T |
|
|
|
|
|
|
| NA | 254 | 88 | 137 | 29 | Reference | – |
| AG | 139 | 51 | 66 | 22 | 2.41
(1.10–5.25) | 0.027a |
| H. pylori
positive |
|
|
|
|
|
|
| NA | 118 | 40 | 70 | 8 | Reference | – |
| AG | 133 | 49 | 62 | 22 | 3.31
(1.35–8.12) | 0.0089a |
|
≤60 |
|
|
|
|
|
|
| NA | 129 | 48 | 64 | 17 | Reference | – |
| AG | 46 | 13 | 23 | 10 | 3.15
(1.03–9.61) | 0.044a |
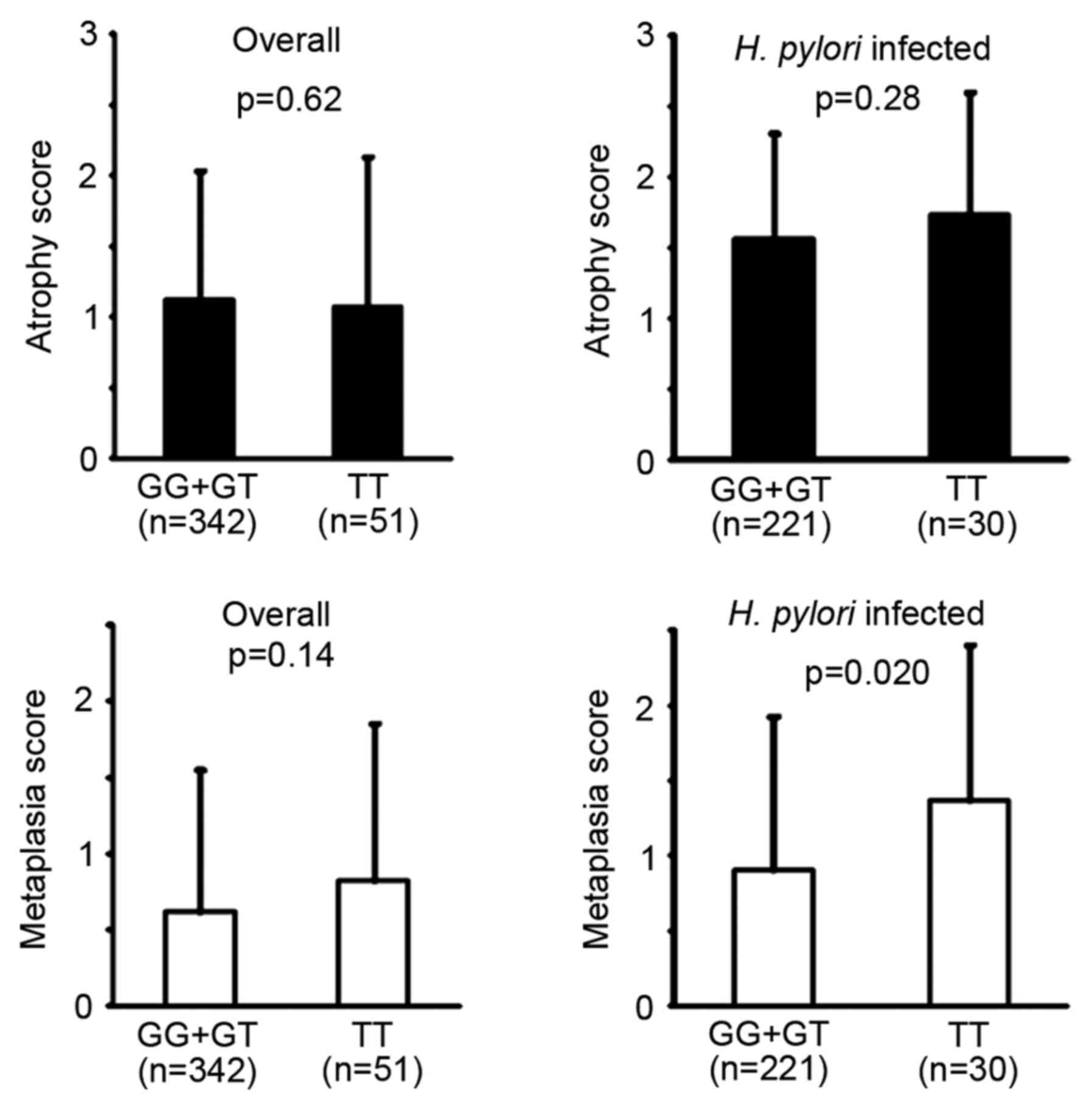
No significant difference was identified between the
GG+GT genotype and TT homozygote for the atrophy and metaplasia
scores in the updated Sydney system (Fig. 3). However, in patients with an
H. pylori infection, the metaplasia score was significantly
higher in the TT homozygote compared with the GG+GT genotype.
Serum PG I/II ratio and rs7521584
genotype
The rs7521584 genotype distribution in 123 patients
whose serum PG were quantified was as follows: GG, 47; GT, 57; and
TT, 19, which did not differ from the distribution observed in all
patients (P=0.63). The H. pylori positivity did not differ
significantly between the GG+GT genotype (61.5%) and TT homozygote
(52.6%).
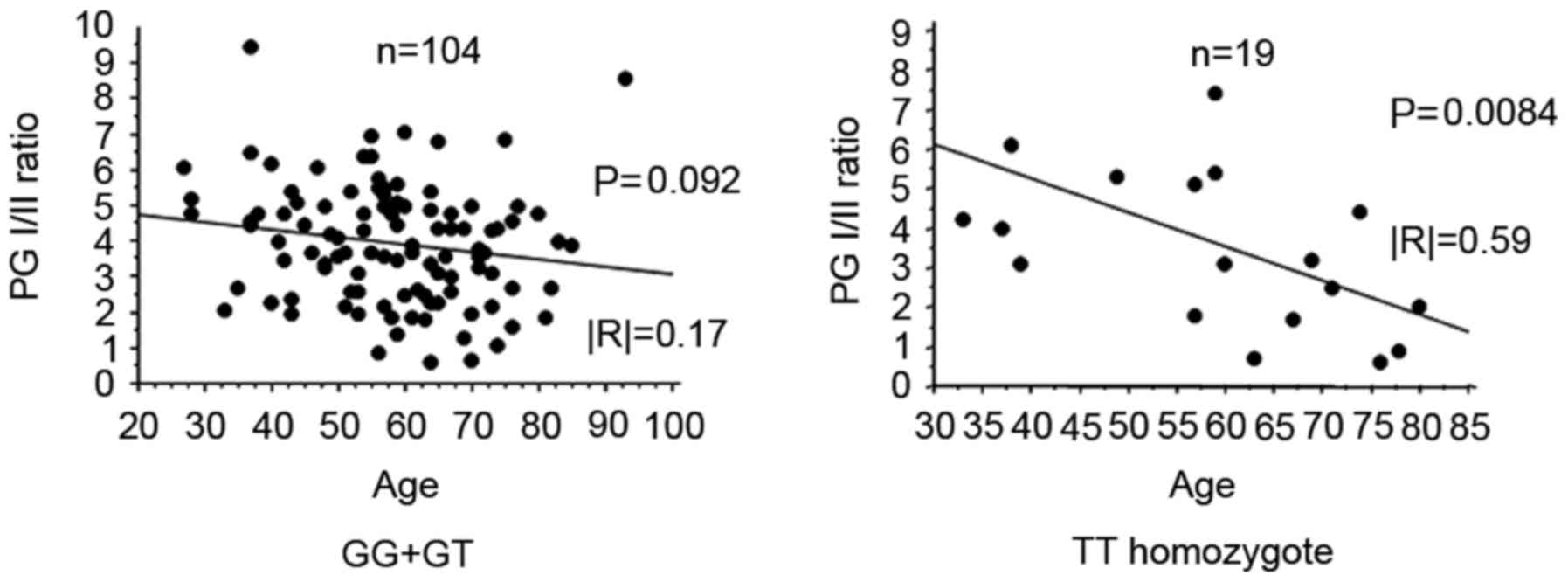
In the rs7521584 TT homozygote, PG I/II ratio
significantly decreased with increasing age (P=0.0084; Fig. 4). However, there was no significant
association between the PG I/II ratio and age in the GG+GT genotype
(Fig. 4).
Discussion
A total of five miRNAs with similar seed sequence
constitute the miR-200 family, located in two distinct genomic
clusters (miR-200a-200b-429 and miR-200c-141 clusters). The
miR-200a-200b-429 cluster is located on chromosome 1. In general,
one mRNA is regulated by numerous miRNAs, and one miRNA may target
numerous mRNAs (17). Therefore,
it is difficult to determine the mechanisms of miRNA in the
pathogenesis of human disorders. However, several previous studies
have indicated that the reduced expression of miR-200 family may
contribute to the development and progression of gastric cancer
(18–20). Chang et al (18) determined that the expression of the
miR-200 family was downregulated in gastric cancer tissues compared
with matched non-cancerous tissues and lower levels of the miR-200
family were associated with the prognosis of the patients with
gastric cancer (18). Zhu et
al (19) has reported that
suppressed expression of miR-429 in the gastric cancer cell line
AGS promoted cancer cell survival mediated by B cell
leukemia/lymphoma 2 against chemotherapy-induced cell death and
that the rescued expression of miR-429 induced the apoptosis of
cancer cells (19). Zhang et
al (20) have indicated that
expression of miR-429 in gastric cancer tissues was downregulated
compared with adjacent normal tissue and have demonstrated that the
expression of fascin-1 (a direct target of miR-429) and miR-429
expression were inversely correlated in vivo (20). These previous findings suggest that
the miR-200 family may act as a tumor suppressor on the development
and progression of gastric cancer. Additionally, the miR-200 family
may also be capable of inhibiting the invasion of carcinoma to the
other organs, including colorectal carcinoma (21–23).
Conversely, Chen et al (24) reported that the overexpression of
miR-200a-3p may be associated with gender and miR-429 with age in
patients with gastric cancer. In addition, it has been previously
reported that upregulated expression of miR-429 in patients with
serous ovarian carcinoma was negatively correlated with survival
(25). Therefore, although the
miR-200 family may have important roles in tumor progression, their
specific functions and the mechanisms involved remain to be fully
elucidated.
The miR-200 family also has an important role in the
inflammation process. Reddy et al (26) identified a proinflammatory function
for the negative feedback loop between miR-200 and zinc finger
e-box-binding homeobox 1 in vascular smooth muscle cells (VSMCs)
under diabetic conditions (26).
They concluded that disruption of this negative feedback loop
enhances the proinflammatory responses of VSMCs, which are
implicated in vascular complications (26). Xiao et al (27) have determined that miR-429 targets
dual specificity protein phosphatase 1 directly, in order to
regulate the activation of p38 mitogen-activated protein kinase and
subsequent production of cytokines in response to
lipopolysaccharide stimulation in alveolar macrophages (27). The miR-200 family may have a tumor
suppressor role by inhibition of the epithelial mesenchymal
transition (28) and a
proinflammatory role in the inflammation process.
In the present study, rs7521584 was significantly
associated with the development of atrophic gastritis, which is
considered as a pre-malignant condition, particularly in patients
with H. pylori infection. Additionally, the significant
progression of atrophic gastritis was observed in patients younger
than 60 years of age. Although no difference was identified in
terms of H. pylori positivity between the two genotypes,
serum PG I/II ratio was reduced with age in the TT homozygote
compared with the GG+GT genotype. The present results suggest that
H. pylori-associated atrophic gastritis may progress more
rapidly in the TT homozygote compared with the other genotypes.
Additionally, the present study indicated that re7521584 is more
closely associated with metaplastic alterations compared with
atrophic alterations. A previous study indicated that the
development of gastric metaplasia may not always be associated with
the progression of gastric mucosal atrophy and H. pylori
infection status (29). Gastric
glandular atrophy, resulting from parietal cell loss due to H.
pylori induced-inflammation, may lead to gastric chief cell
transdifferentiation into spasmolytic polypeptide expressing
metaplasia (SPEM) (30). In the
presence of chronic inflammation, SPEM may progress to more
advanced metaplasia but not to intestinal metaplasia in mice
(31,32). However, in humans, SPEM leads to
intestinal metaplasia, which may progress to cancer (33). Therefore, it is likely that more
advanced factors are required to develop intestinal metaplasia from
atrophic gastric mucosa in humans that have yet to be identified
(30). The rs7521584 polymorphism
may be more closely associated with metaplastic transformation than
atrophic change by H. pylori-induced chronic
inflammation.
The contents of miR-200 families in blood samples
were quantified and no significant differences were identified
among rs7521584 genotypes (data not shown). Therefore, the
influence of rs7521584 on the expression or function of miR-200
families cannot be evaluated. miRNAs may be derived by various
stimulations and the content of miRNA in blood serum may differ
from that in organ tissues, its influence may be difficult to
determine solely from the quantification of the serum sample.
Further investigation using cell assays would be required at this
point. If variation of rs7521584 may influence the expression or
function of one or all miR-200 families; therefore, rs7521584 may
influence the metaplastic transformation in the gastric mucosa,
leading to the later intestinal type of gastric cancer, by
affecting mRNA expression via the alteration of miR-200 family
expression or function.
In conclusion, the present study demonstrated that
the rs7521584 minor allele homozygote was associated with the
development of atrophic gastritis following H.
pylori-induced inflammation, particularly affecting the
severity of metaplastic alterations. The accumulation of similar
findings may lead to an improved understanding of the diverse
clinical course of H. pylori infection.
Acknowledgements
Not applicable.
Funding
Not applicable.
Availability of data and materials
The analyzed datasets generated during the study are
available from the corresponding author on reasonable request.
Author's contributions
TA analyzed the data and was responsible for the
conception of the study and designed the study. TO analyzed the
data and wrote the paper. MN, MO, TN and RH obtained the samples
and the data. WJ and TaS determined the genotypes. TT and ToS
participated in the design of the study.
Ethics approval and consent to
participate
The Ethical Committee of Fujita Health University
and Kanazawa Medical University approved the protocol and written
informed consent was obtained from all patients.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Blaser MJ and Parsonnet J: Parasitism by
the ‘slow’ bacterium Helicobacter pylori leads to altered
gastric homeostasis and neoplasia. J Clin Invest. 94:4–8. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Huang JQ, Sridhar S, Chen Y and Hunt RH:
Meta-analysis of the relationship between Helicobacter
pylori seropositivity and gastric cancer. Gastroenterology.
114:1169–1179. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Correa P: Human gastric carcinogenesis: A
multistep and multifactorial process-first American cancer society
award lecture on cancer epidemiology and prevention. Cancer Res.
52:6735–6740. 1992.PubMed/NCBI
|
|
5
|
Crabtree JE: Gastric mucosal inflammatory
responses to Helicobacter pylori. Aliment Pharmacol Ther. 10
Suppl 1:S29–S37. 1996. View Article : Google Scholar
|
|
6
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Paladini L, Fabris L, Bottai G, Raschioni
C, Calin GA and Santarpia L: Targeting microRNAs as key modulators
of tumor immune response. J Exp Clin Cancer Res. 35:1032016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nakamura K, Sawada K, Yoshimura A, Kinose
Y, Nakatsuka E and Kimura T: Clinical relevance of circulating
cell-free microRNAs in ovarian cancer. Mol Cancer. 15:482016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li X, Nie J, Mei Q and Han WD: MicroRNAs:
Novel immunotherapeutic targets in colorectal carcinoma. World J
Gastroenterol. 22:5317–5331. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tsai MM, Wang CS, Tsai CY, Huang HW, Chi
HC, Lin YH, Lu PH and Lin KH: Potential diagnostic, prognostic and
therapeutic targets of MicroRNAs in human gastric cancer. Int J Mol
Sci. 17:E9452016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mishra PJ and Bertino JR: MicroRNA
polymorphisms: The future of pharmacogenomics, molecular
epidemiology and individualized medicine. Pharmacogenomics.
10:399–416. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Duan R, Pak C and Jin P: Single nucleotide
polymorphism associated with mature miR-125a alters the processing
of pri-miRNA. Hum Mol Genet. 16:1124–1131. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Arisawa T, Tahara T, Shibata T, Nagasaka
M, Nakamura M, Kamiya Y, Fujita H, Hasegawa S, Takagi T, Wang FY,
et al: A polymorphism of microRNA 27a genome region is associated
with the development of gastric mucosal atrophy in Japanese male
subjects. Dig Dis Sci. 52:1691–1697. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dixon MF, Genta RM, Yardley JH and Correa
P: Classification and grading of gastritis: The updated Sydney
system. Am J Surg Pathol. 20:1161–1181. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lim LP, Lau NC, Garrett-Engele P, Grimson
A, Schelter JM, Castle J, Bartel DP, Linsley PS and Johnson JM:
Microarray analysis shows that some microRNAs downregulate large
numbers of target mRNAs. Nature. 433:769–773. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chang L, Guo F, Huo B, Lv Y, Wang Y and
Liu W: Expression and clinical significance of the microRNA-200
family in gastric cancer. Oncol Lett. 9:2317–2324. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhu P, Ahang J, Zhu J, Shi J, Zhu Q and
Gao Y: miR-429 induces gastric carcinoma cell apoptosis through
Bcl-2. Cell Physiol Biochem. 37:1572–1580. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang M, Dong BB, Lu M, Zheng MJ, Chen H,
Ding JZ, Xu AM and Xu YH: miR-429 functions as a tumor suppressor
by targeting FSCN1 in gastric cancer cells. Onco Targets Ther.
9:1123–1133. 2016.PubMed/NCBI
|
|
21
|
Sun Y, Shen S, Liu X, Tang H, Wang Z, Yu
Z, Li X and Wu M: miR-429 inhibits cells growth and invasion and
regulates EMT-related marker genes by targeting Onecut2 in
colorectal carcinoma. Mol Cell Biochem. 390:19–30. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ye ZB, Ma G, Zhao YH, Xiao Y, Zhan Y, Jing
C, Gao K, Liu ZH and Yu SJ: miR-429 inhibits migration and invasion
of breast cancer cells in vitro. Int J Oncol. 46:531–538. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lei W, Liu YE, Zheng Y and Qu L: miR-429
inhibits oral squamous cell carcinoma growth by targeting ZEB1. Med
Sci Monit. 21:383–389. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen Z, Liu X, Hu Z, Wang Y, Liu M, Liu X,
Li H, Ji R, Guo Q and Zhou Y: Identification and characterization
of tumor suppressor and oncogenic miRNAs in gastric cancer. Oncol
Lett. 10:329–336. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim
JH, Kim JW and Kim S: MicroRNA expression profiles in serous
ovarian carcinoma. Clin Cancer Res. 14:2690–2695. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Reddy MA, Jin W, Villeneuve L, Wang M,
Lanting L, Todorov I, Kato M and Natarajan R: Pro-inflammatory role
of microrna-200 in vascular smooth muscle cells from diabetic mice.
Arterioscler Thromb Vasc Biol. 32:721–729. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xiao J, Tang J, Chen Q, Tang D, Liu M, Luo
M, Wang Y, Wang J, Zhao Z, Tang C, et al: miR-429 regulates
alveolar macrophage inflammatory cytokine production and is
involved in LPS-induced acute lung injury. Biochem J. 471:281–291.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Brabletz S and Brabletz T: The ZEB/miR-200
feedback loop-a motor of cellular plasticity in development and
cancer? EMBO Rep. 11:670–677. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Watanabe T, Goto H, Arisawa T, Hase S,
Niwa Y, Hayakawa T and Asai J: Relationship between local immune
response to Helicobacter pylori and the diversity of
disease: Investigation of H. pylori-specific IgA in gastric
juice. J Gastroenterol Hepatol. 12:660–665. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nam KT, Lee HJ, Sousa JF, Weis VG, O'Neal
RL, Finke PE, Romero-Gallo J, Shi G, Mills JC, Peek RM Jr, et al:
Mature chief cells are cryptic progenitors for metaplasia in the
stomach. Gastroenterology. 139:2028–2037.e9. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fox JG, Blanco M, Murphy JC, Taylor NS,
Lee A, Kabok Z and Pappo J: Local and systemic immune responses in
murine Helicobacter felis active chronic gastritis. Infect Immun.
61:2309–2015. 1993.PubMed/NCBI
|
|
32
|
Yoshizawa N, Takenaka Y, Yamaguchi H,
Tetsuya T, Tanaka H, Tatematsu M, Nomura S, Goldenring JR and
Kaminishi M: Emergence of spasmolytic polypeptide-expressing
metaplasia in Mongolian gerbils infected with Helicobacter
pylori. Lab Invest. 87:1265–1276. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Halldórsdóttir AM, Sigurdardóttrir M,
Jónasson JG, Oddsdóttir M, Magnússon J, Lee JR and Goldenring JR:
Spasmolytic polypeptide expressing metaplasia (SPEM) associated
with gastric cancer in Iceland. Dig Dis Sci. 48:431–441. 2003.
View Article : Google Scholar : PubMed/NCBI
|