Introduction
Recurrent pregnancy loss (RPL) was initially defined
as ≥3 consecutive pregnancy losses before 20 weeks of gestation
(1,2), but was later redefined by the
American Society for Reproductive Medicine as ≥2 consecutive
pregnancy losses (3). An estimated
1–5% of women of reproductive age experience a pregnancy loss, and
10–20% of pregnancies end in a miscarriage, which frequently occurs
in the second or third month of pregnancy (2,4,5). RPL
may be caused by genetic disorders or various other factors,
including fetal chromosomal abnormalities, uterine anomalies,
thrombophilia, endocrine, immune and anatomical disorders,
nutritional or environmental factors, psychological stress, or
maternal infections (5). However,
in as many as 50% of women who suffer from recurrent miscarriages,
the causes are considered idiopathic. RPL is an important
reproductive health issue and remains an active field of research
(4). In the present study, the
potential genetic cause of RPL was investigated. Four candidate
genes harboring mutations that have been associated with other
diseases were selected at random as they have not been investigated
in association with RPL, and their association with RPL in Korean
women was assessed.
Frameshift mutations result from insertions or
deletions that alter the reading frame, and affect the subsequent
coding sequence as well as the stop codon. Therefore, frameshift
mutations may result in final polypeptide products with an abnormal
length or in nonsense-mediated mRNA decay (6). This type of mutation has been
implicated in numerous diseases; for example, Crohn's disease is
associated with the nucleotide-binding oligomerization domain 2
3020insC frameshift mutation (7).
Alternatively, abnormalities in alternative splicing have also been
implicated in various diseases, including cancer (8–10).
Next-generation sequencing (NGS) is a more advanced
method of DNA sequencing compared with Sanger sequencing, which
permits the sequencing of the entire human genome in 1 day
(11,12). It offers efficient analysis and
rapid processing time, thereby reducing the labor and expenses
associated with other sequencing options (13,14).
NGS is a powerful tool used to study genetic diseases (13–16),
and was therefore the technique of choice for this study.
In the present study, frameshift mutations in
membrane spanning 4-domains A14 (MS4A14) and solute carrier
family 2 member 7 (SLC2A7), and splice variants in pregnancy
specific β-1-glycoprotein 9 (PSG9) and ATP binding cassette
subfamily B member 5 (ABCB5) were investigated. These were
selected through whole-exome sequencing (WES). To the best of our
knowledge, the following four polymorphisms: MS4A14D>I
(rs3217518), SLC2A7D>I (rs60746313), PSG9C>T
(rs3746297) and ABCB5C>G (rs17143187), have not been
previously studied in RPL. Therefore, the aim of the study was to
examine the association between these four gene polymorphisms and
RPL in Korean women.
Materials and methods
Study population
The study population included 383 female patients
with RPL and 276 control women. Patients with RPL were 22–45 years
old, with an average age of 33.11±4.44 years and a body mass index
(BMI) of 21.48±3.85 kg/m2. Women in the control group
were 20–66 years old, with an average age of 33.09±5.68 years and a
BMI of 21.31±3.30 kg/m2.
Blood samples were collected from patients with RPL
and control women between March 1999 and February 2012 at the
Department of Obstetrics and Gynecology and the Fertility Center of
CHA Bundang Medical Center (Seongnam, Korea). The study was
approved by the Institutional Review Board of CHA Bundang Medical
Center (IRB number: BD2010-123D) and written informed consent was
provided by all patients. Women in the control group had regular
menstrual cycles, a history of ≥1 naturally-conceived pregnancy, no
history of pregnancy losses and a normal karyotype (46,XX). Women
were diagnosed with RPL if they had a history of ≥2 consecutive
spontaneous pregnancy losses. Before week 20 of gestation,
ultrasound and/or physical examinations were performed and human
gonadotropic hormone levels were assessed. The average gestational
age at the time of miscarriage was 7.35±1.93 weeks, and overall
patients had 3.28±1.84 miscarriages.
Patients with a history of smoking or alcohol abuse,
or whose miscarriages were attributed to infection or anatomical,
hormonal, chromosomal, autoimmune or thrombotic causes were
excluded from the study. Anatomical causes of miscarriage were
determined using hysterosalpingography, hysteroscopy, computerized
tomographic scanning or magnetic resonance imaging, to identify
intrauterine adhesion, septate uterus or uterine fibroids. Hormonal
causes of miscarriage included hyperprolactinemia, luteal
insufficiency and thyroid disease; these were determined by
measuring relevant hormone levels in blood samples. The chromosomal
causes for miscarriage were assessed by standard chromosome
analysis using the G-banding method (17). Miscarriages caused by infection
with Ureaplasma urealyticum or Mycoplasma hominis
were identified by bacterial culture. The following autoimmune
diseases: Lupus and antiphospholipid syndrome, were selected for
their strong association with miscarriages; these were identified
by measuring lupus anticoagulant and anticardiolipin antibodies.
Thrombophilia was defined as a thrombotic disorder associated with
miscarriages, and was identified by protein C and protein S
deficiency, and presence of anti-β-2 glycoprotein.
Genotyping
Genomic DNA was extracted from peripheral blood of
each study participant using the G-DEX DNA extraction kit (Intron
Biotechnology, Inc., Seongnam, Korea). Macrogen, Inc. (Seoul,
Korea) was commissioned to perform NGS of 20 patients with RPL. NGS
was conducted using a HiSeq Instrument (Macrogen, Inc., Seoul,
Korea) and paired-end sequences produced by a HiSeq Instrument were
first mapped to the human genome using the mapping program ‘BWA’
(version 0.7.12) (https://sourceforge.net/projects/bio-bwa/files/).
Based on the BAM file previously generated, variant genotyping for
each sample was performed with Haplotype Caller of GATK (Broad
Institute, Cambridge, MA, USA). Then, an in-house program and
SnpEff (snpeff.sourceforge.net/) was applied to filter
additional databases, including ESP6500 (https://esp.gs.washington.edu/), ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/),
and dbNSFP2.9 (https://sites.google.com/site/jpopgen/dbNSFP). WES
statistical analyses were also conducted by Macrogen, Inc. The
frameshift and splice variants genes were identified from the human
genome single nucleotide polymorphism (SNP) database (http://www.ncbi.nlm.nih.gov/snp), and selected
from the WES statistical list. The four polymorphisms selected were
as follows: MS4A14D>I (rs3217518), SLC2A7D>I
(rs60746313), PSG9C>T (rs3746297) and ABCB5C>G
(rs17143187). The four SNPs were genotyped using polymerase chain
reaction-restriction fragment length polymorphism (PCR-RFLP)
analysis; the primers, PCR conditions and restriction enzymes used,
are detailed in Table I (18).
 | Table I.Frameshift and splice variant gene
polymorphisms determined using polymerase chain
reaction-restriction fragment length polymorphism analysis. |
Table I.
Frameshift and splice variant gene
polymorphisms determined using polymerase chain
reaction-restriction fragment length polymorphism analysis.
| Polymorphism | Reference SNP | Chromosome | Position | Function | Primer
sequence | Annealing
temperature (°C) | Restriction
enzyme | Band size (bp) |
|---|
| MS4A14
D>I | rs3217518 | 11 | 60397880 | Frameshift | Forward:
5′-TTGGATGGAGGGAAAGGTGTG-3′ | 55 | TspRI | DD: 229, 140 |
|
|
|
|
|
| Reverse:
5′-TTTTGCCGTGAAGGGAGCT-3′ |
|
| DI: 370, 229,
140 |
|
|
|
|
|
|
|
|
| II: 370 |
| SLC2A7
D>I | rs60746313 | 1 | 9024982 | Frameshift | Forward:
5′-AAGATGGCGGCTACCTTC-3′ | 53 | Hpy166II | DD: 109 |
|
|
|
|
|
| Reverse:
5′-CTACAACCTCTCTGTGGTCA-3 |
|
| DI: 120, 109 |
|
|
|
|
|
|
|
|
| II: 120 |
| PSG9
C>T | rs3746297 | 19 | 43268150 | Splice variant | Forward:
5′-GTAATGGTAGAGGTCCGTCA-3′ | 57 | BfaI | CC: 171, 114 |
|
|
|
|
|
| Reverse:
5′-CGTGTGTGTATCTTCAAGGC-3′ |
|
| CT: 285, 171,
114 |
|
|
|
|
|
|
|
|
| TT: 285 |
| ABCB5
C>G | rs17143187 | 7 | 20626612 | Splice variant | Forward:
5′-GAGAAAGGAAGCAGTTGG-3′ | 52 | DdeI | CC: 115, 32 |
|
|
|
|
|
| Reverse:
5′-TAGTTCCCTCTTTCCCAC-3′ |
|
| CG: 147, 115,
32 |
|
|
|
|
|
|
|
|
| GG: 147 |
The MS4A14D>I polymorphism was amplified
using forward (5′-TTGGATGGAGGGAAAGGTGTG-3′) and reverse
(5′-TTTTGCCGTGAAGGGAGCT-3′) primers, and was amplified by the Solg™
2X h-Taq PCR Pre-mix (SolGent co., Ltd., Daejeon, Korea) under the
following conditions: 95°C for 15 min; 35 cycles of denaturation at
95°C for 30 sec, annealing at 55°C for 30 sec, and extension at
72°C for 30 sec; and a final extension at 72°C for 5 min. The PCR
products were digested with the restriction enzyme TspRI
(New England BioLabs, Inc., Ipswich, MA, USA) at 65°C for 16 h.
The SLC2A7D>I polymorphism was amplified
using forward (5′-AAGATGGCGGCTACCTTC-3′) and reverse
(5′-CTACAACCTCTCTGTGGTCA-3′) primers, and was amplified by PCR
under the following conditions: 95°C for 15 min; 35 cycles of
denaturation at 95°C for 30 sec, annealing at 53°C for 30 sec, and
extension at 72°C for 30 sec; and a final extension at 72°C for 5
min. The genotypes of the amplified products were identified by
electrophoretic separation on a 5% agarose gel using EcoDye™
Nucleic Acid Staining Solution (BioFact Co., Ltd., Daejeon,
Korea).
The PSG9C>T polymorphism was amplified
using forward (5′-GTAATGGTAGAGGTCCGTCA-3′) and reverse
(5′-CGTGTGTGTATCTTCAAGGC-3′) primers, and was amplified by PCR
under the following conditions: 95°C for 15 min; 35 cycles of
denaturation at 95°C for 30 sec, annealing at 57°C for 30 sec, and
extension at 72°C for 30 sec; and a final extension at 72°C for 5
min. The PCR products were digested with the restriction enzyme
BfaI (New England BioLabs, Inc.) at 37°C for 16 h.
The ABCB5C>G polymorphism was amplified
using forward (5′-GAGAAAGGAAGCAGTTGG-3′) and reverse
(5′-TAGTTCCCTCTTTCCCAC-3′) primers, and was amplified by PCR under
the following conditions: 95°C for 5 min; 35 cycles of denaturation
at 95°C for 30 sec, annealing at 52°C for 30 sec, extension at 72°C
for 30 sec; and a final extension at 72°C for 5 min. The PCR
products were digested with the restriction enzyme DdeI
(Enzynomics, Daejeon, Korea) at 37°C for 16 h.
To validate the PCR-RFLP analysis, DNA sequencing
was performed on randomly selected samples (~20% of total samples),
using a BigDye™ Terminator v3.1 Cycle Sequencing kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.), ABI 3730×L DNA
Analyzer (Applied Biosystems; Thermo Fisher Scientific, Inc.).
PCR-RFLP genotyping was 100% concordant with DNA sequencing.
Laboratory tests
Plasma homocysteine, folate, total cholesterol, uric
acid, blood urea nitrogen (BUN) and creatinine were measured in
blood samples collected from patients with RPL after 12 h of
fasting. Homocysteine was measured by IMx fluorescent polarizing
immunoassay using the Abbott IMx analyzer (Abbott Pharmaceutical
Co. Ltd., Lake Bluff, IL, USA). Folic acid was determined with a
radioassay kit (ACS:180; Bayer AG, Leverkusen, Germany). Total
cholesterol, uric acid, BUN and creatinine were measured using
commercially available enzymatic colorimetric tests of the MODULAR
PRE ANALYTICS PLUS system (Roche Diagnostics GmbH, Mannheim.
Germany). Platelet (PLT) and white blood cell (WBC) counts were
measured using the Sysmex XE-2100 Automated Hematology system
(Sysmex Corporation, Kobe, Japan). Prothrombin time (PT) and
activated partial thromboplastin time (aPTT) were measured with the
ACL TOP automated photo-optical coagulometer (Mitsubishi Gas
Chemical Company, Inc., Tokyo, Japan). Blood was collected from the
control group by venipuncture on the second or third day of the
menstrual cycle for the measurement of FSH, LH, E2, TSH, and
prolactin levels. Serum was prepared as previously described
(19) and hormone levels were
determined using either radioimmunoassays [E2 (cat. no. A21854),
TSH (cat. no. IM3712) and PRL (cat. no. IM2121); Beckman Coulter,
Inc., Brea, CA, USA], or enzyme immunoassays using
IMMULITE® 1000 Systems (FSH and LH; Siemens AG, Munich,
Germany) according to the manufacturer's protocols.
Flow cytometric analysis of cluster of
differentiation (CD)56+ natural killer cells
For cell surface marker staining, peripheral blood
samples (50 µl) were mixed with monoclonal anti-CD56+
antibodies labeled with fluorescein isothiocyanate (cat. no.
340417, BD Biosciences, Franklin Lakes, NJ, USA) to a ratio of 5:2,
for 15 min at room temperature. A total of 450 µl 1X BD FACS™
lysing solution (cat. no. 349202, BD Biosciences) was added
followed by gentle vortexing and two washes with 2 ml
fluorescence-activated cell sorting buffer (PBS supplemented with
1% bovine serum albumin and 0.01% sodium azide; Lonza Group, Ltd.,
Basel, Switzerland). Cells were fixed in 200 ml 1% paraformaldehyde
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) at 30 min in 4°C
and washed with BD Perm/Wash™ buffer (cat. no. 554723, BD
Biosciences) prior to acquisition on a BD FACSCalibur (BD
Biosciences) (20,21). The data were analyzed using Cell
Quest software (BD Biosciences).
Statistical analysis
The gene frequencies of MS4A14, SLC2A7, PSG9
and ABCB5 in patients and controls were assessed by logistic
regression, Fisher's exact test and Mann-Whitney test. All genotype
frequencies followed the Hardy-Weinberg equilibrium (HWE). The
association between the MS4A14, SLC2A7, PSG9 and
ABCB5 gene polymorphisms and RPL risk factors were examined
by odds ratio (OR), adjusted odds ratio (AOR) and 95% confidence
intervals (CIs). Data are presented as the means ± standard
deviation. P≤0.05 was considered to indicate a statistically
significant difference. The correlations of each genotype or allele
with the proportion of NK cells and plasma Hcy, folate, total
cholesterol, and uric acid levels were assessed by the
Kruskal-Wallis and Mann-Whitney tests. The false discovery rate
(FDR) was used for multiple comparisons correction. An FDR adjusted
P<0.05 (q-value) was deemed statistically significant (22). MedCalc version 12.1.4 (MedCalc
Software bvba, Ostend, Belgium) or GraphPad Prism version 4.0
(GraphPad Software, Inc., La Jolla, CA, USA) were used for
statistical analysis. The HAPSTAT version 3.0 (www.bios.unc.edu/~lin/hapstat/) was used
to estimate the frequency of the polymorphic haplotype.
Results
Study population
In the present study, patients with RPL and control
women were compared. The clinical characteristics of the two study
groups are summarized in Table
II. There was a significant difference between patients with
RPL and control women for several of the parameters tested. The PLT
values were significantly higher in the RPL patient group than in
the control group. Furthermore, LH values were significantly higher
in the RPL patient group than in the control group.
 | Table II.Clinical characteristics of patients
with RPL vs. controls. |
Table II.
Clinical characteristics of patients
with RPL vs. controls.
| Parameter | Control n=276 (n=in
each parameter) | RPL n=383 | P |
|---|
| Age (years) | 33.09±5.68 | 33.11±4.44 | 0.966a |
| BMI
(kg/m2) | 21.31±3.30 | 21.48±3.85 | 0.304b |
| Previous pregnancy
losses (no.) | NA | 3.28±1.84 |
|
| Live births
(no.) | 1.73±0.72 | NA |
|
| Gestational
weeks | 39.30±1.68 | 7.35±1.93 |
<0.0001a |
| FSH (mIU/ml) | 8.13±2.86
(110) | 7.54±10.57
(195) |
<0.0001b |
| E2 (pg/ml) | 25.87±14.76
(110) | 35.36±29.00
(166) |
0.002b |
| LH (mIU/ml) | 3.28±1.71
(110) | 6.33±12.15
(196) |
<0.0001b |
| TSH (µIU/ml) | – | 2.19±1.56
(211) |
|
| Prolactin
(ng/ml) | – | 15.61±12.92
(206) |
|
| CD56+ NK
cells (%) | – | 18.36±8.02
(131) |
|
| Hematocrit (%) | 35.35±4.24
(214) | 37.31±3.38
(203) |
<0.0001b |
| PLT (103
platelets/µl) | 235.37±64.08
(221) | 256.31±58.83
(203) |
0.001a |
| PT (sec) | 11.58±3.14
(64) | 11.59±0.86
(206) |
<0.0001b |
| aPTT (sec) | 30.79±4.64
(105) | 32.27±4.33
(208) |
0.006a |
| Homocysteine
(µmol/l) | – | 6.96±2.10
(278) |
|
| Folate (ng/ml) | 13.71±8.37
(19) | 14.19±12.01
(220) | 0.865a |
| BUN (mg/dl) | 8.07±2.02 (41) | 9.96±2.74
(196) |
<0.0001b |
| Creatinine
(mg/dl) | 0.69±0.08 (41) | 0.72±0.12
(195) | 0.056b |
| Total cholesterol
(mg/dl) | 239.00±85.19
(15) | 187.59±49.67
(178) |
0.004b |
| Uric acid
(mg/dl) | 4.19±1.44 (10) | 3.80±0.84
(175) | 0.361b |
Combined effects between clinical
factors and gene polymorphism
Interaction analysis was performed to evaluate the
association between adjusted factors, such as age and the different
gene polymorphisms; the results are presented in Table III. The MS4A14 DI+II
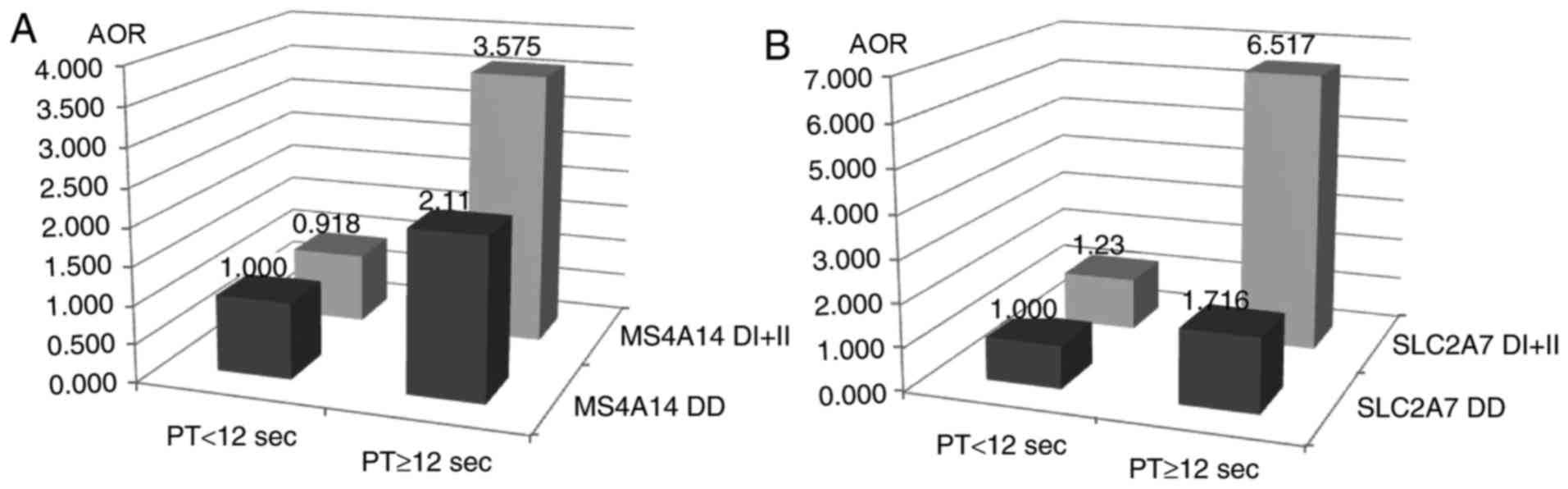
genotype was associated with PT≥12 sec (AOR=3.575; 95%
CI=1.115–11.464) and aPTT≤22.1 sec (AOR=0.453; 95% CI=0.211–0.973).
The SLC2A DI+II genotype was associated with PT≥12 sec
(AOR=6.517; 95% CI=1.783–23.825). The PSG9 CT+TT genotype
was associated with PLT≥279×103/µl (AOR=2.069; 95%
CI=1.079–3.967) and aPTT≤22.1 sec (AOR=0.390; 95% CI=0.172–0.881).
The ABCB5 CG+GG genotype was associated with PT≥12 sec
(AOR=4.224; 95% CI=1.404–12.706). The AOR between the two different
gene polymorphisms and PT are summarized in Fig. 1A and B. Overall, these genotypes
appeared to be associated with coagulation indicators.
 | Table III.Interaction analysis of recurrent
pregnancy loss and coagulation factors. |
Table III.
Interaction analysis of recurrent
pregnancy loss and coagulation factors.
|
| AOR (95% CI) |
|---|
|
|
|
|---|
| Parameter | MS4A14
DD | MS4A14
DI+II | SLC2A7
DD | SLC2A7
DI+II | PSG9 CC | PSG9
CT+TT | ABCB5
CC | ABCB5
CG+GG |
|---|
| Age |
| <33 | 1.000
(reference) | 0.771
(0.483–1.229) | 1.000
(reference) | 0.922
(0.565–1.504) | 1.000
(reference) | 0.866
(0.520–1.442) | 1.000
(reference) | 0.795
(0.490–1.290) |
| ≥33 | 0.852
(0.503–1.443) | 0.802
(0.501–1.285) | 1.018
(0.565–1.834) | 0.884
(0.542–1.443) | 0.912
(0.500–1.662) | 0.860
(0.514–1.438) | 0.853
(0.474–1.533) | 0.821
(0.507–1.329) |
| PLTa |
|
<279×103/µl | 1.000
(reference) | 0.991
(0.625–1.572) | 1.000
(reference) | 0.918
(0.564–1.495) | 1.000
(reference) | 1.047
(0.626–1.750) | 1.000
(reference) | 0.846
(0.520–1.379) |
|
≥279×103/µl | 2.251
(1.083–4.676) | 1.632
(0.873–3.050) | 2.727
(1.126–6.605) | 1.482
(0.800–2.747) | 1.693
(0.737–3.890) | 2.069
(1.079–3.967) | 1.640
(0.664–4.049) | 1.671
(0.909–3.072) |
| PTa |
| <12 sec | 1.000
(reference) | 0.918
(0.476–1.770) | 1.000
(reference) | 1.230
(0.622–2.435) | 1.000
(reference) | 0.612
(0.288–1.302) | 1.000
(reference) | 1.664
(0.842–3.288) |
| ≥12 sec | 2.110
(0.647–6.880) | 3.575
(1.115–11.46) | 1.716
(0.551–5.346) | 6.517
(1.783–23.83) | 1.015
(0.248–4.161) | 2.908
(0.886–9.548) | 4.351
(1.097–17.26) | 4.224
(1.404–12.706) |
| aPTTb |
| >22.1 sec | 1.000
(reference) | 0.957
(0.528–1.735) | 1.000
(reference) | 1.161 (0.630-
2.141) | 1.000
(reference) | 0.990
(0.506–1.936) | 1.000
(reference) | 1.119
(0.609–2.057) |
| ≤22.1 sec | 0.390
(0.171–0.890) | 0.453
(0.211–0.973) | 0.519
(0.250–1.075) | 0.519
(0.250–1.075) | 0.508
(0.188–1.371) | 0.390
(0.172–0.881) | 0.289
(0.088–0.952) | 0.505
(0.248–1.028) |
Genotype frequencies of the VEGF
polymorphisms
The genotype frequencies of the four selected gene
polymorphisms in patients with RPL and normal women are shown in
Table IV. According to the
results, there was no difference in the frequency of these
genotypes between the two study populations, with respect to these
polymorphisms. The genotypes between the two groups did not differ
with respect to the number of pregnancy losses either, regardless
as to whether normal women did not experience pregnancy loss (data
not shown). In addition, to determine if various allele
combinations were associated with the prevalence of RPL, allele
combination analyses were performed with two, three and four allele
combinations. The four-allele combination data are presented in
Table V (data not shown for two
and three allele combinations). Amongst all possible allele
combinations of the four genes,
MS4A14I/SLC2A7D/PSG9T/ABCB5G was
associated with decreased RPL risk (OR=0.448; 95% CI=0.223–0.901;
P=0.033).
 | Table IV.Genotype frequencies in patients with
RPL and controls. |
Table IV.
Genotype frequencies in patients with
RPL and controls.
| Genotype | Control (%)
n=276 | RPL (%) n=383 | AOR (95%
CI)a | Pb | qc |
|---|
| MS4A14
(rs3217518) |
| DD | 92 (33.3) | 141 (36.8) | 1.000
(reference) |
|
|
| DI | 139 (50.4) | 188 (49.1) | 0.882
(0.627–1.242) | 0.473 | 0.785 |
| II | 45 (16.3) | 54 (14.1) | 0.770
(0.478–1.239) | 0.281 | 0.562 |
| Dominant (DD vs.
DI+II) |
|
| 0.855
(0.617–1.183) | 0.344 | 0.582 |
| Recessive (DD+DI
vs. II) |
|
| 0.827
(0.537–1.273) | 0.388 | 0.746 |
| HWE P | 0.535 |
| 0.492 |
|
|
| SLC2A7
(rs60746313) |
| DD | 74 (26.8) | 112 (29.2) | 1.000
(reference) |
|
|
| DI | 148 (53.6) | 202 (52.7) | 0.912
(0.635–1.311) | 0.619 | 0.785 |
| II | 54 (19.6) | 69 (18.0) | 0.854
(0.538–1.356) | 0.504 | 0.672 |
| Dominant (DD vs.
DI+II) |
|
| 0.895
(0.633–1.264) | 0.528 | 0.582 |
| Recessive (DD+DI
vs. II) |
|
| 0.906
(0.610–1.346) | 0.625 | 0.746 |
| HWE P | 0.194 | 0.181 |
|
|
|
| PSG9
(rs3746297) |
| CC | 72 (26.1) | 108 (28.2) | 1.000
(reference) |
|
|
| CT | 149 (54.0) | 195 (50.9) | 0.891
(0.616–1.289) | 0.540 | 0.785 |
| TT | 55 (19.9) | 80 (20.9) | 0.990
(0.627–1.562) | 0.964 | 0.672 |
| Dominant (CC vs.
CT+TT) |
|
| 0.906
(0.639–1.286) | 0.582 | 0.582 |
| Recessive (CC+CT
vs. TT) |
|
| 1.066
(0.724–1.570) | 0.746 | 0.746 |
| HWE P | 0.164 | 0.642 |
|
|
|
| ABCB5
(rs17143187) |
| CC | 74 (26.8) | 113 (29.5) | 1.000
(reference) |
|
|
| CG | 133 (48.2) | 194 (50.7) | 0.950
(0.658–1.371) | 0.785 | 0.785 |
| GG | 69 (25.0) | 76 (19.8) | 0.727
(0.469–1.129) | 0.156 | 0.562 |
| Dominant (CC vs.
CG+GG) |
|
| 0.872
(0.618–1.232) | 0.438 | 0.582 |
| Recessive (CC+CG
vs. GG) |
|
| 0.745
(0.514–1.079) | 0.120 | 0.480 |
| HWE P | 0.551 | 0.658 |
|
|
|
 | Table V.Allele combination analysis in
patients with RPL and controls. |
Table V.
Allele combination analysis in
patients with RPL and controls.
| Allele
combination | Controls
(2na=552; frequency,
%) | RPL (2n=766) | OR (95% CI) | Pb | qc |
|---|
|
MS4A14/SLC2A7/PSG9/ABCB5 |
| D-D-C-C | 51 (9.2) | 82 (10.7) | 1.000
(reference) |
|
|
| D-D-C-G | 39 (7.1) | 58 (7.6) | 0.925
(0.541–1.581) | 0.786 | 0.983 |
| D-D-T-C | 45 (8.1) | 72 (9.4) | 0.995
(0.597–1.659) | 1.000 | 1.000 |
| D-D-T-G | 41 (7.4) | 57 (7.5) | 0.865
(0.508–1.472) | 0.684 | 0.983 |
| D-I-C-C | 49 (8.8) | 52 (6.8) | 0.660
(0.391–1.115) | 0.142 | 0.710 |
| D-I-C-G | 31 (5.6) | 50 (6.5) | 1.003
(0.568–1.771) | 1.000 | 1.000 |
| D-I-T-C | 28 (5.0) | 58 (7.6) | 1.288
(0.728–2.280) | 0.392 | 0.983 |
| D-I-T-G | 40 (7.2) | 40 (5.2) | 0.622
(0.355–1.090) | 0.116 | 0.710 |
| I-D-C-C | 21 (3.9) | 34 (4.5) | 1.007
(0.527–1.923) | 1.000 | 1.000 |
| I-D-C-G | 36 (6.6) | 51 (6.7) | 0.881
(0.508–1.530) | 0.674 | 0.983 |
| I-D-T-C | 37 (6.8) | 54 (7) | 0.908
(0.526–1.566) | 0.781 | 0.983 |
| I-D-T-G | 25 (4.6) | 18 (2.3) | 0.448
(0.223–0.901) | 0.033 | 0.495 |
| I-I-C-C | 31 (5.6) | 44 (5.7) | 0.883
(0.496–1.573) | 0.768 | 0.983 |
| I-I-C-G | 35 (6.3) | 40 (5.2) | 0.711
(0.401–1.260) | 0.246 | 0.923 |
| I-I-T-C | 19 (3.5) | 24 (3.1) | 0.786
(0.392–1.576) | 0.591 | 0.983 |
| I-I-T-G | 24 (4.3) | 32 (4.1) | 0.829
(0.440–1.564) | 0.626 | 0.983 |
Genotype combination analysis
Finally, genotype combination analysis was
performed. The following combinations were significantly associated
with decreased RPL occurrence: MS4A14II/PSG9CT
(OR=0.446; 95% CI=0.200–0.995; P=0.049),
MS4A14II/ABCB5CG (OR=0.397; 95% CI=0.185–0.851;
P=0.018), SLC2A7DI/ABCB5GG (OR=0.485; 95%
CI=0.240–0.981; P=0.044) and SLC2A7II/ABCB5CC
(OR=0.376; 95% CI=0.152–0.932; P=0.035). A summary of the results
is shown in Table VI.
 | Table VI.Gene combination analysis in patients
with RPL and controls. |
Table VI.
Gene combination analysis in patients
with RPL and controls.
| Genotype | Control (%)
n=276 | RPL (%) n=383 | AOR (95% CI) | Pa |
q-valueb |
|---|
|
MS4A14/SLC2A7 |
|
DD/DD | 28 (10.1) | 49 (12.8) | 1.000
(reference) |
|
|
|
DD/DI | 45 (16.3) | 66 (17.2) | 0.828
(0.454–1.510) | 0.538 | 0.613 |
|
DD/II | 19 (6.9) | 26 (6.8) | 0.794
(0.369–1.708) | 0.555 | 0.613 |
|
DI/DD | 33 (12.0) | 46 (12.0) | 0.792
(0.415–1.510) | 0.478 | 0.613 |
|
DI/DI | 80 (29.0) | 111 (29.0) | 0.781
(0.452–1.351) | 0.377 | 0.613 |
|
DI/II | 26 (9.4) | 31 (8.1) | 0.677 (0.335-
1.370) | 0.279 | 0.613 |
|
II/DD | 13 (4.7) | 17 (4.4) | 0.713
(0.295–1.725) | 0.453 | 0.613 |
|
II/DI | 23 (8.3) | 25 (6.5) | 0.607
(0.291–1.269) | 0.185 | 0.613 |
|
II/II | 9 (3.3) | 12 (3.1) | 0.773
(0.285–2.094) | 0.613 | 0.613 |
|
MS4A14/PSG9 |
|
DD/CC | 21 (7.6) | 39 (10.2) | 1.000
(reference) |
|
|
|
DD/CT | 49 (17.8) | 70 (18.3) | 0.782
(0.409–1.496) | 0.457 | 0.713 |
|
DD/TT | 22 (8.0) | 32 (8.4) | 0.786
(0.368–1.681) | 0.535 | 0.713 |
|
DI/CC | 42 (15.2) | 48 (12.5) | 0.612
(0.312–1.201) | 0.153 | 0.408 |
|
DI/CT | 75 (27.2) | 102 (26.6) | 0.759
(0.410–1.404) | 0.380 | 0.713 |
|
DI/TT | 22 (8.0) | 38 (9.9) | 1.034
(0.481–2.223) | 0.933 | 0.933 |
|
II/CC | 9 (3.3) | 21 (5.5) | 1.206
(0.466–3.122) | 0.699 | 0.799 |
|
II/CT | 25 (9.1) | 23 (6.0) | 0.446
(0.200–0.995) | 0.049 | 0.392 |
|
II/TT | 11 (4.0) | 10 (2.6) | 0.476
(0.172–1.318) | 0.153 | 0.392 |
|
MS4A14/ABCB5 |
|
DD/CC | 26 (9.4) | 44 (11.5) | 1.000
(reference) |
|
|
|
DD/CG | 42 (15.2) | 67 (17.5) | 0.955
(0.513–1.780) | 0.886 | 0.974 |
|
DD/GG | 24 (8.7) | 30 (7.8) | 0.732
(0.354–1.514) | 0.400 | 0.640 |
|
DI/CC | 42 (15.2) | 52 (13.6) | 0.715
(0.378–1.351) | 0.301 | 0.640 |
|
DI/CG | 63 (22.8) | 107 (27.9) | 1.010
(0.566–1.801) | 0.974 | 0.974 |
|
DI/GG | 34 (12.3) | 29 (7.6) | 0.503
(0.251–1.007) | 0.052 | 0.208 |
|
II/CC | 6 (2.2) | 17 (4.4) | 1.679
(0.588–4.799) | 0.333 | 0.640 |
|
II/CG | 28 (10.1) | 20 (5.2) | 0.397
(0.185–0.851) | 0.018 | 0.144 |
|
II/GG | 11 (4.0) | 17 (4.4) | 0.912
(0.370–2.245) | 0.841 | 0.974 |
|
SLC2A7/PSG9 |
|
DD/CC | 17 (6.2) | 35 (9.1) | 1.000
(reference) |
|
|
|
DD/CT | 43 (15.6) | 55 (14.4) | 0.676
(0.331–1.380) | 0.282 | 0.437 |
|
DD/TT | 14 (5.1) | 22 (5.7) | 0.836
(0.340–2.056) | 0.696 | 0.696 |
|
DI/CC | 36 (13.0) | 48 (12.5) | 0.657
(0.317–1.361) | 0.258 | 0.437 |
|
DI/CT | 81 (29.3) | 108 (28.2) | 0.652
(0.339–1.252) | 0.199 | 0.437 |
|
DI/TT | 31 (11.2) | 46 (12.0) | 0.770
(0.366–1.622) | 0.492 | 0.562 |
|
II/CC | 19 (6.9) | 25 (6.5) | 0.654
(0.283–1.511) | 0.320 | 0.437 |
|
II/CT | 25 (9.1) | 32 (8.4) | 0.661
(0.300–1.455) | 0.304 | 0.437 |
|
II/TT | 10 (3.6) | 12 (3.1) | 0.601
(0.216–1.668) | 0.328 | 0.437 |
|
SLC2A7/ABCB5 |
|
DD/CC | 20 (7.2) | 37 (9.7) | 1.000
(reference) |
|
|
|
DD/CG | 42 (15.2) | 50 (13.1) | 0.631
(0.319–1.249) | 0.186 | 0.372 |
|
DD/GG | 12 (4.3) | 25 (6.5) | 1.090
(0.450–2.639) | 0.849 | 0.875 |
|
DI/CC | 36 (13.0) | 63 (16.4) | 0.946
(0.477–1.877) | 0.875 | 0.875 |
|
DI/CG | 69 (25.0) | 99 (25.8) | 0.761
(0.406–1.427) | 0.395 | 0.632 |
|
DI/GG | 43 (15.6) | 40 (10.4) | 0.485
(0.240–0.981) | 0.044 | 0.176 |
|
II/CC | 18 (6.5) | 13 (3.4) | 0.376
(0.152–0.932) | 0.035 | 0.176 |
|
II/CG | 22 (8.0) | 45 (11.7) | 1.118
(0.529–2.364) | 0.769 | 0.875 |
|
II/GG | 14 (5.1) | 11 (2.9) | 0.428
(0.164–1.117) | 0.083 | 0.221 |
|
PSG9/ABCB5 |
|
CC/CC | 19 (6.9) | 29 (7.6) | 1.000
(reference) |
|
|
|
CC/CG | 37 (13.4) | 50 (13.1) | 0.865
(0.421–1.777) | 0.693 | 0.892 |
|
CC/GG | 16 (5.8) | 29 (7.6) | 1.206
(0.518–2.805) | 0.665 | 0.892 |
|
CT/CC | 41 (14.9) | 59 (15.4) | 0.915
(0.450–1.860) | 0.806 | 0.892 |
|
CT/CG | 72 (26.1) | 104 (27.2) | 0.956
(0.496–1.840) | 0.892 | 0.892 |
|
CT/GG | 36 (13.0) | 32 (8.4) | 0.569
(0.268–1.209) | 0.143 | 0.892 |
|
TT/CC | 14 (5.1) | 25 (6.5) | 1.259
(0.515–3.077) | 0.613 | 0.892 |
|
TT/CG | 24 (8.7) | 40 (10.4) | 1.091
(0.501–2.376) | 0.827 | 0.892 |
|
TT/GG | 17 (6.2) | 15 (3.9) | 0.606
(0.243–1.513) | 0.284 | 0.892 |
Discussion
In the present study, the association between four
gene polymorphisms, namely MS4A14D>I (rs3217518),
SLC2A7D>I (rs60746313), PSG9C>T (rs3746297) and
ABCB5C>G (rs17143187), and RPL were examined. These
frameshift mutations and splice variants have been associated with
other diseases, such as colorectal cancer (23,24)
and are also implicated in RPL.
The protein encoded by the MS4A14 gene serves
an important role in embryo development and fertilization in rats
(25). SLC2A7, also known
as the glucose transporter 7 gene, catalyzes the cellular uptake of
sugars. During pregnancy, the transplacental nutrient transport of
amino acids, lipids and carbohydrates is important for proper fetal
development, and glucose from the maternal circulation is a
principal source of energy for the fetus (26). The protein encoded by the
PSG9 gene is a member of the pregnancy-specific glycoprotein
(PSG) family. In several studies, it was demonstrated that reduced
serum concentration of PSG was associated with reduced fetal growth
(27,28). The ABCB5 gene is a biomarker
for physiological and pathological stem cells, and is a mediator of
cell fusion, vasculogenesis and drug efflux (29). A successful pregnancy requires the
development of a complex maternal and fetal vascular network that
can support the increasing oxygen and metabolic demands of the
growing fetus (30). Furthermore,
placental development occurs through the vasculogenesis and
angiogenesis stage (31).
Therefore, it may be hypothesized that a mutation in ABCB5
would influence RPL.
Although the individual gene variations examined in
this study were not associated with RPL, gene combinations were
demonstrated to be associated with RPL. Allele combination analysis
(MS4A14I/SLC2A7D/PSG9T/ABCB5G) and
genotype combination analysis (MS4A14II/PSG9CT and
MS4A14II/ABCB5CG) revealed that the I allele affected
RPL when the MS4A14D>I polymorphism was present with
other genes.
The present study also demonstrated that all the
selected gene polymorphisms were associated with RPL when the blood
coagulation factors PLT, PT and aPTT were included in the
interaction analysis. During pregnancy, fibrinolysis and
coagulation must be precisely balanced so that excess fibrin
deposition in placental vessels and intravillous spaces does not
occur, and to ensure fibrin polymerization and stabilization of the
placental basal plate. Defects in this process can have a negative
impact on trophoblast transplantation and placenta development,
ultimately leading to miscarriage (32). Therefore, if blood coagulation
factors deviate from their normal levels this may result in an
unsuccessful pregnancy.
The present study has several limitations. Firstly,
it is unclear whether these polymorphisms can affect gene
transcription and/or translation. Secondly, the study only included
Korean women who visited the CHA Bundang Medical Center, and it
would be useful to validate the findings in a different cohort.
Lastly, the control group size was smaller than the patient group.
Nevertheless, to the best of our knowledge, this study is the first
of its kind to investigate the association between the gene
polymorphisms MS4A14D>I (rs3217518), SLC2A7D>I
(rs60746313), PSG9C>T (rs3746297) and ABCB5C>G
(rs17143187), and RPL. These genes have been implicated in cancer
and some other diseases, but have not been previously studied in
the context of RPL. Therefore, these findings may help to improve
our understanding of frameshift mutations and splice variants in
pregnancy.
In conclusion, the association between RPL and four
gene polymorphisms, MS4A14D>I (rs3217518),
SLC2A7D>I (rs60746313), PSG9C>T (rs3746297) and
ABCB5C>G (rs17143187), was investigated in Korean women.
These four polymorphisms were not associated with RPL individually,
but were associated with RPL when combined with other genes or when
factoring in blood coagulation factors. Notably, the MS4A14
I allele, alongside a PT≥12 sec, may be a potential biomarker for
the diagnosis, prevention and prognosis of RPL. Further studies are
required to clarify the associations between the four gene
polymorphisms and RPL in an ethnically diverse cohort.
Acknowledgements
Not applicable.
Funding
The present study was partly supported by the Basic
Science Research Programs through the National Research Foundation
of Korea funded by the Ministry of Education, Science, and
Technology (grant no. 2009-0093821, 2015R1D1A1A09057432 and
2017R1D1A1B03031542). The present study was also partly supported
by a grant from the Korea Healthcare Technology R&D Project,
Ministry for Health, Welfare & Family Affairs, Republic of
Korea (grant no. HI15C1972010015).
Availability of data and materials
Not applicable.
Authors' contributions
WSL and NKK conceived and designed the experiments.
HAL, CSR, JYL and JOK performed the experiments. EHA, JHK, CSR, JYL
and SHC performed the analysis and interpretation of data for this
study. WSL and NKK. prepared reagents/materials/analytical tools
for experiments and analysis. HAL and EHA wrote the paper. JHK,
SHC, WSL and NKK revised the manuscript for important intellectual
information.
Ethics approval and consent to
participate
The study was approved by the Institutional Review
Board of CHA Bundang Medical Center (IRB number: BD2010-123D) and
written informed consent was provided by all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hefler LA, Tempfer CB, Unfried G,
Schneeberger C, Lessl K, Nagele F and Huber JC: A polymorphism of
the interleukin-1beta gene and idiopathic recurrent miscarriage.
Fertil Steril. 76:377–379. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jang HG, Choi Y, Kim JO, Jeon YJ, Rah H,
Cho SH, Kim JH, Lee WS and Kim NK: Polymorphisms in tumor necrosis
factor-alpha (−863C>A, −857C>T and +488G>A) are associated
with idiopathic recurrent pregnancy loss in Korean women. Hum
Immunol. 77:506–511. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Practice Committee of American Society for
Reproductive Medicine: Definitions of infertility and recurrent
pregnancy loss: a committee opinion. Fertil Steril. 99:632013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Su MT, Lin SH, Lee IW, Chen YC, Hsu CC,
Pan HA and Kuo PL: Polymorphisms of endocrine gland-derived
vascular endothelial growth factor gene and its receptor genes are
associated with recurrent pregnancy loss. Hum Reprod. 25:2923–2930.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kim HS, Lee BE, Jeon YJ, Rah H, Lee WS,
Shin JE, Choi DH and Kim NK: Transcobalamin II (TCN2 67A>G and
TCN2 776C>G) and transcobalamin II receptor (TCblR 1104C>T)
polymorphisms in Korean patients with idiopathic recurrent
spontaneous abortion. Am J Reprod Immunol. 72:337–346. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Altawil AS, Mawlawi HA, Alghamdi KA and
Almijmaj FF: A novel homozygous frameshift mutation in exon 2 of
LEP gene associated with severe obesity: A case report. Clin Med
Insights Pediatr. 10:115–118. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ogura Y, Bonen DK, Inohara N, Nicolae DL,
Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, et
al: A frameshift mutation in NOD2 associated with susceptibility to
Crohn's disease. Nature. 411:603–606. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Camacho Londoño J and Philipp SE: A
reliable method for quantification of splice variants using
RT-qPCR. BMC Mol Biol. 17:82016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Furnham N, Ruffle S and Southan C: Splice
variants: A homology modeling approach. Proteins. 54:596–608. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen J and Weiss WA: Alternative splicing
in cancer: Implications for biology and therapy. Oncogene. 34:1–14.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Orzińska A, Guz K, Mikula M, Kulecka M,
Kluska A, Balabas A, Pelc-Kłopotowska M, Ostrowski J and Brojer E:
A preliminary evaluation of next-generation sequencing as a
screening tool for targeted genotyping of erythrocyte and platelet
antigens in blood donors. Blood Transfus. 16:285–292.
2017.PubMed/NCBI
|
|
12
|
Behjati S and Tarpey PS: What is next
generation sequencing? Arch Dis Child Educ Pract Ed. 98:236–238.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Joensen KG, Engsbro ALØ, Lukjancenko O,
Kaas RS, Lund O, Westh H and Aarestrup FM: Evaluating
next-generation sequencing for direct clinical diagnostics in
diarrhoeal disease. Eur J Clin Microbiol Infect Dis. 36:1325–1338.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hodzic J, Gurbeta L, Omanovic-Miklicanin E
and Badnjevic A: Overview of next-generation sequencing platforms
used in published draft plant genomes in light of genotypization of
immortelle plant (Helichrysium Arenarium). Med Arch. 71:288–292.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bevilacqua J, Hesse A, Cormier B, Davey J,
Patel D, Shankar K and Reddi HV: Clinical utility of a 377 gene
custom next-generation sequencing epilepsy panel. J Genet.
96:681–685. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yoon B, Kim Y-J, Son S-Y, Han K and Park
BC: Whole-exome sequencing in Tricho-rhino-phalangeal syndrome
(TRPS) type I in a Korean family. Genes Genomics. 39:417–422. 2017.
View Article : Google Scholar
|
|
17
|
Barch MJ, Knutsen T and Spurbeck JL: The
AGT Cytogenetics Laboratory Manual. 3rd ed. Lippincott-Raven, New
York: pp. 481–526. 1997
|
|
18
|
Cho H-S, Kim W, Choi M-K, Le MT, Choi HJ,
Kim J-H, Kim K, Soundrarajan N, Park J-K, Lee Y-M, et al: Effects
of natural resistance-associated macrophage protein 1 and toll-like
receptor 2 gene polymorphisms on post-weaning piglet survivability.
Genes Genomics. 38:171–178. 2016. View Article : Google Scholar
|
|
19
|
Choi DH, Kim EK, Kim KH, Lee KA, Kang DW,
Kim HY, Bridges P and Ko CM: Expression pattern of endothelin
system components and localization of smooth muscle cells in the
human pre-ovulatory follicle. Hum Reprod. 26:1171–1180. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rosen HR, Doherty DG, Madrigal-Estebas L,
O'Farrelly C and Golden-Mason L: Pretransplantation CD56(+) innate
lymphocyte populations associated with severity of hepatitis C
virus recurrence. Liver Transpl. 14:31–40. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Carbone T, Nasorri F, Pennino D, Eyerich
K, Foerster S, Cifaldi L, Traidl-Hoffman C, Behrendt H and Cavani
A: CD56highCD16-CD62L- NK cells accumulate in allergic contact
dermatitis and contribute to the expression of allergic responses.
J Immunol. 184:1102–1110. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Stat Soc. 57:289–300. 1995.
|
|
23
|
Yeon SY, Jo YS, Choi EJ, Kim MS, Yoo NJ
and Lee SH: Frameshift mutations in repeat sequences of ANK3,
HACD4, TCP10L, TP53BP1, MFN1, LCMT2, RNMT, TRMT6, METTL8 and
METTL16 genes in colon cancers. Pathol Oncol Res. 24:617–622. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ling Y, Kuang Y, Chen LL, Lao WF, Zhu YR,
Wang LQ and Wang D: A novel RON splice variant lacking exon 2
activates the PI3K/AKT pathway via PTEN phosphorylation in
colorectal carcinoma cells. Oncotarget. 8:39101–39116. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jia XF, Zhou M, Lin JF, Shi WL, Zhang XD
and Shi HJ: Role of SP3111 protein in fertilization and early
embryo development in mice. Zhonghua Nan Ke Xue. 16:14–19.
2010.PubMed/NCBI
|
|
26
|
Stanirowski PJ, Szukiewicz D, Pyzlak M,
Abdalla N, Sawicki W and Cendrowski K: Impact of pre-gestational
and gestational diabetes mellitus on the expression of glucose
transporters GLUT-1, GLUT-4 and GLUT-9 in human term placenta.
Endocrine. 55:799–808. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Moore T and Dveksler GS:
Pregnancy-specific glycoproteins: Complex gene families regulating
maternal-fetal interactions. Int J Dev Biol. 58:273–280. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pihl K, Larsen T, Laursen I, Krebs L and
Christiansen M: First trimester maternal serum pregnancy-specific
beta-1-glycoprotein (SP1) as a marker of adverse pregnancy outcome.
Prenat Diagn. 29:1256–1261. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Volpicelli ER, Lezcano C, Zhan Q, Girouard
SD, Kindelberger DW, Frank MH, Frank NY, Crum CP and Murphy GF: The
multidrug-resistance transporter ABCB5 is expressed in human
placenta. Int J Gynecol Pathol. 33:45–51. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Geva E, Ginzinger DG, Zaloudek CJ, Moore
DH, Byrne A and Jaffe RB: Human placental vascular development:
Vasculogenic and angiogenic (branching and nonbranching)
transformation is regulated by vascular endothelial growth
factor-A, angiopoietin-1, and angiopoietin-2. J Clin Endocrinol
Metab. 87:4213–4224. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kayisli UA, Demir R, Erguler G and Arici
A: Vasodilator-stimulated phosphoprotein expression and its
cytokine-mediated regulation in vasculogenesis during human
placental development. Mol Hum Reprod. 8:1023–1030. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Buchholz T and Thaler CJ: Inherited
thrombophilia: Impact on human reproduction. Am J Reprod Immunol.
50:20–32. 2003. View Article : Google Scholar : PubMed/NCBI
|